Abstract
The phytocystatins of plants are members of the cystatin superfamily of proteins, which are potent inhibitors of cysteine proteases. The Arabidopsis genome encodes seven phytocystatin isoforms (AtCYSs) in two distantly related AtCYS gene clusters. We selected AtCYS1 and AtCYS2 as representatives for each cluster and then generated transgenic plants expressing the GUS reporter gene under the control of each gene promoter. These plants were used to examine AtCYS expression at various stages of plant development and in response to abiotic stresses. Histochemical analysis of AtCYS1 promoter- and AtCYS2 promoter-GUS transgenic plants revealed that these genes have similar but distinct spatial and temporal expression patterns during normal development. In particular, AtCYS1 was preferentially expressed in the vascular tissue of all organs, whereas AtCYS2 was expressed in trichomes and guard cells in young leaves, caps of roots, and in connecting regions of the immature anthers and filaments and the style and stigma in flowers. In addition, each AtCYS gene has a unique expression profile during abiotic stresses. High temperature and wounding stress enhanced the expression of both AtCYS1 and AtCYS2, but the temporal and spatial patterns of induction differed. From these data, we propose that these two AtCYS genes play important, but distinct, roles in plant development and stress responses.
Keywords: Abiotic stress, Cysteine protease inhibitor, Gene expression, GUS staining, Promoter, Transgenic plant
Introduction
Cystatins are a group of proteins that inhibit cysteine (Cys) proteases and are widespread in eukaryotes (Turk and Bode 1991; Margis et al. 1998). Plant cystatins are called phytocystatins (PhyCYSs) and form an independent subfamily in the cystatin superfamily (Margis et al. 1998). Several PhyCYSs have been well characterized and take part in a variety of physiological processes, including programmed cell death (Belenghi et al. 2003), fruit development (Ryan et al. 1998; Neuteboom et al. 2009), and seed development and germination (Abe et al. 1987; Bolter 1993; Misaka et al. 1996; Gaddour et al. 2001; Hong et al. 2007; Hwang et al. 2009). Most studies have focused on the roles played by PhyCYS proteins in defense mechanisms against pathogens and insect attack (Koiwa et al. 1997; Delledonne et al. 2001; Soares-Costa et al. 2002; Urwin et al. 2003; Outchkourov et al. 2004; Yang and Yeh 2005; Christova et al. 2006; Wang et al. 2008). Recently, studies have also suggested that PhyCYSs are involved in responding to abiotic environmental stresses (Gaddour et al. 2001; Diop et al. 2004; Zhang et al. 2008). The expression levels of soybean PhyCYS change during wounding (Botella et al. 1996), and a chestnut PhyCYS is strongly induced in the roots and leaves of plantlets subjected to cold and salt stress, and in the roots after heat stress (Pernas et al. 2000). A cDNA from developing barley endosperm encoding the PhyCYS Hv-CPI (gene Icy) has been reported that its expression in vegetative tissues is affected by cold and anaerobiosis (Gaddour et al. 2001), and its expression in aleurone layers is regulated by two DOF transcription factors (Martínez et al. 2005a). Corn PhyCYS genes are rapidly expressed in response to cold stress and drought (Massonneau et al. 2005). In Arabidopsis, seven PhyCYS genes (AtCYS1–AtCYS7) have been identified (Martínez et al. 2005b). The expression of AtCYSa and AtCYSb (synonymous with AtCYS3 and AtCYS6, respectively) increases resistance to salt and drought stresses, as well as oxidative and cold stresses (Zhang et al. 2008).
Despite the important roles played by PhyCYS proteins in biotic and abiotic stress responses, tissue-specific PhyCYS gene expression patterns or regulation with respect to developmental and environmental cues have not been determined. We examined these facets of two Arabidopsis PhyCYS genes, AtCYS1 and AtCYS2, using β-glucuronidase (GUS; uid gene) expression analysis. Cell- and tissue-specific expression driven by the AtCYS1 and AtCYS2 promoters was monitored at several developmental stages and in response to different abiotic stresses. Our study has determined the specific expression patterns of AtCYS1 and AtCYS2 and establishes a framework for further examination of the physiological roles played by these proteins.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana L. Heynh. ecotype Columbia (Col-0) plants were grown in soil or MS medium (Murashige and Skoog 1962) containing 3% sucrose and 0.25% phyta-gel (pH 5.8), under long-day conditions (16 h of 100 μE s−1 m−2 light and 8 h darkness) at 22°C. To induce synchronous germination, seeds were incubated at 4°C for 3 days in the dark, and then transferred to a growth chamber, as previously described (Lim et al. 2007).
Generation of transgenic Arabidopsis
DNA fragments encompassing the promoter regions of the PhyCYS genes AtCYS1 (At5g12140; −1381 to +30 relative to the ATG translation start codon) and AtCYS2 (At2g31980; −1392 to +30) were PCR-amplified from Arabidopsis genomic DNA using the following primers: AtCYS1 promoter forward (5′-GAA TTC GAG CAA CTG CAA GCT GAG AG-3′), AtCYS1 promoter reverse (5′-GAT CCG ACG ATT GTT CCT GCT TGT TG-3′); AtCYS2 promoter forward (5′-GAA TTC GAG ACT CTT ACG CTT AGG G-3′), and AtCYS2 promoter reverse (5′-GGA TCC TAC AAG AGA GAC CTT CAA CAT GG-3′). The PCR products were cloned into pMD18-T (Takara, Tokyo, Japan) using the TA overhang, and the integrity of the constructs was verified by sequencing. Cloned DNA was digested with EcoRI and BamHI, and the fragments were then inserted into the pCAMBIA1381-GUS plasmid (http://www.cambia.org.au). Each construct was designated AtCYS1P::GUS for AtCYS1 promoter and AtCYS2P::GUS for AtCYS2 promoter. Recombinant plasmids were introduced into Agrobacterium tumefaciens GV3101 and then transferred into Arabidopsis plants using the floral dip method (Clough and Bent 1998). Homozygous T3 lines containing a single T-DNA insertion were used for the analyses, and transgenic plants were maintained under the previously described long-day conditions.
Histochemical GUS assays
Histochemical localization of GUS activity was performed as described by Jefferson et al. (1987). Briefly, wild-type or transgenic Arabidopsis seedlings, organs, and tissues were vacuum-infiltrated in 50 mM sodium phosphate buffer (pH 7.0), 2 mM potassium ferrocyanide (Sigma, St. Louis, MO, USA), 2 mM potassium ferricyanide (Sigma), and 0.2% Triton X-100 (Sigma) containing 1 mM X-GlcA (Duchefa, Haarlem, The Netherlands). The samples were incubated in the dark at 37°C for 12 h and, subsequently, transferred to 70% ethanol to remove the chlorophylls. Digital images were obtained using an Olympus SZX12 stereoscope (Olympus, Tokyo, Japan). GUS staining data are the representatives of at least ten independent transgenic lines for each construct.
Stress treatments for RT-PCR analysis
Arabidopsis plants grown on MS medium at 22°C for 10 days were subjected to various abiotic stresses. Plants were exposed to air (22°C) on filter paper for rapid induction of drought conditions, or placed in a 4 or 37°C chamber in the dark (EYELA, Tokyo, Japan) for thermal stress induction. Mechanical wounding was performed by punching holes in rosette leaves and then incubating the plants in a dark chamber at 22°C. Materials were collected at 0, 1, 3, 6, 12, 24, or 48 h after treatment. Harvested plants were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction.
Total RNA was extracted from 100 mg whole plant tissue using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) was synthesized from 2 μg total RNA using the Revert Aid™ M-MuLV Reverse Transcriptase (Fermentas, Glen Burnie, MD, USA). Each cDNA sample was diluted tenfold, and 1 μl of the diluted cDNA was used for PCR amplification with gene-specific primer sets (AtCYS1 forward, 5′-TCT AGA ATG GCG GAT CAA CAA GCA GG-3′, AtCYS1 reverse, 5′-GGA TCC TTA AAC ATC GTG AAG GTG GTT G-3′; AtCYS2 forward, 5′-TCT AGA ATG GCT ACC ATG TTG AAG GTC-3′, AtCYS2 reverse, 5′-GGT ACC TTA GTA GAC AGG ACT GAC AAC AGG-3′). To minimize inherent RT-PCR errors, each comparison was performed at least three times, using independently isolated RNA samples. The resulting PCR products were sequenced and analyzed by electrophoresis.
Heat and wounding stress treatments of transgenic plants
Heat and wounding stress treatments of transgenic plants carrying an AtCYSP::GUS construct were conducted using plants grown on plates for 10 days. At least three independent transgenic T3 lines of AtCYS1P::GUS and AtCYS2P::GUS were analyzed. For heat stress treatments, plants were placed in a temperature-controlled chamber (EYELA) at 37°C for 48 h in the dark. Wounding experiments were conducted using ticket punches to punch a single hole in each of the rosette leaves. Plants were then placed in a growth chamber in the dark at 22°C. Plant materials were collected 48 h after treatment.
For transverse sections of rosette leaf tissues, GUS-stained samples were dehydrated using a graded ethanol series, stained with 0.1% Eosin Y (Sigma) followed by incubation in a tert-butanol series, and then embedded in paraffin. Embedded tissue sections (12 μm thick) were fixed on poly-lysine-treated slides (Polysciences, PA, USA) at 42°C overnight. Slides were deparaffinized using a clearing agent (Amresco, OH, USA; Yang et al. 2006) and digital images were collected using an Olympus AX-70 microscope (Olympus).
Results
Expression of AtCYS1 and AtCYS2 during plant growth
To monitor the activity of AtCYS promoters during plant development, T3 AtCYS1P::GUS and AtCYS2P::GUS transgenic plants were grown to various stages and then subjected to histochemical staining for GUS activity. In the hypogeal seedling stage [growth stage (gs) 0.1; the gs numbers follow the descriptions of Boyes et al. (2001)], AtCYS1 expression was observed in the cotyledons, whereas AtCYS2 was expressed in the whole seedling (Fig. 1). In epigeal seedlings (gs 0.7–1.0), AtCYS1 expression was restricted to the vasculature, whereas AtCYS2 was expressed in the non-vascular tissue of the cotyledon, hypocotyl and root (gs 0.7). AtCYS2 expression in the cotyledons decreased dramatically as the seedlings matured (gs 1.0). In fully grown plants, AtCYS1 was continuously expressed in the rosette leaves and root vasculature, but AtCYS2 was almost undetectable in whole plants (gs 1.04). AtCYS2 was detected only in unextended young leaves and some root tips. When the transgenic plants started to bolt (gs 5.10), GUS activity was observed in both leaves and roots in AtCYS1P::GUS transgenic plants, but GUS activity was almost undetectable in AtCYS2P::GUS plants. These GUS activity assays clearly show that AtCYS1 expression was low in the seedling stages, but strong in the mature plants, especially in the vasculature. In contrast, AtCYS2 was expressed in the seedling stages, but expression was almost absent in the mature plant stages.
Fig. 1.

Developmentally regulated GUS expression in transgenic Arabidopsis plants carrying the GUS coding region fused to the AtCYS1 (AtCYS1P::GUS) or AtCYS2 (AtCYS2P::GUS) promoter. Histochemical assays show AtCYS1P::GUS and AtCYS2P::GUS expression patterns in representative GUS expression from 1- (gs 0.1), 3- (gs 0.7), 5- (gs 1.0), 10- (gs 1.04), and 28- (gs 5.10) day-old plants grown under normal growth conditions. Scale bars are 500 μm in gs 0.1, 0.7, 1.0, 1.04, and 5 mm in gs 5.10
Expression of AtCYS1 and AtCYS2 in leaves and roots
To explore the tissue-specific expression driven by the AtCYS1 and AtCYS2 promoters, we examined GUS expression in the leaves and roots of gs 1.04 transgenic plants (Fig. 2). In the young rosette leaves of plants, strong expression of AtCYS1 was detected in the vascular tissues and the trichome basal cells, but not in stomata (Fig. 2a–c), whereas AtCYS2 was expressed in the single-celled leaf trichomes and the guard cells (Fig. 2d–f). The expression patterns of AtCYS1 and AtCYS2 in the roots also differed. AtCYS1 expression was observed in the vascular tissue of the primary and lateral roots (Fig. 2g, h), but AtCYS2 expression was restricted to the root cap of the primary root (Fig. 2i, j). These observations indicate that the two AtCYS gene promoters are responsible for the precise transcriptional regulation that determines the tissue-specific and developmental-stage-specific expression of the two AtCYS genes in the leaves and roots of the transgenic Arabidopsis (Hwang et al. 2009).
Fig. 2.
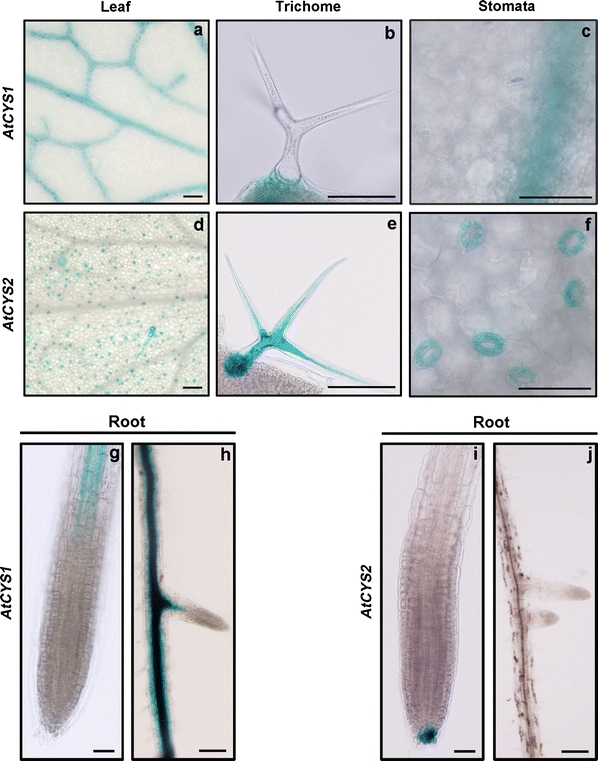
Histochemical localization of AtCYS1P::GUS and AtCYS2P::GUS expression in 10-day-old (gs 1.04) plants. a–f Representative expression patterns in leaf tissues. AtCYS1P::GUS and AtCYS2P::GUS expression in the vascular tissues (a, d), the trichomes (b, e) and the stomata (c, f). Scale bars are 50 μm. g–j Representative expression patterns in root tissues. AtCYS1P::GUS expression in the vascular tissue of the primary (g) and the lateral roots (h). The AtCYS2P::GUS expression in the root caps of the primary roots (i, j). Scale bars are 10 μm in g and i, 50 μm in h and j
Expression of AtCYS1 and AtCYS2 during flowering
Sequential expression of the AtCYS1 and AtCYS2 promoters was observed during flower development (Fig. 3). AtCYS1 was expressed at all flower developmental stages in the vascular tissues of the anther filaments and the sepal, as well as in the regions connecting the stigma and style. AtCYS2 expression was weaker overall than AtCYS1 expression and AtCYS2P::GUS activity was observed in the regions connecting the immature anthers and filaments and the stigma and style at flower development stages (fds) 9–12. After fds 12, AtCYS2 expression declined and was almost undetectable by fds 15. These differences in AtCYS1 and AtCYS2 expression in the floral organs indicate that the AtCYS1 and AtCYS2 promoters control both the developmental and tissue specificity of expression.
Fig. 3.
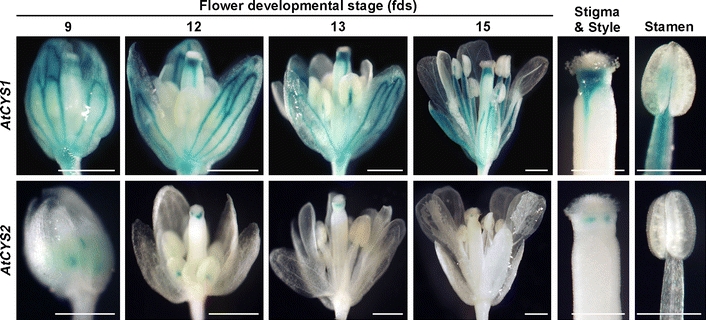
Histochemical localization of AtCYS1P::GUS and AtCYS2P::GUS expression in flowers. The numbers indicate flower development stages (fds). Bars are 500 μm in the main panels and 200 μm in the enlarged images of the carpel and stamen
Semi-quantitative RT-PCR analysis of AtCYS1 and AtCYS2 expression by abiotic stresses
To examine the expression of the AtCYS1 and AtCYS2 genes in relation to abiotic stress, Arabidopsis plants (gs 1.04) were subjected to cold, drought, heat, or wounding stress under dark conditions. By 48 h after the stress treatment, the AtCYS1 expression had increased following cold, drought, heat, and wounding stress. AtCYS2 expression was also induced by drought, heat, and wounding stress, but not by cold stress (Fig. 4a). Additionally, we examined the expression patterns of each gene at various times after heat stress or wounding. As shown in Fig. 4b, AtCYS1 expression started to increase by 1 h after heat stress treatment and reached a maximum at 24 h. However, AtCYS2 expression started to increase 3 h after heat stress treatment and then gradually increased for 12–48 h. To explore the expression of these genes after a wounding stress, hole was punched in the rosette leaves of plants (gs 1.04) and the plants were then incubated in the dark at 22°C. RNA was collected at various times from whole plants as in the heat stress experiments. As shown in Fig. 4c, AtCYS1 and AtCYS2 were both expressed in response to wounding and exhibited expression patterns similar to the heat stress treatments.
Fig. 4.

Expression patterns of AtCYS1 and AtCYS2 in response to abiotic stresses. Semi-quantitative RT-PCR was performed on total RNA using gene-specific primer sets. a Transcript abundance was determined after 10-day-old plants were exposed for 48 h to darkness, heat (37°C), cold (4°C), drought, or wounding. b Transcript levels of AtCYS1 and AtCYS2 genes in response to heat stress (37°C) for the indicated times. c Effects of wounding on AtCYS1 and AtCYS2 transcript accumulation. Total RNA was isolated from whole plants at the indicated time points. The Arabidopsis Actin2 gene (At3g18780) served as a loading control
Computational analysis of the AtCYS1 and AtCYS2 promoter sequences
Generally, stress-responsive cis-acting elements are found in the promoter regions of stress-inducible genes. Since AtCYS1 and AtCYS2 were induced by abiotic stresses (Fig. 4), we performed a motif search using PLACE (Higo et al. 1999; http://www.dna.affrc.go.jp/PLACE/) and PlantCARE (Lescot et al. 2002; http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to identify putative cis-elements in the promoter regions of AtCYS1 and AtCYS2. We identified potential regulatory elements associated with stress-related transcription factor-binding sites including ABA-response elements (ABREs), CCAAT-boxes, heat shock elements (HSEs), stress response elements (STREs) and W-boxes (Fig. 5). The promoter regions of AtCYS1 (−242 to −233) and AtCYS2 (−428 to −420 and −356 to −348) both contain HSEs (Amin et al. 1988; Xiao and Lis 1988). The AtCYS1 promoter contains two perfect CCAAT sequences (−650 to −646 and −607 to −603), while five CCAAT-box sequences were identified in the AtCYS2 promoter (−1293 to −1289, −997 to −993, −524 to −520, −272 to −268, and −143 to −139). These CCAAT enhancer sequences represent binding sites for CCAAT enhancer binding proteins (C/EBP) and act co-operatively with HSEs to increase promoter activation under heat stress conditions (Rieping and Schöffl 1992). STRE elements are important for transcriptional activation in response to a variety of stress conditions and, in particular, heat stress (Siderius and Mager 1997). Two STREs (consensus sequence AGGGG) were identified in the AtCYS2 promoter (−910 to −906 and −903 to −899). The W-box (consensus sequence TTGAC) binds WRKY factors and is responsive to heat and wounding (Ülker and Somssich 2004; Levée et al. 2009; Li et al. 2009). Multiple copies of this element were found in both the AtCYS1 (−1285 to −1282, −1105 to −1102, −779 to −776, −463 to −460, −433 to −430, and −68 to −65) and AtCYS2 (−1341 to 1338, −1303 to −1229, −697 to −693, −550 to 546, and −315 to 311) promoters. Since the promoter regions of these genes contain many cis-elements correlated with responses to heat and wounding stress, the expression of AtCYS1 and AtCYS2 may be controlled by complex regulatory mechanisms that respond to heat and wounding stresses.
Fig. 5.
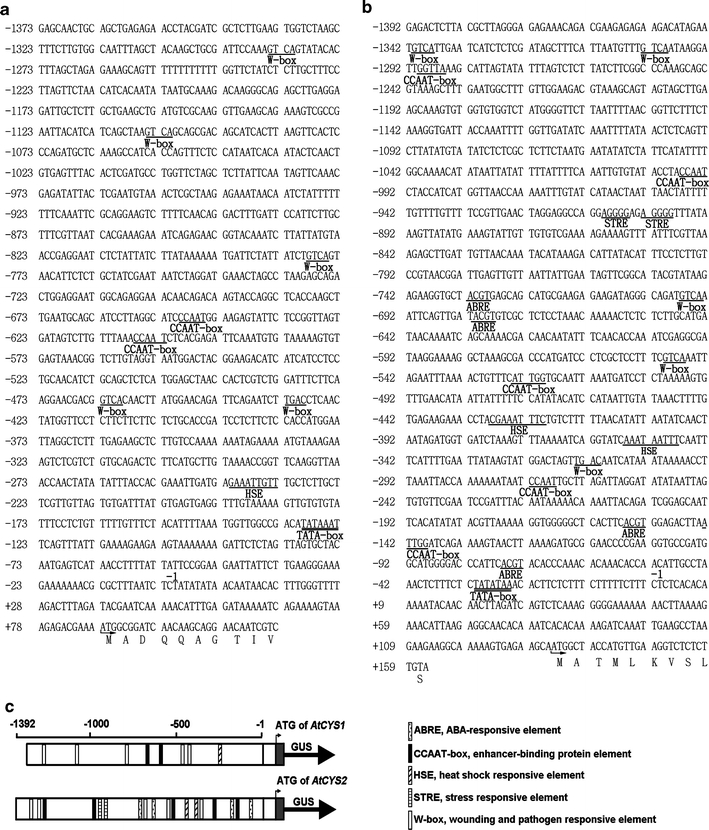
Sequences and structural features of the AtCYS1 and AtCYS2 promoters. The sequences of the 5′-flanking region of AtCYS1 (a) and AtCYS2 (b) are shown together with a partial amino acid sequence. The numbering of nucleotides relative to the putative transcriptional initiation site (−1) is shown above the sequences. The putative TATA box is identified by double underlined. The deduced amino acid sequences of the first exon are indicated by single letter codes in bold. Putative cis-regulatory promoter sequences are underlined. c Schematic representation of the AtCYS1 and AtCYS2 promoters. The locations of abiotic responsive cis-elements of interest were identified using the PLACE and PlantCARE databases. The symbols of each cis-element are annotated below
AtCYS1P::GUS and AtCYS2P::GUS expression in response to heat stress
To precisely define the spatio-temporal expression pattern of AtCYS1 and AtCYS2 under heat stress, we examined the pattern of GUS expression in AtCYS1P::GUS and AtCYS2P::GUS transgenic plants (gs 1.04). As shown in Fig. 6a, heat stress triggers a strong increase in AtCYS1 and AtCYS2 expression in several tissues. However, the pattern of induction differs in roots and leaves between AtCYS1 and AtCYS2 (Fig. 6b, c). In the roots, the expression of AtCYS1 was induced in the vascular bundles, whereas AtCYS2 was detected in the elongation and maturation zone (Fig. 6b). In the lateral roots, AtCYS2 expression increased in the elongation and maturation zones, similar to the primary root, but it was not expressed in the root caps of the lateral roots under heat stress. These data suggest that AtCYS2 may modulate the elongation and maturation of roots during heat stress.
Fig. 6.
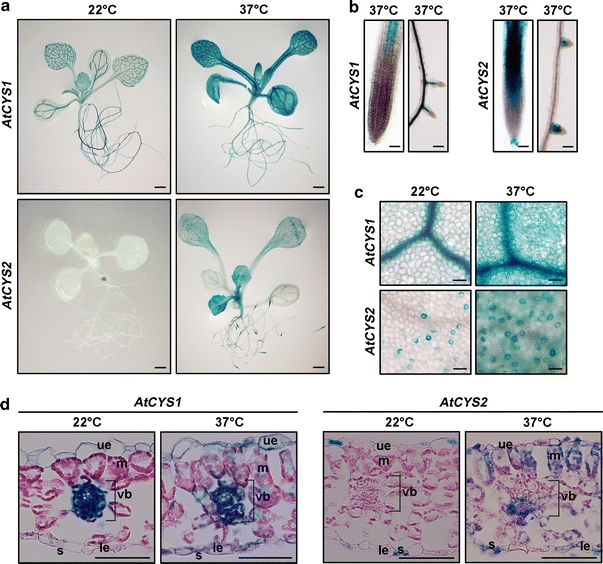
Elevation of GUS expression in transgenic plants following heat stress. a GUS expression after 10-day-old plants was subjected to heat treatment for 48 h at 37°C. Scale bars are 1 mm. b, c GUS activity in the roots (scale bars 15 μm) and leaves (scale bars 12.5 μm). d Transverse sections of leaves from heat-treated seedlings. Scale bars are 50 μm. The abbreviations indicate upper epidermis (ue), mesophyll cells (m), vascular bundle (vb), stoma (s), and lower epidermis (le)
Expression of both AtCYS1 and AtCYS2 was induced in the leaf blade in response to heat stress, with the strongest increase in young leaves in AtCYS2P::GUS expressing plants (Fig. 6c). Transverse sections using the same stage (gs 1.04) leaves used in Fig. 6a were used to examine the expression patterns in detail. As shown in Fig. 6d, AtCYS1 and AtCYS2 expression was consistent with the expression patterns shown in Fig. 6c. Interestingly, AtCYS2 expression was induced in the leaf blade and vascular bundles, where it is not expressed under normal growth conditions. Taken together, the comparisons of AtCYS1 and AtCYS2 expression in response to heat stress have revealed not only differences in expression pattern, but also common expression in some cells and tissues.
AtCYS1P::GUS and AtCYS2P::GUS expression in response to wounding stress
AtCYS1 and AtCYS2 transcripts levels increase in response to wounding stress (Fig. 4a, c), and these data are consistent with previous findings (Belenghi et al. 2003), suggesting that the expression of AtCYS mediates the wounding defense response. In this study, to compare the spatial expression pattern of AtCYS1 and AtCYS2 in response to wounding stress, leaves from AtCYS1P::GUS or AtCYS2P::GUS transgenic plants (gs 1.04) were mechanically wounded with ticket punches. GUS staining patterns were analyzed 48 h after wounding (Fig. 7). Increased GUS activity was detected around the wound sites in both AtCYS1P::GUS and AtCYS2P::GUS transgenic plants (Fig. 7a), but GUS staining was stronger in AtCYS1 transgenic plants. Transverse sections were also cut using the same leaves as in Fig. 7a to examine the expression patterns in detail. As shown in Fig. 7b, AtCYS1 expression was greater in the vascular bundles around the wound and AtCYS2 increased in the lamina and vasculature surrounding the wound. These findings indicate that AtCYS1 and AtCYS2 mediate the wounding defense response in different cells and tissues surrounding the wounded region.
Fig. 7.
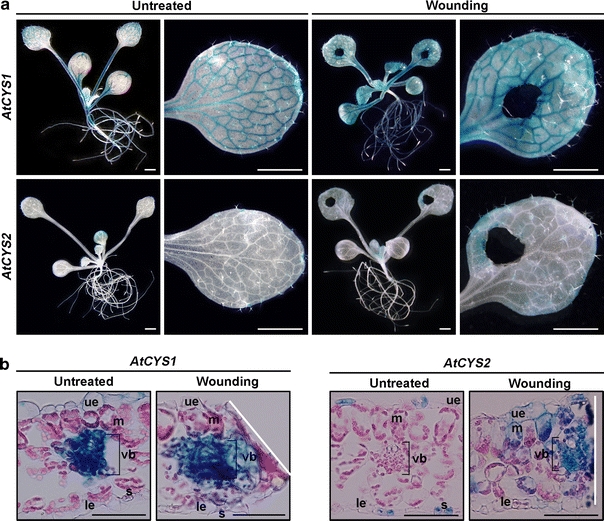
Changes in expression of AtCYS1 and AtCYS2 by wounding stress. a GUS expression 48 h after 10-day-old plants was subjected to wounding. Scale bars are 1 mm. b Transverse sections of wounded and control leaves. Scale bars are 50 μm. White lines indicate the wound sites. The abbreviations indicate upper epidermis (ue), mesophyll cells (m), vascular bundle (vb), stoma (s), and lower epidermis (le)
Discussion
Cys protease and PhyCYS interactions have been repeatedly implicated in the control of plant development and defense mechanisms (Otlewski et al. 2005; Neuteboom et al. 2009). Therefore, to gain a better understanding of the regulatory mechanisms controlling Arabidopsis PhyCYS gene expression, constructs consisting of the GUS reporter gene under the control of either the AtCYS1 or AtCYS2 promoter were introduced into Arabidopsis via Agrobacterium mediated transformation.
In the leaves, AtCYS1 expression was found in the vasculature, whereas AtCYS2 expression was restricted to the stomata and trichomes (Figs. 1, 2). In the roots, AtCYS1 expression was also restricted to the vascular tissues of the primary and lateral roots in transgenic plants. Therefore, AtCYS1 may be involved in regulating vascular development and/or function in roots and leaves, whereas AtCYS2 may play a role in the development of epidermal cells such as trichomes and stomata in young, rapidly growing leaves by interaction with Cys proteases (Reeves et al. 2002). Interestingly, AtCYS2 expression was restricted to the root cap of the primary root (Fig. 2). The root cap is the first point of contact between the growing root system and the soil environment, programmed cell death, and it functions as an important sensory organ (Huh et al. 2002; Jiang et al. 2006). Therefore, we hypothesize that the specific expression of AtCYS2 in the root cap may reflect the involvement of AtCYS2 in root growth and/or cell death in the root cap. Further studies using Cys proteases, which interact with the PhyCYS proteins in the root tips, will be needed to address this possibility. We also identified differences between the tissue-specific expression patterns driven by the AtCYS1 and AtCYS2 promoters in the flower (Fig. 3), based on accurate sampling of floral materials (Smyth et al. 1990). AtCYS1 was strongly expressed in the vasculature of all floral tissues, as in the seedlings and mature plants; however, AtCYS2 was only detected at minimal levels, in the regions connecting the anther and stamen, and the carpel and stigma. The level of AtCYS2 expression rapidly decreased as the anthers matured. These data suggest that AtCYS1 may play roles in development of the floral vasculature in the carpel, sepal, and stamen, whereas AtCYS2 may be involved in regulating growth of the carpel and stamen via interactions with Cys proteases.
Expression of PhyCYS is induced by several abiotic stresses (Pernas et al. 2000; Gaddour et al. 2001; Diop et al. 2004; Massonneau et al. 2005; Zhang et al. 2008) and PhyCYS over-expressing transgenic plants exhibit increased abiotic stress tolerance (Belenghi et al. 2003; Van der Vyver et al. 2003; Huang et al. 2007). These observations support the hypothesis that PhyCYS proteins play a crucial role in plant abiotic stress defense mechanisms. Hence, we assessed the response of AtCYS1 and AtCYS2 to abiotic stresses using RT-PCR (Fig. 4). AtCYS1 expression is induced by cold, drought, heat, and wounding stresses. AtCYS2 expression is induced by drought, heat, and wounding stresses, but not by cold stress. These findings suggest that the selective induction of these genes may be due to the presence of specific promoter elements. Therefore, we analyzed the upstream promoter sequences of the AtCYS1 and AtCYS2 genes to determine whether they contain stress-related cis-elements (Fig. 5). This analysis revealed that AtCYS1 and AtCYS2 contain several interesting putative heat and wounding stress-related cis-elements, including ABREs, CCAAT-boxes, HSEs, STREs, and W-boxes. The ABRE, HSE, and W-box motifs have been identified as binding sites for ABRE-binding factors (ABFs) (Choi et al. 2000), heat shock factors (Hsfs) (Nover et al. 1996), and WRKY transcription factors (Eulgem and Somssich 2007), respectively. ABREs control ABA- and/or heat stress-responsive gene expression, as determined by numerous studies (Giraudat et al. 1994; Gong et al. 1998; Larkindale and Knight 2002). Several WRKY transcription factors are involved in mechanical wounding stress (Levée et al. 2009; Li et al. 2009). The CCAAT-box sequences act cooperatively with HSEs to increase promoter activity (Rieping and Schöffl 1992; Haralampidis et al. 2002). Both motifs were found in the AtCYS1 and AtCYS2 promoter regions and may modulate expression of these genes in response to heat and/or wounding stress.
The cis-elements identified in the AtCYS1 and AtCYS2 promoter sequences prompted us to analyze the transgenic plants carrying the GUS-promoter fusions in heat and wounding stress. The expression of GUS driven by the AtCYS1 and AtCYS2 promoters in transgenic Arabidopsis was induced by heat and wounding stresses (Figs. 6, 7). AtCYS1 expression was induced in most plant tissues, with the exception of root tips, under heat stress (Fig. 6a, b). Heat treatment induced a significant increase in AtCYS2 in young leaves (Fig. 6a), especially in the guard cells (Fig. 6c). This result suggests that AtCYS2 may specifically participate in defense against heat stress via stomatal regulation. In response to wounding stress, both genes exhibited expression patterns similar to heat stress (Fig. 7). AtCYS1 expression was induced in most plant tissues, especially in vasculature around the wounding sites, and AtCYS2 was induced in the mesophyll cells and vasculature around the wounding sites (Fig. 7a, b). In a previous study, wounding caused extensive changes in the synthesis of defense proteins, leading to localized resistance at the site of the lesion (McConn et al. 1997). Expression of both Cys protease inhibitors and Cys proteases is also induced by wounding stress (Kinoshita et al. 1999; Belenghi et al. 2003; Yamada et al. 2004). These observations support the hypothesis that the differential expression of AtCYS1 and AtCYS2 may indicate the presence of different defense mechanisms against wounding, and distinct interactions with target Cys proteases in specific tissues and organs.
The data from our study underscore the importance of comparative analysis in identifying the activities of AtCYS1 and AtCYS2 in each tissue and cell type, as well as in response to different stresses, in order to understand the complexity underlying multiple signaling systems. However, the detailed functions of the entire AtCYS gene family remain to be elucidated, and further experiments are required to define their complete biochemical and physiological functions in response to abiotic stresses.
Acknowledgments
This work was supported by Basic Science Research Program (Grant No. 20100009175) and Environmental Biotechnology National Core Research Center Program (Grant No. 20090091494) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST). Hwang J.E. and Je J. were supported by BK21 fellowship from the MEST, Republic of Korea.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abe K, Emori Y, Kondo H, Suzuki K, Arai S. Molecular cloning of a cysteine proteinase inhibitor of rice (oryzacystatin). Homology with animal cystatins and transient expression in the ripening process of rice seeds. J Biol Chem. 1987;262:16793–16797. [PubMed] [Google Scholar]
- Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988;8:3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenghi B, Acconcia F, Trovato M, Perazzolli M, Bocedi A, Polticelli F, Ascenzi P, Delledonne M. AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur J Biochem. 2003;270:2593–2604. doi: 10.1046/j.1432-1033.2003.03630.x. [DOI] [PubMed] [Google Scholar]
- Bolter CJ. Methyl jasmonate induces papain inhibitor(s) in tomato leaves. Plant Physiol. 1993;103:1347–1353. doi: 10.1104/pp.103.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella MA, Xu Y, Prabha TN, Zhao Y, Narasimhan ML, Wilson KA, Nielsen SS, Bressan RA, Hasegawa PM. Differential expression of soybean cysteine proteinase inhibitor genes during development and in response to wounding and methyl jasmonate. Plant Physiol. 1996;112:1201–1210. doi: 10.1104/pp.112.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/tpc.13.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs, a family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Christova PK, Christov NK, Imai R. A cold inducible multidomain cystatin from winter wheat inhibits growth of the snow mold fungus, Microdochium nivale. Planta. 2006;223:1207–1218. doi: 10.1007/s00425-005-0169-9. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Delledonne M, Allegro G, Belenghi B, Balestrazzi A, Picco F, Levine A, Zelasco S, Calligari P, Confalonieri M. Transformation of white poplar (Populus alba L.) with a novel Arabidopsis thaliana cysteine proteinase inhibitor and analysis of insect pest resistance. Mol Breed. 2001;7:35–42. doi: 10.1023/A:1009605001253. [DOI] [Google Scholar]
- Diop NN, Kidrič M, Repellin A, Gareil M, d’Arcy-Lameta A, Pham Thi AT, Zuily-Fodil Y. A multicystatin is induced by drought-stress in cowpea (Vigna unguiculata (L.) Walp.) leaves. FEBS Lett. 2004;577:545–550. doi: 10.1016/j.febslet.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Gaddour K, Vicente-Carbajosa J, Lara P, Isabel-Lamoneda I, Díaz I, Carbonero P. A constitutive cystatin-encoding gene from barley (Icy) responds differentially to abiotic stimuli. Plant Mol Biol. 2001;45:599–608. doi: 10.1023/A:1010697204686. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Parcy F, Bertauche N, Gosti F, Leung J. Current advances in abscisic acid action and signalling. Plant Mol Biol. 1994;26:1557–1577. doi: 10.1007/BF00016490. [DOI] [PubMed] [Google Scholar]
- Gong M, Li YJ, Chen SZ. Abscisic acid-induced thermotolerance in maize seedlings is mediated by calcium and associated with antioxidant systems. J Plant Physiol. 1998;153:488–496. [Google Scholar]
- Haralampidis K, Milioni D, Rigas S, Hatzopoulos P. Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol. 2002;129:1138–1149. doi: 10.1104/pp.004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JK, Hwang JE, Lim CJ, Yang KA, Jin ZL, Kim CY, Koo JC, Chung WS, Lee KO, Lee SY, Cho MJ, Lim CO. Over-expression of Chinese cabbage phytocystatin 1 retards seed germination in Arabidopsis. Plant Sci. 2007;172:556–563. doi: 10.1016/j.plantsci.2006.11.005. [DOI] [Google Scholar]
- Huang Y, Xiao B, Xiong L. Characterization of a stress responsive proteinase inhibitor gene with positive effect in improving drought resistance in rice. Planta. 2007;226:73–85. doi: 10.1007/s00425-006-0469-8. [DOI] [PubMed] [Google Scholar]
- Huh GH, Damsz B, Matsumoto TK, Reddy MP, Rus AM, Iveas JI, Narasimhan ML, Bressan RA, Hasegawa PM. Salt causes ion disequilibrium-induced programmed cell death in yeast and plants. Plant J. 2002;29:649–659. doi: 10.1046/j.0960-7412.2001.01247.x. [DOI] [PubMed] [Google Scholar]
- Hwang JE, Hong JK, Je JH, Lee KO, Kim DY, Lee SY, Lim CO. Regulation of seed germination and seedling growth by an Arabidopsis phytocystatin isoform, AtCYS6. Plant Cell Rep. 2009;28:1623–1632. doi: 10.1007/s00299-009-0762-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Zhang S, Lee S, Tsai G, Kim K, Huang H, Chilcott C, Zhu T, Feldman LJ. Transcription profile analyses identify genes and pathways central to root cap functions in maize. Plant Mol Biol. 2006;60:343–363. doi: 10.1007/s11103-005-4209-4. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yamada K, Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J. 1999;19:43–53. doi: 10.1046/j.1365-313X.1999.00497.x. [DOI] [PubMed] [Google Scholar]
- Koiwa H, Bressan RA, Hasegawa PM. Regulation of protease inhibitors and plant defense. Trends Plant Sci. 1997;2:379–384. doi: 10.1016/S1360-1385(97)90052-2. [DOI] [Google Scholar]
- Larkindale J, Knight MR. Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 2002;128:682–695. doi: 10.1104/pp.010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levée V, Major I, Levasseur C, Tremblay L, MacKay J, Séguin A. Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol. 2009;184:48–70. doi: 10.1111/j.1469-8137.2009.02955.x. [DOI] [PubMed] [Google Scholar]
- Li S, Fu Q, Huang W, Yu D. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 2009;28:683–693. doi: 10.1007/s00299-008-0666-y. [DOI] [PubMed] [Google Scholar]
- Lim CJ, Hwang JE, Chen H, Hong JK, Yang KA, Choi MS, Lee KO, Chung WS, Lee SY, Lim CO. Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem Biophys Res Commun. 2007;362:431–436. doi: 10.1016/j.bbrc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Margis R, Reis EM, Villeret V. Structural and phylogenetic relationships among plant and animal cystatins. Arch Biochem Biophys. 1998;359:24–30. doi: 10.1006/abbi.1998.0875. [DOI] [PubMed] [Google Scholar]
- Martínez M, Rubio-Somoza I, Fuentes R, Lara P, Carbonero P, Daz I. The barley cystatin gene (Icy) is regulated by DOF transcription factors in aleurone cells upon germination. J Exp Bot. 2005;56:547–556. doi: 10.1093/jxb/eri033. [DOI] [PubMed] [Google Scholar]
- Martínez M, Abraham Z, Carbonero P, Díaz I. Comparative phylogenetic analysis of cystatin gene families from arabidopsis, rice and barley. Mol Genet Genomics. 2005;273:423–432. doi: 10.1007/s00438-005-1147-4. [DOI] [PubMed] [Google Scholar]
- Massonneau A, Condamine P, Wisniewski J-P, Zivy M, Rogowsky PM. Maize cystatins respond to developmental cues, cold stress and drought. Biochim Biophys Acta. 2005;1729:186–199. doi: 10.1016/j.bbaexp.2005.05.004. [DOI] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaka T, Kuroda M, Iwabuchi K, Abe K, Arai S. Soyacystatin, a novel cysteine proteinase inhibitor in soybean, is distinct in protein structure and gene organization from other cystatins of animal and plant origin. Eur J Biochem. 1996;240:609–614. doi: 10.1111/j.1432-1033.1996.0609h.x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Neuteboom LW, Matsumoto KO, Christopher DA. An extended AE-rich N-terminal trunk in secreted pineapple cystatin enhances inhibition of fruit bromelain and is posttranslationally removed during ripening. Plant Physiol. 2009;151:515–527. doi: 10.1104/pp.109.142232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley WB. The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones. 1996;1:215–223. doi: 10.1379/1466-1268(1996)001<0215:THWCAP>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otlewski J, Jelen F, Zakrzewska M, Oleksy A. The many faces of protease–protein inhibitor interaction. EMBO J. 2005;24:1303–1310. doi: 10.1038/sj.emboj.7600611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outchkourov NS, de Kogel WJ, Schuurman-de Bruin A, Abrahamson M, Jongsma MA. Specific cysteine protease inhibitors act as deterrents of western flower thrips, Frankliniella occidentalis (Pergande), in transgenic potato. Plant Biotechnol J. 2004;2:439–448. doi: 10.1111/j.1467-7652.2004.00088.x. [DOI] [PubMed] [Google Scholar]
- Pernas M, Sánchez-Monge R, Salcedo G. Biotic and abiotic stress can induce cystatin expression in chestnut. FEBS Lett. 2000;467:206–210. doi: 10.1016/S0014-5793(00)01157-1. [DOI] [PubMed] [Google Scholar]
- Reeves PH, Murtas G, Dash S, Coupland G. early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development. 2002;129:5349–5361. doi: 10.1242/dev.00113. [DOI] [PubMed] [Google Scholar]
- Rieping M, Schöffl F. Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Mol Gen Genet. 1992;231:226–232. doi: 10.1007/BF00279795. [DOI] [PubMed] [Google Scholar]
- Ryan SN, Laing WA, McManus MT. A cysteine proteinase inhibitor purified from apple fruit. Phytochemistry. 1998;49:957–963. doi: 10.1016/S0031-9422(98)00206-4. [DOI] [PubMed] [Google Scholar]
- Siderius M, Mager WH. General stress response: in search of a common denominator. In: Hohmann S, Mager WH, editors. Yeast stress responses. Heidelberg: Springer; 1997. pp. 213–230. [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-Costa A, Beltramini LM, Thiemann OH, Henrique-Silva F. A sugarcane cystatin: recombinant expression, purification, and antifungal activity. Biochem Biophys Res Commun. 2002;296:1194–1199. doi: 10.1016/S0006-291X(02)02046-6. [DOI] [PubMed] [Google Scholar]
- Turk V, Bode W. The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett. 1991;285:213–219. doi: 10.1016/0014-5793(91)80804-C. [DOI] [PubMed] [Google Scholar]
- Ülker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Urwin PE, Green J, Atkinson HJ. Expression of a plant cystatin confers partial resistance to Globodera, full resistance is achieved by pyramiding a cystatin with natural resistance. Mol Breeding. 2003;12:263–269. doi: 10.1023/A:1026352620308. [DOI] [Google Scholar]
- Van der Vyver C, Schneidereit J, Driscoll S, Turner J, Kunert K, Foyer CH. Oryzacystatin I expression in transformed tobacco produces a conditional growth phenotype and enhances chilling tolerance. Plant Biotechnol J. 2003;1:101–112. doi: 10.1046/j.1467-7652.2003.00010.x. [DOI] [PubMed] [Google Scholar]
- Wang KM, Kumar S, Cheng YS, Venkatagiri S, Yang AH, Yeh KW. Characterization of inhibitory mechanism and antifungal activity between group-1 and group-2 phytocystatins from taro (Colocasia esculenta) FEBS J. 2008;275:4980–4989. doi: 10.1111/j.1742-4658.2008.06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Lis JT. Germline transformation used to define key features of heat-shock response elements. Science. 1988;239:1139–1142. doi: 10.1126/science.3125608. [DOI] [PubMed] [Google Scholar]
- Yamada K, Nishimura M, Hara-Nishimura I. The slow wound-response of γ-VPE is regulated by endogenous salicylic acid in Arabidopsis. Planta. 2004;218:599–605. doi: 10.1007/s00425-003-1131-3. [DOI] [PubMed] [Google Scholar]
- Yang AH, Yeh KW. Molecular cloning, recombinant gene expression, and antifungal activity of cystatin from taro (Colocasia esculenta cv. Kaosiung no. 1) Planta. 2005;221:493–501. doi: 10.1007/s00425-004-1462-8. [DOI] [PubMed] [Google Scholar]
- Yang KA, Lim CJ, Hong JK, Park CY, Cheong YH, Chung WS, Lee KO, Lee SY, Cho MJ, Lim CO. Identification of cell wall genes modified by a permissive high temperature in Chinese cabbage. Plant Sci. 2006;171:175–182. doi: 10.1016/j.plantsci.2006.03.013. [DOI] [Google Scholar]
- Zhang X, Liu S, Takano T. Two cysteine proteinase inhibitors from Arabidopsis thaliana, AtCYSa and AtCYSb, increasing the salt, drought, oxidation and cold tolerance. Plant Mol Biol. 2008;68:131–143. doi: 10.1007/s11103-008-9357-x. [DOI] [PubMed] [Google Scholar]


