Abstract
Fluorescence pulse width can provide size information on the fluorescence-emitting particle, such as the nuclei of propidium iodide-stained cells. To analyze nuclear size in the present study, rather than perform the simple doublet discrimination approach usually employed in flow cytometric DNA content analyses, we assessed the pulse width of the propidium iodide fluorescence signal. The anti-cancer drug etoposide is reportedly cytostatic, can induce a strong G2/M arrest, and results in nuclear enlargement. Based on these characteristics, we used etoposide-treated HCT116 cells as our experimental model system. The fluorescence pulse widths (FL2-W) of etoposide-treated (10 μM, 48 h) cells were distributed at higher positions than those of vehicle control, so the peak FL2-W value of etoposide-treated cells appeared at 400 while those of vehicle control cells appeared at 200 and 270. These results were consistent with our microscopic observations. This etoposide-induced increase in FL2-W was more apparent in G2/M phase than other cell cycle phases, suggesting that etoposide-induced nuclear enlargement preferentially occurred in G2/M phase cells rather than in G0/G1 or S phase cells.
Electronic supplementary material
The online version of this article (doi:10.1007/s10529-010-0277-x) contains supplementary material, which is available to authorized users.
Keywords: Cell cycle, Etoposide, Flow cytometry, Fluorescence, Nuclear size, Pulse width
Introduction
Flow cytometry is a staple methodology in biological research that is frequently employed in the development of cancer chemotherapeutic and chemopreventive agents, chiefly to evaluate biological activities and molecular mechanisms. The basic parameters of flow cytometry include forward (FSC) and side (SSC) scatter, which provide information on the cell size and cell internal complexity, respectively. When cells are stained with a fluorescent dye for DNA content analysis (e.g., propidium iodide, PI), their emitted fluorescence can serve as a pulse signal, and the signal height (FL2-H), width (FL2-W), and area (FL2-A) can be determined (Fig. 1). In the case of cell cycle analysis, FL2-A is the main parameter, and the histogram plot of FL2-A serves as a cell cycle graph. FL2-W is often used to discriminate singlet and aggregated cells through gating on the singlet cell region of the FL2-A vs. FL2-W dot plot (Hoffman 2009; Wersto et al. 2001). However, FL2-W can also be used for other purposes such as size evaluation of the fluorescence-emitting particle. Recently, researchers have examined the calf thymocyte nuclear size using fluorescence pulse width analysis (Hoffman 2009). However, the flow cytometric analysis of fluorescence pulse width for biological particle sizing remains unpopular, with limited published data (Hoffman 2009; Kirkhus et al. 1992; Leary et al. 1979).
Fig. 1.
Schematic diagram of fluorescence pulse signal (height, width, and area)
Etoposide, a famous anti-cancer drug, has been used to treat a wide spectrum of human cancers. Etoposide induces a strong G2/M arrest in various cancer cells (Rudolf and Cervinka 2003; Zhu et al. 2009). Due to the resulting abnormal mitotic features, etoposide reportedly also induces the formation of abnormal giant cells in cervical cancer and Epstein-Barr virus (EBV)-infected lymphocytes (Dedov et al. 2003; Rello-Varona et al. 2006).
In this study, we applied a flow cytometric analysis of fluorescence pulse width to evaluate the nuclear size. This analysis goes beyond the traditional purpose of the method, which is to discriminate between aggregated and singlet cells. We used etoposide-induced HCT116 cell nuclear enlargement as our experimental model system. We also evaluated the effect of the cell cycle phase on etoposide-induced nuclear enlargement through fluorescence pulse width analysis.
Materials and methods
Materials
Etoposide and PI were purchased from Sigma. Etoposide was dissolved in dimethyl sulfoxide (DMSO) for cellular treatment. Antibodies against ATM, p-ATM (Ser1981), α-tubulin, and GAPDH were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibodies against β-actin and lamin B1 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and Abcam (Cambridge, UK), respectively.
Cell culture
HCT116 and DLD-1 cells were obtained from the American Type Culture Collection (Rockville, MD, USA). HCT116 and DLD-1 cells were maintained in minimal essential medium (MEM) supplemented with 10% (v/v) fetal bovine serum, 100 units penicillin/ml, and 100 μg streptomycin/ml. Cells were cultured in a humidified atmosphere (95% air, 5% CO2) at 37°C.
Cell viability assay
HCT116 (2 × 103 cells per well) cells were plated into 96-well plates, incubated at 37°C for 24 h, and treated with etoposide. Cell viability was measured using the EZ-cyTox Cell Viability Assay Kit (Daeil Lab Service, Seoul, Korea) as previously described (Kang et al. 2007).
Cell cycle analysis
HCT116 and DLD-1 cells (5 × 105) were seeded on 60 mm dishes, incubated for 24 h, and then treated with etoposide. The cell cycle was evaluated by flow cytometric DNA content analysis using Modfit LT 3.0 software, as previously described (Kang et al. 2007).
Microscopy and measurement of nuclear size
Cell morphological changes were crudely observed using a phase contrast microscope. For precise observation of the nucleus, we further exploited confocal laser-scanning microscopy. Cells were fixed in 4% formaldehyde for 30 min, and nuclei were stained using a mounting medium supplemented with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). Stained cells were immediately observed using the Leica TCS SP5 system (Leica Microsystems, Heidelberg, Germany). Differential interference contrast (DIC) and DAPI fluorescence images were merged using the LAS AF software.
To measure the nuclear size, detached cells were stained with PI as previously described (Kang et al. 2007). Fluorescence images were obtained using a fluorescence microscope, and the ProgRes CFcool camera system (JENOPTIK, Jena, Germany). Nuclear size (diameter, μm) was determined using the circle measurement algorithm of the ProgRes Capture Pro 2.5 software (JENOPTIK, Jena, Germany).
Western blot analysis
HCT116 (5 × 105) cells were seeded on 60 mm dishes and incubated for 24 h, and then treated with etoposide. Total cell lysates were prepared and subjected to electrophoresis on 10% SDS–PAGE Tris/HCl gels. Additional procedures were performed as previously described (Kang et al. 2009).
Flow cytometry
The fluorescence pulse signal was analyzed using a flow cytometer. Cells were stained with PI as previous described (Kang et al. 2007). FL-2 voltages were adjusted such that the G0/G1, G2 + M peaks had values of 200 and 400, respectively, on FL2-A. A total of 20,000 events were collected for each sample. PI fluorescence pulses were detected using a laser beam at 488 nm and band pass filters at 585/45. For every event, FSC-H, SSC-H, FL2-A, and FL2-W values were measured. The histogram plot and statistics tools of CellQuest Pro software were used for analysis.
Statistical analysis
Data were expressed as means ± SDs. The statistical significance of the differences was determined by Student’s t-test. P values <0.01 were considered statistically significant. Kolmogorov–Smirnov statistics were used to compare sets of two flow cytometric histograms (Young 1977).
Results
Etoposide induces a strong G2/M arrest and formation of giant cell shapes in HCT116 cells
Although it was previously shown that etoposide induces G2/M arrest in various cells and triggers giant cell formation in cervical cancer and EBV-lymphocytes (Dedov et al. 2003; Rello-Varona et al. 2006; Zhu et al. 2009), we reconfirmed these characteristics in HCT116 human colon cancer cells. Etoposide did not significantly inhibit cell proliferation in HCT116 cells. Etoposide treatment (10 μM) for 48 h resulted in only a 30% inhibition of cell viability compared with vehicle control (Fig. 2a). However, treatment for 24 or 48 h triggered a strong, dose-dependent G2/M arrest in HCT116 cells (Fig. 2b). Etoposide triggered formation of giant cell shapes in HCT116 cells, in which HCT116 cells treated with etoposide (10 μM) for 24 and 48 h were much larger than those treated with vehicle control (Fig. 2c). Phase contrast microscopy also suggested that the cell nuclear size increased with etoposide treatment. In addition, we also observed etoposide-induced G2/M arrest and cell enlargement in the p53-mutated colon cancer cell line, DLD-1 (Supplementary Fig. 1a, b).
Fig. 2.
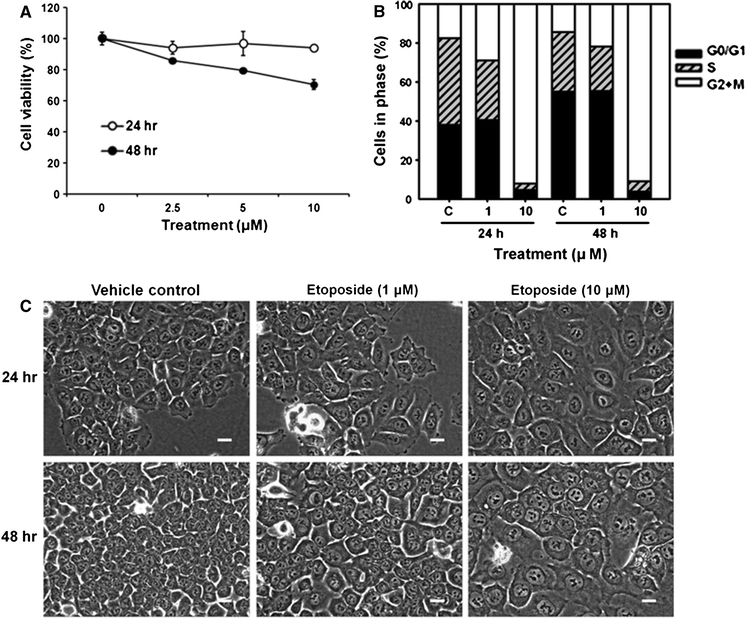
Effects of etoposide on viability, cell cycle distribution, and cellular morphology of HCT116 cells. Cells were treated with etoposide (0–10 μM) for 24 or 48 h. a Cell proliferation was determined using the MTT assay. Data are expressed as the mean ± SD of triplicate experiments. ** P < 0.001 and * P < 0.01 compared to vehicle control. Graph is a representative from three independent experiments. b Cell cycle distribution determined by measuring PI-labeled DNA content using flow cytometry. C vehicle control. Graph is a representative from three independent experiments. c Phase contrast microscope images of HCT116 cells (bar = 20 μm). Representative pictures are shown from at least three independent experiments
Etoposide induces HCT116 cell nuclear enlargement
To more precisely observe the cell nuclei, we performed confocal laser-scanning microscopy after DAPI staining. Etoposide clearly induced the enlargement of both the cell and nuclear sizes (Fig. 3a). We determined the nuclear size quantitatively using fluorescent microscopy after PI staining, according to the circle measurement algorithm of the microscope software, and found that etoposide dose-dependently increased nuclear size at 24 and 48 h of treatment (Fig. 3b). Because etoposide induced nuclear and cell enlargements, we examined the expression levels of cytoskeleton proteins using Western blot analysis. Etoposide triggered the DNA damage signaling pathway in HCT116 cells, as reported previously (Zhu et al. 2009). Etoposide increased both ATM expression and phosphorylation at Ser1981, while the β-actin and α-tubulin expression levels were unaltered by etoposide. Only the expression of lamin B1, which is an important nuclear membrane filament, was decreased by etoposide treatment (Fig. 3c).
Fig. 3.
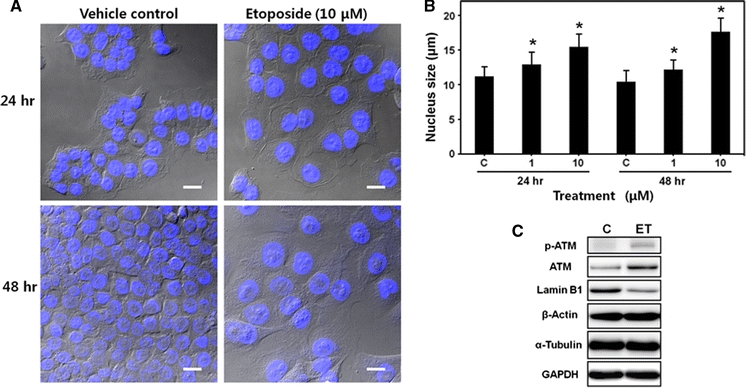
Enlargement of cell and nucleus by etoposide treatment in HCT116 cells. a Confocal microscope images of HCT116 cells treated with etoposide (10 μM) for 24 or 48 h. Cells were fixed and stained with DAPI. Differential interference contrast (DIC) images were superimposed on DAPI fluorescence images (bar = 20 μm). b Measurement of nuclear size. HCT116 cells treated with etoposide (0–10 μM) for 24 or 48 h, fixed, and stained with PI. Nuclei size was determined by fluorescence microscopy and the circle measurement algorithm of the microscope software. Each bar represents the mean ± SD (n = 100). * P < 0.001 compared to vehicle control, C. c Expressions of ATM, lamin B1, β-actin, α-tubulin, and GAPDH in HCT116 cells determined by Western blot analysis. ATM phosphorylation was also measured. Cells were treated with etoposide (ET, 10 μM) for 24 h. C vehicle control. Representative data are shown from two independent experiments
Flow cytometric analysis of PI fluorescence width reveals HCT116 cell nuclear enlargement following etoposide treatment
We used the PI fluorescence pulse width to analyze the nuclear size in HCT116 cells. We divided the region into four portions in the FL2-A vs. FL2-W dot plot of vehicle control cells (R1, singlet cells in G0/G1 to S phases [2N–3N]; R2, aggregated cells such as doublets; R3, singlet cells in S to G2/M phases [3N–4N]; R4, overall region, R1 + R2 + R3) (Supplementary Fig. 2a). We reconfirmed the cell cycle distribution of each gated region in the FL2-A histogram plot (Supplementary Fig. 2b). The FSC-H values of the R3 gated cells were distributed at higher positions than those of the R1 gated cells (Supplementary Fig. 2b, FSC-H histogram plot). The SSC-H value of each gated region was distributed similarly (Supplementary Fig. 2b, SSC-H histogram plot). The FL2-W values of the R3 gated cells were distributed at higher positions than those of the R1 gated cells. The aggregated cells (R2 gated cells) showed a flat peak of FL2-W, with values that were higher than those of the R3 gated cells (Supplementary Fig. 2b, FL2-W histogram plot). These results indicate that the nuclear sizes of cells in the G2/M phase were larger than those of cells in the G0/G1 phase. This may be due to the fact that cells naturally enlarge their cellular components, including DNA and proteins, in preparation for mitosis. Based on this observation, we proposed that the flow cytometric measurement of PI fluorescence could be used to analyze the cell nuclear size.
We further applied the flow cytometric analysis of the PI fluorescence pulse width to evaluate nuclear enlargement by etoposide in HCT116 cells. We divided the region into four portions in the FL2-A vs. FL2-W dot plot based on the cell cycle phase (R1, G0/G1 phase; R2, S phase; R3, G2/M phase; R4, overall region, R1 + R2 + R3) (Fig. 4a). First, we analyzed the FSC-H, SSC-H, FL2-A, and FL2-W values in all cells (R4 gated region) (Fig. 4b). The FSC-H and SSC-H values of the etoposide-treated (10 μM, 48 h) HCT116 cells were 1.4- and 3.2-fold higher, respectively, than those of vehicle control (Fig. 4b, FSC-H and SSC-H histogram plots). The FL2-A histogram plot reconfirmed that etoposide induced G2/M arrest (Fig. 4b, FL2-A histogram plot). The peak FL2-W values of etoposide-treated (10 μM, 48 h) HCT116 cells appeared at about 400, while those of vehicle control cells appeared at about 200 and 270 (Fig. 4b, FL2-W histogram plot). This etoposide-induced increase in FL2-W is consistent with our microscopic observations (Fig. 2a, b), and was also observed at 24 h of etoposide treatment (data not shown) and in DLD-1 cells (Supplementary Fig. 1c).
Fig. 4.
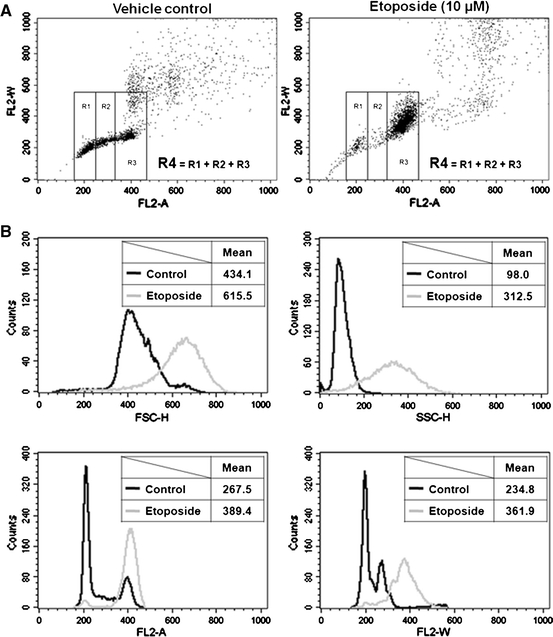
Flow cytometric analysis of PI fluorescence pulse width to evaluate etoposide-induced nuclear enlargement. HCT116 cells were treated with etoposide (10 μM) for 48 h, harvested, fixed in 70% ethanol, and stained with PI. Representative data are shown from three independent experiments. a FL2-A vs. FL2-W dot plot (R1, G0/G1 phase cells; R2, S phase cells; R3, G2 + M phase cells; R4, overall cells, R1 + R2 + R3). b Histogram plots of FSC-H, SSC-H, FL2-A, and FL2-W with gating on R4. Overlay histograms are schematized using vehicle control (dark) and etoposide-treated (48 h) (gray) cell data. Mean values of FSC-H, SSC-H, FL2-A, and FL2-W were calculated using the histogram statistics tool of CellQuest Pro software
Etoposide-induced nuclear enlargement preferentially occurred in G2/M phase cells
To evaluate the effect of the cell cycle phase on etoposide-induced nuclear enlargement, we compared the FL2-W distribution of vehicle control and etoposide-treated cells gated on each cell cycle phase (R1, R2 and R3; as described in Fig. 4a). The FL2-W values of etoposide-treated cells were distributed at higher positions than those of vehicle control with gating on R3 (G2/M phase cells), but were distributed similarly with gating on R1 or R2 (G0/G1 or S phase cells, respectively) (Fig. 5a). These data imply that etoposide-induced nuclear enlargement occurs in the G2/M phase rather than the G0/G1 or S phase. Similar results were also obtained after 24 h of etoposide treatment (data not shown).
Fig. 5.
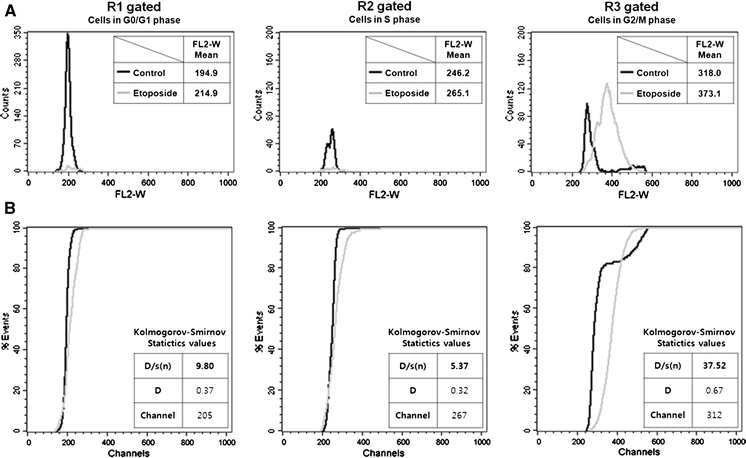
Effects of cell cycle phase on etoposide-induced nuclear enlargement. FL2-W distributions were compared between vehicle control and etoposide-treated cells (10 μM, 48 h). Representative data sets are shown from three independent experiments. a Histogram plots of FL2-W, schematized from cells in each cell cycle phase (left: R1 gated, G0/G1 phase cells; middle: R2 gated, S phase cells; right: R3 gated, G2 + M phase cells; explained in Fig. 4). Overlay histograms schematized using vehicle control (dark) and etoposide-treated (gray) cell data. Mean values of FL2-W are represented in each plot. b Kolmogorov–Smirnov statistics were used to quantitatively compare the FL2-W distribution between vehicle control (dark) and etoposide-treated cells (gray) shown in (a). D/s(n), index of similarity for the two curves. If D/s(n) = 0, the curves are identical. D, greatest difference between the two curves. Channel, channel number of D. P values <0.001 for all data, which is the probability of D being as large as it is assuming that the two selected histogram are from the same population. Each value was calculated using the Kolmogorov–Smirnov statistics tool of CellQuest Pro software
We also quantitatively compared the flow cytometric histograms using Kolmogorov–Smirnov statistics. The D/s(n) value reflects the index similarity for two histograms, such that a higher D/s(n) value indicates a less-similar distribution. The D/s(n) value of cells in the G2/M phase (37.52) was much higher than that of cells in the G0/G1 (9.80) or S phase (5.37) (Fig. 5b). For a more precise comparison, we again calculated the values for vehicle control cells in the G2/M phase after excluding doublet cells (since R3 also contained some doublet cells), and obtained a D/s(n) value of 43.54. Thus, the FL2-W distribution of the etoposide-treated cells was definitely different from that of vehicle control (Supplementary Fig. 3).
Discussion
In flow cytometric cell cycle analysis, the FL2-W value is predominantly used to discriminate between singlet and aggregated cells. However, the pulse width of a fluorescence signal can also provide size information (Hoffman 2009). Here we analyzed the FL2-W values to characterize cell nuclear size, using etoposide-induced nuclear enlargement in HCT116 human colon cancer cells as an experimental model system. This method is convenient and less labor-intensive than microscopic observation, since the pulse width data of DNA fluorescence can be easily obtained after PI staining. Thus, through flow cytometric DNA content analysis, we can simultaneously evaluate both the cell cycle and nuclear size distributions.
We observed that the pulse width of PI fluorescence from etoposide-treated cells was much higher than that of vehicle control, which correlated well with our microscopic observations. The etoposide-induced increase in FL2-W preferentially occurred in G2/M phase cells (Fig. 5), potentially because etoposide induced a strong G2/M phase arrest. Although the cells synthesized DNA and protein for cell division, they could not enter mitosis due to the arrest and were stuck at an abnormal phase with giant nuclei and cell shapes. From the observations of this study, we hypothesize that two properties may be essential for chemically induced nuclear enlargement. First, the chemical should be able to induce cell cycle arrest at late phases such as G2/M, rather than at early phases. Second, the chemical should not induce apoptosis rapidly: it should be cytostatic rather than cytotoxic. For example, although paclitaxel induced G2/M arrest, it also rapidly induced a strong apoptosis in HCT116 cells, and could not induce nuclear enlargement (data not shown). In contrast, etoposide is both cytostatic and induces G2/M arrest. In the future, we plan to evaluate other drugs, such as hydroxyurea and paclitaxel, in association with inhibiting the apoptotic pathway for nuclear size changes.
The FSC-H and SSC-H values were higher when cells were treated with etoposide than in vehicle control cells, indicating that cell size and cellular granularity were increased by etoposide. These findings coincide with a previous report that etoposide induces increased cell mass and protein expression, including that of actin proteins (Dedov et al. 2003; Perez et al. 1997; Rello-Varona et al. 2006). The results also suggest that the increased presence and reorganization of cytoskeletal proteins might contribute to the cell size enlargement. Western blot analyses showed that α-tubulin and β-actin were not changed by etoposide treatment, perhaps because the Western blot showed the ratio of the protein expression in the overall cells. The expression of proteins, including cytoskeletal proteins, in one cell should be increased by etoposide treatment (Min et al. 2006). Lamin B1 expression was noticeably decreased by etoposide. Lamins are the major architectural proteins of nuclear envelope, and are critical for nuclear integrity and assembly (Parnaik 2008), implying that etoposide might inhibit nuclear membrane formation due to the uncontrolled enlargement of the nucleus. Another possibility is that the decreased lamin B1 expression might have been triggered by the preparation of apoptosis induction, similar to its reported response in apoptotic human leukemia cells (Fadeel et al. 1999).
In conclusion, we measured the PI fluorescence pulse width, typically presented as FL2-W, by flow cytometry to analyze the nuclear size. The etoposide-induced nuclear enlargement in HCT116 cells was used as the experimental model system, because etoposide induces cytostasis and a strong G2/M arrest. The PI fluorescence width of the etoposide-treated cells was increased compared to vehicle control cells, in agreement with our microscopic observations. Furthermore, the etoposide-induced nuclear enlargement preferentially occurred in G2/M phase cells. Our findings provide insights into the mechanisms underlying etoposide-induced nuclear enlargement. We suggest that our modified fluorescence pulse width analysis method could be used for nuclear size evaluation, as a useful tool for the functional study of cancer therapies and prevention agents.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by an intramural grant (2Z03300) from Korea Institute of Science and Technology, Gangneung Institute.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Dedov VN, Dedova IV, Nicholson GA. Inhibition of topoisomerase II overrides the G2/M check points of the cell cycle in EBV-lymphocytes. Apoptosis. 2003;8:399–406. doi: 10.1023/A:1024133304637. [DOI] [PubMed] [Google Scholar]
- Fadeel B, Hassan Z, Hellstrom-Lindberg E, Henter JI, Orrenius S, Zhivotovsky B. Cleavage of Bcl-2 is an early event in chemotherapy-induced apoptosis of human myeloid leukemia cells. Leukemia. 1999;13:719–728. doi: 10.1038/sj/leu/2401411. [DOI] [PubMed] [Google Scholar]
- Hoffman RA (2009) Pulse width for particle sizing. Curr Protoc Cytom Chapter 1:Unit 1.23 [DOI] [PubMed]
- Kang K, Lee HJ, Kim CY, Lee SB, Tunsag J, Batsuren D, Nho CW. The chemopreventive effects of Saussurea salicifolia through induction of apoptosis and phase II detoxification enzyme. Biol Pharm Bull. 2007;30:2352–2359. doi: 10.1248/bpb.30.2352. [DOI] [PubMed] [Google Scholar]
- Kang K, Lee SB, Jung SH, Cha KH, Park WD, Sohn YC, Nho CW. Tectoridin, a poor ligand of estrogen receptor alpha, exerts its estrogenic effects via an ERK-dependent pathway. Mol Cells. 2009;27:351–357. doi: 10.1007/s10059-009-0045-8. [DOI] [PubMed] [Google Scholar]
- Kirkhus B, Glaso M, Clausen OP. Multivariate flow cytometry of epidermal regeneration provoked by a skin irritant and a tumor promoter. Cytometry. 1992;13:267–274. doi: 10.1002/cyto.990130308. [DOI] [PubMed] [Google Scholar]
- Leary JF, Todd P, Wood JC, Jett JH. Laser flow cytometric light scatter and fluorescence pulse width and pulse rise-time sizing of mammalian cells. J Histochem Cytochem. 1979;27:315–320. doi: 10.1177/27.1.374594. [DOI] [PubMed] [Google Scholar]
- Min DJ, Moskowitz NP, Brownstein C, Lee H, Horton TM, Carroll WL. Diverse pathways mediate chemotherapy-induced cell death in acute lymphoblastic leukemia cell lines. Apoptosis. 2006;11:1977–1986. doi: 10.1007/s10495-006-0081-1. [DOI] [PubMed] [Google Scholar]
- Parnaik VK. Role of nuclear lamins in nuclear organization, cellular signaling, and inherited diseases. Int Rev Cell Mol Biol. 2008;266:157–206. doi: 10.1016/S1937-6448(07)66004-3. [DOI] [PubMed] [Google Scholar]
- Perez C, Vilaboa NE, Garcia-Bermejo L, de Blas E, Creighton AM, Aller P. Differentiation of U-937 promonocytic cells by etoposide and ICRF-193, two antitumour DNA topoisomerase II inhibitors with different mechanisms of action. J Cell Sci. 1997;110:337–343. doi: 10.1242/jcs.110.3.337. [DOI] [PubMed] [Google Scholar]
- Rello-Varona S, Gamez A, Moreno V, Stockert JC, Cristobal J, Pacheco M, Canete M, Juarranz A, Villanueva A. Metaphase arrest and cell death induced by etoposide on HeLa cells. Int J Biochem Cell Biol. 2006;38:2183–2195. doi: 10.1016/j.biocel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Rudolf E, Cervinka M. Topoisomerases and tubulin inhibitors: a promising combination for cancer treatment. Curr Med Chem Anticancer Agents. 2003;3:421–429. doi: 10.2174/1568011033482242. [DOI] [PubMed] [Google Scholar]
- Wersto RP, Chrest FJ, Leary JF, Morris C, Stetler-Stevenson MA, Gabrielson E. Doublet discrimination in DNA cell-cycle analysis. Cytometry. 2001;46:296–306. doi: 10.1002/cyto.1171. [DOI] [PubMed] [Google Scholar]
- Young IT. Proof without prejudice: use of the Kolmogorov–Smirnov test for the analysis of histograms from flow systems and other sources. J Histochem Cytochem. 1977;25:935–941. doi: 10.1177/25.7.894009. [DOI] [PubMed] [Google Scholar]
- Zhu H, Miao ZH, Huang M, Feng JM, Zhang ZX, Lu JJ, Cai YJ, Tong LJ, Xu YF, Qian XH, Ding J. Naphthalimides induce G(2) arrest through the ATM-activated Chk2-executed pathway in HCT116 cells. Neoplasia. 2009;11:1226–1234. doi: 10.1593/neo.09986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


