Abstract
While recurrent gene fusions involving ETS family transcription factors are common in prostate cancer, their products are considered “undruggable” by conventional approaches. Recently, rare “targetable” gene fusions (involving the ALK kinase), have been identified in 1–5% of lung cancers1, suggesting that similar rare gene fusions may occur in other common epithelial cancers including prostate cancer. Here we employed paired-end transcriptome sequencing to screen ETS rearrangement negative prostate cancers for targetable gene fusions and identified the SLC45A3-BRAF and ESRP1-RAF1 gene fusions. Expression of SLC45A3-BRAF or ESRP1-RAF1 in prostate cells induced a neoplastic phenotype that was sensitive to RAF and MEK inhibitors. Screening a large cohort of patients, we found that although rare (1–2%), recurrent rearrangements in the RAF pathway tend to occur in advanced prostate cancers, gastric cancers, and melanoma. Taken together, our results emphasize the importance of RAF rearrangements in cancer, suggest that RAF and MEK inhibitors may be useful in a subset of gene fusion harboring solid tumors, and demonstrate that sequencing of tumor transcriptomes and genomes may lead to the identification of rare targetable fusions across cancer types.
Recurrent gene fusions characterized by 5′ genomic regulatory elements (most commonly controlled by androgen) fused to members of the ETS family of transcription factors are present in at least half of all prostate cancers2,3. Unfortunately, such rearrangements involving oncogenic transcription factors are considered poor therapeutic targets by conventional pharmaceutical approaches, unlike rearrangements involving protein kinases. The recent identification of rearrangements involving a protein kinase (EML4-ALK) in a rare subset of non-small cell lung carcinomas, and preclinical and phase I/II clinical data suggesting that these patients respond to investigational ALK inhibitors1,4, demonstrates that rare “druggable” rearrangements may exist in small subsets of patients across common solid tumors.
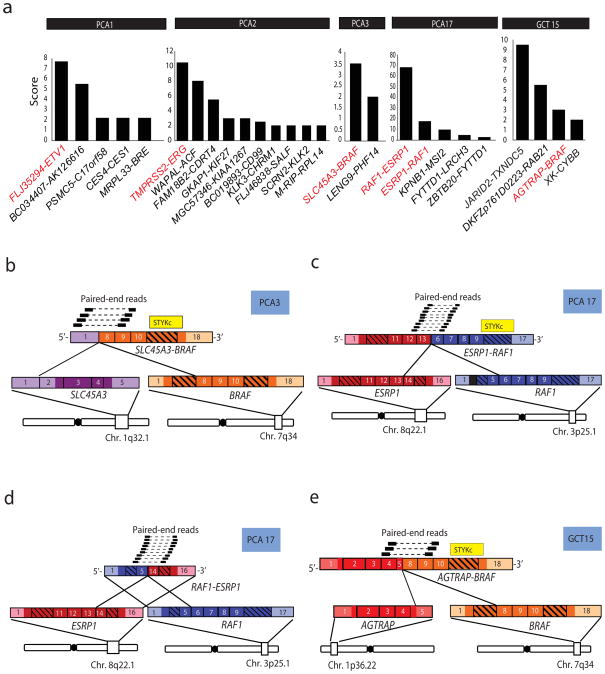
To search for such “druggable” rearrangements in prostate cancer, we employed paired-end, massively parallel transcriptome sequencing to prioritize candidate gene fusions in prostate tumors. We developed a prioritization strategy, which generates a score derived from the quantity of mate-pair reads that meet a series of computational filters implemented to reduce potential false positive chimera nominations5. As shown in Fig. 1a, prioritization histograms for two ETS rearrangement positive prostate cancers, PCA1 and PCA2, which harbor FLJ35294-ETV1 and TMPRSS2-ERG gene fusions, respectively, demonstrate that the ETS gene fusion had the highest score in each sample, as we have reported previously5,6.
Fig. 1. Discovery of the SLC45A3-BRAF and ESRP1-RAF1 gene fusions in prostate cancer by paired-end transcriptome sequencing.
a, Histograms of gene fusion nomination scores in clinically localized prostate tumor samples PCA1, PCA2, PCA3, and PCA17 harboring FLJ35294-ETV1, TMPRSS2-ERG, SLC45A3-BRAF, ESRP1-RAF1 and RAF1-ESRP1, respectively, and a gastric cancer sample GCT15 harboring AGTRAP-BRAF. Co-occurring fusions in each sample are also indicated. Data from ETV1 and ERG fusions are provided as controls derived from paired-end transcriptome data presented in a previous study5. b, Schematic representation of reliable paired-end reads supporting the inter-chromosomal gene fusion between SLC45A3 (purple) and BRAF (orange). The protein kinase domain in the BRAF gene (yellow) remains intact following the fusion event. Respective exons are numbered. c, d, As in b, except showing the fusions between ESRP1 (red) and RAF1 (blue), resulting in reciprocal fusion genes ESRP1-RAF1 and RAF1-ESRP1. e, As in b, except showing the fusion between AGTRAP (red) and BRAF (orange).
In this study, we sequenced 5 ETS gene fusion positive and 10 ETS gene fusion negative prostate cancers (ETS gene fusion status was determined by Fluorescence In Situ Hybridization (FISH) and/or qRT-PCR and found that two ETS negative samples, PCA3 and PCA17, each prioritized a fusion involving BRAF and RAF1 genes, key serine/threonine kinase components of the RAF signaling pathway (Fig. 1a).
While activating somatic mutations in the RAF kinase pathway, such as BRAFV600E are common in melanoma, thyroid, colon and ovarian cancers7–10, activating gene fusions of pathway members have been reported less frequently and found in subsets of relatively rare cancers11,12,13. Importantly, the RAF kinase pathway is druggable, with multiple approved and investigational agents in late stage development. Sorafenib, an FDA approved drug, was originally identified as a RAF kinase inhibitor, but was subsequently found to target other kinases such as VEGFR-2, VEGFR-3 and PDGFR-β14. An emerging lead drug candidate, PLX-4032, appears to be highly selective for the BRAFV600E mutation and is being evaluated in patients with advanced melanoma15. Thus, we proceeded to characterize and validate the potentially druggable gene fusions we identified in prostate tumors PCA3 and PCA17.
The first case, PCA3, revealed an inter-chromosomal rearrangement resulting in the fusion of untranslated exon 1 of SLC45A3 with exon 8 of BRAF (Fig. 1b). Importantly, SLC45A3 is a prostate-specific, androgen responsive gene which has been found fused to ERG16,17, ETV118, ETV519 and ELK420,21 in a subset of prostate tumors. The predicted open reading frame encodes for 329 amino acids of the C-terminal portion of BRAF (Supplementary Fig. 1a), retaining the kinase domain but losing the N-terminal RAS binding domain, suggesting that the mutant protein may be constitutively active. Having inherited promoter regulatory elements from SLC45A3, this BRAF fusion is likely under androgen regulation (Supplementary Fig. 2). Consistent with this, the C-terminal exons of BRAF (8–18) present in the fusion are over-expressed in PCA3 relative to benign prostate and other prostate cancers (Supplementary Fig. 3a,b). The second case, PCA17, revealed two highly expressed gene fusions involving ESRP1 and RAF1 (Fig. 1c,d) presumably formed by a balanced reciprocal translocation. ESRP1 is a splicing factor that regulates the formation of epithelial cell-specific isoforms of mRNA22, while RAF1 (or CRAF) is a serine/threonine protein kinase.
The ESRP1-RAF1 fusion transcript involves the fusion of exon 13 of ESRP1 to exon 6 of RAF1 (Fig. 1c). The predicted open reading frame encodes a 120 kDa fusion protein comprised of the majority of ESRP1, including its 3 RNA recognition motifs, fused to the C-terminal kinase domain of RAF1 (Supplementary Fig. 1c). Loss of the RAS-binding domain of RAF1 suggests that this fusion protein may be constitutively active, while the significance of the RNA binding domains of ESRP1 is unclear.
In addition to ESRP1-RAF1, we also detected the reciprocal gene fusion RAF1-ESRP1, produced from the same genomic rearrangement in PCA17. The RAF1-ESRP1 transcript involves the fusion of exon 5 of RAF1 with exon 14 of ESRP1 (Fig. 1d) which encodes a predicted 30kDa protein comprised of the RAS binding domain of RAF1 fused to 194 amino acids from the C-terminus of ESRP1 (Supplementary Fig. 1c). Unlike SLC45A3-BRAF, ESRP1-RAF1 is predicted not to be regulated by androgen since wild-type ESRP1 is not androgen regulated (Supplementary Fig. 2).
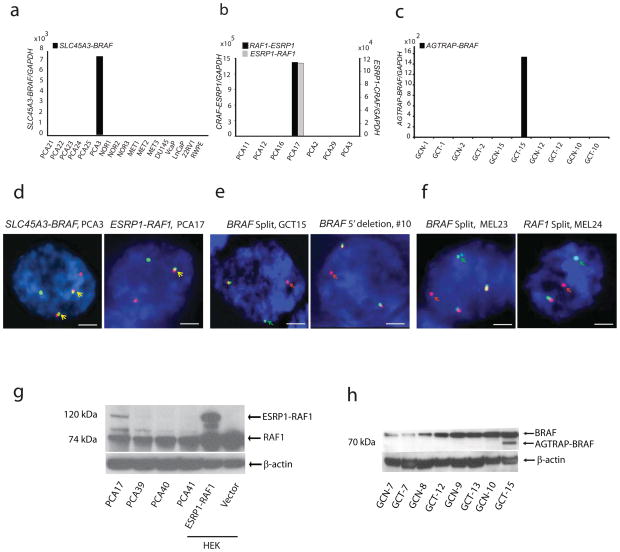
Next, the SLC45A3-BRAF fusion was validated by fusion specific qPCR in PCA3 (Fig. 2a). Rearrangement at the DNA level was validated by FISH and confirmed the presence of two copies of rearranged chromosomes by break apart (Supplementary Fig. 4a) and fusion assays (Fig. 2d, left). Expression of the SLC45A3-BRAF fusion gene in HEK293 cells and stable expression in RWPE prostate epithelial cells generated a 37kDa protein (Supplementary Fig. 5a,b).
Fig. 2. Experimental validation of the SLC45A3-BRAF, ESRP1-RAF1 and RAF1-ESRP1 and AGTRAP-BRAF gene fusions.
qRT-PCR validation of a) SLC45A3-BRAF gene fusion in PCA3, b) ESRP1-RAF1 and RAF1-ESRP1 fusions in PCA17, and c) AGTRAP-BRAF fusion in GCT15. d, FISH validation of SLC45A3-BRAF (left) and ESRP1-RAF1 (right) gene fusions in PCA3 and PCA17, respectively. The individual green and red signals indicate the normal chromosomes 1 and 7 (SLC45A3 and BRAF, respectively) in PCA3 and chromosomes 8 and 3 (ESRP1 and RAF1, respectively) in PCA17. Co-localizing green and red signals (yellow signal and arrow) indicate the fusion event. Tumor PCA3 displays two copies of the rearranged chromosome. e, FISH validation of the BRAF rearrangement in GCT15. Individual green and red signals (arrows) indicate rearrangement and co-localizing yellow signals indicate the normal chromosome. A second case (#10) showed deletion of the 5′ region probe with an intact 3′ probe. f. FISH validation of the BRAF rearrangement in melanoma cases MEL23 and RAF1 rearrangement in melanoma case MEL24. Individual green and red signals (arrows) indicate the rearrangement and co-localizing yellow signals indicate the normal chromosome. Scale bars indicate 2μm. g, Expression of the 120kDa ESRP1-RAF1 fusion protein in the index case PCA17. ESRP1-RAF1 fusion was detected by an antibody against C-terminus of RAF1. HEK293 cells expressing ESRP1-RAF1 fusion served as a positive control. β-actin serves as a loading control. H, Expression of a 70kDa AGTRAP-BRAF fusion protein in GCT15.
Similarly, ESRP1-RAF1 and RAF1-ESRP1 was validated by qRT-PCR (Fig. 2b) in the index case PCA17. FISH confirmed the DNA level rearrangement and fusion of the ESRP1 and RAF1 loci (Fig. 2d, right, Supplementary Fig. 4b). Expression of a 120 kDa ESRP1-RAF1 fusion protein was observed in PCA17, and upon over-expression in HEK293 (Fig. 2g) and RWPE cells (Supplementary Fig. 5c).
BRAF and RAF1 rearrangement frequencies in three independent prostate cancer clinical cohorts were estimated by FISH on tissue microarrays (TMAs) using break-apart probes. Out of 349 prostate cancer cases that were evaluable by FISH, 6 cases displayed an aberration at the BRAF locus (5 rearrangements and 1 deletion of the 5′ probe) and 4 of 450 cases displayed rearrangement at the RAF1 locus (1 rearrangement and 3 deletions of the 3′ probe). Other than the index cases PCA3 and PCA17, these cases did not display rearrangement of the SLC45A3 or ESRP1 loci suggesting fusions involving multiple 5′ partners, similar to ETV1 fusions in prostate cancer18. Due to the lack of availability of frozen tissue we were unable to characterize the 5′ fusion partners in these specific cases. Importantly, a majority of the cases that were positive for rearrangements of BRAF or RAF1 had aggressive features including high Gleason score and exhibited castration-resistance. All the cases were negative for ETS gene rearrangement (except MET37 which had an ERG rearrangement), suggesting that these aberrations occur predominantly in ETS negative prostate cancers (Supplementary Table 1a).
We extended the analysis of BRAF and RAF1 rearrangements to other solid tumors using break apart FISH probes on TMAs of breast (n=49), endometrial (n=26), gastric (n=85), melanoma (n=131), and liver tumors (n= 42). Similar to prostate cancer we found a 1–2% incidence of BRAF aberrations in gastric cancer (2/105) (Fig. 2e) and one case each of BRAF and RAF1 rearrangement in melanoma (2/131) (Fig. 2f). In the gastric cancer index case GCT-15, paired-end transcriptome sequencing revealed that exon 8 of the BRAF gene was fused with exon 5 of AGTRAP (Fig. 1e). We validated the AGTRAP-BRAF fusion transcript by qRT-PCR (Fig. 2c) and the DNA level rearrangement by FISH analysis (Fig. 2f). The AGTRAP-BRAF fusion resulted in the formation of a 597aa fusion protein with the C-terminal kinase domain of BRAF fused to the N-terminal angiotensin II type 1 receptor associated domain of AGTRAP (Supplementary Fig. 1d). Expression of the predicted AGTRAP-BRAF fusion protein was confirmed by immunoblot analysis of the index tumor GCT-15 (Fig. 2h). The 5′ partner gene for the second gastric case with BRAF rearrangement and BRAF and RAF1 rearrangement positive melanoma cases were not confirmed due to lack of frozen tissue (Supplementary Table 1b).
Considering the prevalence of oncogenic mutations in BRAF in different cancer types, we screened for the BRAFV600E mutation by pyrosequencing in 274 prostate samples, 23 gastric cancer samples, 2 gastroesophageal cancer samples and 34 melanoma samples. We found 20/34 (59%) melanoma samples, 1/25 gastroesophageal cancers and 0/274 prostate samples were positive for the BRAFV600 mutation. Importantly none of the RAF pathway gene rearrangement positive prostate cancers, gastroesophageal cancers and melanomas identified herein harbored the V600 mutations, suggesting genomic rearrangement, rather than mutation, as a mechanism for RAF gene activation in a subset of solid tumors. In an Asian cohort, 10% of prostate cancer cases have been reported to be positive for BRAFV600E mutations23. We were unable to find any BRAFV600 mutations in our prostate cohorts which is consistent with a recently published study (0/95 prostate cancers were positive for V600E)24. This discrepancy could reflect differences between the ethnic backgrounds of the various cohorts studied.
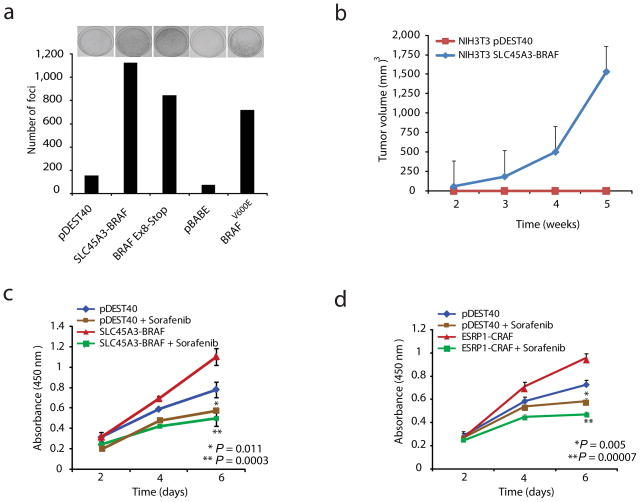
We next examined the functional relevance of these fusions involving RAF pathway members in prostate cancer. First, we examined the SLC45A3-BRAF fusion in NIH3T3 cells, a system classically used to study RAS/RAF biology25. Over-expression of SLC45A3-BRAF (Supplementary Fig. 1b) or mutant BRAFV600E showed a dramatic increase in the number of foci as compared to vector controls (Fig. 3a). The foci assay data was further validated by automated colony counting (Supplementary Fig. 6a). NIH3T3 cells over-expressing SLC45A3-BRAF formed rapidly growing tumors in nude mice (Fig. 3b); however NIH3T3 cells over-expressing ESRP1-RAF1 did not form tumors (data not shown), which may reflect signaling differences between the different fusion products.
Fig. 3. Oncogenic properties of SLC45A3-BRAF and ESRP1-RAF1 gene fusions.
a, Foci formation by SLC45A3-BRAF, BRAFV600E and vector controls (pDEST40 and pBABE) constructs in NIH3T3 cells. Representative plates are shown for each sample above the respective quantification of foci formation (from two independent experiments). b, Over-expression of SLC45A3-BRAF fusion transcript in NIH3T3 cells induces tumor formation in nude mice. Stable polyclonal NIH3T3 cells expressing SLC45A3-BRAF (5×106) were implanted subcutaneously into nude mice. Tumor growth was monitored weekly up to 5 weeks. The c) SLC45A3-BRAF and d) ESRP1-RAF1 fusions promote cell proliferation in RWPE prostate cells. Stable RWPE cells were treated with sorafenib (0.25μM) or DMSO vehicle and proliferation was monitored by WST-1 assay at indicated times. Error bars represent SEM. P-values represent Student’s t-test and the comparison was stable pDEST40 control cells. Sorafenib-treated SLC45A3-BRAF or ESRP1-RAF1 cells were compared with vehicle control.
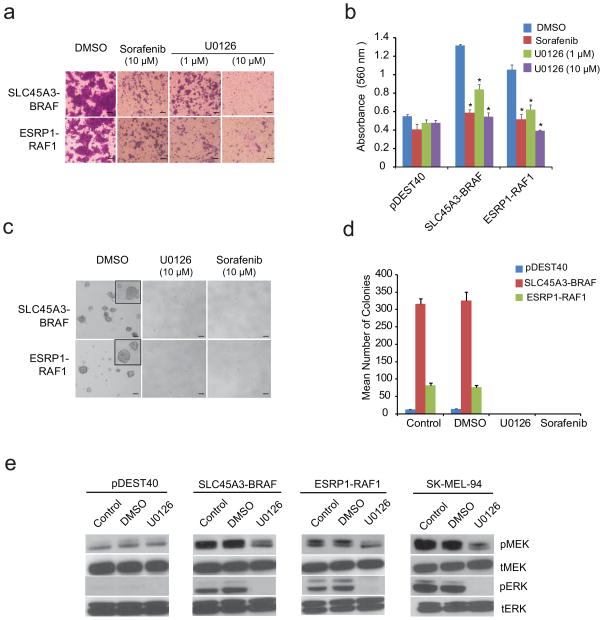
To examine the role of these fusions in the prostate, we over expressed SLC45A3-BRAF or ESRP1-RAF1 in RWPE cells (benign immortalized prostate epithelial cells), which both resulted in increased cell proliferation that was sensitive to the RAF kinase inhibitor sorafenib (Fig. 3c,d). We also observed a dramatic increase in cell invasion in RWPE cells expressing either SLC45A3-BRAF or ESRP1-RAF1, which was sensitive to sorafenib or the MEK inhibitor U0126 (Fig. 4a,b). Furthermore, RWPE cells expressing either SLC45A3-BRAF or ESRP1-RAF1 formed anchorage independent colonies in soft agar, which were again sensitive to RAF and MEK inhibitors (Fig. 4c,d). Finally, RWPE cells stably expressing SLC45A3-BRAF formed small tumors in immunodeficient mice, which regressed after 4 weeks (Supplementary Fig. 6b).
Fig. 4. RAF and MEK inhibitors block SLC45A3-BRAF or ESRP1-RAF1 gene fusion mediated oncogenic phenotypes.
a, SLC45A3-BRAF or ESRP1-RAF1 mediated cell invasion in RWPE prostate cells is sensitive to sorafenib (10μM) or the MEK inhibitor U0126 (1 or 10μM). Cells were photographed after invasion through Matrigel and stained with crystal violet or b) quantitated by absorbance. pDEST40 represents the empty vector control. Error bars and P values were calculated as in Fig 3d. c, Photomicrographs or d) quantitation of SLC45A3-BRAF or ESRP1-RAF1 induced anchorage independent colony growth in soft agar, which was sensitive to sorafenib or U0126. Scale bars indicate 50μm. e, Evaluation of the downstream signaling pathways activated by the SLC45A3-BRAF or ESRP1-RAF1 gene fusions in RWPE prostate cells. SLC45A3-BRAF or ESRP1-RAF1 expressing pooled populations and vector controls were treated with U0126 (10 μM) for two hours and immunoblotted for phosphor (p)- and total (t) MEK-1/2 and ERK-1/2. SK-MEL-94, a BRAFV600E positive melanoma cell line, was included as a positive control.
The RAF family is known to play a pivotal role in transducing signals from RAS to downstream kinases, mitogen activated protein kinase (MAPK) and extracellular signal regulated kinase (ERK) kinase (MEK)-1/2 and ERK-1/226. As expected, over-expression of SLC45A3-BRAF or ESRP1-RAF1 in RWPE cells induced MEK/ERK phosphorylation, sensitive to treatment with a MEK inhibitor (Fig. 4e). The MEK inhibitor also decreased MEK-1/2 and ERK-1/2 phosphorylation in a control BRAFV600E mutation positive melanoma cell line, SK-MEL-94, consistent with previous data27. We also found an increase in mRNA expression of feedback effectors (DUSP6 and SPRY227) in stable RWPE cells expressing SLC45A3-BRAF or ESRP1-RAF1, and the expression of these feedback effectors was decreased upon MEK inhibitor treatment (Supplementary Fig. 7).
Our results emphasize the importance of the RAF pathway in prostate cancer development and progression. Although rare (possibly non-existent) in human prostate tumors, activation of the BRAF pathway via the V600E mutation in genetically-engineered mice was shown to cooperate with other lesions to initiate the development of invasive prostate cancer28. This model may now have greater clinical significance for the study of human prostate cancer. Finally, ETS transcription factors, including ETV1, have been shown to be downstream targets activated by the RAS-RAF-MAPK signaling pathway29,30, suggesting a possible common pathway.
Sequencing tumor transcriptomes and genomes may identify rare targetable fusions across cancer types. Screening for RAF kinase fusions may be useful in identifying cancer patients that may benefit from RAF kinase inhibitors, similar to what is already being considered clinically for ALK fusions in lung cancer. The identification of RAF pathway gene rearrangements in 1–2% of prostate cancers, gastric cancers, and melanomas (and earlier work by others in rarer cancers7–13) supports the general principle that cancers should be classified by driving molecular event(s), rather than organ site, in the context of rationale targeted therapy.
METHODS
Samples and paired end library preparation for Illumina sequencing
Prostate cancer tissues negative for ETS family gene rearrangements were selected for paired-end sequencing from the University of Michigan cohort. Total RNA was isolated with Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Quality assessment of RNA was performed using Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). Paired end libraries (n=15) for sequencing with Illumina Genome Analyzer II were prepared according to the protocol provided by Illumina with minor modifications using mRNA-seq sample prep kit (Illumina). Sequence analysis was carried out by the Illumina data analysis pipeline.
Nomination of prostate gene fusions
Mate pair transcriptome reads were mapped to the human genome (hg18) and Refseq transcripts, allowing up to two mismatches, using ELAND (Efficient Alignment of Nucleotide Databases) pair within the Illumina Genome Analyzer Pipeline software. Sequence alignments were subsequently processed to nominate gene fusions using described methodology20. In brief, mate pairs were processed to identify any that either ‘encompassed’ or ‘spanned’ the fusion junction. Encompassing mate pairs refer to those in which each read aligned to an independent transcript, thereby encompassing the fusion junction. In contrast, spanning mate pairs refer to those in which one sequence read aligns to a gene and its mate spans the fusion junction. Both categories undergo a series of filtering steps to remove putative false positive before being merged together to generate the final chimera nominations.
Cloning of full length fusion transcript
The full length fusion transcripts of SLC45A3-BRAF and ESRP1-RAF1 were cloned into pCR8/GW/TOPO Entry vector (Invitrogen, USA) by TA cloning. All entry vector clones were sequence confirmed and recombined into the Gateway pcDNA-DEST40 mammalian expression vector (Invitrogen, USA) and pAd/CMV/V5-DEST Adenoviral expression system (Invitrogen, USA) by LR Clonase II. Plasmids with N-terminus FLAG tag and C-terminus V5 tags were generated for initial verification of protein expression in HEK293 cells.
Invasion and WST-1 assays
Equal amount of cells were plated into 96-well plates and WST-1 proliferation assay was performed using the manufacturer’s protocol (Roche, Indianapolis, IN, USA). For Boyden chamber matrigel invasion assay, equal numbers of cells were plated into each matrigel coated transwell in the presence of sorafenib or U0126 or DMSO. Invasion assays were performed as described previously 31, 32.
In vitro soft agar growth
For in vitro growth in soft agar, 2 ml of 0.6% SeaPlaque GTG Agarose (Cambrex, ME, Rockland) dissolved in complete Keratinocyte-SFM was poured into 6-well dishes. After polymerization, a second layer containing 2 ml of 0.4% agar in complete Keratinocyte-SFM and RWPE cells stably expressing SLC45A3-BRAF or ESRP1-RAF1 (1 × 104 cells per well) was poured on top. The next day, cells were treated with sorafenib or U0126 (10 μM) in 1 ml of supplement-free keratinocyte media. Soft agar assay plates were incubated for 14 days at 37°C. MEK inhibitor was changed once a week. Each experimental condition was done in triplicate. On the 14th day, colonies larger than 40μm were counted.
Fluorescence in situ hybridization (FISH)
FISH hybridizations were performed on tissue microarrays (TMA) of indicated cancer types. Rearrangement positive cases identified from TMA were further validated on individual formalin fixed and paraffin-embedded (FFPE) sections. BAC clones were selected from UCSC genome browser and purchased through BACPAC resources (Children’s Hospital, Oakland, CA). Following colony purification, midi prep DNA was prepared using QiagenTips-100 (Qiagen, USA). DNA was labeled by nick translation with biotin-16-dUTP and digoxigenin-11-dUTP (Roche, USA). Probe DNA was precipitated and dissolved in hybridization mixture containing 50% formamide, 2XSSC, 10% dextran sulphate, and 1% Denhardt’s solution. Approximately 200ng of labeled probe was hybridized to normal human chromosomes to confirm the map position of each BAC clone. FISH signals were obtained using anti digoxigenin-fluorescein and alexa fluor594 conjugate, to obtain green and red colors, respectively. Fluorescence images were captured using a high resolution CCD camera controlled by ISIS image processing software (Metasystems, Germany).
MEK/ERK Signaling Pathway analysis
Stable pooled population of RWPE cells expressing SLC45A3-BRAF or ESRP1-RAF1 were maintained in Keratinocyte SFM medium without supplements for two hours. Whereas for MEK inhibitor treatments, U0126 (10μM) was added in the supplement-free keratinocyte media for two hours. MEK and ERK activation was assessed by Western blot analysis using phospho-MEK or ERK and total MEK or ERK antibodies (Cell Signaling Technologies, USA).
Statistical analyses
All data are presented as means ± SEM, and significance was determined by two-tailed Student’s t test.
Additional methods
Detailed methodology is described in the supplementary methods.
Supplementary Material
Acknowledgments
We thank Dr. R.B. Jenkins for providing prostate cancer tissues with BRAF over expression for FISH evaluation. R. Morey for assistance in paired-end sequencing, and Illumina for technical support. T. Barrette, R. Lonigro, M. Quist, C. Quist and S. Begley for hardware support, sample database maintenance, data curation and maintenance of paired-end sequence data and useful discussions; D. Sanders, M. Vinco, for their assistance in providing gastric cancer samples; D. Kim, R. Mehra and R. Varambally for providing prostate and melanoma tissue microarray and clinical information from the University of Michigan cohort. D.F. Fries, X. Jiang, L. Wang, R. Jagirdar, N. Kitabayashi and X. Jing for technical assistance. K. Giles for critical reading of the manuscript. The Biobank at Weill Cornell Medical College and A.K. Tewari for providing PCa samples for mutation analysis. This work is supported in part by the National Institutes of Health (R01CA132874), Early Detection Research Network (EDRN) UO1 CA111275, Prostate SPORE P50CA69568, National Center for Integrative Bioinformatics (U54 DA21519-01A1), the National Center for Functional Genomics supported by the Department of Defense (A.M.C) and R01 CA125612-01 (M.A.R and F.D). S.A.T. is supported by a Young Investigator Award from the Prostate Cancer Foundation. A.M.C. is supported by the Doris Duke Charitable Foundation Clinical Scientist Award, a Burroughs Welcome Foundation Award in Clinical Translational Research and the Prostate Cancer Foundation. A.M.C. is an American Cancer Society Research Professor. N.P. supported by the development award from Melanoma Research Alliance. C.A.M. currently derives support from the American Association of Cancer Research Amgen Fellowship in Clinical/Translational Research and the Canary Foundation and American Cancer Society Early Detection Postdoctoral Fellowship. B.A. is supported by a Genentech Foundation Postdoctoral Fellowship and Young Investigator Award from Expedition Inspiration. T.A.B. is supported by funding from the Prostate Cancer Foundation and Young Investigator Award and CIHR (Canadian Institute of Health Research).
Footnotes
Accession numbers
Nucleotide sequences for the ESRP1-RAF1, SLC45A3-BRAF and AGTRAP-BRAF fusion genes have been deposited at GenBank with accession numbers GU149302, GU149303, and HM053972 respectively.
Author Contributions
N.P., B.A., S.A.T., C.M., A.M.C. designed experiments and wrote the manuscript. N.P. performed paired-end transcriptome library preparation, FISH probe design, preparation and analysis. S.K., C.A.M performed bioinformatics analysis for gene fusion nominations. B.A., K.R., S.S., Q.C., S.M.D., S.V., S.A.T conducted experiments including design and generation of expression constructs, in vitro assays, western blot, qRT PCR validation, data analysis and interpretation. B.A. performed in vivo experiments. N.P., B.H., K.S., D.P., performed FISH assays on cancer tissue microarrays. X.C performed sequencing. C.K. performed BRAF mutation analysis by pyrosequencing. Y.C, R.E, S.B, C.J.L., J.S., F.D., P.M., T.A.B., R.K., and M.A.R provided prostate cancer specimens and performed FISH assays. D.R.F., J.K.G., T.M.J, provided melanoma tissue microarrays. T.J.G and P.T provided gastric cancer tissue microarray. A.M.C. directed the study.
References
- 1.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 2.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 3.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koivunen JP, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maher CA, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X-S. Integrative analyses reveal functional and genetic associations of gene fusions in cancer. Nature Biotechnology. 2009 [Google Scholar]
- 7.Cohen Y, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 8.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, et al. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer research. 2003;63:5209–5212. [PubMed] [Google Scholar]
- 10.Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer. 2005;12:245–262. doi: 10.1677/erc.1.0978. [DOI] [PubMed] [Google Scholar]
- 11.Ciampi R, et al. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. The Journal of clinical investigation. 2005;115:94–101. doi: 10.1172/JCI23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones DT, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer research. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dessars B, et al. Chromosomal translocations as a mechanism of BRAF activation in two cases of large congenital melanocytic nevi. J Invest Dermatol. 2007;127:1468–1470. doi: 10.1038/sj.jid.5700725. [DOI] [PubMed] [Google Scholar]
- 14.Wilhelm SM, et al. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol Cancer Ther. 2008;7:3129–3140. doi: 10.1158/1535-7163.MCT-08-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sala E, et al. BRAF silencing by short hairpin RNA or chemical blockade by PLX4032 leads to different responses in melanoma and thyroid carcinoma cells. Mol Cancer Res. 2008;6:751–759. doi: 10.1158/1541-7786.MCR-07-2001. [DOI] [PubMed] [Google Scholar]
- 16.Esgueva R, et al. Prevalence of TMPRSS2-ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Mod Pathol. :539–546. doi: 10.1038/modpathol.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han B, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer research. 2008;68:7629–7637. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 19.Helgeson BE, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer research. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 20.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rickman DS, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer research. 2009;69:2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho NY, et al. BRAF and KRAS mutations in prostatic adenocarcinoma. Int J Cancer. 2006;119:1858–1862. doi: 10.1002/ijc.22071. [DOI] [PubMed] [Google Scholar]
- 24.MacConaill LE, et al. Profiling critical cancer gene mutations in clinical tumor samples. PloS one. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garte SJ, Currie DD, Troll W. Inhibition of H-ras oncogene transformation of NIH3T3 cells by protease inhibitors. Cancer research. 1987;47:3159–3162. [PubMed] [Google Scholar]
- 26.Hoeflich KP, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15:4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 27.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong JH, et al. BRAF activation initiates but does not maintain invasive prostate adenocarcinoma. PloS one. 2008;3:e3949. doi: 10.1371/journal.pone.0003949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janknecht R. Analysis of the ERK-stimulated ETS transcription factor ER81. Mol Cell Biol. 1996;16:1550–1556. doi: 10.1128/mcb.16.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bosc DG, Janknecht R. Regulation of Her2/neu promoter activity by the ETS transcription factor, ER81. J Cell Biochem. 2002;86:174–183. doi: 10.1002/jcb.10205. [DOI] [PubMed] [Google Scholar]
- 31.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Q, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.