Abstract
The intraocular lens contains high levels of both cholesterol and sphingolipids, which are believed to be functionally important for normal lens physiology. The aim of this study was to explore the spatial distribution of sphingolipids in the ocular lens using mass spectrometry imaging (MSI). Matrix-assisted laser desorption/ionization (MALDI) imaging with ultra high resolution Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) was used to visualize the lipid spatial distribution. Equatorially-cryosectioned, 12 μm thick slices of tissue were thaw-mounted to an indium-tin oxide (ITO) glass slide by soft-landing to an ethanol layer. This procedure maintained the tissue integrity. After the automated MALDI matrix deposition, the entire lens section was examined by MALDI MSI in a 150 μm raster. We obtained spatial- and concentration-dependent distributions of seven lens sphingomyelins (SM) and two ceramide-1-phosphates (CerP), which are important lipid second messengers. Glycosylated sphingolipids or sphingolipid breakdown products were not observed. Owing to ultra high resolution MS, all lipids were identified with high confidence, and distinct distribution patterns for each of them are presented. The distribution patterns of SMs provide an understanding of the physiological functioning of these lipids in clear lenses and offer a novel pathophysiological means for understanding diseases of the lens.
Keywords: ocular lens, sphingolipid, matrix-assisted laser desorption/ionization
Spatial localization in tissue sections has traditionally focused on protein distribution and relied on the use of immunohistochemistry. The assessment of lipid spatial distribution has been even more difficult as immunohistochemical methods are not available. Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) has been developed and successfully used for spatial localizations of proteins, peptides, and lipids (1, 2). MSI generates two-dimensional molecular maps from particular ions, which are directly desorbed/ionized from the surface of the tissue (2, 3). This gives an opportunity to detect molecules of interest at multiple points in whole tissue sections. MSI allows for precise localization of chemical information in a tissue section, and this is the main reason the technique has become a popular tool in bioanalytical, biological, and medicinal chemistry. Several MS-based methods have been developed for molecular imaging of lipids in tissues. The main focus has been on traditional ionization techniques based on desorption/ionization in a vacuum, mainly secondary ion mass spectrometry (SIMS) (4, 5) and MALDI (6). However, ambient ionization techniques, such as desorption electrospray (DESI) (7–9) or desorption atmospheric pressure photo ionization (DAPPI) (7), were also introduced for lipid imaging of tissue sections.
MALDI MSI has been used for molecular imaging of proteins in ocular lenses. To the best of our knowledge, only a few studies discuss the spatial distribution of lens proteins, namely, the integral lens protein aquaporin and its truncation products (10, 11) and α-crystallin (1, 12). Interestingly, it seems that protein modifications, such as phosphorylation and protein truncation, occur at distinct areas of the lens, suggesting that these modifications may affect global lens functioning (10). We are not aware of any published studies that have used MSI for mapping lipids in lenses. Rujoi et al. (13), however, performed MALDI-time of flight (TOF) MS analysis of neutral phospholipids in selected spots of the lens tissue.
The avascular intraocular lens is composed of concentric layers of fiber cells. Newly formed fiber cells lose their internal organelles and are translocated from the equatorial plane of the lens toward its center. Consequently, cells are successively layered, forming an onion-like structure where the oldest fiber cells are positioned at the innermost segment (nucleus) of the lens, and the younger cells are located in the cortical region (cortex) of the lens. This process begins in early embryogenesis and continues throughout an organism's life. The plasma membrane becomes practically the only membrane of the lens because of the loss of internal organelles in lens fiber cells. It is unique because it contains high concentrations of aquaporin 0 (AQP0), a 26 kDa water channel protein that lacks polyunsaturated phospholipids and contains unusually high concentrations of sphingomyelins (SM) and cholesterol (14, 15). The cholesterol content of the lens is the highest of all tissues. The presence of high concentrations of SMs and cholesterol are likely to increase the rigidity of fiber cell plasma membranes and may contribute to cataractogenesis.
Previously, we and others used MALDI-TOF MS to identify the major lipid components of the porcine lens (13, 16). We found that the major phospholipid components were long chain phosphatidylcholines (PC) and SMs. The long chain PCs and SMs likely caused ordering of the lens fiber membranes and allowed for more interactions between phospholipids and cholesterol (17).
We have extended our previous studies to provide spatial organization of SMs and other identified sphingolipid species in clear porcine lenses using ultra high resolution MALDI imaging coupled with Fourier transform ion-cyclotron resonance mass spectrometry (FT-ICR MS). An advantage of FT-ICR MS over TOF MS is the superior resolution and mass accuracy of the FT-ICR instrument, which allows for clear determinations of exact masses and, consequently, more confident identification of lipid species.
MATERIALS AND METHODS
Tissue preparation
Porcine eyes from ∼2-year-old animals were obtained from a local abattoir and transferred to the laboratory within 6 h of death. Lenses were extracted immediately upon arrival by excision with an anterior approach and stored on ice during transportation in a physiological saline solution. Animals were treated in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals and Ethical Committees. These organizations approved this research. Tissues were shock-frozen in liquid nitrogen and stored in a freezer at −80°C. Lenses were left to adapt to the cryostat cutting temperature (−20°C) for 2 h prior to cutting (Leica CM 1950, Leica Biosystem, Germany). Lenses were frozen-glued on a cryostat steel plate using plain water and were then cut into 12 µm sections from the equatorial plane. Single slices were thaw-mounted onto an indium tin oxide (ITO) glass slide (conductive glass slides for MALDI imaging, Bruker Daltonics, Bremen, Germany) with a thin layer of ethanol (2 µL, Merck, Germany). The tissue slice on the ITO glass slide was placed in a desiccator for 10 min and then immediately covered with a MALDI matrix using an ImagePrep deposition device (Bruker Daltonics, Germany). The MALDI matrix was α-cyano-4-hydroxycinnamic acid (CHCA) (Sigma-Aldrich, Germany) and was prepared at a 7 mg/ml concentration in 50:50 (v/v) acetonitrile:trifluoroacetic acid (0.2%) (Sigma-Aldrich, St. Louis, MO, USA). The tissue was analyzed immediately after the matrix deposition. The fixed tissue on ITO glass was imaged with a Nikon scanner (Coolscan VED, Nikon, Tokyo, Japan) before and after matrix deposition.
Mass spectrometry imaging
MALDI MSI of porcine lens was performed using an APEX Ultra 9.4 T FT-ICR (dual ion source ESI/MALDI, Bruker Daltonics, Germany) mass spectrometer. MALDI imaging was conducted using a smart beam laser (200 Hz) with a diameter spot of 50 µm. The mass spectra data set was acquired from the whole area of porcine lens with a 150 µm raster step size and 600 laser shots per spot. One spectrum acquired per pixel was sufficient for satisfactory signal intensity over the whole porcine lens. Mass spectra were acquired in positive ion mode within a mass range of 200–1100 m/z and 512,000 data points collected. Mass resolution at 400 m/z was 66,000. Time fill for an ICR cell was 1,400 μs. Spectra were apodized using square sin apodization with one zero fill.
Data analysis
Spatial distribution of lipids of interest was visualized using FlexImaging software (Version 2.1, Bruker Daltonics, Germany). Data were normalized to total ion count, and each lipid was displayed within an interval of ±0.002 amu. Mass spectra were open in Data Analysis (Version 4.0, Bruker Daltonics, Germany) for qualitative analysis. Internal calibrations were conducted with CHCA matrix using a 4-point linear trend. The peak list was exported into the publicly accessible search algorithm LIPID MAPS, which is available on the Internet (18). The LIPID MAPS database was searched with a tight tolerance of 0.01 amu for protonated ions and sodium and potassium adducts. Resulting lipids were cross-checked in DataAnalysis for matching molecular formulas and isotopic patterns.
ImageJ software (version 1.43i 64-bit on Mac OS X) (19) was used to measure the local abundance of lipids from MS images. Colored images of lipid distribution from FlexImaging were transferred to ImageJ and converted into grayscale images. The edge of the nucleus was first marked, and the cortex area and Mean Gray Values were assessed directly. The Mean Gray Value of the cortex was obtained by subtracting the Mean Gray Value of nucleus from the Mean Gray Value of the entire lens. Two default area patterns, including one for the nucleus and a second for the entire tissue, were created using the software and applied to all lens images to measure the local gray channel gain. Measured local intensities of lipids were visualized from changes in the Mean Gray Value. Results were further processed in Microsoft Excel (Microsoft, Version 2007).
RESULTS AND DISCUSSION
Sample preparation
One of the key steps in MALDI imaging is sample preparation, which strongly affects the quality of the MS image. Important factors in sample preparation include the cutting temperature, thickness of the slice, consistency of the tissue, matrix deposition, and the skills of the operator. The onion-like structure of the lens is especially prone to damage during cutting, and careless handling may result in undesirable loss of tissue parts. This may in turn lead to a loss of chemical information from the missing area of the tissue section. To preserve the tissue integrity, the slice was attached to an ITO glass slide using ethanol-assisted soft-landing. Similar approaches with methanol (10, 11) and ethanol (1) have previously been used to fix animal lenses on MALDI surfaces for MSI of proteins. Unlike proteins, lipids are soluble in ethanol. Therefore, we deduced that a more polar solvent could possibly prevent deterioration of the spatial distribution of lipids in the tissue. However, using water to fix the ocular lens on the ITO glass did not help to preserve the tissue structure. The tissue, which was cut and transferred onto the ITO glass at −20°C, did not establish good contact with the ice. This improper fixing caused ruptured tissue during the MALDI matrix deposition. The same result was observed with untreated tissue thaw-mounted on the ITO glass. To minimize the possible dilution of analytes with ethanol after soft-landing, minimum amounts of ethanol were spread on the ITO surface. Excess ethanol was evaporated within a couple of seconds of tissue attachment by placing a finger on the other side of the glass slide. Visual observation of the ethanol soft-landing procedure showed that the solvent did not penetrate through the tissue, which suggests that the bottom side was fixed to the ITO surface, while the upper part was left intact.
The matrix deposition in MALDI is another crucial factor during sample preparation. The matrix should be applied on the tissue surface to create a uniform layer of crystals that dissolves the analytes and facilitates the desorption/ionization process during laser irradiation. In addition, deposition of the matrix solution should not dissolve the analytes excessively to prevent the deterioration of their spatial distribution in the tissue, which is usually accomplished by repeating matrix deposition and drying. However, such matrix deposition procedures may affect the tissue stability. If the tissue slice is not properly affixed to the MALDI surface, alternating the wetting and drying during MALDI matrix deposition may cause tissues to flake. In our experiment, six sliced lenses were attached on an ITO glass slide. However, usually only one or two slices stayed intact after MALDI matrix deposition and were usable for further MALDI MSI analysis. This outcome is not surprising considering that others have reported difficulties with attaching fragile lenses to surfaces (13). The importance and necessity of using a fixative agent for MSI of the ocular lens is displayed in Fig. 1. Plain thaw-mounting of the ocular lens to ITO glass was not sufficient for preserving the tissue integrity during the MALDI matrix deposition. In these cases, the tissue ruptured into flakes. In contrast, the application of ethanol soft-landing preserved the entire area of the tissue.
Fig. 1.
Comparison of two different sample preparation procedures. In procedure A, the ocular lens was attached to ITO glass by common thaw-mounting. This procedure did not preserve the fragile structure of the tissue during the MALDI matrix deposition, and some tissue parts flaked off (left scan). In procedure B, a thin layer of ethanol was used for soft-landing the ocular lens to the ITO glass (right scan). MS images in the lower row compare molecular images deteriorated by unsuitable sample preparation (left) to the successful ethanol-soft landing procedure (right). Black void spots with no lipid signal indicate places where the tissue was removed (observed using procedure A). ITO, indium-tin oxide; MALDI, matrix-assisted laser desorption/ionization; SM, sphingomyelin.
Previous MALDI-TOF MS studies aimed to obtain information about the regional distribution of lipids in ocular lens. These studies were based either on the analysis of separate extracts of the nucleus and cortex (20) or on the direct analysis of the tissue with the manual collection of spectra from several spots (13). The compound, 2,5-dihydroxybenzoic acid (DHBA), which is a common MALDI matrix, was used in the first study; however, the second study claimed that DHBA caused poor signal-to-noise ratios for porcine lenses. Therefore, a matrix based on 4-nitroaniline (PNA) was used instead. Nevertheless, both methods produced valuable MALDI-MS spectra of lipids. In the current study, CHCA was used. Previous MSI experiments in our laboratory have proven that CHCA matrix is advantageous for MALDI analysis of small molecules in tissue sections. In addition, CHCA, which forms smaller crystals on the tissue surface than DHBA, helps to maintain a good lateral resolution.
MALDI MSI
In MALDI MSI, the laser performs a raster scan across the tissue section of the entire lens within a set raster pattern. The intensity of the m/z signal is plotted against its position to construct a molecular image. The spatial distribution of a molecule of interest is usually overlaid with an optical image to correlate chemical composition with visible structure. The advantage of MSI experiments conducted in this study is that the entire lens was examined to reveal the distribution of individual lipids or molecules of interest. In this study, we have analyzed the regional distribution of sphingolipid species in porcine lens.
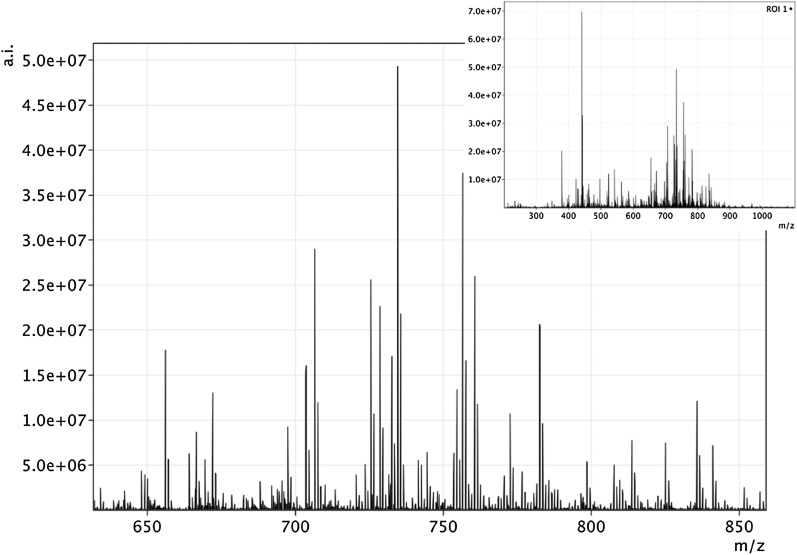
Figure 2 shows an average MALDI FT-ICR MS spectrum obtained from a single spot of a porcine lens tissue section. The spectrum shows a pattern of peaks between m/z 600 and 900 that were attributed mostly to phospholipid species. This work focuses on sphingolipids, but several other lipid classes were identified with high confidence. We were not able to detect the cholesterol ion nor its dehydrated fragment ion, probably due to the strong integration of cholesterol molecules in the lens or because cholesterol is partly crystallized in the lens. The localization of cholesterol in tissues was performed instead using SIMS (4, 21, 22) and DESI (7, 23).
Fig. 2.
Average mass spectrum obtained from the entire area of porcine lens. The spectrum is zoomed to the mass range relevant for the lipid envelope. The insert displays the full mass range spectrum. Peaks at the following nominal m/z are due to the matrix: 379, 401, 417, and 568.
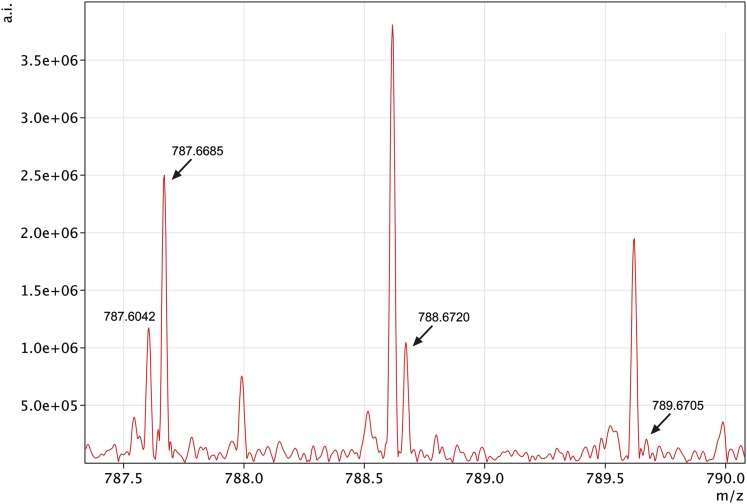
The benefit of ultra high resolution MS in MSI is demonstrated for a lipid with a nominal mass of 787 (Fig. 3). While the isotopic peak at exact m/z of 787.6685 represents the SMs (22:0) in its protonated form, the neighboring peak at m/z 787.6042 is an unknown ionized molecule. MS instruments with lower resolution analyzers would not be able to resolve the two peaks, and their overlap would deteriorate qualitative, quantitative, and spatial distribution analyses.
Fig. 3.
The benefit of high-resolution MS demonstrated by mass 787.6685 and its isotopes (788.6720 and 789.6705). The mono-isotopic mass 787.6685 was identified as a protonated SM (18:1/22:0), while the 787.6042 represents an unknown impurity. SM, sphingomyelin.
We identified a number of SM species in the porcine lens (Table 1), consistent with previous studies (13, 16, 24). The protonated ions as well as the sodium and potassium adducts of all identified SMs were observed. The sodium adduct was the most abundant, followed by the protonated ion (40–75% abundance relative to Na adducts) and the potassium adduct (21–28% abundance relative to Na adducts). Exact masses of all adducts were analyzed for the molecular formula match, with an accuracy <2 ppm in most cases. Spectra were checked for isotopic pattern agreement, which was positive in all cases. Ultra high resolution MS analyses allow for the identification of molecular formulas with high mass accuracy. This attribute increases our confidence that correct molecular formulas are assigned to analytes of interest. However, resolving isobaric compounds may represent a limitation for the current method, particularly for low-intensity ions, such as SMs, which are difficult to subject to tandem MS. An exact mass of 787.6680 was identified in the database as a protonated ion of SM (18:1/22:0) or SM (18:0/22:1). Structural information could not be mined from the tandem MS experiment due to the low intensity of the parent ion. This ion has been previously identified in animal and human ocular lenses as SM (18:1/22:0) (13, 24). In this study, we followed previous analyses and present the molecular ion lateral distribution based on an exact mass assignment by FT-ICR MS. However, identification and discovery of new biomarkers and metabolites requires analysis complementary to MSI, which is usually performed by LC-MS of the tissue extract (25).
TABLE 1.
Lipid molecular formulas, measured m/z values for all identified ions, and deviations of measured m/z values from theoretical values
| Compound | Molecular Formula | Measured (m/z) | Ion | Molecular Formula Match (ppm) |
|---|---|---|---|---|
| SM (18:1/14:0) | C37H75N2O6P | 675.5434 | [M+H]+ | 0.2 |
| 697.5242 | [M+Na]+ | 0.7 | ||
| 713.4989 | [M+K]+ | 1.8 | ||
| SM (18:1/16:0) | C39H79N2O6P | 703.5746 | [M+H]+ | 0.3 |
| 725.5568 | [M+Na]+ | 0.2 | ||
| 741.5303 | [M+K]+ | 0.5 | ||
| SM (18:1/18:0) | C41H83N2O6P | 731.6054 | [M+H]+ | 1.1 |
| 753.5878 | [M+Na]+ | 0.4 | ||
| 770.5665 | [M+K]+ | 1.1 | ||
| SM (18:1/20:0) | C43H87N2O6P | 759.6365 | [M+H]+ | 0.5 |
| 781.6188 | [M+Na]+ | 0.1 | ||
| 797.5920 | [M+K]+ | 1.6 | ||
| SM (18:1/22:0) | C45H91N2O6P | 787.6680 | [M+H]+ | 1.0 |
| 809.6505 | [M+Na]+ | 0.2 | ||
| 825.6253 | [M+K]+ | 0.8 | ||
| SM (18:1/22:1) | C45H89N2O6P | 785.6519 | [M+H]+ | 1.5 |
| 807.6338 | [M+Na]+ | 1.5 | ||
| 823.6081 | [M+K]+ | 1.0 | ||
| SM (18:1/24:1) | C47H93N2O6P | 813.6828 | [M+H]+ | 2.0 |
| 835.6664 | [M+Na]+ | 1.8 | ||
| 851.6374 | [M+K]+ | 3.4 | ||
| CerP (18:1/18:1) | C36H70NO6P | 666.4830 | [M+Na]+ | 0.5 |
| CerP (18:1/20:0) | C38H76NO6P | 696.5307 | [M+Na]+ | 0.6 |
Abbreviations: CerP, ceramide-1-phosphate; SM, sphingomyelin.
No glycosphingolipid species were detected in this analysis. Interestingly, ceramides were not observed with the exception of a (18:1/18:1) and (18:1/20:0) ceramide-1-phosphates (CerP) (Table 1). In contrast to SMs, these ceramide-1-phosphates were detected only as the sodium adduct. High abundance of sodium adducts could be the result of the tendency for phosphates to form salts, and sodium salts were introduced during our matrix deposition. Conversely, SMs have high proton affinities due to the presence of two nitrogens; thus, protonated forms of SMs were also observed in relatively high abundance. Another factor that can contribute to the elevated abundance of sodium adducts is higher stability of alkali metal adducts than proton adducts in the gas phase, which is the MALDI environment during the desorption/ionization process.
Figure 4 shows the two-dimensional distribution of all identified SM species and two ceramide-1-phosphates in an equatorially sliced porcine lens. For each lipid, the sodium adduct was chosen for visualization. The spatial distributions of all protonated molecules and adducts were identical. Black spots indicate points without a detectable signal (below the respective limit of detection). In the MS images, the white line defines the contour of the porcine lens. The distribution of SMs and ceramide-1-phosphates correlate with the structure of the ocular lens, and the nucleus is distinctly visible as being apart from the cortex. Some lipids are also present outside of the lens, which was probably caused by the ethanol used to fix the tissue to the ITO glass slide. Ethanol dissolved some compounds from the bottom part of the tissue. This dissolution is particularly visible at the top of some images as a sharp change in the lipid intensity toward the outside of the edge of the rastered area. In the case of dissolving lipids from the upper part of the tissue, transitions in lipid intensity at the edge of the tissue would be continuous. This transition demonstrates that the upper part of the tissue stayed intact without contacting the ethanol, and the lateral distribution of lipids, which was examined by a laser, stayed preserved. The quality of the SM spatial distribution in the lens tissue was maintained, and ethanol can be used as a fixative agent for the MSI of lipids. The optical scan displayed in Fig. 4 shows the lens after deposition of the MALDI matrix. Its uniform distribution is one of the conditions for obtaining an MS signal from each point of the MSI image.
Fig. 4.
Distribution of individual SM and ceramide-1-phosphates in porcine ocular lens along the equatorial plane together with optical images of porcine eye lens covered with the MALDI matrix. In MS images, white lines show the size of the ocular lens tissue slice. In the optical image, the inner black line defines the size of the ocular lens tissue, and the red line shows the area selected for the MS imaging experiment, including an area for reading MS background (on the left hand side of the optical image). CerP, ceramide-1-phosphate; MALDI, matrix-assisted laser desorption/ionization; SM, sphingomyelin.
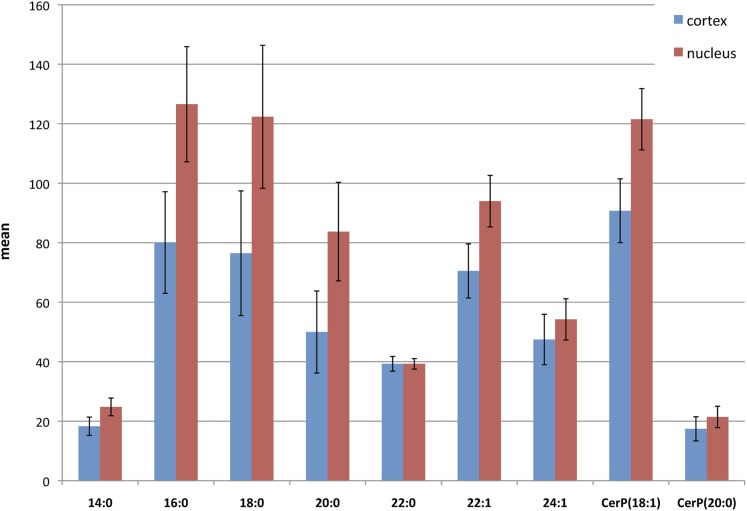
Figure 5 shows compositional changes between the lens nucleus and cortical layer for each identified SM and ceramide-1-phosphate species. All SMs are more abundant in the nucleus than cortex, and the difference is 5% for 14:0, 37% for 16:0, 36% for 18:0, 27% for 20:0, 19% for 22:1, and 5% for 24:1, as normalized to the most abundant SM (16:0). However, SM 22:0 had the same mean intensity in both the nucleus and cortex. Rujoi et al. (13) determined regional changes of SMs and PCs based on the average of three-to-four laser shots collected within 50 µm of each other across different parts of the tissue. Regional changes of SMs in the nucleus and cortex using this approach were in relatively good agreement with our MSI results. In our MSI experiments, whole areas of the nucleus and cortex were selected for measurement of the mean intensity of each SM, which resulted in more precise information. Similarly to SMs, both ceramide-1-phosphates are more abundant in the nucleus than in the cortex, and the difference is 3% for CerP (18:1) and 4% for CerP (20:0). Lipids are distributed in the lens in concentric layers, and so reproducible quantitative information can be obtained from a small part of the tissue. This was probably the reason that we obtained outcomes similar to the study by Rujoi et al. (13). Chemometric standards have not yet been established for MSI, mainly because of the unique properties of each biological tissue, making assessment of the reproducibility ambiguous, and the very high time demands for the experimental time. The data presented in Fig. 5 were obtained from two sets of experiments of individual lenses. The relatively high reproducibility is satisfactory considering high number of steps in the sample preparation.
Fig. 5.
The comparison of intensities of local SM and ceramide-1-phosphates distributions in the cortex and nucleus (absolute mean values). The error bars represent standard deviation. CerP, ceramide-1-phosphate; SM, sphingomyelin.
The lipid composition of the lens is unusual compared with other mammalian cellular membranes because it has high levels of cholesterol and sphingolipids, especially SMs and dihydrosphingomyelins (DHSM), which may constitute half of the total phospholipid composition (14). Conversely, polyunsaturated lipids are almost absent. Surprisingly, we did not observe any DHSMs in this study, but this may be explained by the substantial variation in DHSM levels across mammals (26–28). It is believed that the loss of functional integrity of fiber cell plasma membranes is an early step in the development of cataracts (29). Levels of almost all identified SMs increase toward the nucleus, and the cholesterol content increases toward the lens nucleus (30, 31). This observation suggests the parallel regulation of levels of these lipids. We suggest that this relationship is necessary for providing an optimal lipid environment for integral membrane protein regulation.
CONCLUSIONS
We identified and provided the spatial distribution of seven SMs and two ceramide-1-phosphates in an equatorial slice of the porcine lens using MALDI MSI. SMs were identified as sodium and potassium adducts and protonated ions, while the ceramide-1-phosphates were measured as the sodium adduct alone. Ceramides were not observed. FT-ICR MS provided ultra high mass resolution that minimized undesirable mass peak overlaps and facilitated accurate mass determinations of molecular formulas (high confidence, <2 ppm). MSI of the entire lens slice allowed for the precise measurement of compositional changes between the lens nucleus and the cortical layer of each SM and ceramide-1-phosphate identified. All investigated lipids were more abundant in the nucleus than in the cortex, except for SM 22:0, which exhibited the same abundance in both examined lens areas. This work focuses on sphingolipids and the more recently discovered ceramide-1-phosphate in porcine lens (32). However, several other lipid classes were identified with high confidence and will be discussed elsewhere. The results presented here are important to increasing our understanding of the role of lipids in healthy and diseased lenses.
Footnotes
Abbreviations:
- CerP
- ceramide-1-phosphate
- FT-ICR MS
- Fourier transform ion cyclotron resonance mass spectrometry
- ITO
- indium-tin oxide
- MALDI
- matrix-assisted laser desorption/ionization
- MSI
- mass spectrometry imaging
This work was supported by funding from the Sigrid Juselius Foundation (J.M.H.); the Finnish Eye Foundation (J.M.H.); the Academy of Finland Projects 122354 (J.P.) and 114292 (S.K.W.); the University of Helsinki Project 2105060 (S.K.W.); the Ministry of Education, Youth, and Sports of the Czech Republic (LC07017 and MSM 6198959216); the Institute of Microbiology, Academy of Sciences of the Czech Republic (AV0Z50200510); and a Marie Curie International Reintegration grant (M.V.).
REFERENCES
- 1.Han J., Schey K. L. 2006. MALDI tissue Imaging of ocular lens alpha-crystallin. Invest. Ophthalmol. Vis. Sci. 47: 2990–2996. [DOI] [PubMed] [Google Scholar]
- 2.Caprioli R. M., Farmer T. B., Gile J. 1997. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 69: 4751–4760. [DOI] [PubMed] [Google Scholar]
- 3.Chaurand P., Norris J. L., Cornett D. S., Mobley J. A., Caprioli R. M. 2006. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J. Proteome Res. 5: 2889–2900. [DOI] [PubMed] [Google Scholar]
- 4.Börner K., Malmberg P., Månsson J-E., Nygren H. 2007. Molecular imaging of lipids in cells and tissues. Int. J. Mass Spectrom. 260: 128–136. [Google Scholar]
- 5.Amaya K. R., Monroe E. B., Sweedler J. V., Clayton D. F. 2007. Lipid imaging in the zebra finch brain with secondary ion mass spectrometry. Int. J. Mass Spectrom. 260: 121–127. [Google Scholar]
- 6.Murphy R. C., Hankin J. A., Barkley R. M. 2009. Imaging of lipid species by MALDI mass spectrometry. J. Lipid Res. 50: S317–S322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pól J., Vidová V., Kruppa G., Kobliha V., Novák P., Lemr K., Kotiaho T., Kostiainen R., Havlíček V., Volný M. 2009. Automated ambient desorption-ionization platform for surface imaging integrated with a commercial Fourier transform ion cyclotron resonance mass spectrometer. Anal. Chem. 81: 8479–8487. [DOI] [PubMed] [Google Scholar]
- 8.Wiseman J. M., Ifa D. R., Song Q. Y., Cooks R. G. 2006. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew. Chem. Int. Ed. Engl. 45: 7188–7192. [DOI] [PubMed] [Google Scholar]
- 9.Dill A. L., Ifa D. R., Manicke N. E., Zheng O. Y., Cooks R. G. 2009. Mass spectrometric imaging of lipids using desorption electrospray ionization. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2883–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grey A. C., Schey K. L. 2009. Age-related changes in the spatial distribution of human lens alpha-crystallin products by MALDI imaging mass spectrometry. Invest. Ophthalmol. Vis. Sci. 50: 4319–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grey A. C., Chaurand P., Caprioli R. M., Schey K. L. 2009. MALDI imaging mass spectrometry of integral membrane proteins from ocular lens and retinal tissue. J. Proteome Res. 8: 3278–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grey A. C., Schey K. L. 2008. Distribution of bovine and rabbit lens alpha-crystallin products by MALDI imaging mass spectrometry. Mol. Vis. 14: 171–179. [PMC free article] [PubMed] [Google Scholar]
- 13.Rujoi M., Estrada R., Yappert M. C. 2004. In situ MALDI-TOF MS regional analysis of neutral phospholipids in lens tissue. Anal. Chem. 76: 1657–1663. [DOI] [PubMed] [Google Scholar]
- 14.Byrdwell W. C., Borchman D., Porter R. A., Taylor K. G., Yappert M. C. 1994. Separation and characterization of the unknown phospholipid in human lens membranes. Invest. Ophthalmol. Vis. Sci. 35: 4333–4343. [PubMed] [Google Scholar]
- 15.Borchman D., Delamere N., McCauley L., Paterson C. 1989. Studies on the distribution of cholesterol, phospholipid, and protein in the human and bovine lens. Lens Eye Toxic. Res. 6: 703–724. [PubMed] [Google Scholar]
- 16.Sane P., Tuomisto F., Wiedmer S. K., Nyman T., Vattulainen I., Holopainen J. M. 2010. Temperature-induced structural transition in-situ in porcine lens—changes observed in void size distribution. Biochim. Biophys. Acta. 1798: 958–65. [DOI] [PubMed] [Google Scholar]
- 17.Holopainen J. M., Metso A. J., Mattila J. P., Jutila A., Kinnunen P. K. J. 2004. Evidence for the lack of a specific interaction between cholesterol and sphingomyelin. Biophys. J. 86: 1510–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipid Maps. Nature Publishing Group and LIPID MAPS Consortium. Available from: http://www.lipidmaps.org.
- 19.Rasband W. S., Image J. 1997–2009. U. S. National Institutes of Health. Available from: http://rsb.info.nih.gov/ij/.
- 20.Estrada R., Yappert M. C. 2004. Regional phospholipid analysis of porcine lens membranes by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 39: 1531–1540. [DOI] [PubMed] [Google Scholar]
- 21.Altelaar A. F. M., van Minnen J., Jimenez C. R., Heeren R. M. A., Piersma S. R. 2005. Direct molecular imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal. Chem. 77: 735–741. [DOI] [PubMed] [Google Scholar]
- 22.Nygren H., Malmberg P., Kriegeskotte C., Arlinghaus H. F. 2004. Bioimaging TOF-SIMS: localization of cholesterol in rat kidney sections. FEBS Lett. 566: 291–293. [DOI] [PubMed] [Google Scholar]
- 23.Wu C., Ifa D. R., Manicke N. E., Cooks R. G. 2009. Rapid, direct analysis of cholesterol by charge labeling in reactive desorption electrospray ionization. Anal. Chem. 81: 7618–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yappert M. C., Rujoi M., Borchman D., Vorobyov I., Estrada R. 2003. Glycero-versus sphingo-phospholipids: correlations with human and non-human mammalian lens growth. Exp. Eye Res. 76: 725–734. [DOI] [PubMed] [Google Scholar]
- 25.Burnum K. E., Cornett D. S., Puolitaival S. M., Milne S. B., Myers D. S., Tranguch S., Brown H. A., Dey S. K., Caprioli R. M. 2009. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J. Lipid Res. 50: 2290–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byrdwell W. C. 1998. Dual parallel mass spectrometers for analysis of sphingolipid, glycerophospholipid and plasmalogen molecular species. Rapid Commun. Mass Spectrom. 12: 256–272. [DOI] [PubMed] [Google Scholar]
- 27.Iwata J. L., Bardygulanonn L. G., Greiner J. V. 1995. Interspecies comparisons of lens phospholipids. Curr. Eye Res. 14: 937–941. [DOI] [PubMed] [Google Scholar]
- 28.Greiner J. V., Auerbach D. B., Leahy C. D., Glonek T. 1994. Distribution of membrane phospholipids in the crystalline lens. Invest. Ophthalmol. Vis. Sci. 35: 3739–3746. [PubMed] [Google Scholar]
- 29.Kinoshita J. H. 1974. Mechanisms initiating cataract formation (Proctor Lecture). Invest. Ophthalmol. Vis. Sci. 13: 713–724. [PubMed] [Google Scholar]
- 30.Li L. K., So L., Spector A. 1987. Age-dependent changes in the distribution and concentration of human lens cholesterol and phospholipids. Biochim. Biophys. Acta. 917: 112–120. [DOI] [PubMed] [Google Scholar]
- 31.Li L. K., So L., Spector A. 1985. Membrane cholesterol and phospholipid in consecutive concentric sections of human lenses. J. Lipid Res. 26: 600–609. [PubMed] [Google Scholar]
- 32.Estrada R., Puppato A., Borchman D., Yappert M. C. 2010. Reevaluation of the phospholipid composition in membranes of adult human lenses by 31P NMR and MALDI MS. Biochim. Biophys. Acta. 1798: 303–311. [DOI] [PubMed] [Google Scholar]