Abstract
The conversion of lysophosphatidic acid (LPA) to phosphatidic acid is carried out by the microsomal enzymes 1-acylglycerol-3-phosphate-O-acyltransferases (AGPATs). These enzymes are specific for acylating LPA at the sn-2 (carbon 2) position on the glycerol backbone and are important, because they provide substrates for the synthesis of phospholipids and triglycerides. At least, mutations in one isoform, AGPAT2, cause near complete loss of adipose tissue in humans. We cloned a cDNA predicted to be an AGPAT isoform, AGPAT11. This cDNA has been recently identified also as lysophosphatidylcholine acyltransferase 2 (LPCAT2) and lyso platelet-activating factor acetyltransferase. When AGPAT11/LPCAT2/lyso platelet-activating factor acetyltransferase cDNA was expressed in CHO and HeLa cells, the protein product localized to the endoplasmic reticulum. In vitro enzymatic activity using lysates of Human Embryonic Kidney-293 cells infected with recombinant AGPAT11/LPCAT2/lyso platelet-activating factor-acetyltransferase cDNA adenovirus show that the protein has an AGPAT activity but lacks glycerol-3-phosphate acyltransferase enzymatic activity. The AGPAT11 efficiently uses C18:1 LPA as acyl acceptor and C18:1 fatty acid as an acyl donor. Thus, it has similar substrate specificities for LPA and acyl-CoA as shown for AGPAT9 and 10. Expression of AGPAT11 mRNA was significantly upregulated in human breast, cervical, and colorectal cancer tissues, indicating its adjuvant role in the progression of these cancers. Our enzymatic assays strongly suggest that the cDNA previously identified as LPCAT2/lyso platelet-activating factor-acetyltransferase cDNA has AGPAT activity and thus we prefer to identify this clone as AGPAT11 as well.
Keywords: LPAAT, phospholipids, lipodystrophy
The biosynthesis of glycerophospholipids and triglycerides in eukaryotic cells initiates with the successive esterification of the hydroxyl groups with various fatty acids on the glyercol-3-phosphate backbone. These acylations are enzymatic processes catalyzed sequentially by glycerol-3-phosphate acyltransferases (GPATs), 1-acylglycerol-3-phosphate-O-acyltransferases (AGPATs), and diacylglycerol acyltransferases (DGATs) (1, 2). It is known that phospholipids also undergo remodeling. That is, after the initial synthesis, the fatty acids are removed by phospholipases and reacylated by acyltransferases (3–5). To accomplish this, several phospholipases and acyltransferases are now known that could carry out this task. We have been interested in identifying novel sn-2 acyltransferases (AGPATs) to determine their role in adipocyte biology ever since we discovered that mutations in AGPAT2 result in a near-complete loss of adipose tissue in patients with congenital generalized lipodystrophy, type 1 (6). We have previously reported enzymatic properties and tissue expression of AGPAT isoforms 8-10 (7–9). As more sequences became available and based on a multiple sequence alignment of all the previously known AGPATs, it became apparent that additional isoforms of AGPAT might also exist. This report describes the cloning and enzymatic activity of an isoform we identified as AGPAT11. Because the same cDNA has been recently reported to acylate lysophosphatidyl choline and platelet-activating factor (10, 11), we now identify the clone as AGPAT11/ lysophosphatidylcholine acyltransferase 2 (LPCAT2)/lyso platelet-activating factor acetyltransferase cDNA.
METHODS
Generation of V5-epitope-tagged wild-type AGPAT11 recombinant adenovirus
The strategy for generating recombinant adenovirus using the AdEasy adenoviral system (Stratagene, La Jolla, CA) has been described previously (9). Briefly, the full-length open reading frame (ORF) AGPAT11 cDNA was tagged with V5-epitope at the amino-terminus by amplifying the cDNA from a plasmid containing the ORF for AGPAT11 (the details for the generation of the cDNA for human AGPAT11 are described in the supplementary material provided with this manuscript), with primers 5′- GCGTCGAC-ATG GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACG AGCCGGTGCGCCCAG -3′; the V5 epitope is underlined, and 5′-CGCTCGAGTCAGTCATCTTTTTTGTCTGAGGTAC-3′. The amplified product was cloned in TA-cloning vector, sequenced, restriction digested with XhoI, and cloned in the pShuttleCMV vector at the same site. The orientation of the insert was confirmed by restriction digest and sequencing of the plasmid. To generate recombinant adenovirus, the pShuttle-CMV-AGPAT11 plasmid was digested with PmeI and cotransformed with pAdEasy-1 into BJ5183 cells. The recombinant AdEasy-1-AGPAT11 plasmid was digested with PacI and transfected in AD293 cells using Lipofectamine-2000. Several viral pools were further propagated in the same AD293 cells. The viral pool that showed most enzymatic activity was selected for further amplification and purification using the Virabind adenovirus purification kit (Cell Biolabs Inc., San Diego, CA). The generation of recombinant adenovirus-β-galactosidase has been described previously (9).
Generation of EGFP-tagged wild-type AGPAT11 expression vector
The construction of the wild-type AGPAT11-GFP fusion protein was carried out using the AGPAT11 cloned in TA cloning vector. The ORF for AGPAT11 was amplified using wild-type plasmid as the template and primer pair 13 and 14 (supplementary Table I). The amplified product and the plasmid pEGFP-N3 (Clontech, Mountain View, CA) were restricted with XhoI and BamHI and cloned in the similar sites. The expression plasmid was sequenced to ascertain the orientation and the correctness for the junction sequences.
AGPAT enzymatic activity of AGPAT11 in the cell lysate
For all enzymatic assays, the adenoviral expressed recombinant protein was used. The AD293 cells were infected with AGPAT11 recombinant adenovirus at a multiplicity of infection of 150. It is to be noted that the method employed for the virus purification yielded low level of infectious viral particle. After 48 h, the infected cells were collected and washed once with PBS. The viral pellet was resuspended in the lysis buffer (100 mM Tris, pH 7.4, 10 mM NaCl containing protease inhibitor cocktail, Roche, Indianapolis, IN). The cells were lysed in two different ways: first with three freeze-thaw cycles and second via sonication which was carried out at 65% output (which generates energy of 20 J) for three 6 s bursts with intermittent cooling on ice (Sonics Vibra Cell, Danbury, CT). The cell lysate was centrifuged at 3,000 g for 10 min at 4°C to remove the cell debris. Protein concentration was determined by a commercially available colorimetric assay (Bio-Rad Laboratories, Hercules, CA).
The enzyme activity was determined by measuring the conversion of tritiated lysophosphatidic acid [3H]LPA to tritiated phosphatidic acid [3H]PA as described previously (7, 12). Briefly, the enzymatic reaction was assembled in 200 μl of 100 mM Tris-HCl buffer, pH 7.4, containing the following: 10 μmol/L LPA (oleoyl- sn1-glycerol-3-phosphate, Avanti Polar Lipids, Alabaster, AL), 50 μmol/L oleoyl CoA (Sigma, St. Louis, MO), 1 μl of [3H] oleoyl-LPA (specific activity 30-60 Ci/mmol; PerkinElmer Life and Analytic Sciences, Boston, MA), and 1 mg/ml fatty acid free BSA. The reaction was started by adding 30 μg of cell lysate followed by incubation for 10 min at 37°C. The reaction was terminated by adding 0.5 ml of 1-butanol containing 1 NHCl and to extract phospholipids. The butanol extract was dried under vacuum and the LPA and PA were resolved using the solvent system: chloroform-methanol-acetic acid-water (85:12.5:12.5:3). Radioactive spots were identified by co-migration with unlabeled LPA and PA standard and visualized in iodine vapors. The [3H]LPA and [3H]PA spots were scraped and counted for radioactivity (Tri-Carb Liquid Scintillation Counter 3100TR; PerkinElmer Life Sciences, Boston, MA).
The acyltransferase activity for additional lysophospholipids, LPE, LPG, LPI, and LPS, all containing the C18:1 fatty acid at the sn-1 position, were determined by substituting these lysophospholipids for LPA.
Acyl-CoA specificity
The specificity of esterification of sn-2 position of [3H]LPA (sn-1-oleoyl-2-hydroxy-sn-glycerol-3-phosphate) was determined by substituting the fatty acids with the following acyl-CoAs: octoyl (C8:0)-, decanoyl (C10:0)-, lauroyl (C12:0)-, tridecanoyl (C13:0)-, myristoyl (C14:0)-, pentadecanoyl (C15:0)-, palmitoyl (C16:0)-, heptadecanoyl (C17:0)-, stearoyl (C18:0)-, oleoyl (C18:1)-, linoleoyl (C18:2)-, linolenoyl (C18:3;n-3)-, γ-linolenoyl (C18:3;n-6)-, nonadecanoyl (C19:0)-, arachidoyl (C20:0)-, arachidonoyl (C20:4;n-6)-, heneicosanoyl (C21:0)-, behenoyl (C22:0)-, docosahexaenoyl (C22:6;n-3)-, tricosanoyl (C23:0)-, lignoceroyl (C24:0)-, nervonoyl (C24:1;n-9), pentacosanoyl (C25:0)-, and hexacosanoyl (C26:0)-CoA. The assay conditions remained the same as described for the AGPAT activity.
sn-1-Acyl-lysophosphaditic acid specificity
To determine the specificity of various LPAs for the enzymatic activity, the following LPAs with various fatty acids at the sn-1 position were used for the assay: palmitoyl (C16:0)-, heptadecanoyl (C17:0)-, stearoyl (C18:0)-, and arachidonoyl (C20:4). The enzymatic activity of these LPA species was compared with that of sn-1-oleoyl (C18:1)-2-hydroxy-sn-glycerol-3-phosphate. The enzymatic assay was assembled as before except that unlabeled LPAs and [14C]oleoyl-CoA were used as substrates, and the activity was determined by formation of [14C]PA.
GPAT assay
GPAT activity was determined as described earlier (9). Briefly, Huh7 cells depleted of AGPAT2 expression by shRNA were infected with recombinant adenoviruses expressing either GPAT1 or AGPAT11 at a multiplicity of infection of 150, and the cell lysate was prepared as described above. The enzyme reaction was assembled by adding 60 μM acyl-CoA, 150–800 μM glycerol-3-phosphate (G-3-P), spiked with either [14C] glycerol-3-phosphate (American radiolabels) or [14C]oleoyl CoA (Perkin Elmer) in a buffer mix consisting of 75 mM Tris, pH 7.4, 2 mg/ml BSA, 4 mM MgCl2, and 2 mM DTT in total volume of 200 μl. The enzymatic reaction was started by adding 100 μg of cell lysate and further incubated for 20 min at room temperature. The lipid extraction procedure was the same as for the acyltransferase assay. The product formed (LPA) was resolved on TLC using the following solvent system: chloroform:methanol:acetic acid:H2O (50:25:8:4). G-3-P and LPA spots were identified by exposure to iodine, scraped, and quantified. In some assays, PA spots were also counted for erroneous AGPAT activity.
Western blot analysis
Cells overexpressing V5-tagged AGPAT11 were collected and lysed in RIPA buffer containing protease inhibitor cocktail (Roche, Indianapolis, IN). Lysates were centrifuged at 1,000 g for 10 min to remove cellular debris to obtain the fraction containing mitochondria, microsomes, and cytosol (WCL). The supernatant was further fractionated into mitochondria, microsomes, and cytosol by differential centrifugation. Protein concentrations were determined using Bio-Rad DC protein assay (BioRad, Hercules, CA). Total protein (50 μg) from each fraction was resolved on 10% SDS-PAGE followed by transfer onto nylon membrane (Millipore, Billerica, MA). The membrane was blocked with 5% non-fat milk containing 0.2% Tween-20 and then incubated with V5 antibody (dilution 1:5,000) conjugated to horseradish alkaline phosphatase (Invitrogen) for 2 h at room temperature. The membrane was washed and then incubated with ECL reagent (GE Healthcare Piscataway, NJ) and exposed to X-ray film to visualize the immunoreactive protein. The same blots were stripped using the restore, a Western blot stripping buffer (Pierce, Rockford, IL), according to the manufacturer's protocol, reprobed with anti-GAPDH antibody at a dilution of 1:5,000, and detected with IgG conjugated with horseradish peroxidase at a dilution of 1:5,000.
Immunofluorescence microscopy
Stably expressing AGPAT11-EGFP CHO cells were grown on cover slips 1 day before the experiment. HeLa cells were plated on cover slips in a 6-well plate and transiently transfected with 1.5 μg of AGPAT11-EGFP 24 h before the experiment. Cells were fixed/permeabilized by incubation with methanol (−20°C) for 20 min. Cells were washed 3 × 5 min with PBS and then incubated with primary antibody (Sec61β at a diluted of 1:200, Upstate, Lake Placid, NY) for 30 min at 37°C in a humidified chamber. Cells were then washed 3 × 5 min with PBS and incubated with AlexaFluor 598 coupled fluorescent secondary antibody (Invitrogen) for 30 min at 37°C in a humidified chamber. After incubation, cells were washed 3 × 5 min with PBS, counter stained with 4’-6-diamidino-2-phenylindole during the washes and mounted on a glass slide with Aqua Poly/Mount (Polysciences, Inc., Warrington, PA). Cells were observed with DeltaVision RT Deconvolution Microscope (Applied Precision, LLC; Issaquah, WA).
Quantitative real-time PCR in human tissue panel
Quantitative PCR was performed using the TaqMan primer and probes and designed using primer express software, ABI PRISM 7700 sequence detection system, and PRIMER EXPRESS analysis software (13) using the human cDNA panel from Clontech (Palo Alto, CA). To amplify AGPAT11, 100 pg of cDNA was added to the forward (5′-CACTCTTTGACAGGAACCATGATG-3′) and reverse (5′-ACAGCCAGGCCAATCACATAC-3′) primers along with fam-labeled probe (CAGCATTGACTTCCG) and universal mix containing AmpliTaq and appropriate buffers. The PCR was followed for 40 cycles of 94°C for 15 s and 60°C for 30 s. The cDNA was amplified in duplicate along with G3PDH as an internal control. The ΔCt value for each tissue was calculated as ΔCt = [Ct (tissue) − Ct (G3PDH)] where necessary.
Human tissues and cell lines
To determine the expression of AGPAT11 in various carcinomas, we used cultured cells derived from the human colon, breast, and cervix cancer tissues, as well as human carcinoma tissues. The HEK293, T84, HT-29, SW480, SW620, CaCo-2, MCF-7, and MDAMB231 were from ATCC (Manassas, VA). All cells were cultured in DMEM supplemented with 10% serum and antibiotics, except HT-29, which was cultured in McCoy's 5a medium, SW480, and SW620 in Leibovitz's L-15 medium and T84 in DMEM/F12 1:1 medium. The human tissues were obtained through The Cooperative Human Tissue Network, Southern Division, University of Alabama at Birmingham, Birmingham, AL.
Total RNA isolation and reverse transcription
The cultured cells were lysed in RNA Stat-60 (Tel-Test, Inc., Friendswood, TX). Similarly, the normal and carcinoma tissues were homogenized in RNA Stat-60 to obtain total RNA as suggested by the manufacturer. A known quantity of total RNA (1 μg) was treated with DNase I to remove residual DNA, and the RNA was reverse transcribed with omniscript RT kit following the manufacturer's protocol (Qiagen) in a total volume of 20 μl. The expression of AGPAT11 was quantified using Assay on Demand TaqMan Master Mix from ABI (primer/probe set Hs00215225_ml).
Statistical analysis
All quantitative enzymatic data are shown as mean ± SD. P-values were obtained using a Student's t-test. Statistical analyses were performed using the SAS version 9.13 (SAS Institute, Cary, NC).
RESULTS
Tissue distribution and quantitative expression in human tissue panel
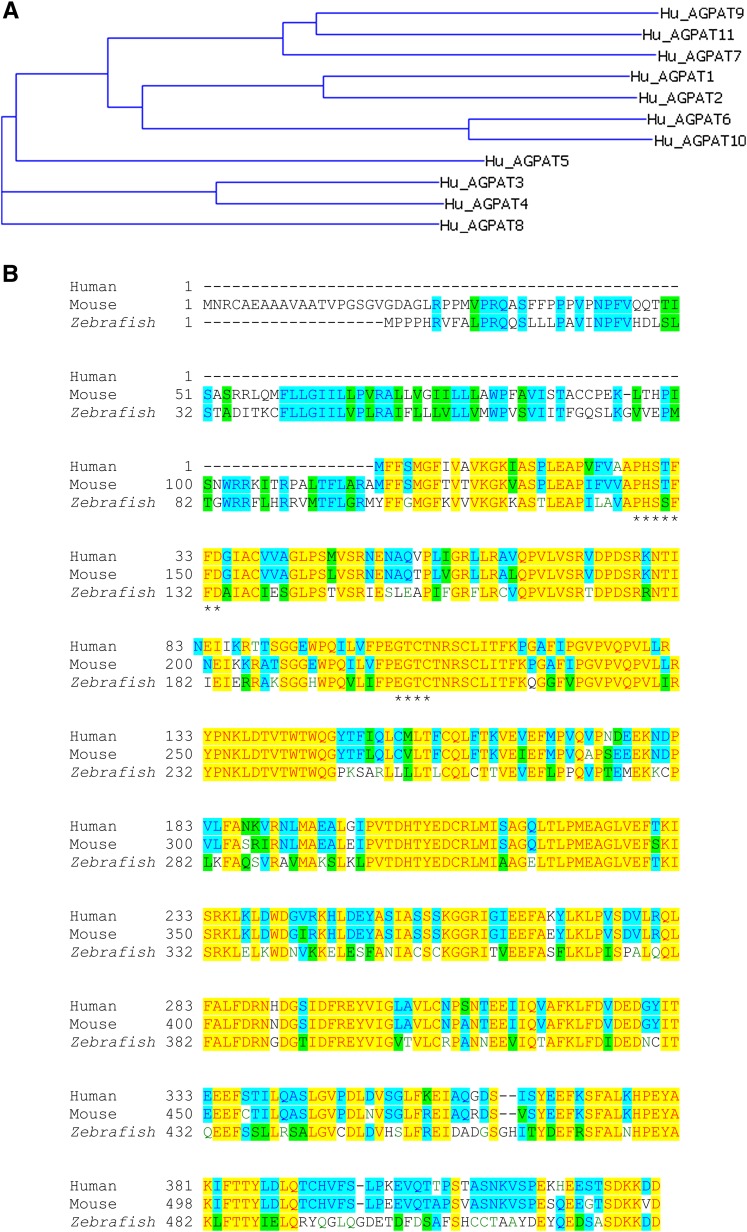
The northern blot analysis detected two mRNA of approximate sizes close to 4 and 5 kb in heart, skeletal muscle, and peripheral leukocyte (supplementary Fig. I). We expanded our search for expression in human tissues by using cDNA and quantified using real-time PCR. Shown in Table 1 are the ΔCt values for various tissues after being normalized to the housekeeping gene G3PDH. In this tissue panel, the highest expression was detected in the lung and the leukocytes, with slightly decreased expression in the spleen and adipose tissue. The multiple protein alignment of known AGPATs reveals that human AGPAT11 is very close to the human AGPAT9 (Fig. 1A). The mRNA expression data for the two isoforms, AGPAT9/LPCAT1, which we reported earlier (8), and AGPAT11/LPCAT2 (this study), are also similar to what we reported previously. Quantitative real-time PCR revealed that the two isoforms have very similar expression patterns for most tissues in humans, except that AGPAT11/LPCAT2 is expressed 1- to 2-fold less than AGPAT9/LPCAT1. However, in the lung, spleen, and leukocytes, AGPAT11 is expressed 5- to 6-fold less than the AGPAT9.
TABLE 1.
Tissue distribution of human AGPAT11 as quantified by TaqMan real-time PCR
| Tissue | Ct values | Tissue | Ct values |
|---|---|---|---|
| Heart | 11.00 | Prostate | 8.85 |
| Brain | 9.82 | Testis | 8.39 |
| Placenta | 8.06 | Ovary | 8.60 |
| S. muscle | 14.81 | S. intestine | 8.64 |
| Kidney | 9.41 | Colon | 8.80 |
| Pancreas | 8.12 | Leukocyte | 6.98 |
| Lung | 6.99 | Adipose | 7.84 |
| Spleen | 7.31 | Liver | 11.14 |
| Thymus | 10.31 |
Shown are the mean ΔCt values, performed in duplicate and corrected for internal control, G3PDH. S. muscle, skeletal muscle; S. intestine, small intestine.
Fig. 1.
Multiple sequence alignment. A: Evolutionary dendrogram and conserved protein motifs among AGPATs. The dendrogram was computed from the aligned protein sequences using the ClustalW algorithm and VectorNTi's phylogenetic tree program with default settings. B: Alignment of amino acid sequences of AGPAT11 from the human, mouse, and zebrafish. The two conserved motifs, N/PHX4D and EGTR/C, are marked with asterisks.
Comparison with other known AGPATs
Multiple sequence alignment of the known human AGPATs revealed that AGPAT11 was more similar to AGPAT9 than any other acyltransferases within the family (Fig. 1A). The two isoforms are 58% homologous and 41.5% identical at the amino acid level. The signature motifs involved in catalytic and substrate binding, NHX4D and EGTR, are conserved in all the phospholipid acyltransferases. However, alignment of AGPAT11 human isoform with mouse and zebrafish showed that the arginine residue in the EGTR motif is substituted by cysteine, and in motif NHX4D, the asparagine is substituted with proline (Fig. 1B).
Subcellular localization of AGPAT11-GFP
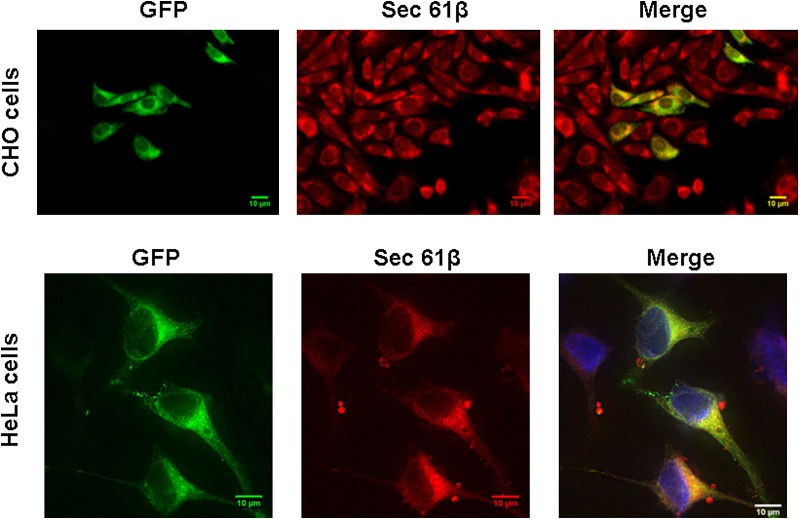
In CHO cells stably expressing the human AGPAT11-GFP fusion protein, fluorescence microscopy showed an endoplasmic reticulum (ER)-like expression pattern. This subcellular localization was confirmed by colocalizing AGPAT11-GFP with the ER-specific protein sec61-β (Fig. 2). The subcellular localization was further confirmed in HeLa cells, except that the cells were transiently transfected with the AGPAT11-GFP construct, which again localized to ER (Fig. 2).
Fig. 2.
Localization of AGPAT11-GFP to ER in cultured cells. Upper panel: CHO cells overexpressing AGPAT11-GFP were fixed in methanol and incubated with antibody, specific for ER, sec61 and imaged for green and red fluorescence using fluorescence microscopy. Shown are representative images for sec61β (red fluorescence), AGPAT11 (green fluorescence), 4’-6-diamidino-2-phenylindole (blue fluorescence), and the merge. Lower panel: The HeLa cells were transiently transfected with AGPAT11-GFP construct. The cells were fixed as above after 48 h of transfection and processed as for CHO cells.
AGPAT activity
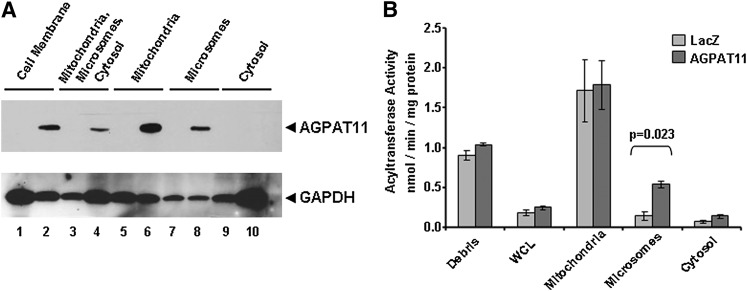
We initially created stable CHO cells expressing AGPAT11. However, we were unable to detect any significant AGPAT activity compared with those expressing β-galactosidase. Most likely, this was due to excessive background (data not shown). We then generated recombinant adenovirus expressing AGPAT11 and expressed it in the AD293 cells. To determine the expression of AGPAT11 protein, the cell lysates were fractionated into subcellular fractions and subjected to Western blot analysis. As shown in Fig. 3A the expression of AGPAT11 could be recovered in both the mitochondria and microsomes, but not in the cytosol fraction. Immunofluorescence localization of AGPAT11 did not reveal that the protein localizes to the mitochondria other than the ER. This could be due to a couple of reasons: it could be an artifact of the overexpression of the recombinant adenovirus in AD293 cells, or, alternatively, it could be the feature of the host cells. When the enzymatic activity was determined in various cellular fractions, only the microsomal fraction showed significant AGPAT activity (Fig. 3B).
Fig. 3.
Western blot of wild-type human AGPAT11 and AGPAT enzymatic activity in AD293 cells. A: The Western blot for the recombinant AGPAT11 protein from the whole cell lysate and various subcellular fractions, as probed with V5-antibody. LacZ cell lysates were loaded as a negative control. Even-numbered lanes represent recombinant AGPAT11 and odd-numbered represent β-galactosidase (control). The same blot was stripped and reprobed with full form G3PDH to demonstrate protein loading. B: The AGPAT activity determined as conversion of 3H-LPA to 3H-PA in the presence of oleoyl-CoA and expressed as product (3H-PA) formed per minute per milligram protein. The LPA to PA conversion by recombinant adenovirus β-galactosidase was used as a control. Not shown is the conversion of substrate in the absence of enzyme. Each bar represents mean ± SD from two independent experiments carried out in triplicate. WCL, whole cell lysate. P-value is shown above the bars.
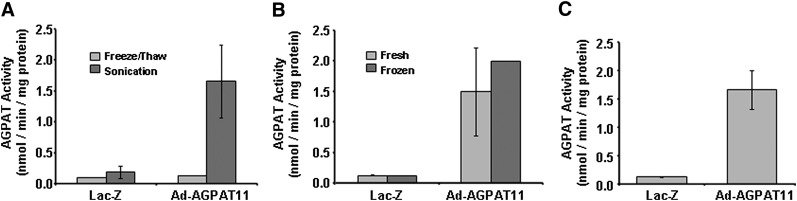
Because we observed AGPAT activity for AGPAT11 in microsomes and its detection in mitochondria by Western blot, for all further enzymatic assays we employed postnuclear fraction. We then performed preliminary optimization for the AGPAT enzymatic activity. First, the cells were subjected to either freeze-thaw or only sonication of the cellular lysate and determined the AGPAT activity. As shown in Fig. 4A, the sonicated lysate retained much more AGPAT activity than the freeze-thaw. We then determined enzymatic activity of the cell lysate obtained by sonicating either freshly prepared or frozen cell lysate. In our hands, there was no significant difference between the two processing methods (Fig. 4B). Therefore, for all the enzymatic activity assays, we used cellular lysate obtained by sonication of frozen lysates for ease of the experiments (Fig. 4C).
Fig. 4.
Optimization of AGPAT11 enzymatic activity in cellular lysates. A: AGPAT activity in cell lysates upon processing of the cells either by three cycles of freeze-thaw or by sonication. B: The AGPAT activity when determined either with freshly prepared lysates or frozen and thawed again. C: AGPAT activity obtained from the cellular lysate frozen and used for the enzymatic activity. All data are expressed as mean ± SD from two independent experiments in triplicate.
Substrate specificity of AGPAT11
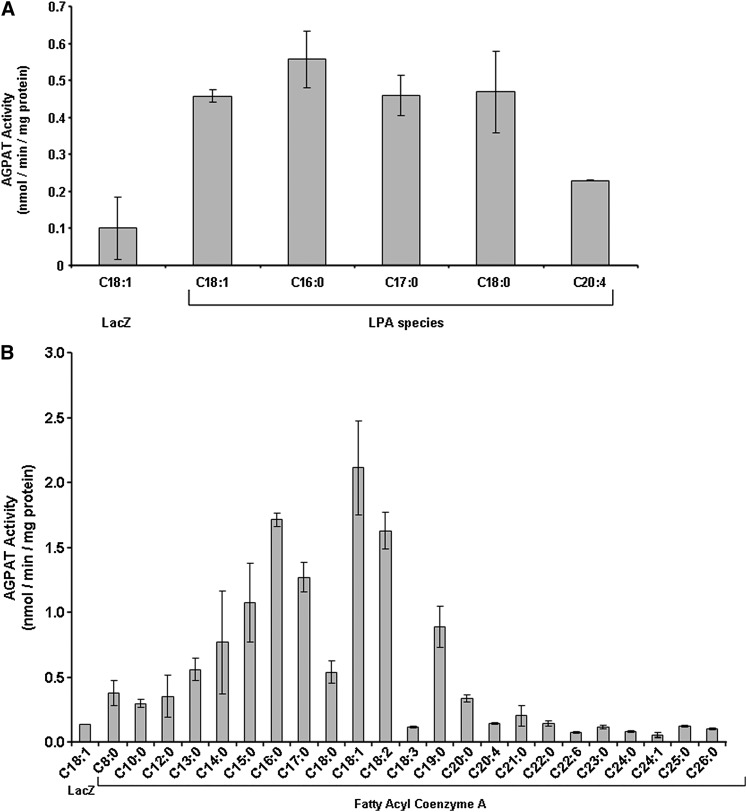
Because human AGPAT11 revealed AGPAT activity, we then determined its acyl-CoA and LPA specificity. We employed a panel of LPAs with various long-chain fatty acids at the sn-1 position and using oleoyl-CoA to determine the AGPAT activity as shown in Figure 5A. AGPAT11 appears to have broader preferences for the LPA containing saturated fatty acids C16:0-C18:0, including LPA containing monounsaturated fatty acid (C18:1). LPA with fatty acid C20:4 is acylated at only about one-half maximal rate compared with other LPAs.
Because the LPA with C18:1 fatty acid retained significant enzymatic activity, we used this LPA as the acyl acceptor to determine the specificity of various acyl-CoA. The panel consists of fatty acids from C8:0 to C26:0, including some with unsaturated fatty acid. The preferred acyl donor was in the order of C18:1 > C16:0 > C18:2 > C17:0. Fatty acids consisting of short-chain carbon: C8:0 to C13:0 and very long-chain from C20:0 to C26:0 retained no enzymatic activity (Fig. 5B). There also appeared to be an upper limit for the C18 fatty acid unsaturation. The acyl-CoA (C18:3) consisting of three double bonds did not acylate the LPA to PA.
Fig. 5.
Specificity of AGPAT11 for various LPA and Acyl-CoA. A: LPA specificity of the recombinant human AGPAT11 expressed in AD293 cells. Specificity of human recombinant AGPAT11 for various species of sn-1-LPA acceptors was determined using oleoyl-CoA as donor. Various species of LPA with long-chain fatty acids at sn-1 position were used as acceptors. Activity was expressed as product (3H-PA) formed per minute per milligram protein. All enzymatic activities were determined in two independent experiments in triplicate. Bar represents mean ± SD. B: Acyl-CoA specificity of recombinant human AGPAT11 expressed in AD293 cells. Specificity of human recombinant AGPAT11 for acyl-CoA donors were determined using sn-1-oleoyl-LPA as an acceptor and various short-, medium-, and long-chain fatty acyl-CoA as donors. All enzymatic activities were determined in two independent experiments in triplicate. Bar represents mean ± SD.
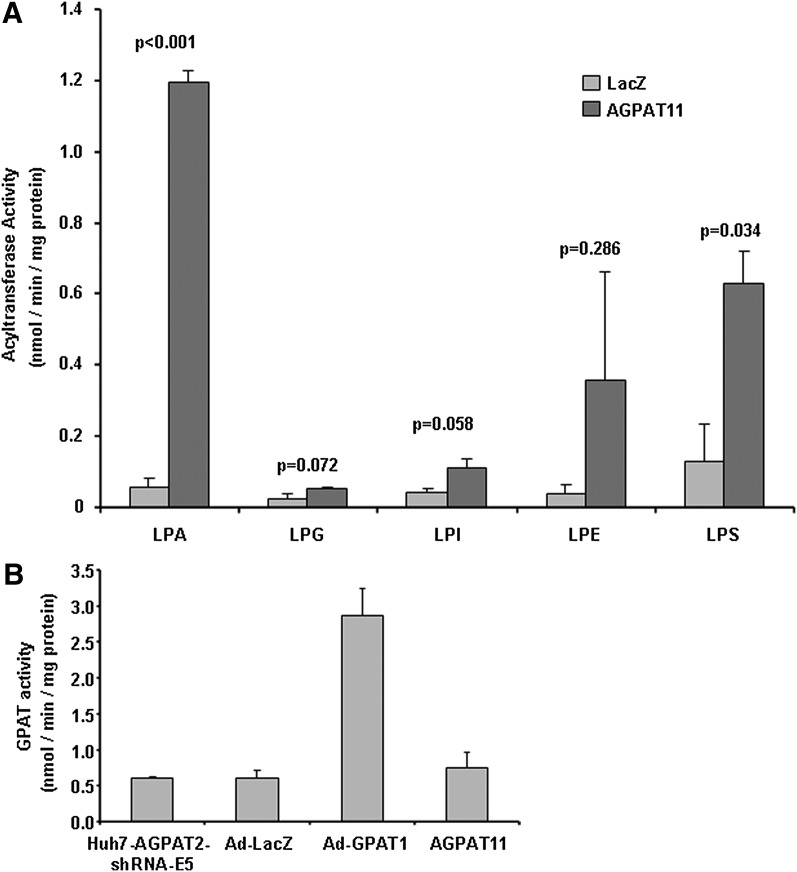
We employed various other lysophospholipids, LPS, LPE, LPI, and LPG, to ascertain whether AGPAT11 could esterify these phospholipids. AGPAT11 did not show any significant acyltransferase activity above the background for LPG, LPE, or LPI. LPS is the only other lysophospholipid for which AGPAT11 retained some enzymatic activity, but is not as robust as when LPA is the acyl acceptor (Fig. 6A). We then investigated if the AGPAT11 has any additional acyltransferase activities. It did not show GPAT activity (Fig. 6B).
Fig. 6.
Specificity for acyltransferase activity of AGPAT11 for lysophospholipids LPA, LPG, LPI, LPE, and LPS and for glycerol-3-phosphate. A: The enzymatic activity was determined as the conversion of various lysophospholipids to their corresponding [3H]phospholipids in the presence of [3H] oleoyl-CoA and expressed as product [3H]phospholipids formed per minute per milligram protein. The recombinant adenovirus β-galactosidase was used as a control. Not shown is the conversion of substrate in the absence of enzyme. Shown are the mean values from two independent assays carried out in triplicate. Bar represents mean ± SD. P-values are shown above the bars. B: GPAT enzymatic activity of AGPAT11 in Huh-7 cells depleted for AGPAT2 via shRNA (HuH7-AGPAT2-shRNA-E5) cell lysate. The GPAT activity was determined by incubating [14C]glycerol-3-phosphate with an acyl-CoA. Recombinant adenovirus expressing human GPAT1 was included as a positive control. The recombinant adenovirus β-galactosidase was used as a control. Not shown is the conversion of substrate in the absence of enzyme. Bar represents mean ± SD.
Expression of AGPAT11 in human cancer tissues and cultured cell lines
Because human AGPAT11 has significant homology to human AGPAT9/LPCAT1 that is shown to be upregulated in human colorectal cancer (CRC), we investigated whether the expression of AGPAT11 was also modulated in CRC. We examined the expression of AGPAT11 in cancer cell lines (Fig. 7A) compared with HEK293 cell line (which does not represent a cancer cell). The human breast cancer cells MCF-7 and MDAMB231 did not overexpress AGPAT11 compared with the HEK293 cells. The expression of AGPAT11 in HeLa cells, which are derived from human cervical cancer, doubled compared with HEK293 cells. In the CRC cell lines HT29, SW480, and CaCo-2, AGPAT11 showed 2-fold, 5-fold, and 3- to 5-fold increased expression, respectively; however, T84 and SW620 cells did not. It is interesting to note that SW620 is derived from SW480 but seems to lose its ability to overexpress AGPAT11. We also examined the expression of AGPAT11 in pathological tissues. In human cancer tissues, there is a distinct upregulation of AGPAT11 in breast cancer tissue compared with normal breast (Fig. 7B). Because in normal breast, AGPAT11 expression is undetectable, it is not possible to present any fold increase in the expression of the AGPAT11 mRNA. The case for cervical cancer is similar (Fig. 7C). In CRC tissues, there is at least a 3- to 4-fold increase in the expression of AGPAT11 (Fig. 7D).
Fig. 7.
Expression of AGPAT11 mRNA in cultured tumor cells and human tumor tissues. A: Shown are the quantitative expression of AGPAT11 in cultured cells representing CRC (T84, HT29, SW480, SW620, and CaCo-2), breast cancer (MCF-7 and MDAMB231), and cervical cancer (HeLa). The expression is normalized to 18S housekeeping gene and expressed relative to HEK293 cells. B: Expression of AGPAT11 in normal and cancer tissues from breast. The quantitative PCR values are expressed as an arbitrary unit where 1 represents Ct = 40 and 100 represents Ct = 1. Arbitrary units were obtained from two independent determinations. Because of undetectable expression in normal breast, the expression data are not normalized to 18S. The lower panel shows the same PCR analyzed on 2% agarose gel stained with ethidium bromide. Lane 1, normal breast tissue with fibrocystic changes; lanes 2–4, breast cancer tissues with varying degrees of tumor. C: Expression of AGPAT11 in normal and cervical cancer tissues from cervix. The quantitative PCR values are expressed as an arbitrary unit where 1 represents Ct = 40 and 100 represents Ct = 1. Each bar represents the mean Ct values from two independent determinations not normalized to 18S because of undetectable levels of AGPAT11 in normal tissues. The lower panel shows agarose gel analyses: lanes 1 and 2, normal cervical tissues; lanes 3–5, the cervical cancer tissues. D: Expression of AGPAT11 in normal and tumor tissues from CRC. In one out of three normal colorectal tissues, the expression of AGPAT11 was undetectable, and in two out of three colorectal cancerous tissues its expression increased compared with normal tissues. The quantitative PCR values are expressed as an arbitrary unit where 1 represents Ct = 40 and 100 represents Ct = 1. The lower panel shows the agarose gel analyses: lanes 1–3 are from normal and lanes 4–6 are from cancerous tissues. For details on cancer tissues pathology, see supplementary Table III.
DISCUSSION
Our goal has been to identify new isoforms for the AGPAT enzymes and characterize their tissue expression patterns and enzymatic activities in the hope that one of these isoforms, in addition to AGPAT2, is also highly expressed in the adipose tissue, which might provide additional genetic loci for patients with lipodystrophy. We have earlier reported mutations in the AGPAT2 gene that lead to the loss of adipose tissue in humans (6). This observation has now been corroborated in Agpat2 null mice (14).
In this study, we demonstrate that the recombinant AGPAT11 protein had AGPAT enzymatic activity when expressed in AD293 cells and it localizes to the ER. This enzyme prefers LPA with oleic acid at its sn-1 position as an acyl acceptor as well as oleoyl-CoA as the acyl donor. This substrate specificity is very similar to AGPAT9 and 10, which we reported earlier (8, 9). We also showed that AGPAT11 lacks enzymatic activity toward other lysophospholipids, such as LPE, LPG, and LPI. However, we did observe reduced but statistically significant acyltransferase enzymatic activity for LPS, which was only 50% of that for LPA. The significance of this substrate specificity remains unclear from this study. The AGPAT11 has no GPAT activity. Other investigators have shown that this cDNA has LPCAT activity as well (10, 11). Although we could show robust AGPAT activity, the LPCAT activity did not reach statistical significance (data not shown). It remains unclear why we were unable to detect LPCAT activity. Thus, the results of ours and others require additional experiments to verify these discrepancies.
The human AGPAT11 is located on chromosome 16q.13 and consists of 14 exons (supplementary Table II). When aligned with other known AGPATs, AGPAT11 shows significant homology to AGPAT9/LPCAT1 (Fig. 1A) (15). The human AGPAT11 is highly conserved with the mouse and zebrafish (Fig. 1B). However, there is no ortholog for this gene to be found in D. melanogaster or C. elegans. Tissue expression patterns, using quantitative RT-PCR analysis, revealed that AGPAT11 is highly expressed in the lung, spleen, and leukocytes and also present in most other tissues. It is least expressed in the heart, liver, and thymus. This tissue expression pattern is also very similar to that of human AGPAT9, reported earlier, except for qualitative differences (8).
The expression of AGPAT2 is upregulated in ovarian cancer (16). Patients with ovarian cancer have higher levels of circulating LPA, and the ascites fluid is enriched in LPA level (17). Because the human AGPAT9/LPCAT1 is reported to be upregulated in CRC (18), and with the close homology of this isoform to AGPAT11, we examined the human tumor tissues for the expression of AGPAT11. We looked at the cultured cells as well as those from the tumor tissue. Compared with the normal human breast tissue (with some fibrocystic changes), breast cancer tissue (with 50–100% tumor tissue) showed significantly increased expression of AGPAT11. We also observed upregulation of AGPAT11 in cervical cancer tissue (tumor grade 15–90%). The expression of AGPAT11 was reflective to the degree of tumor tissue present. The higher the tumor grade, the greater was the expression of AGPAT11. In CRC tissues, the expression was increased for AGPAT11. However, we observed that in normal colon, AGPAT11 is expressed as well, but less than that observed in the tumor tissue. Thus, we show that in breast and cervical cancers, AGPAT11 is activated, because normal breast and cervical tissue do not reveal significant expression of AGPAT11. An overexpression of AGPAT11 in these tumor tissues would indicate an elevated level of phosphatidic acid, which itself is a signaling molecule. PA can activate and amplify Ras signaling, resulting in mitogen-activated protein kinase and PI3K/AKT for survival pathways for neoplastic anchorage-independent survival of tumors (19–21). It is also important to mention that a specific PA species might be generated by the overexpression of AGPAT11, required for the activation of downstream target(s) genes for neoplastic cell proliferation and survival. AGPAT9/LPCAT1 is not upregulated in the human breast and cervical cancer tissues (18).
With this report, there are now at least 11 isoenzymes possessing AGPAT activity. An obvious question is why so many of these isoforms are present in the genome, all of which localize to ER, except AGPAT3, which also seems to be present in the Golgi apparatus of the cell (22). It is possible that despite having similar in vitro enzymatic activity, the specificity is accorded in conjunction with additional factor(s) still undetermined. A possibility also exists that some of these isoforms are expressed in specific ER domains providing AGPAT activity in generating specific lipid molecules either for localized cellular function or localized membrane structure. Alternatively, some of these enzymes function in concert with phospholipases to remodel the newly synthesized phospholipids via the Lands cycle (3, 4). It is important to mention in this context that until now, the colocalization of the two enzymes, AGPATs and phospholipases, has not been demonstrated and none of these enzymes has been shown to possess transacylase enzymatic activity.
PA and diacylglycerol, a dephosphorylated product of PA, are precursors for the biosynthesis for various phospholipids and triacylglycerol. It is possible that all actions for these enzymes are within the membrane where they are localized. The phospholipids are integral components of all the cellular membranes, including those of mitochondria, ER, peroxisomes, and plasma membrane. For ER and mitochondrial biogenesis, there is local demand for specific phospholipid species. AGPAT isoforms might fulfill this location-specific role. To understand the functions of various AGPATs, an antibody to each isoform is required to determine their subcellular localization instead of employing GFP-fusion protein. Future experiments shall be directed toward understanding the specific role of each of these AGPATs in tissue models to determine specific subcellular localization (microdomain) and by developing mouse models by homologous gene deletion to understand their function in whole animals. In this context, it is important to mention that when Agpat2 null mice were generated and analyzed, an unexpected role of monoacylglycerol acyltransferase 1 in the biosynthesis of triglycerides in liver was discovered (14).
Supplementary Material
Acknowledgments
We thank Katie Tunison, Robert Barnes, Ruth Giselle Huet, and Diana Varghese for technical assistance, Daniel Linden, AstraZeneca, Sweden, for recombinant adenovirus-GPAT1, and Richard Auchus, MD. Ph.D. for critical review of the manuscript.
Footnotes
Abbreviations:
- AGPAT
- 1-acylglycerol-3-phosphate-O-acyltransferase
- CHO
- chinese hamster ovary cell
- GPAT
- glycerol-3-phosphate acyltransferase
- HeLa
- human (Henrietta Lacks) cervical cancer cell
- LPA
- lysophosphatidic acid
- LPCAT2
- lysophosphatidylcholine acyltransferase 2
- LPI
- lysophosphatidylinositol
- LPE
- lysophosphatidylethanolamine
- LPG
- lysophosphatidylglycerol
- LPS
- lysophosphatidylserine
- ORF
- open reading frame
- PA
- phosphatidic acid
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure and one table.
This work was supported by the National Institutes of Health grants R01-DK54387 and by the Southwestern Medical Foundation. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Coleman R. A., Lee D. P. 2004. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43: 134–176. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal A. K., Garg A. 2003. Congenital generalized lipodystrophy: significance of triglyceride biosynthetic pathways. Trends Endocrinol. Metab. 14: 214–221. [DOI] [PubMed] [Google Scholar]
- 3.Lands W. E. 1960. Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin. J. Biol. Chem. 235: 2233–2237. [PubMed] [Google Scholar]
- 4.Lands W. E., Merkl I. 1963. Metabolism of glycerolipids. III. Reactivity of various acyl esters of coenzyme A with alpha’-acylglycerophosphorylcholine, and positional specificities in lecithin synthesis. J. Biol. Chem. 238: 898–904. [PubMed] [Google Scholar]
- 5.Merkl I., Lands W. E. 1963. Metabolism of glycerolipids. IV. Synthesis of phosphatidylethanolamine. J. Biol. Chem. 238: 905–906. [PubMed] [Google Scholar]
- 6.Agarwal A. K., Arioglu E., de Almeida S., Akkoc N., Taylor S. I., Bowcock A. M., Barnes R. I., Garg A. 2002. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat. Genet. 31: 21–23. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal A. K., Barnes R. I., Garg A. 2006. Functional characterization of human 1-acylglycerol-3-phosphate acyltransferase isoform 8: cloning, tissue distribution, gene structure and enzymatic activity. Arch. Biochem. Biophys. 449: 64–76. [DOI] [PubMed] [Google Scholar]
- 8.Agarwal A. K., Sukumaran S., Bartz R., Barnes R. I., Garg A. 2007. Functional characterization of human 1-acylglycerol-3-phosphate-O-acyltransferase isoform 9: cloning, tissue distribution, gene structure, and enzymatic activity. J. Endocrinol. 193: 445–457. [DOI] [PubMed] [Google Scholar]
- 9.Sukumaran S., Barnes R., Garg A., Agarwal A. 2009. Functional characterization of the human 1-acylglycerol-3-phosphate-O-acyltransferase isoform 10 / glycerol-3-phosphate acyltransferase isoform 3 (AGPAT10/GPAT3). J Mol Endocrinol. 42: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shindou H., Hishikawa D., Nakanishi H., Harayama T., Ishii S., Taguchi R., Shimizu T. 2007. A single enzyme catalyzes both platelet-activating factor production and membrane biogenesis of inflammatory cells. Cloning and characterization of acetyl-CoA:LYSO-PAF acetyltransferase. J. Biol. Chem. 282: 6532–6539. [DOI] [PubMed] [Google Scholar]
- 11.Harayama T., Shindou H., Ogasawara R., Suwabe A., Shimizu T. 2008. Identification of a novel noninflammatory biosynthetic pathway of platelet-activating factor. J. Biol. Chem. 283: 11097–11106. [DOI] [PubMed] [Google Scholar]
- 12.Haque W., Garg A., Agarwal A. K. 2005. Enzymatic activity of naturally occurring 1-acylglycerol-3-phosphate-O-acyltransferase 2 mutants associated with congenital generalized lipodystrophy. Biochem. Biophys. Res. Commun. 327: 446–453. [DOI] [PubMed] [Google Scholar]
- 13.Livak K. J. 1997. ABI Prism 7700 Sequencer detection system. User Bulletin 2. PE Applied Biosystems, Foster City, CA: 1–36. [Google Scholar]
- 14.Cortes V. A., Curtis D. E., Sukumaran S., Shao X., Parameswara V., Rashid S., Smith A. R., Ren J., Esser V., Hammer R. E., et al. 2009. Molecular mechanisms of hepatic steatosis and insulin resistance in the AGPAT2-deficient mouse model of congenital generalized lipodystrophy. Cell Metab. 9: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakanishi H., Shindou H., Hishikawa D., Harayama T., Ogasawara R., Suwabe A., Taguchi R., Shimizu T. 2006. Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J. Biol. Chem. 281: 20140–20147. [DOI] [PubMed] [Google Scholar]
- 16.Diefenbach C. S., Soslow R. A., Iasonos A., Linkov I., Hedvat C., Bonham L., Singer J., Barakat R. R., Aghajanian C., Dupont J. 2006. Lysophosphatidic acid acyltransferase-beta (LPAAT-beta) is highly expressed in advanced ovarian cancer and is associated with aggressive histology and poor survival. Cancer. 107: 1511–1519. [DOI] [PubMed] [Google Scholar]
- 17.Burton A. 2006. LPAAT-beta identifies aggressive ovarian cancer. Lancet Oncol. 7: 893. [DOI] [PubMed] [Google Scholar]
- 18.Mansilla F., da Costa K. A., Wang S., Kruhoffer M., Lewin T. M., Orntoft T. F., Coleman R. A., Birkenkamp-Demtroder K. 2009. Lysophosphatidylcholine acyltransferase 1 (LPCAT1) overexpression in human colorectal cancer. J. Mol. Med. 87: 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Athenstaedt K., Daum G. 1999. Phosphatidic acid, a key intermediate in lipid metabolism. Eur. J. Biochem. 266: 1–16. [DOI] [PubMed] [Google Scholar]
- 20.Yalcin A., Clem B., Makoni S., Clem A., Nelson K., Thornburg J., Siow D., Lane A. N., Brock S. E., Goswami U., et al. 2010. Selective inhibition of choline kinase simultaneously attenuates MAPK and PI3K/AKT signaling. Oncogene. 29: 139–149. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Du G. 2009. Phosphatidic acid signaling regulation of Ras superfamily of small guanosine triphosphatases. Biochim. Biophys. Acta. 1791: 850–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt J. A., Brown W. J. 2009. Lysophosphatidic acid acyltransferase 3 regulates Golgi complex structure and function. J. Cell Biol. 186: 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.