Abstract
The effect of apolipoprotein (apo) E genotype on apoB-100 metabolism was examined in three normolipidemic apoE2/E2, five type III hyperlipidemic apoE2/E2, and five hyperlipidemic apoE3/E2 subjects using simultaneous administration of 131I-VLDL and 125I-LDL, and multi-compartmental modeling. Compared with normolipidemic apoE2/E2 subjects, type III hyperlipidemic E2/E2 subjects had increased plasma and VLDL cholesterol, plasma and VLDL triglycerides, and VLDL and intermediate density lipoprotein (IDL) apoB concentrations (P < 0.05). These abnormalities were chiefly a consequence of decreased VLDL and IDL apoB fractional catabolic rate (FCR). Compared with hyperlipidemic E3/E2 subjects, type III hyperlipidemic E2/E2 subjects had increased IDL apoB concentration and decreased conversion of IDL to LDL particles (P < 0.05). In a pooled analysis, VLDL cholesterol was positively associated with VLDL and IDL apoB concentrations and the proportion of VLDL apoB in the slowly turning over VLDL pool, and was negatively associated with VLDL apoB FCR after adjusting for subject group. VLDL triglyceride was positively associated with VLDL apoB concentration and VLDL and IDL apoB production rates after adjusting for subject group. A defective apoE contributes to altered lipoprotein metabolism but is not sufficient to cause overt hyperlipidemia. Additional genetic mutations and environmental factors, including insulin resistance and obesity, may contribute to the development of type III hyperlipidemia.
Keywords: apoE2/E2 genotype, kinetics, Type III hyperlipidemia
Apolipoprotein (apo) E is a 34.2 kDa glycoprotein synthesized by the liver and to a lesser extent by peripheral tissues (1). It is associated in plasma with triglyceride-rich lipoproteins (TRL) and HDL particles (2). ApoE plays a central role in mediating hepatic recognition and uptake of TRL and their remnants by acting as a ligand for lipoprotein binding to the LDL receptor, LDL receptor-related protein, and glycosaminoglycans (3). ApoE may also activate enzymes involved in lipoprotein metabolism, including hepatic lipases, cholesteryl ester transfer protein, and lecithin cholesteryl:acytransferase (4). The apoE gene is polymorphic, with three common alleles coding for three major isoforms identified as E2, E3, and E4 (5).
Type III hyperlipidemia, also known as dysbetalipoproteinemia, is a rare form of genetic dyslipidemia characterized by elevated plasma cholesterol and triglycerides and the presence of cholesterol-enriched remnant particles, collectively known as β-VLDL (6). The accumulation of β-VLDL particles may be associated with impaired TRL catabolism (7). Type III hyperlipidemic subjects may have severe xanthomatosis and significantly increased risk of developing premature atherosclerosis (8). The majority of type III hyperlipidemic subjects are homozygotes for the apoE2 allele of the apoE (APOE) gene. However, <10% of apoE2 homozygotes develop hyperlipidemia, and most apoE2/E2 subjects are either normolipidemic or hypocholesterolemic (6). The onset of hyperlipidemia may be associated with additional genetic factors and/or environmental factors such as obesity, diabetes, and menopausal status (6). The regulation of lipoprotein transport in subjects with the apoE2/E2 genotype, in particular those without type III hyperlipidemia, is not well known. A better understanding of the mechanisms underlying the lipid and lipoprotein changes in apoE2/E2 subjects is potentially of clinical importance.
In this study, we investigated the kinetics of VLDL, intermediate density lipoprotein (IDL), and LDL apoB using simultaneous administration of 131I-VLDL and 125I-LDL and multi-compartmental modeling in apoE2/E2 subjects with or without type III hyperlipidemia, and a group of hyperlipidemic subjects heterozygous for the apoE allele.
METHODS
Subjects
Thirteen Caucasian subjects (eight males and five females) were studied. Eight of these subjects had the apoE2/E2 genotype and five had the apoE3/E2 genotype. Of the eight with the apoE2/E2 genotype, three were normolipidemic and the remaining five were classified with type III hyperlipidemia (9). Patient 6 was subsequently shown to be heterozygous for E2 (Arg 158→Cys) and E2 Christchurch (Arg 136→Ser) (10). The presence of other apoE2 mutations, including E2 Christchurch and E2 Dunedin (Arg 228 →Cys) (11), were examined and excluded in subjects with the apoE3/E2 genotype. None of these subjects had a history of diabetes or other metabolic disorder, cardiovascular disease, renal dysfunction (macroproteinuria and/or serum creatinine >150 μmol/L), alcohol consumption >30 g alcohol/day, or were taking lipid-modifying agents at the time of the study. All subjects provided informed written consent, and the study was approved by the Ethics Committee of the North Canterbury Hospital Board.
Preparation of labeled lipoproteins
Blood samples were collected from subjects using 0.1% EDTA following a 14 h overnight fast. Plasma was separated immediately by centrifugation. VLDL (density < 1.006 g/ml) and LDL (1.019 < density < 1.063 g/ml) were isolated by ultracentrifugation at 115,000 g for 17 h at 4°C in a Kontron TFT 45.6 fixed angle rotor (Watford, Hertfordshire, UK). VLDL and LDL were washed and concentrated by recentrifugation for 17 h at 4°C at background densities of 1.006 g/ml and 1.06 3g/ml, respectively. LDL was dialyzed extensively against 0.15M NaCl, pH 7.4, to remove sodium bromide prior to labeling.
VLDL and LDL were labeled with 131I and 125I (obtained from the Radiochemical Center, Amersham, UK), respectively, using the iodine monochloride method of McFarlane (12). Unbound iodine was removed by passage of the sample through Sephadex G10 eluted with sterile saline. Residual free iodine was removed by dialysis against sterile saline containing 0.001% EDTA. 131I-VLDL was dialyzed for 2 h and then centrifuged at d = 1.006 g/ml for 15 h at 115,000 g. 125I-LDL was dialyzed for 16 h with two changes of buffer. Prior to injection, the labeled VLDL and LDL were each made up to a volume of 8 ml with sterile saline and sterilized by 0.45 μm Millipore filtration (Millipore, New Zealand). The lipid labeling was 6–13% in the 131I-VLDL and 1-13% in the 125I-LDL.
Study protocol
Subjects were admitted to the metabolic ward and fasted 14 h overnight prior to injection of the labeled lipoproteins. They received a bolus injection of labeled lipoproteins, 20–30 μCi 131I-VLDL and 10–20 μCi 125I-LDL, administered within 96 h of the initial blood collection via an indwelling catheter placed in a forearm vein. A total of 10 blood samples were collected within the first 24 h and a further 8–10 samples over the next 56–72 h. Subjects were discharged from the metabolic ward 72 h postinjection. Additional daily fasting blood samples were collected until 14 days postinjection. In addition, 24 h urine samples were collected daily for the 14 day period.
During the initial 48 h postinjection, subjects consumed a 1,500 kcal diet containing <5 g of fat/day to minimize intestinal production of chylomicrons. During the first 48 h of the kinetic study, plasma triglycerides remained in steady state, varying by <10% of the mean value. After the initial 48 h, subjects adhered to their usual low animal fat diet (30% of calories from fat, 15% from protein, and 55% from carbohydrates). Potassium iodide (50 mg/day) was given to prevent thyroidal uptake of radioiodine until the completion of the study.
Analysis of samples
VLDL, IDL, and LDL were isolated by sequential ultracentrifugation at densities of 1.006, 1.019, and 1.063 g/ml, respectively. ApoB radioactivity in VLDL, IDL, and LDL was determined by precipitation of apoB using redistilled tetramethyl urea (TMU) (Sigma Chemical Co. Ltd., St Louis, MO) followed by extraction of the remaining lipids with organic solvents as previously described (13). In brief, equal volumes of VLDL and TMU were mixed and incubated at 37°C for 10 min. Insoluble apoB and lipid was separated by centrifugation at 8,000 g for 20 min, and the TMU soluble material was removed. Lipid was extracted overnight at −20°C in 3 ml of ethanol:ether (3:2, v/v) followed by two further ether extractions. The apoB precipitate dried under nitrogen was dissolved in 1 ml of 0.5M NaOH. Aliquots of the solubilized apoB were taken for counting and for protein determination (14). ApoB-specific radioactivity was then calculated as counts per minute per milligram of apoB protein. For kinetic analysis, VLDL, IDL, and LDL apoB activity data were expressed as a fraction of the injected dose per 1 ml of plasma.
Determination of apoB pool sizes
Aliquots of the VLDL samples were pooled and washed by recentrifugation at d = 1.006 g/ml. On these respun samples, VLDL apoB concentration was determined as the difference between total VLDL protein measured by the method of Lowry et al. (14) using BSA as the standard curve and the TMU soluble protein concentration (15). Correction for incomplete recovery was determined by the measurement of triglyceride concentrations in the original and respun VLDL samples. The apoB concentration was multiplied by a factor of 0.77 to correct for the difference in chromogenicity between apoB and BSA (16). Aliquots of LDL samples were similarly pooled, respun at density = 1.063 g/ml, and LDL apoB concentration was determined by the method of Lowry et al. Correction for incomplete recovery was based on cholesterol measurements in the original and respun LDL samples. VLDL, IDL, and LDL apoB pool sizes were derived by multiplying their concentrations by plasma volume; plasma volume was estimated as 4.5% of body weight.
Additional analyses
ApoE isoforms were isolated from delipidated VLDL using isoelectric focusing with Pharmacia ampholytes (Sci-Med Ltd., New Zealand). Lipoprotein electrophoresis was performed according to the Helena Laboratories method on cellulose acetate strips (Chemac Laboratories Ltd., New Zealand) (9–11, 17).
Kinetic analysis
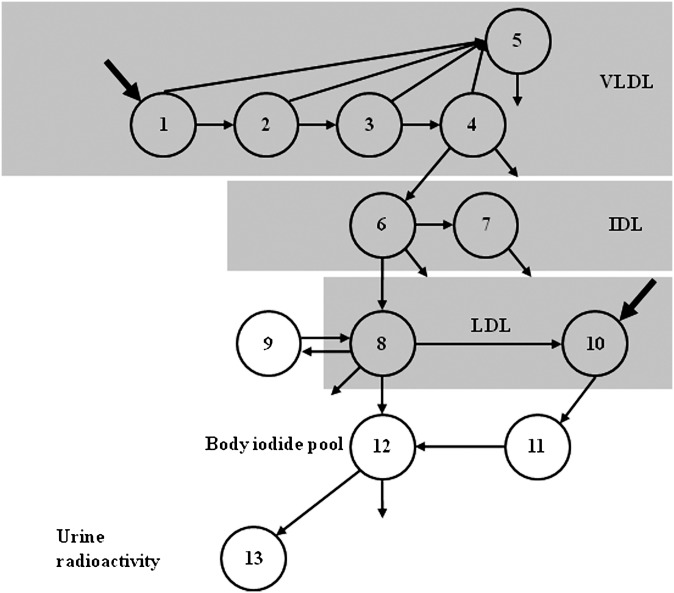
The SAAM II program was used for modeling the data. The 131I and 125I-derived tracer data were modeled simultaneously. This approach permitted the integration of all tracer data into a single model (Fig. 1). Five compartments are used to describe the kinetics of apoB in VLDL, which takes into account the delipidation cascade (compartments 1–4) and a slowly turning over VLDL pool (compartment 5). The residence time in each compartment in the chain is the same. Evidence for the existence of a slowly turning over population of particles can be observed in the terminal slope of the VLDL decay curve. VLDL particles can be converted to IDL or removed directly from plasma. While IDL (and potentially LDL) particles may be derived from the slowly turning over VLDL pool (compartment 5), the coefficient of variation associated with such pathways exceeded 100%. Although such conversion may exist, the present study does not permit us to resolve this, and additional data would be required to accurately define such pathways. Plasma IDL kinetics are described by two compartments (compartments 6 and 7), where one represents a slowly turning over pool of IDL (compartment 7), as observed in the tail of the IDL radioactivity curve. The turnover rate of compartment 7 was set to equal that of compartment 5, as the terminal slopes of the VLDL and IDL curves were parallel. The material in compartments 5 and 7 may represent a pool of particles that spans the VLDL and IDL density fraction. Despite evidence to suggest that there is a significant direct input of apoB into the IDL [Svedberg flotation rate (SF) 12-60] in type III hyperlipidemia (18), this input appears confined to the small VLDL (Sf 20-60) fraction (19). It was, therefore, assumed that all IDL was derived from the VLDL fraction. IDL particles can be converted to LDL (compartment 8) or removed directly from plasma.
Fig. 1.
Compartment model describing VLDL, IDL, and LDL apoB kinetics. Five compartments are used to describe the kinetics of apoB in VLDL. Compartments 1–4 represent the VLDL delipidation cascade and compartment 5, a slowly turning over VLDL pool. VLDL particles can be converted to IDL or removed directly from plasma. Plasma IDL kinetics are described by compartments 6 and 7, where compartment 7 represents a slowly turning over pool of IDL. IDL particles can be converted to LDL (compartment 8) or removed directly from plasma. LDL kinetics are described by two independent plasma compartments (compartments 8 and 10) that account for the heterogeneous nature of LDL particles. Compartment 8 exchanges with compartment 9, an extravascular compartment. LDL particles in compartment 8 can be removed directly from plasma and the 125I-label, originally associated with the LDL tracer, can be transported to the body iodide pool (compartment 12). LDL particles in compartment 10 were converted to compartment 11, and subsequently the iodine associated with the LDL was converted to compartment 12. Compartment 11 acts as a delay between compartment 10 and the body iodide pool. Compartment 13 represents the urine 125I pool.
The kinetics of the LDL fraction were estimated from the 125I-LDL co-injected with the VLDL tracer. In general, LDL kinetics are not accurately determined from tracer appearance and disappearance originating from VLDL/IDL into the LDL fraction. The administration of labeled LDL particles, coupled with the collection of blood and/or urine for 10 or more days, overcomes this limitation. LDL kinetics are described by two independent plasma compartments (compartments 8 and 10) that account for the heterogeneous nature of LDL particles. It was assumed that LDL derived from the conversion of VLDL and LDL entered the LDL fraction via compartment 8. LDL particles not derived from this pathway were assumed to enter the LDL fraction via compartment 10 (20). Both inputs of particles into the LDL fraction account for total LDL production. Compartment 8 was assumed to exchange with compartment 9, an extravascular compartment. LDL particles in compartment 8 can be removed directly from plasma and the 125I-label, originally associated with the LDL tracer, can be transported to the body iodide pool (compartment 12) where the turnover rate was fixed at 2.5 pools/day (21). LDL particles in compartment 10 were converted to compartment 11, and subsequently the iodine associated with the LDL was converted to compartment 12. Compartment 11 acts as a delay between compartment 10 and the body iodide pool. The model also included compartments 13, a compartment that accounted for the appearance of iodine in urine derived from the catabolism of the LDL tracer.
Statistical analysis
Data are reported as mean ± SEM unless specified. Skewed variables were logarithmically transformed where appropriate. The statistical significance of differences between mean values was assessed using independent t-tests. Associations between study variables were examined using simple and stepwise regression analyses. Statistical significance was defined at P < 0.05. All data were analyzed using SPSS 15.0 (SPSS, Chicago, IL) software. The number of subjects included in the study was small. As such, the use of Bonferroni adjustments of P-values was not applied.
RESULTS
Table 1 shows the individual clinical and biochemical characteristics of the study subjects. Compared with normolipidemic apoE2/E2 subjects, apoE/E2 subjects with type III hyperlipidemia and apoE3/E2 hyperlipidemic subjects were older and had significantly higher plasma cholesterol, triglyceride, VLDL cholesterol, and VLDL triglyceride concentrations (Table 1). There were no significant differences between apoE2/E2 subjects with type III hyperlipidemia and apoE3/E2 hyperlipidemic subjects.
TABLE 1.
Clinical and biochemical characteristics of study subjects
| Subject Group | Subject No. | Sex | Age (years) | Weight (kg) | BMI (kg/m ) | Cholesterol (mmol/L) | Triglycerides (mmol/L) | LDL Cholesterol (mmol/L) | HDL Cholesterol (mmol/L) | VLDL Cholesterol (mmol/L) | VLDL Triglyceride (mmol/L) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E2/E2 normolipidemic | |||||||||||
| G1 | 1 | M | 32 | 64 | 21.6 | 4.06 | 1.35 | 1.10 | 1.31 | 0.39 | 0.78 |
| G1 | 2 | F | 30 | 55 | 21.6 | 4.75 | 1.20 | 1.44 | 1.83 | 0.65 | 0.57 |
| G1 | 3 | F | 37 | 65 | 27.9 | 4.75 | 2.83 | 1.92 | 0.86 | 1.15 | 1.78 |
| Mean | 33 | 61.7 | 23.7 | 4.52 | 1.79 | 1.49 | 1.33 | 0.73 | 1.04 | ||
| SEM | 1.70 | 2.57 | 1.69 | 0.19 | 0.42 | 0.19 | 0.23 | 0.18 | 0.30 | ||
| E2/E2-hyperlipidemic | |||||||||||
| G2 | 4 | M | 46 | 72 | 28.6 | 5.51 | 4.12 | 1.23 | 0.69 | 2.01 | 2.54 |
| G2 | 6 | M | 47 | 79 | 26.8 | 8.93 | 5.26 | 1.73 | 0.56 | 4.98 | 4.09 |
| G2 | 6 | M | 40 | 59 | 19.3 | 7.64 | 3.39 | 2.52 | 0.84 | 2.00 | 2.41 |
| G2 | 7 | M | 55 | 87 | 26.4 | 10.89 | 4.34 | 3.07 | 1.12 | 2.91 | 2.70 |
| G2 | 8 | M | 38 | 69 | 23.7 | 6.97 | 2.66 | 2.39 | 1.04 | 1.88 | 1.51 |
| Mean | 45* | 73.2 | 25.0 | 7.99* | 3.95* | 2.19 | 0.85 | 2.76* | 2.65* | ||
| SEM | 2.99 | 4.74 | 1.63 | 0.91 | 0.44 | 0.32 | 0.10 | 0.59 | 0.42 | ||
| E3/E2-hyperlipidemic | |||||||||||
| G3 | 9 | M | 57 | 83 | 27.1 | 7.71 | 9.41 | 1.81 | 0.60 | 3.68 | 5.47 |
| G3 | 10 | F | 65 | 61 | 25.9 | 7.84 | 3.58 | 4.34 | 1.08 | 1.61 | 2.46 |
| G3 | 11 | F | 58 | 84 | 35.3 | 10.54 | 3.45 | 6.62 | 1.03 | 1.52 | 2.04 |
| G3 | 12 | M | 49 | 70 | 25.5 | 7.92 | 2.21 | 4.53 | 0.53 | 1.11 | 1.38 |
| G3 | 13 | F | 58 | 67 | 26.5 | 7.46 | 5.74 | 2.45 | 0.99 | 2.32 | 3.79 |
| Mean | 57* | 72.9 | 28.1 | 8.29* | 4.88* | 3.95* | 0.85 | 2.05* | 3.03* | ||
| SEM | 2.54 | 4.47 | 1.83 | 0.57 | 1.27 | 0.85 | 0.12 | 0.45 | 0.73 | ||
P < 0.05 compared with E2/E2 normolipidemic.
P < 0.05 compared with E2/E2-hyperlipidemic.
Table 2 shows the concentrations and kinetics of VLDL, IDL, and LDL apoB for the three groups of subjects. Compared with normolipidemic apoE2/E2 subjects, apoE2/E2 subjects with type III hyperlipidemia had a significantly higher VLDL apoB concentration and decreased VLDL apoB fractional catabolic rate (FCR). There was no significant difference in VLDL apoB production rate (PR). The proportion of VLDL apoB in the slowly turning over VLDL compartment was significantly increased (Fig. 1, compartment 5; 26.0 ± 2.95% vs. 9.80 ± 1.86% of VLDL apoB pool; P < 0.01). The apoE2/E2 subjects with hyperlipidemia also had a higher IDL apoB concentration (P = 0.016) and a trend toward decreased IDL apoB FCR (P = 0.059) compared with normolipidemic apoE2/E2 subjects. LDL apoB concentrations were not significantly different between apoE2/E2 subjects, with or without type III hyperlipidemia. The apoE2/E2 subjects with type III hyperlipidemia, however, had a significantly lower LDL apoB FCR, and this was accompanied by a commensurate decrease in direct LDL synthesis (85.8 ± 5.23% vs. 70.3 ± 3.72%; P < 0.05).
TABLE 2.
VLDL, IDL, and LDL apoB kinetic parameters
| Subject Group |
P |
|||||
|---|---|---|---|---|---|---|
| Parameter | E2/E2-N | E2/E2-H | E3/E2-H | E2/E2-N vs. E2/E2-H | E2/E2-N vs. E3/E2-H | E2/E2-H vs. E3/E2-H |
| ApoB concentrations (mg/dl) | ||||||
| VLDL | 4.01 ± 0.39 | 18.3 ± 3.76 | 18.3 ± 5.81 | 0.005 | 0.012 | 0.998 |
| IDL | 5.10 ± 1.17 | 14.8 ± 3.05 | 7.73 ± 0.91 | 0.016 | 0.093 | 0.040 |
| LDL | 28.0 ± 6.08 | 38.3 ± 6.56 | 84.6 ± 18.4 | 0.316 | 0.019 | 0.026 |
| FCR (pools/day) | ||||||
| VLDL | 2.57 ± 0.47 | 1.13 ± 0.20 | 1.52 ± 0.17 | 0.022 | 0.044 | 0.175 |
| IDL | 1.15 ± 0.20 | 0.67 ± 0.11 | 0.83 ± 0.08 | 0.059 | 0.118 | 0.275 |
| LDL | 0.70 ± 0.07 | 0.41 ± 0.06 | 0.38 ± 0.09 | 0.025 | 0.040 | 0.762 |
| PR (mg/kg/day) | ||||||
| VLDL | 4.78 ± 1.18 | 8.24 ± 1.29 | 11.5 ± 2.51 | 0.102 | 0.073 | 0.372 |
| IDL | 2.78 ± 1.03 | 3.73 ± 0.30 | 2.83 ± 0.37 | 0.201 | 0.701 | 0.075 |
| LDL | 8.88 ± 1.94 | 6.73 ± 1.03 | 12.8 ± 2.33 | 0.386 | 0.288 | 0.036 |
| Conversion rate (%) | ||||||
| VLDL to IDL | 65.6 ± 22.1 | 47.7 ± 4.26 | 31.2 ± 8,58 | 0.504 | 0.132 | 0.123 |
| IDL to LDL | 43.8 ± 8.25 | 50.3 ± 4.52 | 80.1 ± 7.80 | 0.473 | 0.021 | 0.009 |
| VLDL to LDL | 28.4 ± 11.7 | 23.3 ± 0.50 | 23.6 ± 5.03 | 0.702 | 0.672 | 0.949 |
| VLDL apoB in slowly turning over compartment (%) | 9.80 ± 1.86 | 26.0 ± 2.95 | 7.69 ± 2.75 | 0.008 | 0.609 | 0.002 |
| Direct LDL synthesis (%) | 85.8 ± 5.23 | 70.3 ± 3.72 | 80.2 ± 4.08 | 0.047 | 0.427 | 0.111 |
Data presented as mean ± SEM. Bold text indicates statistical significance a P < 0.05. N: normolipidemic; H: hyperlipidemic.
As shown in Table 2, VLDL apoB kinetic parameters were comparable between apoE2/E2 subjects with type III hyperlipidemia and subjects heterozygous for the apoE2 allele (apoE3/E2), with the exception for the proportion of VLDL apoB in the slowly turning over VLDL compartment (Fig. 1, compartment 5; 26.0 ± 2.95% vs. 7.69 ± 2.75%; P < 0.01), which was significantly greater in apoE2/E2 type III hyperlipidemic subjects. Compared with apoE3/E2 hyperlipidemic subjects, apoE2/E2 type III hyperlipidemic subjects had a higher IDL apoB concentration and lower LDL apoB concentration and PR (Table 2). The conversion of IDL to LDL apoB was also significantly decreased in these subjects. No difference was observed in the conversion of VLDL to IDL and VLDL to LDL apoB.
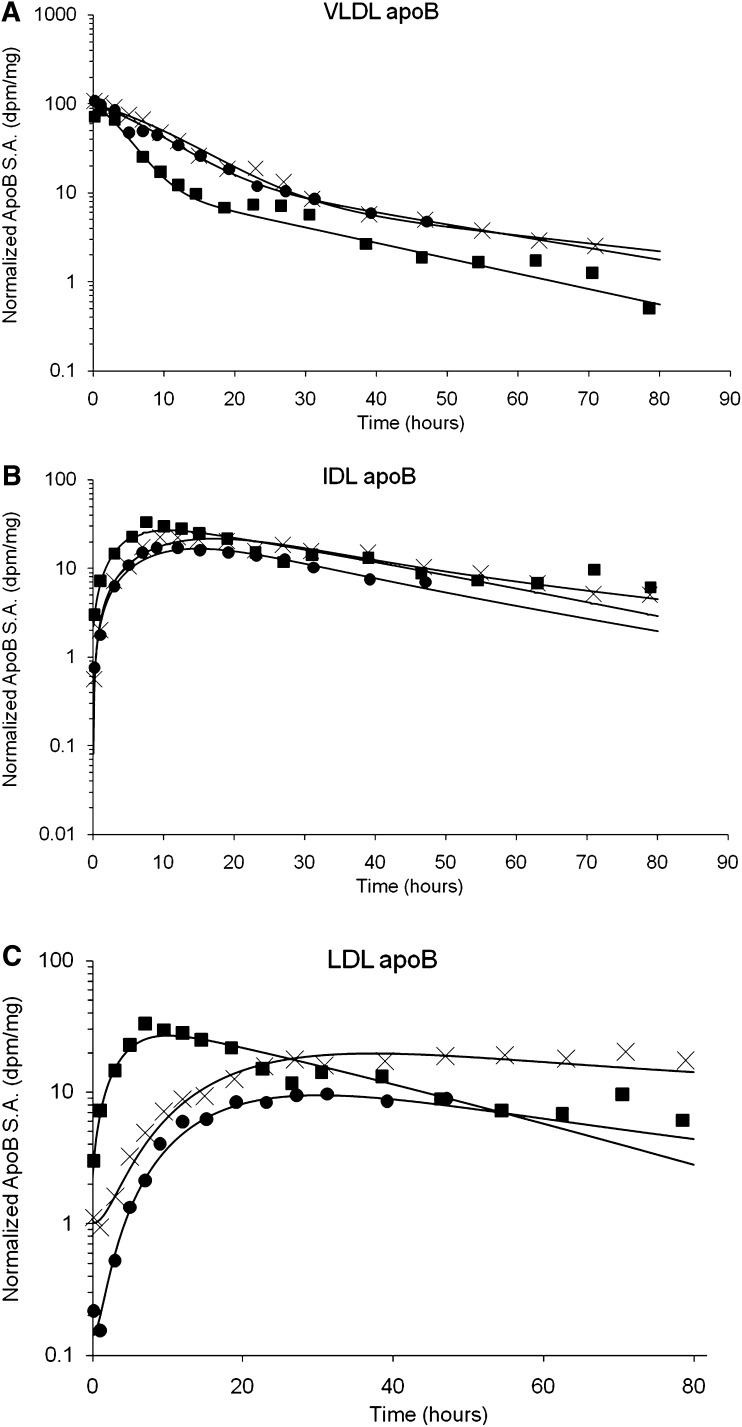
Figure 2 shows the normalized VLDL, IDL, and LDL apoB-specific activity-time curves for a representative subject from each group. The normalized VLDL apoB-specific activity-time curve decayed faster in normolipidemic apoE2/E2 subjects compared with hyperlipidemic apoE2/E2 and apoE3/E2 subjects (Fig. 2A). The IDL apoB- specific activity-time curve decayed faster in normolipidemic apoE2/E2 and hyperlipidemic apoE3/E2 subjects compared with type III hyperlipidemic subjects (Fig. 2B). The normalized LDL apoB-specific activity-time curve decayed faster in normolipidemic apoE2/E2 subjects compared with hyperlipidemic apoE2/E2 and apoE3/E2 subjects (Fig. 2C).
Fig. 2.
Normalized specific activity-time curves for in a representative subject in E2/E2 normolipidemic (filled square), E2/E2 type III hyperlipidemic (X), and E3/E2 hyperlipidemic (filled circle) for VLDL apoB (A), IDL apoB (B), and LDL apoB (C).
Table 3 shows the associations between VLDL cholesterol and VLDL triglyceride concentrations with kinetic parameters of VLDL, IDL, and LDL apoB in a pooled analysis after adjusting for subject group. The adjustment was performed to exclude subject group driving associations between VLDL cholesterol and VLDL triglycerides with study variables. VLDL cholesterol concentration was positively associated with VLDL and IDL apoB concentrations and the proportion of VLDL apoB in the slowly turning over VLDL compartment (compartment 5) (P < 0.05), and negatively associated with VLDL apoB FCR (P < 0.05). VLDL triglyceride concentration was positively associated with VLDL apoB concentrations and the production rate of VLDL and IDL apoB (P < 0.05). In a stepwise regression model that included VLDL apoB residence time and subject group, VLDL apoB concentration and the proportion of VLDL apoB in the slowly turning over compartment were both independent predictors of VLDL cholesterol concentrations (adjusted r2 = 0.81, P < 0.001). In a stepwise regression model that included VLDL and IDL apoB PR and subject group, VLDL apoB concentration was the only significant predictor of VLDL triglycerides (adjusted r2 = 0.67, P < 0.001).
TABLE 3.
Associations of VLDL cholesterol and VLDL triglycerides concentrations with kinetic parameters of VLDL, IDL, and LDL apoB
| VLDL Cholesterol |
VLDL Triglycerides |
|||
|---|---|---|---|---|
| Parameter | R | P | r | P |
| ApoB concentrations (mg/dl) | ||||
| VLDL | 0.845 | 0.001 | 0.783 | 0.003 |
| IDL | 0.687 | 0.014 | 0.514 | 0.087 |
| LDL | −0.461 | 0.131 | −0.480 | 0.114 |
| FCR (pools/day) | ||||
| VLDL | -0.659 | 0.020 | −0.549 | 0.065 |
| IDL | −0.430 | 0.163 | −0.064 | 0.844 |
| LDL | 0.156 | 0.628 | 0.145 | 0.654 |
| PR (mg/kg/day) | ||||
| VLDL | 0.514 | 0.087 | 0.603 | 0.038 |
| IDL | 0.437 | 0.156 | 0.632 | 0.027 |
| LDL | −0.174 | 0.588 | −0.145 | 0.654 |
| Conversion rate (%) | ||||
| VLDL to IDL | −0.216 | 0.501 | −0.055 | 0.865 |
| IDL to LDL | −0.207 | 0.519 | 0.019 | 0.953 |
| VLDL to LDL | −0.272 | 0.393 | −0.036 | 0.911 |
| VLDL apoB in slowly turning over compartment (%) | 0.611 | 0.035 | 0.395 | 0.203 |
| Direct LDL synthesis (%) | −0.328 | 0.298 | −0.466 | 0.127 |
Bold text indicates statistical significance at P < 0.05.
DISCUSSION
We provide new data on the effects of apoE genotype on the metabolism of apoB-containing lipoproteins in normolipidemic and hyperlipidemic subjects. We demonstrate for the first time that compared with normolipidemic apoE2/E2 subjects, apoE2/E2 subjects with type III hyperlipidemia have increased VLDL and IDL apoB concentrations, primarily as a consequence of decreased VLDL and IDL apoB FCR. We also showed that compared with hyperlipidemic subjects heterozygous for the apoE allele, type III hyperlipidemic apoE2/E2 subjects have significantly increased IDL apoB concentrations and decreased conversion of IDL to LDL particles. Both normolipidemic and type III hyperlipidemic apoE2/E2 subjects had decreased conversion of IDL to LDL particles compared with subjects heterozygous for the apoE allele.
Few studies have examined the metabolism of apoB-containing lipoproteins in subjects with type III hyperlipidemia. Stalenhoef et al. (7) showed that type III hyperlipidemia is characterized by delayed catabolism of apoB-100- and apoB-48-containing lipoproteins, with little conversion to IDL and LDL particles. Turner et al. (22) showed that the production of IDL is increased in these subjects and that the conversion of IDL to LDL apoB is delayed by 5-fold. Studies of chylomicron metabolism have shown that type III hyperlipidemia is associated with delayed catabolism of chylomicrons and their remnants (23, 23, 24, 24). Trembley et al. (25) recently showed that in patients with coexisting type III hyperlipidemia and heterozygous familial hypercholesterolemia, VLDL and IDL apoB FCR were decreased and LDL apoB FCR increased compared with familial hypercholesterolemia heterozygous and normolipidemic subjects. We extend previous reports by examining the metabolism of apoB-containing lipoproteins in normolipidemic apoE2/E2 and type III hyperlipidemic apoE2/E2 subjects using dual-tracers and multi-compartmental modeling. We also investigated a group of subjects heterozygous for the apoE allele with hyperlipidemia similar to that seen in type III hyperlipidemic subjects.
Consistent with previous studies, type III hyperlipidemic apoE2/E2 subjects had elevated levels of plasma cholesterol, triglycerides, VLDL cholesterol, and VLDL triglycerides (22–24). They also had higher VLDL and IDL apoB concentrations due, primarily, to reduced VLDL and IDL apoB FCR compared with normolipidemic apoE2/E2 subjects. The reduction in VLDL and IDL apoB FCR may be consequent on the inhibitory effect of apoE2 on LPL-mediated lipolysis (26) and decreased binding affinity of apoE2 to hepatic clearance receptors (27), resulting in the accumulation of remnants particles in the circulation (27). Type III hyperlipidemic subjects also had a significantly greater proportion of VLDL particles in the slowly turning VLDL pool. Earlier studies suggest that particles in this pool may correspond to β-VLDL particles (28, 29). The greater proportion of VLDL particles in the slowly turning over VLDL pool, exclusively in apoE2/E2 subjects with type III hyperlipidemia, suggest that this may be a key mechanism in the expansion of plasma β-VLDL pool (30). Furthermore, the increased proportion of VLDL apoB in the slowly turning over VLDL pool is an independent determinant, together with VLDL apoB concentrations, of VLDL cholesterol levels. Collectively, these kinetic defects may account for the dyslipoproteinemia seen in our apoE2/E2 type III hyperlipidemic subjects. Whether downregulation of VLDL receptors or other extrahepatic receptors (31, 32), or the inhibition of lipolytic enzymes by other apoproteins, such as apoC-III (33, 34), contributes further to the impaired catabolism and accumulation of β-VLDL particles in circulation requires further investigation.
Of interest, normolipidemic apoE2/E2 subjects, like the type III hyperlipidemic apoE2/E2 subjects (7, 22, 35), have decreased conversion of IDL to LDL apoB. Furthermore, VLDL apoB FCR was, on average, 66% slower in the normolipidemic apoE2/E2 subjects than that reported in normolipidemic subjects without the apoE2/E2 genotype [mean (range): 2.57 (1.68–3.30) vs. 7.70 (4.30–13.9), pools/day] (36). These kinetic defects are consistent with the presence of apoE2 (7, 22, 35). Despite this, these subjects had lipid and lipoprotein profiles that are within the normal range (29, 36–38). To explain this, there was a commensurate decrease in VLDL apoB production (∼64%) in normolipidemic apoE2/E2 subjects compared with normolipidemic subjects without the apoE2/E2 genotype (36). The mechanism for this is unclear and warrants further investigation. Normolipidemic apoE2/E2 subjects also had an elevated direct LDL apoB synthesis, coupled with a compensatory increase in LDL apoB FCR. As such, LDL apoB concentration remained low in these subjects. The exact mechanism for the increase in direct LDL apoB synthesis is unclear and requires further study. The increased LDL apoB FCR may relate to upregulation of hepatic LDL receptors, in part, a function of decreased uptake of chylomicron remnants and associated cholesterol by the liver (35). Furthermore, there may be reduced competition for LDL receptor-mediated removal of VLDL remnants, which is apoE-dependent (32). Our observation supports the notion that apoE2 homozygosity contributes to altered lipoprotein metabolism but is not sufficient to cause the development of type III hyperlipidemia (6, 32), and that additional genetic polymorphisms (39, 40), such as polymorphisms in proteins that regulate lipolysis of lipoproteins (34) and/or other environmental factors leading to insulin resistance and obesity (41), contribute to the expression and severity of type III hyperlipidemia.
The apoE2/E2 type III hyperlipidemic and apoE3/E2 hyperlipidemic subjects exhibited comparable hyperlipidemia. However, different metabolic mechanisms underlie the hyperlipidemia observed. The inhibitory effects of apoE2 on LPL-mediated lipolysis, in part through displacement of apoC-II, and the reduced ability of apoE2 to enhance hepatic lipase-mediated lipolysis may explain these differences (6). In vitro studies have demonstrated that the addition of apoE3 but not apoE2 restored LPL-mediated conversion of hepatic β-VLDL into LDL particles (42, 43). Consistent with this, subjects with the apoE3/E2 genotype in this study had higher rates of conversion of IDL to LDL and production of LDL apoB that was coupled with higher LDL apoB levels compared with apoE2/E2 homozygotes, with or without type III hyperlipidemia. Whether the presence of additional genetic mutations where type III hyperlipidemia may be inherited in a dominant fashion (E3 Leiden and E1 Harrisburg) (44, 45) may explain the hyperlipidemia in the apoE3/E2 subjects requires further investigation.
The study has limitations. The isolation of VLDL and LDL particles by ultracentrifugation may remove apoE from these particles. However, apoE is rapidly exchanged between VLDL and HDL particles (46). As such, it is anticipated that VLDL particles will acquire apoE from endogenous VLDL and HDL particles shortly after reinjection. It is also important to acknowledge the possibility that the LDL particles (or a component of this fraction) isolated represents the spillover of particles from the VLDL and IDL density fractions (47). However, the kinetics of the particles in the LDL fraction are distinct from those of the particles in the VLDL and IDL fractions. The kinetics of apoB-48 were not examined in our subjects. Previous studies have demonstrated that in apoE2/E2 normolipidemic and type III hyperlipidemic subjects, the catabolism of apoB-48-containing lipoproteins were impaired (7). The measurement of lipid transfer proteins, including cholesterol ester transfer protein and phospholipid transfer protein, which may contribute to particle remodeling, may have clarified the mechanism for altered lipoprotein composition and kinetics (48). The sample size is small, and larger numbers may have demonstrated increased rates of VLDL apoB secretion, in addition to reduced VLDL apoB FCR, in hyperlipidemic subjects. This study, however, provides important insights into the effect of apoE genotype on apoB metabolism and is hypothesis generating.
In conclusion, the present data demonstrate that plasma lipid and lipoprotein abnormalities in apoE2/E2 subjects with type III hyperlipidemia are due to a combination of VLDL and IDL apoB catabolic defects and increased proportion of VLDL in the slowly turning over VLDL pool. The abnormalities may account for the increase in risk of premature atherosclerosis in type III hyperlipidemia. Our data support the notion that a defective apoE is essential but not sufficient to cause overt hyperlipidemia. Secondary factors, such as additional genetic mutations and environmental factors, including obesity and insulin resistance, may play contributing roles to the development of type III hyperlipidemia. Our results also reveal the differing mechanisms underlying comparable hyperlipidemias and reinforce the need for targeted interventions on lipoprotein metabolism in different states of dyslipidemia for better cardiovascular risk management.
Acknowledgments
We are grateful for the assistance of the late Sally Collins and the staff of the Medical Unit, The Princess Margaret Hospital, Christchurch, New Zealand.
Footnotes
This study was funded in part by a research grant from the National Heart Foundation of New Zealand. E.M.M.O. is supported by an A & A Saw Medical Research Fellowship and a National Health and Medical Research Council (NHMRC) Postdoctoral Research Fellowship. P.H.R.B. is an NHMRC Senior Research Fellow.
Abbreviations:
- apo
- apolipoprotein
- FCR
- fractional catabolic rate
- IDL
- intermediate density lipoprotein
- PR
- production rate
- TMU
- tetramethyl urea
- TRL
- triglyceride-rich lipoprotein
REFERENCES
- 1.Weisgraber K. H. 1994. Apolipoprotein E: structure-function relationships. Adv. Protein Chem. 45: 249–302. [DOI] [PubMed] [Google Scholar]
- 2.Gibson J. C., Brown W. V. 1988. Effect of lipoprotein lipase and hepatic triglyceride lipase activity on the distribution of apolipoprotein E among the plasma lipoproteins. Atherosclerosis. 73: 45–55. [DOI] [PubMed] [Google Scholar]
- 3.Mahley R. W., Huang Y. 1999. Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr. Opin. Lipidol. 10: 207–217. [DOI] [PubMed] [Google Scholar]
- 4.Greenow K., Pearce N. J., Ramji D. P. 2005. The key role of apolipoprotein E in atherosclerosis. J. Mol. Med. 83: 329–342. [DOI] [PubMed] [Google Scholar]
- 5.Davignon J., Gregg R. E., Sing C. F. 1988. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 8: 1–21. [DOI] [PubMed] [Google Scholar]
- 6.Mahley R. W., Huang Y., Rall S. C., Jr 1999. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia). Questions, quandaries, and paradoxes. J. Lipid Res. 40: 1933–1949. [PubMed] [Google Scholar]
- 7.Stalenhoef A. F., Malloy M. J., Kane J. P., Havel R. J. 1986. Metabolism of apolipoproteins B-48 and B-100 of triglyceride-rich lipoproteins in patients with familial dysbetalipoproteinemia. J. Clin. Invest. 78: 722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins P. N., Wu L. L., Hunt S. C., Brinton E. A. 2005. Plasma triglycerides and type III hyperlipidemia are independently associated with premature familial coronary artery disease. J. Am. Coll. Cardiol. 45: 1003–1012. [DOI] [PubMed] [Google Scholar]
- 9.Janus E. D., Grant S., Lintott C. J., Wardell M. R. 1985. Apolipoprotein E phenotypes in hyperlipidaemic patients and their implications for treatment. Atherosclerosis. 57: 249–266. [DOI] [PubMed] [Google Scholar]
- 10.Wardell M. R., Brennan S. O., Janus E. D., Fraser R., Carrell R. W. 1987. Apolipoprotein E2-Christchurch (136 Arg—-Ser). New variant of human apolipoprotein E in a patient with type III hyperlipoproteinemia. J. Clin. Invest. 80: 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wardell M. R., Rall S. C., Jr, Brennan S. O., Nye E. R., George P. M., Janus E. D., Weisgraber K. H. 1990. Apolipoprotein E2-Dunedin (228 Arg replaced by Cys): an apolipoprotein E2 variant with normal receptor-binding activity. J. Lipid Res. 31: 535–543. [PubMed] [Google Scholar]
- 12.McFarlane A. S. 1958. Efficient trace-labelling of proteins with iodine. Nature. 182: 53. [DOI] [PubMed] [Google Scholar]
- 13.Janus E. D., Nicoll A. M., Turner P. R., Magill P., Lewis B. 1980. Kinetic bases of the primary hyperlipidaemias: studies of apolipoprotein B turnover in genetically defined subjects. Eur. J. Clin. Invest. 10: 161–172. [DOI] [PubMed] [Google Scholar]
- 14.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193: 265–275. [PubMed] [Google Scholar]
- 15.Kane J. P. 1973. A rapid electrophoretic technique for identification of subunit species of apoproteins in serum lipoproteins. Anal. Biochem. 53: 350–364. [DOI] [PubMed] [Google Scholar]
- 16.Margolis S., Langdon R. G. 1966. Studies on human serum beta-1-lipoprotein. I. Amino acid composition. J. Biol. Chem. 241: 469–476. [PubMed] [Google Scholar]
- 17.Wardell M. R., Suckling P. A., Janus E. D. 1982. Genetic variation in human apolipoprotein E. J. Lipid Res. 23: 1174–1182. [PubMed] [Google Scholar]
- 18.Reardon M. F., Poapst M. E., Steiner G. 1982. The independent synthesis of intermediate density lipoproteins in type III hyperlipoproteinemia. Metabolism. 31: 421–427. [DOI] [PubMed] [Google Scholar]
- 19.Packard C. J., Clegg R. J., Dominiczak M. H., Lorimer A. R., Shepherd J. 1986. Effects of bezafibrate on apolipoprotein B metabolism in type III hyperlipoproteinemic subjects. J. Lipid Res. 27: 930–938. [PubMed] [Google Scholar]
- 20.Beltz W. F., Kesaniemi Y. A., Howard B. V., Grundy S. M. 1985. Development of an integrated model for analysis of the kinetics of apolipoprotein B in plasma very low density lipoproteins, intermediate density lipoproteins, and low density lipoproteins. J. Clin. Invest. 76: 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman M. 1979. Kinetic analysis of turnover data. Prog. Biochem. Pharmacol. 15: 67–108. [PubMed] [Google Scholar]
- 22.Turner P. R., Cortese C., Wootton R., Marenah C., Miller N. E., Lewis B. 1985. Plasma apolipoprotein B metabolism in familial type III dysbetalipoproteinaemia. Eur. J. Clin. Invest. 15: 100–112. [DOI] [PubMed] [Google Scholar]
- 23.Haffner S. M., Kushwaha R. S., Hazzard W. R. 1989. Metabolism of chylomicrons in subjects with dysbetalipoproteinaemia (type III hyperlipoproteinaemia). Eur. J. Clin. Invest. 19: 486–490. [DOI] [PubMed] [Google Scholar]
- 24.Redgrave T. G., Watts G. F., Martins I. J., Barrett P. H., Mamo J. C., Dimmitt S. B., Marais A. D. 2001. Chylomicron remnant metabolism in familial dyslipidemias studied with a remnant-like emulsion breath test. J. Lipid Res. 42: 710–715. [PubMed] [Google Scholar]
- 25.Tremblay A. J., Lamarche B., Ruel I. L., Hogue J. C., Deshaies Y., Gagne C., Couture P. 2006. Effects of fenofibrate on apolipoprotein kinetics in patients with coexisting dysbetalipoproteinemia and heterozygous familial hypercholesterolemia. Atherosclerosis. 188: 203–212. [DOI] [PubMed] [Google Scholar]
- 26.Jong M. C., Dahlmans V. E., Hofker M. H., Havekes L. M. 1997. Nascent very-low-density lipoprotein triacylglycerol hydrolysis by lipoprotein lipase is inhibited by apolipoprotein E in a dose-dependent manner. Biochem. J. 328: 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisgraber K. H., Innerarity T. L., Mahley R. W. 1982. Abnormal lipoprotein receptor-binding activity of the human E apoprotein due to cysteine-arginine interchange at a single site. J. Biol. Chem. 257: 2518–2521. [PubMed] [Google Scholar]
- 28.Kushwaha R. S., Haffner S. M., Foster D. M., Hazzard W. R. 1985. Compositional and metabolic heterogeneity of alpha 2- and beta-very-low-density lipoproteins in subjects with broad beta disease and endogenous hypertriglyceridemia. Metabolism. 34: 1029–1038. [DOI] [PubMed] [Google Scholar]
- 29.Berman M., Hall M., III, Levy R. I., Eisenberg S., Bilheimer D. W., Phair R. D., Goebel R. H. 1978. Metabolsim of apoB and apoC lipoproteins in man: kinetic studies in normal and hyperlipoproteininemic subjects. J. Lipid Res. 19: 38–56. [PubMed] [Google Scholar]
- 30.Vega G. L., East C., Grundy S. M. 1988. Lovastatin therapy in familial dysbetalipoproteinemia: effects on kinetics of apolipoprotein B. Atherosclerosis. 70: 131–143. [DOI] [PubMed] [Google Scholar]
- 31.Crawford D. C., Nord A. S., Badzioch M. D., Ranchalis J., McKinstry L. A., Ahearn M., Bertucci C., Shephard C., Wong M., Rieder M. J., et al. 2008. A common VLDLR polymorphism interacts with APOE genotype in the prediction of carotid artery disease risk. J. Lipid Res. 49: 588–596. [DOI] [PubMed] [Google Scholar]
- 32.Rall S. C., Jr, Weisgraber K. H., Innerarity T. L., Mahley R. W. 1983. Identical structural and receptor binding defects in apolipoprotein E2 in hypo-, normo-, and hypercholesterolemic dysbetalipoproteinemia. J. Clin. Invest. 71: 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ooi E. M., Barrett P. H., Chan D. C., Watts G. F. 2008. Apolipoprotein C–III: understanding an emerging cardiovascular risk factor. Clin. Sci. (Lond.). 114: 611–624. [DOI] [PubMed] [Google Scholar]
- 34.Henneman P., van der Sman-de Beer F., Moghaddam P. H., Huijts P., Stalenhoef A. F., Kastelein J. J., van Duijn C. M., Havekes L. M., Frants R. R., van Dijk K. W., et al. 2009. The expression of type III hyperlipoproteinemia: involvement of lipolysis genes. Eur. J. Hum. Genet. 17: 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demant T., Shepherd J., Packard C. J. 1988. Very low density lipoprotein apolipoprotein B metabolism in humans. Klin. Wochenschr. 66: 703–712. [DOI] [PubMed] [Google Scholar]
- 36.Kesaniemi Y.A., Vega G.L., Grundy S.M. 1985. ApoB kinetics in normal vs. hyperlipidemic man: review of current data. J. Clin. Invest. 76: 586–595.3861622 [Google Scholar]
- 37.Egusa G., Beltz W. F., Grundy S. M., Howard B. V. 1985. Influence of obesity on the metabolism of apolipoprotein B in humans. J. Clin. Invest. 76: 596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kesaniemi Y. A., Beltz W. F., Grundy S. M. 1985. Comparisons of metabolism of apolipoprotein B in normal subjects, obese patients, and patients with coronary heart disease. J. Clin. Invest. 76: 586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans D., Seedorf U., Beil F. U. 2005. Polymorphisms in the apolipoprotein A5 (APOA5) gene and type III hyperlipidemia. Clin. Genet. 68: 369–372. [DOI] [PubMed] [Google Scholar]
- 40.Geisel J., Bunte T., Bodis M., Oette K., Herrmann W. 2002. Apolipoprotein E2/E2 genotype in combination with mutations in the LDL receptor gene causes type III hyperlipoproteinemia. Clin. Chem. Lab. Med. 40: 475–479. [DOI] [PubMed] [Google Scholar]
- 41.de Beer F., Stalenhoef A. F., Hoogerbrugge N., Kastelein J. J., Gevers Leuven J. A., van Duijn C. M., Havekes L. M., Smelt A. H. 2002. Expression of type III hyperlipoproteinemia in apolipoprotein E2 (Arg158 → Cys) homozygotes is associated with hyperinsulinemia. Arterioscler. Thromb. Vasc. Biol. 22: 294–299. [DOI] [PubMed] [Google Scholar]
- 42.van Dijk K. W., Hofker M. H., Havekes L. M. 1999. Dissection of the complex role of apolipoprotein E in lipoprotein metabolism and atherosclerosis using mouse models. Curr. Atheroscler. Rep. 1: 101–107. [DOI] [PubMed] [Google Scholar]
- 43.Chung B. H., Segrest J. P. 1983. Resistance of a very low density lipoprotein subpopulation from familial dysbetalipoproteinemia to in vitro lipolytic conversion to the low density lipoprotein density fraction. J. Lipid Res. 24: 1148–1159. [PubMed] [Google Scholar]
- 44.Mann W. A., Gregg R. E., Sprecher D. L., Brewer H. B., Jr 1989. Apolipoprotein E-1Harrisburg: a new variant of apolipoprotein E dominantly associated with type III hyperlipoproteinemia. Biochim. Biophys. Acta. 1005: 239–244. [DOI] [PubMed] [Google Scholar]
- 45.Havekes L., de Wit E., Leuven J. G., Klasen E., Utermann G., Weber W., Beisiegel U. 1986. Apolipoprotein E3-Leiden. A new variant of human apolipoprotein E associated with familial type III hyperlipoproteinemia. Hum. Genet. 73: 157–163. [DOI] [PubMed] [Google Scholar]
- 46.Cohn J. S., Batal R., Tremblay M., Jacques H., Veilleux L., Rodriguez C., Mamer O., Davignon J. 2003. Plasma turnover of HDL apoC-I, apoC-III, and apoE in humans: in vivo evidence for a link between HDL apoC-III and apoA-I metabolism. J. Lipid Res. 44: 1976–1983. [DOI] [PubMed] [Google Scholar]
- 47.Murdoch S. J., Boright A. P., Paterson A. D., Zinman B., Steffes M., Cleary P., Edwards K., Marcovina S. S., Purnell J. Q., Brunzell J. D. 2007. LDL composition in E2/2 subjects and LDL distribution by Apo E genotype in type 1 diabetes. Atherosclerosis. 192: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang X. C., Zhou H. W. 2006. Plasma lipid transfer proteins. Curr. Opin. Lipidol. 17: 302–308. [DOI] [PubMed] [Google Scholar]