Abstract
Cholesterol 7α-hydroxylase (CYP7A1) plays a critical role in regulation of bile acid synthesis in the liver. CYP7A1 mRNAs have very short half-lives, and bile acids destabilize CYP7A1 mRNA via the 3′-untranslated region (3′-UTR). However, the underlying mechanism of translational regulation of CYP7A1 mRNA remains unknown. Screening of a human micro RNA (miRNA) microarray has identified five differentially expressed miRNAs in human primary hepatocytes treated with chenodeoxycholic acid, GW4064, or fibroblast growth factor (FGF)19. These compounds also significantly induced the expression of miR-122a, a liver-specific and the predominant miRNA in human hepatocytes. The putative recognition sequences for miR-122a and miR-422a were localized in the 3′-UTR of human CYP7A1 mRNA. The miR-122a and miR-422a mimics inhibited, whereas their inhibitors stimulated CYP7A1 mRNA expression. These miRNAs specifically inhibited the activity of the CYP7A1-3′-UTR reporter plasmids, and mutations of miRNA binding sites in 3′-UTR abrogated miRNA inhibition of reporter activity. These results suggest that miR-122a and miR-422a may destabilize CYP7A1 mRNA to inhibit CYP7A1 expression. However, these miRNAs did not play a role in mediating FGF19 inhibition of CYP7A1 transcription. Under certain conditions, miRNA may reduce CYP7A1 mRNA stability to inhibit bile acid synthesis, and the miR-122a antagomirs may stimulate bile acid synthesis to reduce serum cholesterol and triglycerides.
Keywords: bile acid synthesis, FGF19, nuclear receptors, FXR, lipid metabolism
Bile acids are physiological detergents that facilitate absorption, transport, and distribution of dietary fats, sterols, and lipid-soluble vitamins, and disposal of toxic metabolites and xenobiotics. Bile acids also are signaling molecules that activate cell signaling pathways to regulate lipid, glucose, and energy homeostasis (1). Bile acid synthesis is feedback inhibited by bile acids returning to the liver via enterohepatic circulation of bile acids to inhibit cholesterol 7α-hydroxylase (CYP7A1) and sterol 12α-hydroxylase (CYP8B1) expression (2). Many studies suggest that expression of CYP7A1 and CYP8B1 is mainly regulated at gene transcriptional levels by various factors, including nuclear receptors and cell signaling pathways (1). It has been proposed that bile acid-activated farnesoid X receptor (FXR) inhibits CYP7A1 and CYP8B1 transcription by two mechanisms. In the liver, FXR induces the small heterodimer partner to inhibit CYP7A1 and CYP8B1 transcription. In the intestine, FXR induces fibroblast growth factor (FGF)15, which activates FGF15 receptor 4 (FGFR4) signaling in hepatocytes to inhibit CYP7A1 expression (3, 4). FGF19, a human ortholog of mouse FGF15, has been identified in human sera (5). Feeding chenodeoxycholic acid (CDCA) increases serum FGF19 levels, whereas feeding cholestyramine reduces serum FGF19 levels. Interestingly, serum FGF19 levels exhibit two peaks about 2 h following the peaks of serum bile acid levels in human patients. This is consistent with the increased bile acid flux during the postprandial state to induce intestine FGF19, which inhibits hepatic bile acid synthesis. We have reported recently that CDCA induces FGF19 expression in primary human hepatocytes (PHHs), and FGF19 strongly inhibits CYP7A1 mRNA expression by activating the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK)1/2 pathway (6). Our results suggest that FXR induces FGF19 in hepatocytes to inhibit CYP7A1 by an autocrine mechanism. This is confirmed by a recent report that FGF19 levels were increased in the liver of patients with extrahepatic cholestasis (7). However, the mechanism underlying FGF19 inhibition of CYP7A1 expression remains unknown.
Earlier studies have shown that CYP7A1 mRNA, protein, and activity are induced by cholestyramine, and bile acids inhibited CYP7A1 expression in vivo (8). It has been determined that CYP7A1 mRNA has a very short half-life of about 30 min (9–11). The 3′-untranslated region (3′-UTR) of CYP7A1 mRNA contains multiple AUUUA elements that are present in short-lived mRNA (12, 13). However, the posttranscriptional regulation of CYP7A1 has not been explored. It has been reported that bile acids reduce CYP7A1 mRNA stability via sequences in the 3′-UTR (11, 13).
Recently, a new class of small noncoding RNAs, microRNAs (miRNAs), has been discovered in animals and plants. MiRNAs are 19- to 25-nucleotide RNAs that are able to bind to complementary sequences in the 3′-UTR of mRNAs to guide an RNA-induced silencing complex to mRNAs to repress protein translation, cleave targeted messages, and degrade mRNAs (14, 15). More than 700 miRNAs have been identified in the human genome. A potential role of miRNAs in the regulation of cholesterol, fatty acid, and lipid metabolism in mice has been reported recently (16, 17).
We have screened a human miRNA microarray and identified miRNAs that are differentially expressed by CDCA, FGF19, and GW4064 (a specific FXR agonist). We have identified the complementary sequences in the 3′-UTR of human CYP7A1 mRNA that bind miR-122a and miR-422a. Reporter assays and mutagenesis analyses have confirmed the function of these two miRNAs in regulation of CYP7A1 mRNA expression in human hepatocytes. This study has revealed a novel posttranscriptional mechanism for regulation of CYP7A1 gene expression in human hepatocytes.
MATERIALS AND METHODS
Cell culture
PHHs were obtained from the Liver Tissue and Cell Distribution System of the National Institute of Diabetes and Digestive and Kidney Diseases (LTCDS, N01-DK-7-0004/HHSN267200700004C, S. Strom, University of Pittsburgh, PA). The human hepatoma cell lines, HepG2, were obtained from the American Type Culture Collection (Manassas, VA). Huh7 cells were kindly provided by Dr. Jin Ye (University of Texas Southwestern Medical Center, Dallas, TX). Cells were maintained as described previously (18).
Reagents
Recombinant FGF19 was from R and D Systems (Minneapolis, MN). GW4064 was a generous gift from Dr. C. Kremoser (Phenex Pharma AG, Ludwigshafen, Germany).
RNA isolation and quantitative real-time PCR
PHHs were treated with CDCA (25 µM), GW4064 (1 µM), or FGF19 (40 ng/ml) for 24 h, and total RNA was isolated using QIAGEN miRNeasy isolation kit (catalog no. 217004, QIAGEN) according to the manufacturer's instruction. Reverse-transcription and quantitative real-time PCR were performed to detect CYP7A1, CYP8B1, and solute carrier 7A1 (SLC7A1) mRNAs as described previously (18). The expression of mature miRNAs was assayed using Taqman MicroRNA Assays (Applied Biosystems, Foster City, CA). cDNA was synthesized from total RNA using gene-specific primers according to the TaqMan MicroRNA Assay protocol. The Taqman MicroRNA Assay for RNU48 was used to normalize the relative abundance of miRNA.
The miRNA mimics, miRNA mimic negative control, miRNA inhibitors, and miRNA inhibitor negative controls for miR-122a and miR-422a were purchased from Dharmacon Research (Lafayette, CO) and transfected into HepG2 cells and PHHs using Lipofectin (Invitrogen). Forty-eight hours after transfection, cells were extracted and analyzed for relative mRNA expression of CYP7A1, CYP8B1, and SLC7A1by quantitative real-time PCR.
miRNA microarray
Total RNA was isolated from PHHs treated with CDCA (25 μM), GW4064 (1 μM), or FGF19 (40 ng/ml) for 24 h. RNA quality control, labeling, hybridization, and scanning were performed by Exiqon (Denmark) using LNA miRCURY™ array (version 9.2). The miRNA expression levels that were modulated with a log2 ratio (treated/control) ≥0.5 were considered differentially expressed (Table 1; supplementary Fig. II). The miRNA microarray analysis data were deposited to ArrayExpress (accession no. E-TABM-691).
TABLE 1.
Microarray analysis of differentially expressed miRNAs in PHHs treated with CDCA (25 μM), GW4064 (1 μM), or FGF19 (40 ng/ml)
| MicroRNA Expression (log 2 ratio) | |||
|---|---|---|---|
| Annotation | CDCA | GW4064 | FGF19 |
| miR-637 | 0.60 | 0.10 | 0.10 |
| miR-422a | 0.16 | 0.51 | 0.63 |
| miR-622 | 0.55 | 0.75 | 0.72 |
| miR-487b | −0.9 | −0.76 | −0.41 |
| miR-597 | −0.65 | −0.57 | −0.44 |
| miR-122a | 0.14 | 0.39 | 0.31 |
* Log2 (treated sample/control) ratios: >50% differential expression in one or more treatment samples, P < 0.05, n = 3. The most abundant miR-122a in human hepatocytes is also included.
Analysis of miRNA target sites
The analysis of 3′-UTR of human CYP7A1 mRNA for miRNA target prediction was determined using the TargetScan (19), miRanda (20), or miRBase (21). The TargetScan score for miR-422a on CYP7A1 3′-UTR was −0.28. The TargetScan score and miRanda score for miR-422a on CYP8B1 3′-UTR were −0.09 and 16.2684, respectively. The imperfect miR-122a target site on CYP7A1 3′-UTR was identified based on miR-122a consensus target sequence using MacVector software.
Luciferase reporter assays
The human CYP7A1 3′-UTR sequence from the first nucleotide after the stop codon to nt 200 was amplified by PCR using the primers 5′-GACTAGTATACATGGCTGGAATAAGAGG-3′ (CYP7A1-3′-UTR-F1) and 5′-CCCAAGCTTTTTTCACTAGCAGAGTCTGTG-3′ (CYP7A1-3′-UTR-R200) and cloned downstream of the luciferase gene in pMir REPORT luciferase vector (Ambion). The human CYP7A1 3′-UTR from the first nucleotide after the stop codon to nt 1,298 was amplified by PCR using CYP7A1-3′-UTR-F1 and 5′-AGATTCATACTTTTATTTCTGAA-3′ (CYP7A1-3UTR-R1298) and cloned into the pGEM-T vector (Promega). This plasmid was cut by restriction enzymes SPEI and HindIII to generate a fragment containing sequences from −203 to 982 to insert downstream of the luciferase gene in the pMir REPORT luciferase vector. The 3′-UTR fragment from nt 73 to 326 of the CYP8B1 was PCR amplified using primers: 5′-ACTAGTCCTCCCTCTGGTCCTGTGG (F) and 5′-AAGCTTGGAACCAGTCTCTGTACTCAGGACTT (R) and inserted downstream of the pMir REPORT vector. CYP7A1 3′-UTR reporter constructs containing mutations in the miR-122a and miR-422a target sequences were generated using a Quick Change Mutagenesis kit (Stratagene). For luciferase reporter assays, HepG2 cells were plated in 24-well plates and transiently transfected with pMir REPORT, pMir-CYP7A1-1-200, or pMir-CYP7A1-203-982 together with pMir REPORT β-gal control plasmid and miRNA mimics of miR-122a or miR-422a, using lipofectin. When studying the effect of FGF19, cells were serum starved for 24 h before treatment. Luciferase activities are expressed as relative luciferase unit/β-galactosidase activity as described previously (18). Each reporter assay was repeated three times in triplicate.
Nuclei isolation and nuclear run-on assay
PHHs cultured in T75 flasks were treated with FGF19 (40 ng/ml) for 24 h. Nuclei were isolated from cells with a Chemicon Nuclear Extract kit (Millipore, MA). The nuclei were resuspended in nuclei buffer containing 40% glycerol, 50 mM Tris-HCl, 5 mM MgCl2, and 0.1 mM EDTA. About 5 × 106 nuclei were incubated in 50 μl reaction buffer (5 mM Tris-HCl, pH 8.0, 2.5 mM MgCl2, 150 mM KCl, 1 mM each of ATP, GTP, and CTP) and 1 mM biotin-16-UTP at 30°C for 45 min. The reaction was stopped by the addition of 1/10 vol of DNase I 10× buffer and 200 U RNase-free DNase I and incubated for 10 min at 37°C. The nuclei were then lysed by the addition of 1/10 vol of 10× proteinase K buffer (10× buffer: 10% SDS, 10 mM Tris-HCL, pH 7.4, 50 mM EDTA) and 10 μl of proteinase K (10 mg/ml) for 30 min at 45°C. RNA was extracted with 1 ml TRIzol and resuspended in 100 μl RNase-free H2O. Biotinylated RNA was purified by adding streptavidin magnetic particle beads (Promega), followed by 1 h incubation at 25°C with rotation. Beads were washed twice with 2× SSC for 5 min and eluted by incubating the beads in 95% formamide/10 mM EDTA solution at 90°C for 10 min. Supernatant was collected and RNA was precipitated. Then 10 μl RNA was used for reverse-transcription with random decamer and CYP7A1 transcripts were detected with real-time PCR. A reaction without the addition of biotin-16-UTP was used as background control.
Statistical analyses
Statistical analyses were performed by one-way ANOVA followed by Dunnett's test to determine which groups were significantly different from the control group. Comparison of two groups was performed using an unpaired Student's t-test. A P-value of <0.05 was considered significantly different.
RESULTS
Identification of differentially expressed miRNAs by bile acid, GW4064, and FGF19 in PHHs
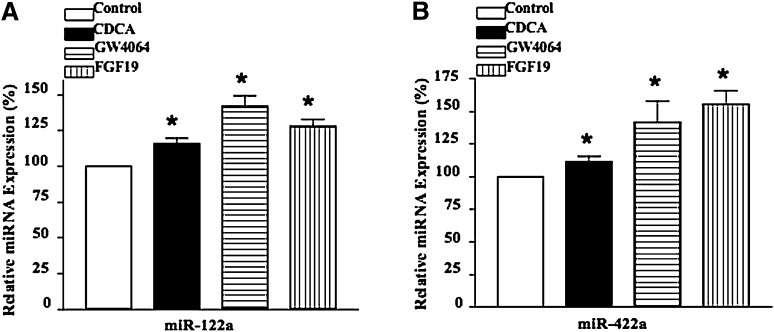
It has been reported that bile acids regulate CYP7A1 mRNA stability through the 3′-UTR sequences. MiRNAs are known to bind to the complementary sequences in the 3′-UTR of mRNA transcripts to induce mRNA degradation and reduce mRNA stability. We screened a human miRNA microarray using RNA isolated from PHHs treated with CDCA (25 μM), FGF19 (40 ng/ml), or GW4064 (1 μM) for 24 h. Of more than 500 human miRNAs in the microarray, only 5 (miR-422a, miR-622, miR-637, miR-487b, and miR-597) were found to be differentially expressed with a log2 ratio (treated/control) of >0.5 (Table 1; supplementary Fig. I). When converted to percent of changes, CDCA induced miR-622 and miR-637 by ∼50%, but inhibited miR-487b by ∼50% and miR-597 by ∼40%. On the other hand, GW4064, a specific and potent FXR agonist, and FGF19 had a stronger effect on induction of miR-422a and miR-622 and inhibition of miR-487b and miR-597. We found that miR-122a, which is a liver-specific and the predominant miRNA expressed in human liver (more than 70% of total miRNA) (22, 23), was induced ∼30% by GW4064, ∼24% by FGF19, and ∼10% by CDCA. FGF19 and GW4064 have similar effects on the expression of these miRNAs. We further confirmed the effect of CDCA, GW4064, and FGF19 on the expression of miR-122a and miR-422a in four different PHH preparations using quantitative RT-PCR. Results in Fig. 1 show similar levels of induction of miR-122a and miR-422a expression by these reagents as obtained from miRNA microarray analysis (Table 1).
Fig. 1.
CDCA, GW4064, and FGF19 induce miR-122a and miR-422a mRNA levels in PHHs. PHHs were treated with CDCA (25 µM), GW4064 (1 µM), or FGF19 (40 ng/ml) for 24 h. Total RNA was isolated for quantitative real-time PCR analysis of miR-122a (A) and miR-422a (B) expression levels. miRNA expression is relative to vehicle-treated control.
We then performed analysis of potential binding sequences for these six miRNAs in the 3′-UTR of human CYP7A1 mRNA and identified one putative binding site each for miR-122a and miR-442a (Fig. 2). The “seed match“ sequence of miR-122a matched 7/8 nucleotides in positions 62–69. The seed match sequence is important in guiding miRNA to specifically bind to the 3′ complementary sequences in mRNA, and some upstream sequences may be also involved in binding. The seed match sequences of miR-422a matched perfectly (7/7 in positions 243–249). A miR-422a site, but not miR-122a, was identified in the 3′-UTR of CYP8B1 and matched 7/7 nucleotides (positions 237–243). The other four miRNA binding sites in CYP7A1 and CYP8B1 have not been identified or listed in the miRNA databases. We thus focused our study on miR-122a and miR-422a.
Fig. 2.
Putative miR-122a and miR-422a target sites in the 3′-UTR of human CYP7A1 mRNA. Complementation of miR-122a and miR-422a sequences to the putative target sequences in the 3′-UTR of human CYP7A1 mRNA and miR-422a to the 3′-UTR of human CYP8B1 mRNA are shown. Nucleotide numbers are counted from the stop codon of human CYP7A1 mRNA. The nucleotides 2–8 in the 5′-end of the miRNA is referred to as seed match sequences that are responsible for binding of the miRNA and the 3′-UTR of target mRNA. The seed match sequences are indicated by bold type.
Differential expression of miRNAs, CYP7A1, and FGF19 mRNAs in PHHs and HepG2 and Huh7 hepatoma cells
We used quantitative real-time PCR assays to analyze mRNA expression levels of miR-122a, miR-422a, CYP7A1, CYP8B1, FGF19, and several nuclear receptors involved in the regulation of bile acid synthesis in PHH, HepG2, and Huh7 cells (supplementary Fig. II.). Because the ΔΔCt method cannot be used to compare mRNA expression in different cells, tissues, or species, we used the Ct values (the threshold cycle numbers for quantitative real-time PCR amplification of mRNAs) to compare the relative expression levels of mRNA transcripts in different human cells. Higher Ct values indicate lower mRNA expression levels in different cells. It has been reported that miR-122a is the most abundant miRNA expressed in human hepatocytes, but not in HepG2 cells (24). We confirmed that miR-122a is highly expressed in PHHs with a Ct value of about 21, which is about 100-fold higher than in Huh7 cells (Ct = 28) and 10,000-fold higher than in HepG2 (Ct = 35), assuming exponential amplification in different cells (internal standards RNU48 and UBC are similar). We thus chose PHHs for loss-of-function and HepG2 for gain-of-function studies of miR-122a. MiR-422a levels are much lower (Ct = ∼29–32) than miR-122a in all three types of hepatocytes. CYP7A1 mRNA expression levels are low but detectable in PHH (Ct ∼32) and HepG2 cells (Ct = ∼30), but undetectable (Ct = ∼40, the limit of detection) in Huh7 cells. In PHH, CYP8B1 levels (Ct = ∼26 in PHHs and ∼29 in HepG2) are about 100-fold higher than CYP7A1. On the other hand, FGF19 is highly expressed in Huh7 cells (Ct = 22), but expressed at very low levels in PHH (Ct = ∼31) and HepG2 (Ct = ∼33) cells. FXR levels are higher in Huh7 cells than in PHH and HepG2 cells. The relatively high levels of FGF19, FXR, and maybe also FGFR4 (Ct = ∼20) may contribute to the low levels of CYP7A1 mRNA in Huh7 cells. Similarly, relatively low levels of FGF19, FGFR4, and β-Klotho (a coactivator of FGFR4) may contribute to higher levels of CYP7A1 mRNA expression in PHH and Hep2 cells than in Huh7 cells.
Effects of miR-122a and miR-422a on human CYP7A1 gene expression
Because PHHs express high levels and HepG2 cells express low levels of endogenous miR-122a and miR-422a, we chose PHHs for loss-of-function and HepG2 for gain-of-function studies of miR-122a and miR-422a on CYP7A1 mRNA levels. The miRNA-mimics (Mir) are double-stranded RNA oligonucleotides designed to mimic the function of endogenous miRNA. The miRNA mimic negative controls are scrambled miRNA mimic sequences. We also used miR-122a and miR-422a inhibitors to antagonize their effect on CYP7A1 mRNA expression in PHHs, which express both miR-122a and miR-422a. The miRNA inhibitors (I-Mir) are single-stranded, chemically enhanced oligonucleotides designed to inhibit the function of endogenous miRNA. The miRNA inhibitor negative controls are scrambled miRNA inhibitor sequences.
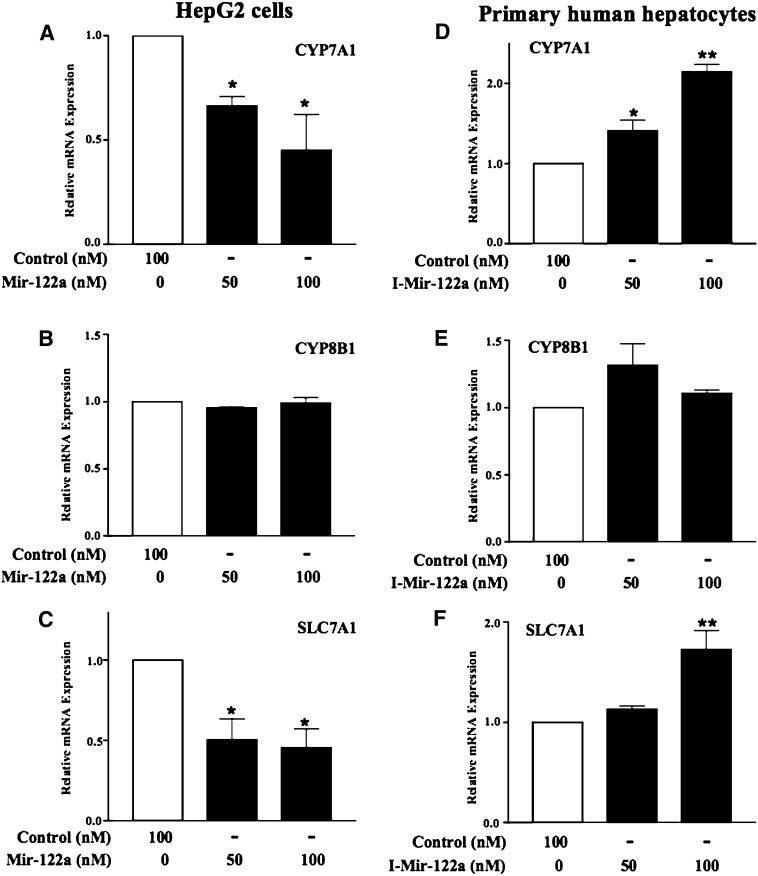
Transfection of miR-122a-mimic and miR-422a-mimic dose-dependently increased the abundance of miR-122a and miR-422a in HepG2 cells (supplementary Fig. III). As shown in Fig. 3A, transfection of a miR-122a-mimic into HepG2 cells significantly repressed CYP7A1 mRNA expression by 50% but had no effect on CYP8B1 (Fig. 3B). A miR-122a-mimic negative control (control) had no effect on CYP7A1 mRNA expression (Fig. 3A, open bar, set as 1). We also assayed the effect of Mir-122a on mRNA expression of SLC7A1, a well-characterized target of miR-122a in hepatocytes (17). SLC7A1 mRNA was significantly repressed by Mir-122a (Fig. 3C).
Fig. 3.
MiR-122a inhibits CYP7A1 mRNA expression. A–C: HepG2 cells were transfected with the miR-122a mimic (Mir-122) or miR-122a mimic control (Control). mRNAs were isolated for quantitative real-time PCR analysis. The effect of miR-122a mimic on SLC7A1 mRNA expression was assayed as a positive control. D–F: PHHs were used to study the effect of a miR-122a inhibitor (I-Mir-122a) on CYP7A1, CYP8B1, and SLC7A1 mRNA expression. SLC7A1 mRNA expression was assayed as a positive control. Data represent the mean ± SD of at least three individual experiments. * P < 0.05 and ** P < 0.001 indicate statistically significant difference of MiR-122a or I-Mir-122a versus their respective negative control (Control).
To assess further whether inactivation of miR-122a would increase CYP7A1 mRNA levels, we transfected a miR-122a inhibitor (I-Mir-122a) into PHHs for 48 h. I-Mir-122a significantly increased the CYP7A1 (Fig. 3D) and SLC7A1 (Fig. 3F) mRNA levels by 2-fold but had no effect on CYP8B1 (Fig. 3E). The miR-122a inhibitor negative control (Control) had no effect on CYP7A1 mRNA expression (Fig. 3D, open bar, set as 1).
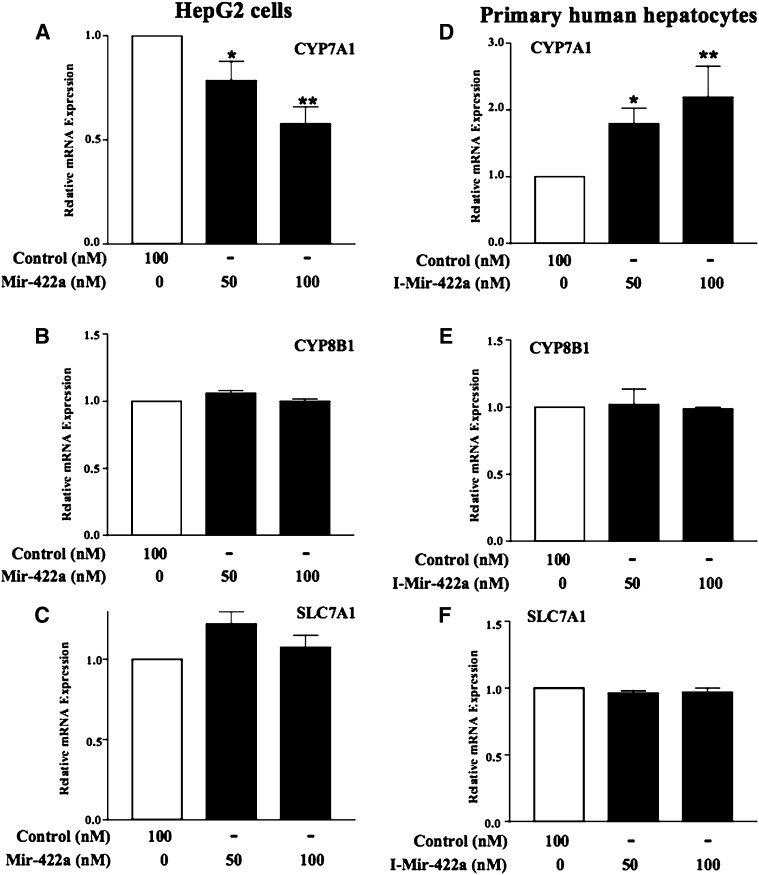
We also studied the effect of miR-422a-mimic on CYP7A1 mRNA expression in HepG2 cells and PHHs. Figure 4A shows that miR-422a-mimic (Mir-422a) inhibited CYP7A1 mRNA expression levels by 50% at 100 nM but had no effect on CYP8B1 (Fig. 4B) and SLC7A1 (Fig. 4C) mRNA expression levels. On the other hand, a miR-422a inhibitor (I-Mir-422a) increased CYP7A1 mRNA expression levels by 2-fold in PHHs (Fig. 4D) but had no effect on CYP8B1 (Fig. 4E) and SLC7A1 (Fig. 4F) mRNA expression. These miRNA overexpression and inhibition assays demonstrated that miR-122a inhibited CYP7A1 mRNA expression, but not CYP8B1 mRNA, which lacks a binding site for miR-122a. On the other hand, miR-422a specifically inhibited CYP7A1 mRNA expression, but had no effect on CYP8B1 mRNA despite the presence of a putative miR-422a site in CYP8B1 mRNA.
Fig. 4.
MiR-422a inhibits CYP7A1 mRNA expression. A–C: HepG2 cells were transfected with the miR-422a mimic (Mir-422) or miR-422a mimic control (Control). MRNAs were isolated for quantitative real-time PCR analysis. D–F: PHHs were used to the assay effect of a miR-422a inhibitor (I-Mir-422a) on CYP7A1, CYP8B1, and SLC7A1 mRNA expression. Data represent the mean ± SD of at least three individual experiments. The * P < 0.05 and ** P < 0.001 indicates statistically significant difference of MiR-422a or I-Mir-422a versus their respective negative control (Control).
Target validation of miR-122a and miR-422a on the 3′-UTR of human CYP7A1 mRNA
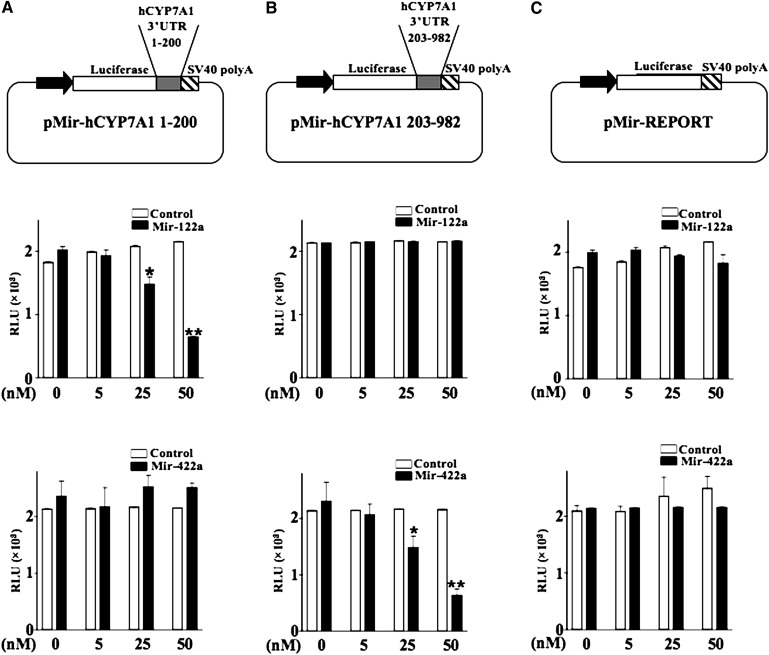
To investigate whether the predicted miR-122a and miR-422a target sequences in the 3′-UTR of CYP7A1 mRNA are functional, we cloned these miRNA target sequences downstream of the luciferase reporter gene to assay the effect of the miR-122a and miR-422a on the luciferase reporter activity. Figure 5A shows that miR-122a-mimic (Mir-122a) strongly inhibited the reporter activity of pMir-CYP7A1-1-200, which contains the miR-122a target sequence in the 3′-UTR of human CYP7A1 mRNA (nt 1–200), whereas miR-422a-mimic (Mir-422a) had no effect on this reporter activity. Figure 5B shows that MiR-422a strongly inhibited the reporter activity of pMir-CYP7A1-203-982, which contains a miR-422a target sequence in the 3′-UTR of human CYP7A1 mRNA (nt 203–982), whereas MiR-122a had no effect on this reporter activity. Reporter activity of the empty plasmid, pMir-REPORT, was neither affected by MiR-122a nor by MiR-422a (Fig. 5C). The miR-mimic negative controls (Control) for miR-122a and miR-422a had no effect on the luciferase activity of these reporters (Fig. 5A–C, open bars). Furthermore, mutations of Mir-122a and Mir-422a target sequences abolished the miR-122a mimic and miR-422a mimic effect on the reporter activities (Fig. 6A, B). Despite the presence of a perfect seed match sequence for miR-422a in the 3′-UTR of CYP8B2 transcript (Fig. 2C), miR-422a mimic did not affect a reporter construct containing the miR-422a binding sequence in the CYP8B1-3′-UTR (supplementary Fig. IV). The CYP8B1-3′-UTR does not have a miR-122a binding site, and miR-122a mimic did not affect the reporter activity as expected. These results suggest that miR-122a and miR-422a binding sites identified in the 3′-UTR of CYP7A1 mRNA are functional and specifically recognized by miR-122a and miR-422a to exert their inhibitory effects on CYP7A1 expression. On the other hand, the miR-422a site in the 3′-UTR of CYP8B1 transcript is not functional, and these miRNAs do not regulate CYP8B1 expression.
Fig. 5.
MiR-122a-mimic and miR-422a-mimic inhibit CYP7A1 reporter activity via the 3′-UTR of human CYP7A1 mRNA. Schematic diagram of human CYP7A1 3′-UTR luciferase reporter constructs are illustrated. A: Luciferase reporter assay of a reporter plasmid pMir-hCYP7A1 1-200 containing the putative miR-122a target sequence derived from nucleotides 1–200 of the human CYP7A1 3′-UTR. B: Luciferase reporter assay of a reporter plasmid pMir-hCYP7A1 203-982 containing the putative miR-422a target sequence derived from nucleotide 203–982 of the human CYP7A1 3′-UTR. C: Luciferase reporter assay of an empty plasmid pMir-REPORT. Reporter activities were assayed in HepG2 cells transfected with miR-122a-mimic (Mir-122a) or miR-422a-mimic (Mir-422a) or their respective negative control (Control). All experiments were done in triplicates and data represent the mean ± SD of three individual experiments. The * P < 0.05 and ** P < 0.001 indicate statistically significant difference of Mir-122a or MiR-422a versus their respective negative control (Control).
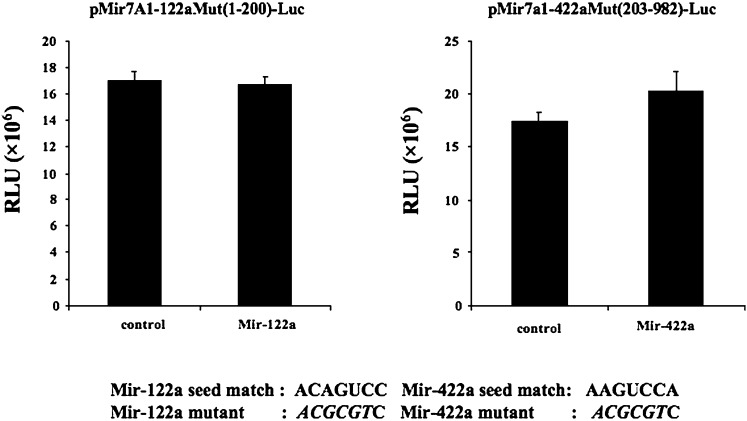
Fig. 6.
Mutagenesis analysis of miR-122a and miR-422a binding sites on regulation of CYP7A1 3′-UTR reporter activity. HepG2 cells were transfected with a reporter plasmid pMir-hCYP7A1-122aMut (1-200)-Luc containing mutant miR-122a target sequences (A) or luciferase reporter plasmid pMir-hCYP7A1-422aMut (203-982)-Luc containing mutant miR-422a target sequence (B), and cotransfected with 50 nM miR-122a-mimic, miR-422a-mimic, or miRNA controls as indicated for 48 h. Mutations (italic, lower case) of the seed match sequences are illustrated. All experiments were done in triplicates and data represent the mean ± SD.
Inhibition of miR-122a and miR-422a did not reverse the inhibitory effect of FGF19 on CYP7A1 mRNA expression
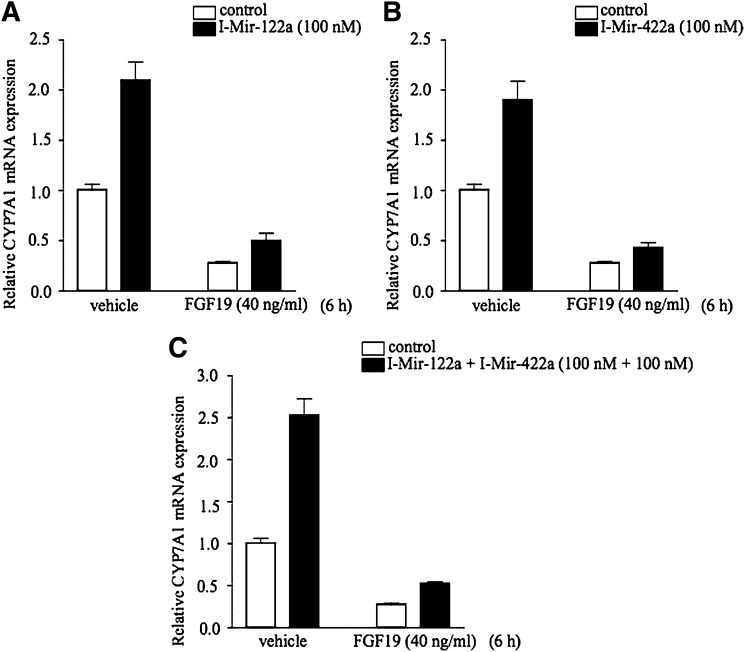
We reported previously that FGF19 signaling activates the MAPK/ERK1/2 K pathway to inhibit CYP7A1 gene transcription (6). However, the downstream factor (s) involved in direct regulation of CYP7A1 gene expression is not known. Results from this study may imply that FGF19 may inhibit CYP7A1 gene transcription via induction of miR-122a and miR-422a expression in hepatocytes. The endogenous miR-122a and miR-422a in PHHs were inhibited by I-Mir-122a and/or I-Mir-422, and then FGF19 was added to test if the inhibitory effect of FGF19 is attenuated. Figure 7 shows that FGF19 strongly inhibited CYP7A1 mRNA expression as expected. However, the inhibitory effect of FGF19 on CYP7A1 mRNA expression was not attenuated in hepatocytes treated with I-Mir-122a (Fig. 7A), I-Mir-422a (Fig. 7B), or both I-Mir-122a and Mir-422a (Fig. 7C). These results suggest that the inhibitory effect of FGF19 is not mediated through miR-122a and miR-422a.
Fig. 7.
MiR-122a and miR-422a do not play a role in mediating FGF19 inhibition of CYP7A1 mRNA expression in PHH. PHH cells were treated with FGF19 (40 mg/ml) and 100 nM of I-Mir-122a (A), I-Mir-422a (B), or both I-Mir-122a and I-Mir-422a (C) for 6 h. mRNAs were isolated for real time PCR analysis of CYP7A1 mRNA expression levels.
FGF19 inhibited the rate of CYP7A1 gene transcription
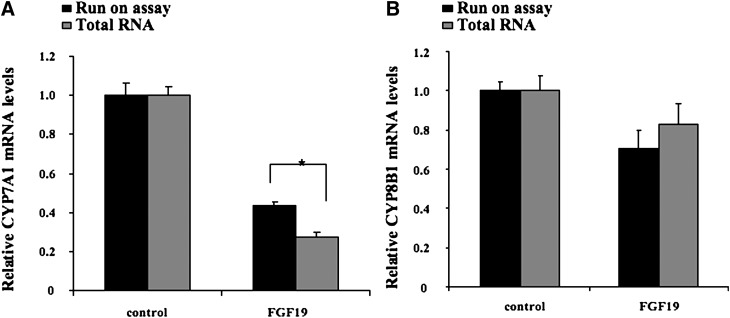
To ascertain if the inhibitory effect of FGF19 on CYP7A1 mRNA expression is through transcriptional and/or posttranscriptional mechanisms, we performed a nuclear run-on assay of the effect of FGF19 on the rate of CYP7A1 gene transcription. Figure 8A shows that FGF19 repressed the rate of CYP7A1 gene transcriptional by ∼60% in 24 h in primary hepatocytes. Real-time PCR assays show that FGF19 reduced the steady-state CYP7A1 mRNA levels by 75%. These results suggest that FGF19 inhibited CYP7A1 gene expression mainly by transcriptional mechanism, and posttranscriptional mechanisms played only a minor role. Interestingly, FGF19 had no significant effect on the CYP8B1 gene transcriptional rate and steady-state mRNA levels in human hepatocytes (Fig. 8B). These results further suggest that FGF19 signaling differentially regulates CYP7A1 and CYP8B1 expression in human hepatocytes.
Fig. 8.
Nuclear run-on assay of FGF19 effects on CYP7A1 mRNA transcription in PHH cells. PHH cells were treated with FGF19 at 40 ng/ml for 24 h. An aliquot of cells were used for total RNA preparation and reverse transcription. The rest of the cells were used for nuclei isolation and nuclear run-on assay as described in the “Materials and Methods” section. The CYP7A1 and CYP8B1 mRNA transcripts from total RNA and run-on assays were measured using real-time PCR and normalized to internal control UBC mRNA. Results were expressed as mean ± SD. N = 3. A P-value of <0.05 is considered as statistical significance.
DISCUSSION
This is the first report that CDCA, GW4064, and FGF19 modulate the expression of a small number of miRNAs in human hepatocytes. GW4064 and FGF19 have similar effects on the expression of miRNAs, suggesting that GW4064 specifically activates FXR to induce FGF19, which may modulate the expression of these miRNAs in human hepatocytes. In this study, we focused on miR-122a and miR-422a, because putative binding sites for those two miRNAs are localized in the 3′-UTR of CYP7A1 mRNA. Inhibition of CYP7A1 mRNA expression by miR-122a-mimic and miR-422a-mimic and stimulation by their inhibitors confirmed the inhibitory effect of these two miRNAs on CYP7A1 mRNA expression. Furthermore, miR-122a-mimic and miR-422a-mimic inhibit the activity of luciferase reporters containing miR-122a and miR-422a target sequences found in the 3′-UTR of human CYP7A1 mRNA. These results clearly demonstrated that these two miRNAs were functional in inhibiting CYP7A1 expression. In contrast, FGF19 did not affect CYP8B1 gene transcription and mRNA expression levels in human hepatocytes. The 3-UTR of CYP8B1 mRNA does not have a miR-122a binding site. A putative miR-422a targeting sequence was located in the 3′-UTR of human CYP8B1 mRNA but was not functional. We conclude that human miR-122a and miR-422a differentially regulate CYP7A1 and CYP8B1 expression in human hepatocytes. Accumulating evidences from recent studies have shown that the CYP7A1 and CYP8B1 genes are differentially regulated (25–27). The FXR/FGF19/FGFR4 pathway may be more important in the regulation of CYP7A1, whereas the FXR/small heterodimer partner pathway may be more important in the regulation of CYP8B1.
It has been determined that CYP7A1 mRNA has a very short half-life of about 30 min (9–11). The 3′-UTR of CYP7A1 mRNA contains multiple AUUUA elements (AT-rich sequences) that are present in short-lived mRNA (12, 13). Bile acids reduce CYP7A1 mRNA stability via sequences in the 3′-UTR (11, 13). It is possible that miR-122a may mediate bile acid inhibition by translational control of CYP7A1 expression in human hepatocytes. It is interesting that the miR-122a seed match sequence (nt 63–69) in the 3′-UTR of human CYP7A1 mRNA is located very close to an 84AUUUA88 element, which may bind apolipoprotein B editing enzyme (Apobec-1). It has been reported recently that Apobec-1 binds to CYP7A1-3′-UTR to stabilize CYP7A1 expression, and Apobec-1 null mice are susceptibility to diet-induced gallstone formation (28). It is possible that miR-122a and Apobec-1 may counteract each other to regulate the steady-state CYP7A1 expression levels in hepatocytes.
Several recent studies report that miR-122a plays a key role in regulating cholesterol biosynthesis and lipid metabolism in mice (16, 17, 29). Inhibition of miR-122a expression in mice using antagomirs or antisense oligonucleotides have uncovered the phenotypes of reduced serum cholesterol and triglyceride levels, increased fatty acid oxidation, and decreased hepatic fatty acid and cholesterol synthesis rate (16, 17, 29). Inhibition of miR-122a resulted in reducing plasma cholesterol and improving steatosis in a diet-induced obesity mouse model (17). In nonalcoholic steatohepatitis patients, miR-122a expression was reduced 63%, implicating a role of miR-122a in inhibiting fatty acid synthesis and preventing inflammation, proliferation, and steatosis (30). The mechanism of miR-122a in regulation of cholesterol metabolism is unknown. The miR-122a antagomirs do not affect mRNA expression, and the miR-122a binding site is not present in the 3′-UTR of mRNAs encoding 3-hydroxy-3-methyl- glutaryl-CoA (HMG-CoA) reductase and LDL receptor in mouse livers. These investigators speculated that miR-122a might indirectly induce these genes by inhibiting the expression of a repressor for these genes. Results from this study provide a plausible mechanism that miR-122a antagomirs may induce CYP7A1, which converts cholesterol to bile acids, and result in reducing serum cholesterol and triglyceride levels. Bile acids are known to reduce plasma triglyceride levels (1).
Interestingly, analysis of human miRNA databases revealed that a putative miR-122a binding sequence was present in the 3′-UTR of the FGF19 mRNA and a miR-597 site in human FGFR4 and bile salt export pump (BSEP) or ATP-binding cassette B11 (ABCB11) mRNAs. If these miRNA binding sites are functional, FGF19 may induce miR-122a as a negative feedback control of FGF19 expression. FGF19 may also inhibit miR-597 as a feed forward activation of FGFR4 signaling to inhibit CYP7A1 and induce BSEP to stimulate biliary bile acid excretion. Further studies are needed to verify these miRNA target genes involved in bile acid homeostasis.
CYP7A1 expression is mainly regulated by gene transcription, although posttranscriptional regulation of CYP7A1 has been reported [review in (1)]. Recent studies have provided very strong evidence that bile acid feedback inhibition of CYP7A1 gene transcription is mediated through FGF19/FGFR4 signaling, which activates the MAPK/ERK1/2 pathway to inhibit CYP7A1 gene transcription (6). However, the downstream factors that are activated by FGF19 signaling to inhibit CYP7A1 gene transcription are not known. Induction of miR-122a and Mir-422a by FGF19 reported in this study may imply that these miRNAs may be the downstream factors mediating the inhibitory effect of FGF19. Our nuclear run-on assay and miRNA inhibitor assay did not support the role of miRNA-122a and miR-422a in mediating posttranscriptional regulation of CYP7A1 by FGF19. Further studies are needed to elucidate the underlying mechanism of FGF19 inhibition of CYP7A1 gene transcription and to identify the physiological conditions that activate miRNAs to regulate CYP7A1 mRNA stability. It is well documented that CYP7A1 expression exhibits a distinct diurnal rhythm and is highly regulated by many stimuli, including inflammatory cytokines, growth factors, and insulin (1). In response to these stimuli, miRNA may play a role in regulation of CYP7A1 mRNA stability and the rate of bile acid synthesis. Future studies are needed to identify these conditions.
In conclusion, this is the first study that revealed a novel posttranscriptional regulation of CYP7A1 expression by miRNA in human hepatocytes. miRNAs may play a role in translational control of the steady-state levels of CYP7A1 mRNA to regulate bile acid synthesis, maintain cholesterol homeostasis, and protect hepatocytes from cholestatic liver injury. The miR-122a antagomirs may increase CYY7A1 mRNA stability to stimulate bile acid synthesis and reduce serum cholesterol and triglycerides. The miR122a antagomirs may be promising therapeutic drugs for treating hyperlipidemia and nonalcoholic fatty liver diseases.
Supplementary Material
Footnotes
Abbreviations:
- Apobec
- apolipoprotein B editing enzyme
- CDCA
- chenodeoxycholic acid
- CYP7A1
- cholesterol 7α-hydroxylase
- CYP8B1
- sterol 12α-hydroxylase
- ERK
- extracellular signal-regulated kinase
- FGF
- fibroblast growth factor
- FGFR4
- fibroblast growth factor 15 receptor 4
- FXR
- farnesoid X receptor
- I-mir
- miRNA inhibitor
- MAPK
- mitogen-activated protein kinase
- miRNA
- micro RNA
- miR
- miRNA mimic
- PHH
- primary human hepatocyte
- SLC7A1
- solute carrier 7A1
- 3′-UTR
- 3′-untranslated region
This study is supported by the National Institutes of Health grants DK44442 and DK58379. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figure.
REFERENCES
- 1.Chiang J. Y. 2009. Bile acids: regulation of synthesis. J. Lipid Res. 50: 1955–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang J. Y. 2002. Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr. Rev. 23: 443–463. [DOI] [PubMed] [Google Scholar]
- 3.Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., et al. 2003. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 17: 1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225. [DOI] [PubMed] [Google Scholar]
- 5.Lundasen T., Galman C., Angelin B., Rudling M. 2006. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J. Intern. Med. 260: 530–536. [DOI] [PubMed] [Google Scholar]
- 6.Song K-H., Chiang J. Y. L. 2009. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7α-hydoxylase gene expression. Hepatology. 49: 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaap F. G., van der Gaag N. A., Gouma D. J., Jansen P. L. 2009. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology. 49: 1228–1235. [DOI] [PubMed] [Google Scholar]
- 8.Chiang J. Y., Miller W. F., Lin G. M. 1990. Regulation of cholesterol 7α-hydroxylase in the liver. Purification of cholesterol 7α-hydroxylase and the immunochemical evidence for the induction of cholesterol 7α-hydroxylase by cholestyramine and circadian rhythm. J. Biol. Chem. 265: 3889–3897. [PubMed] [Google Scholar]
- 9.Noshiro M., Nishimoto M., Okuda K. 1990. Rat liver cholesterol 7 alpha-hydroxylase. Pretranslational regulation for circadian rhythm. J. Biol. Chem. 265: 10036–10041. [PubMed] [Google Scholar]
- 10.Pandak W. M., Stravitz R. T., Lucas V., Heuman D. M., Chiang J. Y. 1996. Hep G2 cells: a model for studies on regulation of human cholesterol 7α-hydroxylase at the molecular level. Am. J. Physiol. 270: G401–G410. [DOI] [PubMed] [Google Scholar]
- 11.Baker D. M., Wang S. L., Bell D. J., Drevon C. A., Davis R. A. 2000. One or more labile proteins regulate the stability of chimeric mRNAs containing the 3′-untranslated region of cholesterol-7α-hydroxylase mRNA. J. Biol. Chem. 275: 19985–19991. [DOI] [PubMed] [Google Scholar]
- 12.Li Y. C., Wang D. P., Chiang J. Y. L. 1990. Regulation of cholesterol 7α-hydroxylase in the liver: cDNA cloning, sequencing and regulation of cholesterol 7α-hydroxylase mRNA. J. Biol. Chem. 265: 12012–12019. [PubMed] [Google Scholar]
- 13.Agellon L. B., Cheema S. K. 1997. The 3′-untranslated region of the mouse cholesterol 7α-hydroxylase mRNA contains elements responsive to post-transcriptional regulation by bile acids. Biochem. J. 328: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miska E. A. 2005. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 15: 563–568. [DOI] [PubMed] [Google Scholar]
- 15.Khvorova A., Reynolds A., Jayasena S. D. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell. 115: 209–216. [DOI] [PubMed] [Google Scholar]
- 16.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 438: 685–689. [DOI] [PubMed] [Google Scholar]
- 17.Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3: 87–98. [DOI] [PubMed] [Google Scholar]
- 18.Song K. H., Ellis E., Strom S., Chiang J. Y. 2007. Hepatocyte growth factor signaling pathway inhibits cholesterol 7α-hydroxylase and bile acid synthesis in human hepatocytes. Hepatology. 46: 1993–2002. [DOI] [PubMed] [Google Scholar]
- 19.Lewis B. P., Burge C. B., Bartel D. P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 20.Enright A. J., John B., Gaul U., Tuschl T., Sander C., Marks D. S. 2003. MicroRNA targets in Drosophila. Genome Biol. 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36: D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 309: 1577–1581. [DOI] [PubMed] [Google Scholar]
- 23.Appel N., Bartenschlager R. 2006. A novel function for a micro RNA: negative regulators can do positive for the hepatitis C virus. Hepatology. 43: 612–615. [DOI] [PubMed] [Google Scholar]
- 24.Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M. A., Xu C., Mason W. S., Moloshok T., Bort R., et al. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 1: 106–113. [DOI] [PubMed] [Google Scholar]
- 25.Kim I., Ahn S. H., Inagaki T., Choi M., Ito S., Guo G. L., Kliewer S. A., Gonzalez F. J. 2007. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 48: 2664–2672. [DOI] [PubMed] [Google Scholar]
- 26.Mataki C., Magnier B. C., Houten S. M., Annicotte J. S., Argmann C., Thomas C., Overmars H., Kulik W., Metzger D., Auwerx J., et al. 2007. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol. Cell. Biol. 27: 8330–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y. K., Schmidt D. R., Cummins C. L., Choi M., Peng L., Zhang Y., Goodwin B., Hammer R. E., Mangelsdorf D. J., Kliewer S. A. 2008. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol Endocrinol. 22: 1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie Y., Blanc V., Kerr T. A., Kennedy S., Luo J., Newberry E. P., Davidson N. O. 2009. Decreased expression of cholesterol 7α-hydroxylase and altered bile acid metabolism in apobec-1−/− mice lead to increased gallstone susceptibility. J. Biol. Chem. 284: 16860–16871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elmen J., Lindow M., Silahtaroglu A., Bak M., Christensen M., Lind-Thomsen A., Hedtjarn M., Hansen J. B., Hansen H. F., Straarup E. M., et al. 2008. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 36: 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung O., Puri P., Eicken C., Contos M. J., Mirshahi F., Maher J. W., Kellum J. M., Min H., Luketic V. A., Sanyal A. J. 2008. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 48: 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.