Abstract
The aim of the present study was to examine whether pretreatment with different fatty acids, as well as the liver X receptor (LXR) agonist T0901317, could modify metabolic switching of human myotubes. The n-3 FA eicosapentaenoic acid (EPA) increased suppressibility, the ability of glucose to suppress FA oxidation. Substrate-regulated flexibility, the ability to increase FA oxidation when changing from a high glucose, low fatty acid condition (“fed”) to a high fatty acid, low glucose (“fasted”) condition, was increased by EPA and other n-3 FAs. Adaptability, the capacity to increase FA oxidation with increasing FA availability, was enhanced after pretreatment with EPA, linoleic acid (LA), and palmitic acid (PA). T0901317 counteracted the effect of EPA on suppressibility and adaptability, but it did not affect these parameters alone. EPA per se accumulated less, however, EPA, LA, oleic acid, and T0901317 treatment increased the number of lipid droplets (LD) in myotubes. LD volume and intensity, as well as mitochondrial mass, were independent of FA pretreatment. Microarray analysis showed that EPA regulated more genes than the other FAs and that specific pathways involved in carbohydrate metabolism were induced only by EPA. The present study suggests a favorable effect of n-3 FAs on skeletal muscle metabolic switching and glucose utilization.
Keywords: lipid metabolism, metabolic flexibility, adaptability, suppressibility, substrate-regulated flexibility
Skeletal muscle in lean, healthy individuals is characterized by metabolic flexibility; the ability to switch from predominantly lipid oxidation during fasting conditions to suppression of lipid oxidation and increased glucose oxidation in response to insulin (1, 2). Loss of this capacity to switch easily between glucose and lipid oxidation was termed metabolic inflexibility by Kelley and Mandarino (3). Obesity, insulin resistance, and type 2 diabetes (T2D) are associated with reduced lipid oxidation during fasting, impaired postprandial switch from lipid to glucose oxidation, and reduced capacity to increase lipid oxidation during exercise (4–7). Furthermore, a reduced postprandial switch from lipid to glucose oxidation has been observed in men with impaired glucose tolerance (4), suggesting that inflexibility plays a role in the early development of T2D. Ukropcova et al. described metabolic switching in vitro in human myotubes as suppressibility, defined as the ability of acutely added glucose to suppress fatty acid oxidation, and as adaptability, defined as the capacity of the cell to increase fatty acid oxidation upon increased fatty acid availability (8). In vitro suppressibility was found to be inversely correlated with insulin sensitivity and metabolic flexibility in vivo, whereas adaptability was positively correlated with these parameters, indicating that metabolic switching is an intrinsic characteristic of skeletal muscle cells (8). However, metabolic inflexibility could be due to both intrinsic and induced factors. Clinical studies have shown that postprandial impairments in metabolic flexibility can be improved by weight loss (4, 9, 10) and that exercise improves the ability of skeletal muscle to oxidize fatty acids during exercise and fasting (10–12). Still, the effect of dietary fat quality, such as different fatty acids, on metabolic flexibility is unknown.
A growing amount of evidence shows different effects of distinct fatty acids on lipid metabolism and insulin sensitivity. In vivo in rats, a diet rich in saturated fatty acids caused insulin resistance, whereas a diet rich in n-6 polyunsaturated fatty acids (PUFA) resulted in increased insulin sensitivity (13). Storlien et al. showed that replacement of the n-6 fatty acid linoleic acid (LA) with n-3 fatty acids prevented the development of insulin resistance in high-fat fed rats (14). Increased intake of fish oil or n-3 PUFA eicosapentaenoic acid (EPA) reduced insulin resistance in some but not all human studies (15–19). Haugaard et al. showed that changes in insulin resistance were inversely correlated to changes in membrane concentrations of EPA, docosahexaenoic acid, and total n-3 fatty acids, as well as the ratio of PUFA n-3/n-6 in human skeletal muscle (20). In vitro studies have shown that incubation of myotubes with the saturated fatty acid palmitic acid (PA) increased the cellular pool of diacylglycerol (DAG) and ceramides, as well as induced insulin resistance (21, 22). By contrast, incubation of myotubes with the unsaturated fatty acid oleic acid (OA) increased triacylglycerol (TAG) accumulation without inhibiting insulin action (21, 22). Pretreatment of human myotubes with EPA has been shown to promote increased uptake of fatty acids and increased TAG accumulation, also compared with pretreatment with OA, without impairing insulin-stimulated glucose uptake (23, 24). Wensaas et al. also showed that the level of total acyl-CoA was reduced after EPA pretreatment versus OA (24). The mechanism by which unsaturated fatty acids exert positive effects on skeletal muscle insulin sensitivity is unclear, but it may involve changes in glucose and fatty acid metabolism as well as changes in the level of different lipid intermediates in skeletal muscle. Furthermore, the effect may be mediated by changes in gene expression via specific nuclear receptors that function as fatty acid sensors. PUFAs are shown to act as antagonists on liver X receptors (LXR) (25, 26) and PUFAs as well as other FAs act as agonists on peroxisome proliferator-activated receptors (PPAR) (27–29). LXRs and PPARs are important regulators of energy metabolism in the body and might contribute to the regulation of metabolic switching. However, the roles for LXR and PPAR as well as the cross-talk between them in nutritional regulation are complex and not fully understood.
According to the Randle cycle (30), FAs reduce glucose oxidation, whereas glucose reduces FA oxidation in what is often called reverse Randle cycle (31, 32). While the mechanistic basis for the Randle cycle is relatively well understood, the molecular mechanism underlying metabolic inflexibility largely remains unknown. Furthermore, it is not known whether it is possible to change metabolic flexibility through dietary changes. The purpose of the present study was to test whether treatment with different fatty acids, including n-3 FAs and LXR activation by T0901317, could modify metabolic switching of myotubes to get insight into the cellular mechanisms behind a potential effect. Moreover, we wanted to use previously suggested parameters (8) and suggest a new parameter to describe energy metabolism in cell cultures.
MATERIALS AND METHODS
Materials
Dulbecco's modified Eagle's medium (DMEM-Glutamax™), DMEM without phenol red, nonheat-inactivated fetal calf serum (FCS), and penicillin-streptomycin-amphotericin B were purchased from Gibco Invitrogen (Gibco, Life Technologies Paisley, UK). BSA (essentially fatty acid-free), L-carnitine, and Dulbecco's phosphate-buffered saline (DPBS; with Mg2+ and Ca2+), oleic acid (OA), palmitic acid (PA), eicosapentaenoic acid (EPA), linoleic acid (LA), docosahexaenoic acid (DHA), and α-linolenic acid (ALA), extracellular matrix (ECM) gel, glycogen, and HEPES were obtained from Sigma (St Louis, MO). Ultroser G was purchased from Ciphergen (Cergy-Saint-Christophe, France), insulin (Actrapid®) from NovoNordisk (Bagsvaerd, Denmark). [1-14C]oleic acid (55 mCi/mmol), [1-14C]palmitic acid (57.5 mCi/mmol), [1-14C]linoleic acid (56 mCi/mmol), [1-14C]eicosapentaenoic acid (55 mCi/mmol), and D-[14C(U)]glucose (5 mCi/mmol) were from American Radiolabeled Chemicals (St Louis, MO). T0901317 was purchased from Cayman Chemical Co. (Ann Arbor, MI). Ecoscint A scintillation solution was from National diagnostics (Hessle, England, UK). Corning® CellBIND® tissue culture plates were obtained from Corning Life-Sciences (Schiphol-Rijk, The Netherlands). Glass bottom plates were from MatTek (Ashland, MA). OptiPhase Supermix, UniFilter®-96 GF/B and ScintiPlate®-96 TC plates were delivered by PerkinElmer (Shelton, CT). Protein assay reagens was purchased from BioRad (Copenhagen, Denmark). Phospho-Akt (Ser473) and Akt antibodies were from Cell Signaling Technology (Beverly, MA). RNeasy minikit was from Qiagen (Venlo, The Netherlands). NuGO human Genechip arrays were obtained from Affymetrix (Santa Clara, CA). Bodipy 493/503 (4.4-difluoro-1.3.5.7.8- pentamethyl-4-bora-3a.4a-diaza-s-indacene), MitoTracker® Red FM, and Hoechst 33258 were obtained from Molecular Probes, Invitrogen (Carlsbad, CA). All other chemicals used were of standard commercial high-purity quality.
Cell culture and fatty acid incubation
Satellite cells were isolated as previously described (33) from the M. obliquus internus abdominis of eight healthy donors. Both males and females were included, age 50 ± 10 years, body mass index 24.0 ± 3.2 kg/m2, fasting glucose 6.0 ± 1.0 mmol/l, insulin, plasma lipids and blood pressure within normal range, and no family history of diabetes. The biopsies were obtained with informed consent and approval by the National Committee for Research Ethics, Oslo, Norway.
The cells were cultured on 96-well plates (CellBIND®) with DMEM-Glutamax™ (5.5 mM glucose), 2% FCS, 2% Ultroser G, P/S, and amphotericin B. At 70%–80% confluence, the growth medium was replaced by DMEM-Glutamax™ supplemented with 2% FCS, P/S, 1.25 µg/ml amphotericin B, and 25 pM insulin to induce differentiation. The cells were cultured in humidified 5% CO2 atmosphere at 37°C, and the medium was changed every 2–3 days. After 5 days of differentiation, the cells were pretreated with 1 µM T0901317 or vehicle (DMSO) for 24 h, and then preincubated with different fatty acids (OA, PA, EPA, or LA; 100 µM) or BSA (40 µM, control) in combination with T0901317 (for EPA) or vehicle for another 24 h. Stock solutions of fatty acid sodium salts (6 mM) and BSA (2.4 mM) were heated to 45°C and rapidly mixed (molar ratio of 2.5:1). Only optically clear solutions were used. There were no differences between the myotubes after preincubation with fatty acids or T0901317 as evaluated by microscopic inspection searching for floating cells. The myotubes had similar protein content independent of preincubation medium.
Substrate oxidation assay
The muscle cells were cultured on 96-well CellBIND® microplates as described above. Substrate, [1-14C]oleic acid (1 µCi/ml, 100 µM), with and without glucose (5 mM) present, or D-[14C(U)]glucose (1 µCi/ml, 200 µM), was given in DPBS with 10 mM HEPES and 1 mM L-carnitine. A 96-well UniFilter®-96 GF/B microplate was mounted on top of the CellBIND® plate as described before (34), and the cells were incubated for 4 h at 37°C. The CO2 trapped in the filter was counted by liquid scintillation in a 1450 MicroBeta TriLux scintillation counter (PerkinElmer). The remaining cell-associated radioactivity was also assessed by liquid scintillation, and the sum of CO2 and cell-associated radioactivity was considered as total substrate utilization. Protein content in each well was determined by use of Coomassie reagent (35).
Scintillation proximity assay
Radiolabeled substrates taken up and accumulated by adherent cells will be concentrated close to the scintillator embedded in the plastic bottom of each well (ScintiPlate®-96 TC, PerkinElmer) and provide a stronger signal than the radiolabel dissolved in the medium alone. Measurements of fatty acid uptake by scintillation proximity assay (SPA) were performed in medium as described above without phenol red, with an additional 100 µM OA, PA, LA, or EPA (1 µCi/ml [1-14C]FA), bound to BSA (40 µM) at a ratio of 2.5:1, and were monitored at 0, 1, 2, 3, 4, 6, 8, and 24 h during the incubation. After the 24 h experiment, measurement of acid-soluble metabolites (ASM) was performed with a method modified from Skrede et al. (36). Incubation media (30 µl) were transferred to a new multiwell plate, precipitated with 100 µl HClO4 (1 M) and 10 µl BSA (6%), and centrifuged at 2100 g for 10 min at 4°C. Then, 30 µl of the supernatant was counted by liquid scintillation. ASMs consist mainly of tricarboxylic acid cycle metabolites and reflect incomplete oxidation of fatty acids.
Lipid distribution
Myotubes were incubated with 100 µM fatty acids (OA, PA, LA, and EPA) supplemented with a trace amount of [1-14C]oleic acid (0.5 µCi/ml) for 24 h. Myotubes were washed twice with PBS (1 ml), harvested into a tube with two additions of 250 µl distilled water, and frozen at −20°C. Cells were later assayed for protein (35), and cellular lipids were extracted (37). Briefly, homogenized cell fractions were extracted, lipids were separated by thin-layer chromatography, and radioactivity was quantified by liquid scintillation. The amount of neutral lipids was calculated by using protein levels for standardization.
Glycogen synthesis
After preincubation with fatty acids and T0901317, the cells were incubated for 3 h with D-[14C(U)]glucose (2 µCi/ml, 5.5 mM) with and without 1 µM insulin. The cells were washed twice with PBS and harvested in 1 M KOH. Protein content in each well was determined by use of Coomassie reagent (35). The rest of the sample was added glycogen (10 mg/ml) and incubated for 20 min at 80°C on a heat block. After cooling down, the samples were added to ice-cold 100% ethanol and centrifuged (10500 g, 20 min, 4°C). The pellets were washed once with ice-cold 70% ethanol, centrifuged (10500 g, 20 min, 4°C), air-dried, and resuspended in distilled water. Radioactivity was measured by scintillation counting, and the glycogen synthesis was calculated by using protein levels for standardization.
Live imaging of lipid droplets and mitochondria
Myotubes were cultured and pretreated as described above, with the exception of using 12-well glass bottom plates coated with ECM. Myotubes were incubated with Bodipy 493/503 (2 µg/ml), which is a lipophilic dye that enters the nonpolar lipid droplet (LD) core, for 5 min and washed with PBS. Next, the cells were incubated for 15 min with MitoTracker®Red FM (100 nM) to stain mitochondria and washed with PBS. Finally, the cells were incubated for 15 min with Hoechst 33258 (2.5 µg/ml) diluted in PBST-BSA to stain nuclei and washed with PBS. The Olympus Scan^R platform was used for automated image acquisition and subsequent analysis. The basis of this platform is an Olympus IX81 inverted fluorescence microscope with a computer-controlled motorized stage that uses autofocus to find the z-position of interest. The microscope has a temperature and a CO2 enrichment incubator for long-term live imaging. Epi-fluorescence illumination was from a MT20 arc burner (Olympus) coupled into the microscope through an optical fiber. We used a 20× objective and standard filter sets with excitation windows for Hoechst 33258 (350 nm), Bodipy 493/503 (488 nm), and MitoTracker®Red FM (594 nm), and a triple band filter for emission.
Images were made at 25 positions per well. We used the background-subtracted maximal intensity projection from 12 images taken in z-direction (1 µm apart) for each color channel at each position. The images were analyzed and quantified using Scan^R analysis software, which uses an edge detection algorithm for object segmentation. We segmented and quantitatively analyzed the nuclei, LDs, and mitochondria in the appropriate color-channels. Parameters measured were the total intensity of MitoTracker®Red FM signal per image (mitochondrial mass), the number of LDs per frame, the number of nuclei per frame, as well as the diameter and intensity of each LD. After gating out obviously artifactual image abnormalities, the images were quantified.
Microarray
Human myotubes were preincubated with different fatty acids (OA, PA, EPA, or LA; 100 µM) or BSA (40 µM) for 24 h. Thereafter, cells were harvested, and total RNA was prepared from primary myotubes from three donors per treatment using RNeasy mini kit according to the supplier's protocol (Qiagen). RNA was used individually, and RNA integrity was checked on chip analysis (Agilent 2100 bioanalyzer, Agilent Technologies, Amsterdam, The Netherlands) according to the manufacturer's instructions. RNA was judged as suitable for array hybridization only if samples exhibited intact bands corresponding to the 18S and 28S rRNA subunits and displayed no chromosomal peaks or RNA degradation products (RNA Integrity Number > 8.0). Ten micrograms of RNA were used for one cycle cRNA synthesis (Affymetrix, Santa Clara, CA). Hybridization, washing, and scanning of Affymetrix human NuGO Genechip arrays were according to standard Affymetrix protocols. Probe sets were defined according to Dai et al. (38). In this method, probes are annotated using up-to-date databases and assigned to unique gene identifiers, in this case Entrez IDs. The probes present on the human NuGO arrays (Gene Expression Omnibus platform number GPL7020) represent 16483 Entrez IDs. After quality control, three arrays per treatment were considered suitable for further analysis. Arrays were normalized with the Robust Multi-array Average (RMA) method (39, 40). Only probe sets with normalized signal intensities above 20 on at least two arrays were selected for further analysis. Individual fold changes (FC) for each donor were calculated as log2-transformed expression level after fatty acid treatment divided by log2-transformed expression level after BSA treatment (control). Mean fold changes were calculated based on the log2-transformed individual fold changes. Differentially expressed probe sets were identified using intensity-based moderated paired t-statistics (41). P values were corrected for multiple testing using a false discovery rate (FDR) method (42). Probe sets that satisfied the criterion of FDR < 20% (q-value < 0.2) were considered to be significantly regulated. Gene set enrichment analysis (GSEA) was performed for functional analysis of changes in gene expression. GSEA is focused on predefined gene sets, that is, groups of genes that share biological function, chromosomal location, or regulation (43). The “functional catalogue” constructed by Subramanian et al. (43) was modified to contain only 505 well-defined biochemical, metabolic, and signal pathways compiled from the following publicly available, curated databases: Biocarta, GenMAPP (44), Kyoto Encyclopedia of Genes and Genomes (KEGG) (45), Sigma-Aldrich pathways, and Signal Transduction Knowledge Environment. The analysis was run using 1,000 permutations per gene set. Gene sets with an FDR < 0.2 were considered significantly regulated. The advantage of this method is that it is unbiased because a score is calculated based on all genes in a gene set. All array data have been submitted to Gene Expression Omnibus (accession number GSE18589).
Real-time RT-PCR
Total RNA was isolated using RNeasy minikit (Qiagen, Venlo, The Netherlands) according to manufacturer's instructions. RNA sample concentrations were determined using a NanoDrop ND-1000 spectrophotometer (Isogen, Maarssen, The Netherlands). One microgram of total RNA was reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Veenendaal, The Netherlands). Real-time PCR was performed with platinum Taq polymerase (Invitrogen, Breda, The Netherlands) and SYBR green on an iCycler PCR machine (Bio-Rad Laboratories). Melt curve analysis was included to assure a single PCR product was formed. PCR primer sequences were taken from the PrimerBank and ordered from Eurogentec (Seraing, Belgium). Sequences of the primers used are available upon request.
Immunoblotting
Total cell lysates prepared in Laemmli buffer were electrophoretically separated on 10% (w/v) polyacrylamide gels (acrylamide/NVNV-bis-methylene acrylamide 5 30:0.8) followed by immunoblotting overnight with antibody recognizing Akt1 [protein kinase B (PKB)] phosphorylated at Ser473 and Akt2 and Akt3 when phosphorylated at equivalent sites, as well an antibody detecting total level of Akt1, Akt2, and Akt3 (Cell Signaling Technology, Beverly, MA). Immunoreactive bands were visualized with enhanced chemiluminescence and quantified with Gel-Pro Analyzer (version 2.0) software.
Calculations of metabolic parameters
Suppressibility is the ability of the cells to decrease OA oxidation by acutely added glucose: [(1-(oxidation of OA at 5 mM glucose / oxidation of OA at no glucose added)) × 100%]. Adaptability is ability to increase the OA oxidation with increasing OA concentration: [oxidation of 100 µM OA / oxidation of 5 µM OA]. Substrate-regulated flexibility is the ability to increase the OA oxidation while changing from the “fed” (low fatty acid, high glucose) to the “fasted” (high fatty acid, no glucose added) condition: [oxidation of 100 µM OA without glucose added / oxidation of 5 µM OA at 5 mM glucose].
Presentation of data and statistics
All values are reported as means ± SEM. The value n represents the number of different donors used. Linear mixed models (LMM) were used to compare effects of different treatments. The statistical analyses were performed with SPSS version 12 (SPSS Inc., Chicago, IL). A P value < 0.05 was considered significant. Bonferroni correction was applied for multiple comparisons.
RESULTS
Fatty acid uptake and effects on insulin-stimulated glucose metabolism in myotubes
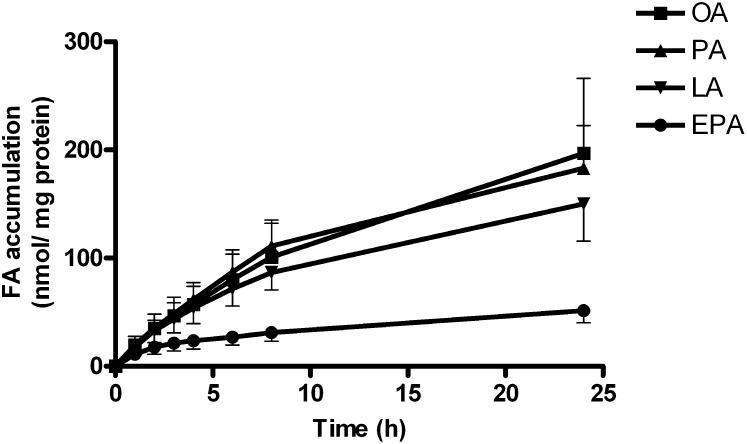
Real-time determination of accumulation of 100 µM of [14C]oleic acid (OA, 18:1, n-9), [14C]palmitic acid (PA, 16:0), [14C]linoleic acid (LA, 18:2, n-6), and [14C]eicosapentaenoic acid (EPA, 20:5, n-3) in differentiated myotubes during 24 h showed that EPA accumulated significantly less than LA, OA, and PA (P < 0.05, Fig. 1). However, the oxidation of [14C]EPA to ASMs was 2.5-fold increased compared with OA (P = 0.02, data not shown).
Fig. 1.
Real-time accumulation of [14C]oleic acid (OA), [14C]palmitic acid (PA), [14C]linoleic acid (LA) and [14C]eicosapentaenoic acid (EPA) during 24 h in differentiated myotubes. Myotubes were incubated with [14C]fatty acids (100 µM, 1 µCi/ml) and measurements of cell-associated radioactivity were monitored by SPA as described in “Materials and Methods” at 0, 1, 2, 3, 4, 6, 8, and 24 h during the incubation. Results represent means ± SEM from 3 different donors.
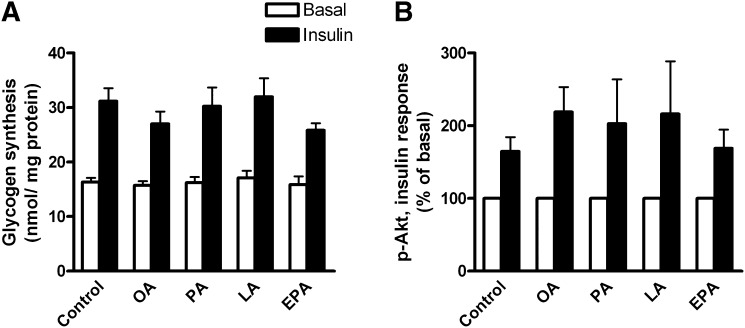
Preincubation of myotubes with 100 µM fatty acids (FAs) for 24 h had no negative influence on glucose metabolism and insulin action. Myotubes were pretreated for 24 h with 100 µM OA, PA, LA, EPA or 40 µM BSA (BSA, fatty acid-free control). Pretreatment with fatty acids did not affect basal or insulin-stimulated glycogen synthesis (Fig. 2A, average insulin response 84%, P < 0.001). Furthermore, insulin sensitivity, measured as phosphorylation of Akt, adjusted for the level of total Akt, was not changed after preincubation with 100 µM fatty acids (Fig. 2B).
Fig. 2.
Insulin responses. (A) Glycogen synthesis. Myotubes were pretreated for 24 h with 100 µM OA, EPA, LA, PA, or 40 µM BSA (control), before 3 h incubation with D-[14C(U)]glucose (5.5 mM, 2 µCi/ml) in the presence or absence of 1 µM insulin. Glycogen synthesis was measured as described in “Materials and Methods.” Results represent means ± SEM for 4 different donors. (B) Akt phosphorylation. Myotubes were pretreated for 24 h with 100 µM OA, EPA, LA, PA, or 40 μM BSA (control), before 15 min incubation in the presence or absence of 1 µM insulin. Cells were harvested and immunoblot analysis was performed with antibodies against phosphorylated Akt and total Akt as described in “Materials and Methods.” Optical density of phosphorylated Akt was adjusted for total Akt. Results represent means relative to basal level ± SEM for 3–4 different donors. EPA, eicosapentaenoic acid; LA, linoleic acid; OA, oleic acid; PA, palmitic acid.
Effect of FA pretreatment on glucose uptake and oxidation
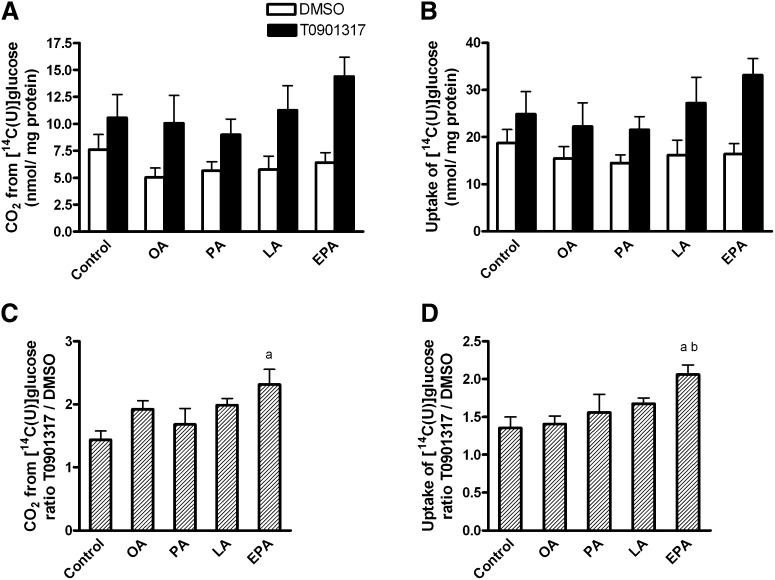
The effect of pretreatment with fatty acids and T0901317 on glucose metabolism was further examined. Myotubes were pretreated for 24 h with 1 µM T0901317 or vehicle (DMSO), and then for another 24 h with 100 µM OA, EPA, LA, PA, or 40 µM BSA (control) in the presence or absence of T0901317. Pretreatment with T0901317 for 48 h increased glucose oxidation by 44-132% (average 87%, P < 0.001, Fig. 3A) and glucose uptake, which is the sum of recovered CO2 and cell-associated glucose, with 36%–106% (average 61%, P < 0.001, Fig. 3B). When combined with EPA, the T0901317-induced augmentation of glucose oxidation was enhanced compared with control (P < 0.05, Fig. 3C). Also the T0901317-mediated augmentation in glucose uptake was larger when T0901317 was used in combination with EPA than with OA or control (P < 0.05, Fig. 3D). Since the effect of T0901317 was more pronounced only when combined with EPA, we chose to further examine this combination.
Fig. 3.
Glucose uptake and oxidation. Myotubes were pretreated for 24 h with 1 µM T0901317 or vehicle (DMSO), and then for another 24 h with 100 µM OA, EPA, LA, PA, or 40 µM BSA (control) in the presence or absence of T0901317. Thereafter the cells underwent CO2 trapping for 4 h with D-[14C(U)]glucose (200 µM, 1 µCi/ml) as described in “Materials and Methods.” Effect of T0901317 treatment on (A) glucose oxidation T0901317 versus DMSO (P < 0.001) and (B) glucose uptake, which is the total recovered CO2 and cell-associated glucose T0901317 versus DMSO (P < 0.001). Pretreatment with EPA affected the T0901317 effect on (C) glucose oxidation and (D) glucose uptake. Same raw data as in (A) and (B), but in (C) and (D) ratio T0901317:DMSO for each fatty acid treatment was calculated. The results represent means ± SEM (n = 6). Bonferroni correction was applied for multiple comparisons. aP < 0.05 versus control; bP < 0.05 versus OA. EPA, eicosapentaenoic acid; LA, linoleic acid; OA, oleic acid; PA, palmitic acid; T, T0901317.
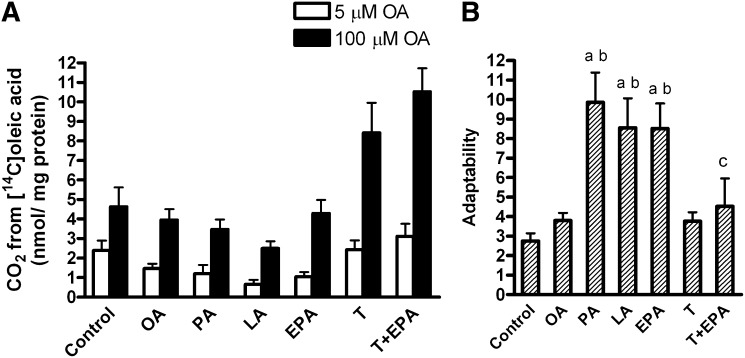
Oxidation of labeled OA after pretreatment with FAs and the effect of acute glucose
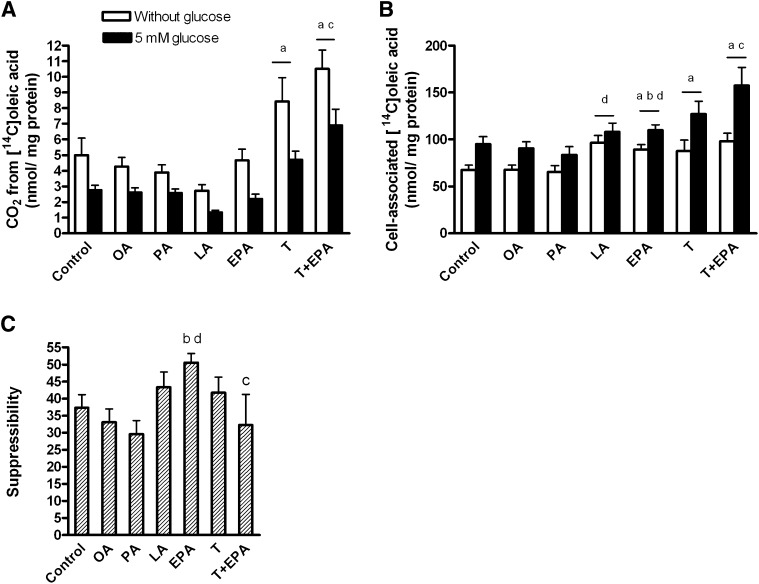
The myotubes were pretreated for 24 h with 1 µM T0901317 or vehicle (DMSO), and then for another 24 h with 100 µM OA, EPA, LA, PA, 40 µM BSA (control), T0901317 plus BSA, or a combination of T0901317 and EPA. Thereafter, myotubes were treated acutely for 4 h with [14C]OA (100 µM) in the absence or presence of 5 mM glucose. The inhibitory effect of acute glucose on [14C]OA oxidation was highly significant (43% inhibition, overall effect P < 0.001, Fig. 4A), and the presence of glucose increased the cellular accumulation of [14C]OA by 32% (overall effect P < 0.001, Fig. 4B). Activation of LXR by T0901317 significantly increased OA oxidation (P < 0.05, Fig. 4A) and OA accumulation (P < 0.01, Fig. 4B) in the myotubes. Pretreatment with EPA increased the cellular accumulation of OA compared with pretreatment with BSA (control) and OA (P < 0.05, Fig. 4B).
Fig. 4.
Suppressibility by glucose. Myotubes were pretreated for 24 h with 1 µM T0901317 or vehicle (DMSO), and then for another 24 h with 100 µM OA, EPA, LA, PA, 40 µM BSA (control), T0901317 plus BSA, or a combination of T0901317 and EPA, and thereafter underwent CO2 trapping for 4 h with 100 µM [14C]oleic acid (OA) in the presence or absence of 5 mM glucose as described in “Materials and Methods.” Overall effect of glucose on [14C]OA oxidation (A) P < 0.001 and [14C]OA accumulation (B) P < 0.001. Suppressibility (C), the ability of the cells to decrease OA oxidation by glucose, was calculated as [(1-(oxidation of OA at 5 mM glucose/oxidation of OA at no glucose added)) × 100%]. Results represent means ± SEM for n = 6–17. Bonferroni correction was applied for multiple comparisons. aP < 0.05 versus control; bP < 0.05 versus OA; cP < 0.05 versus EPA; dP < 0.05 versus PA. EPA, eicosapentaenoic acid; LA, linoleic acid; OA, oleic acid; PA, palmitic acid; T, T0901317.
To study metabolic switching of the myotubes after fatty acid and T0901317 pretreatment, three different parameters were defined; suppressibility and adaptability according to Ukropcova et al. (8) and a new parameter we called substrate-regulated flexibility.
Suppressibility was defined as the capacity of the cells to decrease OA oxidation by acute addition of glucose (5 mM). Pretreatment with EPA significantly increased suppressibility of the myotubes compared with pretreatment with OA (P = 0.007) and PA (P = 0.004), while PA and LA showed minimal effect (Fig. 4C). Exposure to T0901317 in combination with EPA blunted the increase in suppressibility (P = 0.03) seen with EPA alone, and T0901317 had no effect on suppressibility per se (Fig. 4C).
Myotubes were able to increase fatty acid oxidation by 64% upon increased availability of OA (overall effect, P < 0.001, Fig. 5A). The adaptability of the myotubes, defined as the capacity to increase acute fatty acid oxidation with increasing OA availability, was significantly increased after pretreatment with EPA, LA, and PA compared with control and OA pretreatment (P < 0.05, Fig. 5B). Treatment with T0901317 did not affect the adaptability, but it counteracted the effect of EPA (P = 0.007).
Fig. 5.
Adaptability. Myotubes were pretreated for 24 h with 1 µM T0901317 or vehicle (DMSO), and then for another 24 h with 100 µM OA, EPA, LA, PA, 40 µM BSA (control), T0901317 plus BSA, or a combination of T0901317 and EPA, and thereafter underwent CO2 trapping for 4 h with 5 or 100 µM [14C]OA in the absence of glucose as described in “Materials and Methods.” (A) Oxidation of 5 and 100 µM OA. Fatty acid oxidation increased by 64% upon increased availability of OA (overall effect P < 0.001). (B) Adaptability was defined as the ability to increase the acute OA oxidation with increasing OA concentration and calculated as follows: [oxidation of 100 µM OA / oxidation of 5 µM OA]. Results represent means ± SEM for n = 6–12. Bonferroni correction was applied for multiple comparisons. aP < 0.05 versus control; bP < 0.05 versus OA; cP < 0.05 versus EPA. EPA, eicosapentaenoic acid; LA, linoleic acid; OA, oleic acid; PA, palmitic acid; T, T0901317.
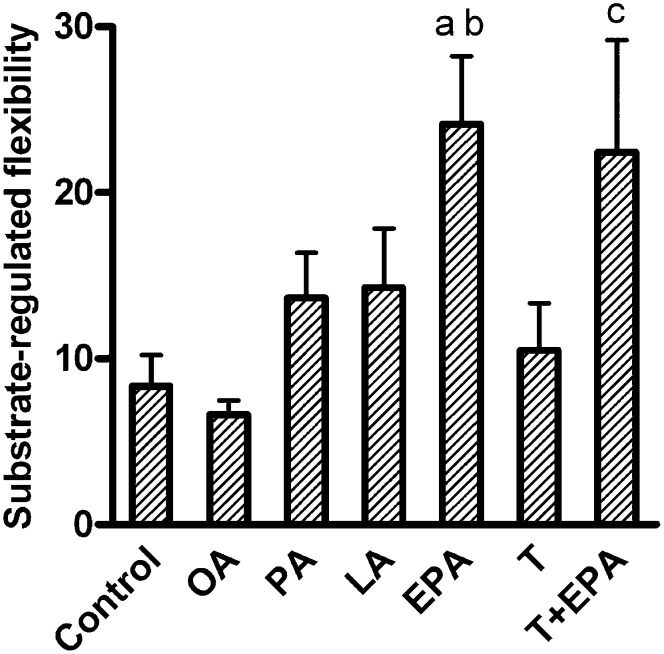
Substrate-regulated flexibility, defined as the ability of the myotubes to increase the acute fatty acid oxidation while changing from a “fed” (low fatty acid and high glucose concentration) to a “fasted” condition (high fatty acid and no glucose added), was increased after pretreatment with EPA compared with control and OA pretreatment (P < 0.05, Fig. 6). Exposure to PA and LA had minimal effect on this parameter. Treatment with T0901317 alone did not affect the substrate-regulated flexibility of the myotubes, nor did it interfere with the effect of EPA (Fig. 6). Pretreatment with the PPARδ agonist GW501516 (10 nM) for 96 h increased OA oxidation as previously reported (34), but it did not change any of the parameters of metabolic switching (data not shown). To investigate whether the effects of EPA were specific to EPA or were a general quality of n-3 fatty acids, additional experiments with ALA (18:3, n-3) and DHA (22:6, n-3) were performed. Pretreatment with ALA and DHA increased substrate- regulated flexibility significantly (P < 0.05 versus control, data not shown) and to the same extent as pretreatment with EPA (fold change 2.8, 3.1, and 2.7 for ALA, DHA, and EPA, respectively).
Fig. 6.
Substrate-regulated flexibility. Myotubes were pretreated for 24 h with 1 µM T0901317 or vehicle (DMSO), and then for another 24 h with 100 µM OA, EPA, LA, PA, 40 µM BSA (control), T0901317 plus BSA, or a combination of T0901317 and EPA, and thereafter underwent CO2 trapping for 4 h with 100 µM [14C]OA in the absence of glucose or with 5 µM [14C]OA in the presence of 5 mM glucose as described in “Materials and Methods.” Substrate-regulated flexibility was defined as the ability to increase the acute OA oxidation while changing from the “fed” (low fatty acid and high glucose concentration) to the “fasted” (high fatty acid and no glucose added) state and calculated as: [oxidation of 100 µM OA without glucose / oxidation of 5 µM OA in the presence of 5 mM glucose]. Results represent means ± SEM for n = 6–12. Bonferroni correction was applied for multiple comparisons. aP < 0.05 versus control; bP < 0.05 versus OA; cP < 0.05 versus T0901317. EPA, eicosapentaenoic acid; LA, linoleic acid; OA, oleic acid; PA, palmitic acid; T, T0901317.
Pretreatment with FAs and T0901317 affected the number LDs in myotubes
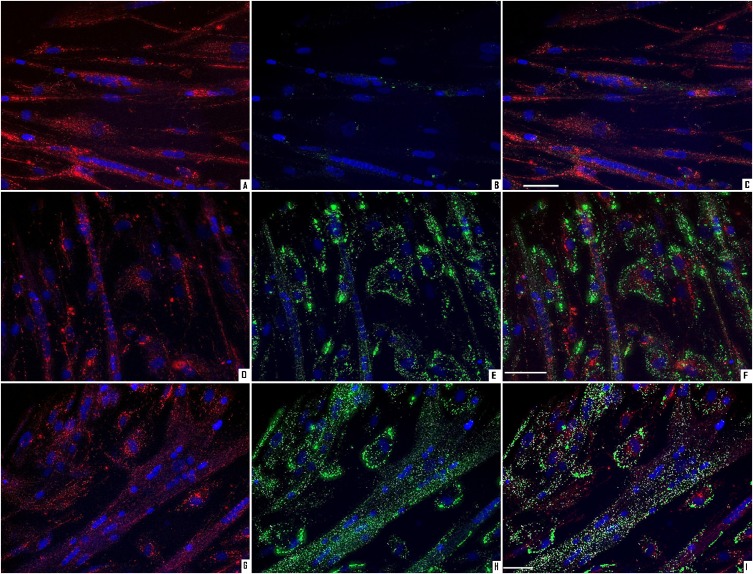
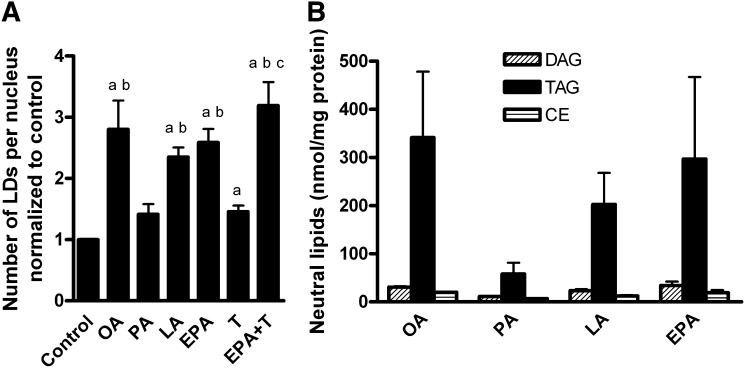
The myotubes were pretreated as described above. Thereafter, the cells were stained for nuclei, LDs, and mitochondria (Fig. 7). Pretreatment with OA, LA, EPA, T0901317, and the combination of EPA and T0901317 increased the number of LDs per nucleus compared with control (P < 0.05, Fig. 8A). PA also tended to increase the number of LDs per nucleus (P = 0.09) but to a smaller extent than the other FAs. The absolute values of LDs per nucleus were in the range 26–260, with 52 ± 7, 164 ± 24, and 136 ± 42 as mean values after control, OA, and EPA exposure, respectively. The average LD volume was measured to be about 1.1 μm3 and was not affected by pretreatment with FAs (data not shown). Density of the LDs was measured as fluorescence intensity per LD. The average fluorescence intensity per LD was not significantly different after pretreatment with various FAs (data not shown). Mitochondrial mass, measured as total intensity of MitoTracker®Red FM per nucleus, was also independent of the different pretreatments (data not shown).
Fig. 7.
Live imaging of lipid droplets and mitochondria. Myotubes were pretreated for 24 h with 1 µM T0901317 or vehicle (DMSO), and then for another 24 h with 100 µM OA, EPA, LA, PA, 40 µM BSA (control), T0901317, or a combination of T0901317 and EPA. The cells were stained for nuclei (blue), mitochondria (red) and LDs (green) as described in “Materials and Methods.” Representative images are presented for control (A–C), pretreatment with EPA (D–F), and EPA + T0901317 (G–I). Images A, D, and G show mitochondria and nuclei; B, E, and H show LDs and nuclei; and C, F, and I show mitochondria, LDs, and nuclei. Scale bar is 50 µm. EPA, eicosapentaenoic acid; LA, linoleic acid; LD, lipid droplet; OA, oleic acid; PA, palmitic acid; T, T0901317.
Fig. 8.
Number of lipid droplets per nucleus and lipid distribution. (A) Number of LDs per nucleus. The myotubes were pretreated for 24 h with 1 µM T0901317 or vehicle (DMSO), and then for another 24 h with 100 µM OA, EPA, LA, PA, 40 µM BSA (control), T0901317, or a combination of T0901317 and EPA. The cells were stained for LDs, mitochondria, and nuclei as described in “Materials and Methods.” Results represent means normalized to control ± SEM for 4 different donors, quantified from in total 200 images where one image contained in average 37 ± 3 nuclei. aP < 0.05 versus control; bP < 0.05 versus PA; cP < 0.05 versus T0901317. (B) Lipid distribution into neutral lipids. Myotubes were incubated with 100 µM OA, PA, LA and EPA supplemented with a trace amount of [1-14C]oleic acid (0.5 µCi/ml) for 24 h. Myotubes harvested and assayed for protein. Cellular lipids were extracted and separated by thin-layer chromatography, and radioactivity was quantified by liquid scintillation as described in “Materials and Methods.” Results represent means (nmol per mg cell protein) ± SEM for 3 different donors. CE, cholesteryl ester; DAG, diacylglycerol; EPA, eicosapentaenoic acid; LA, linoleic acid; LD, lipid droplet; OA, oleic acid; PA, palmitic acid; T, T0901317; TAG, triacylglycerol.
To investigate lipid distribution, myotubes were incubated with 100 µM OA, PA, LA, or EPA supplemented with a trace amount of [1-14C]oleic acid for 24 h. We investigated the content of neutral lipids, as LDs are generally composed of neutral lipids, mostly TAGs and cholesteryl esters (CE) (46). Extraction and separation of cellular lipids showed that in average TAG constitutes 80%, diacylglycerol (DAG) 12%, and CE 8% of neutral lipids. The amount of neutral lipids after different pretreatments followed the same pattern as the number of LDs per nucleus (Fig. 8B). However, the distribution of neutral lipids into different lipid classes was not changed by pretreatment with distinct FAs.
Gene expression and pathway analysis after FA pretreatment
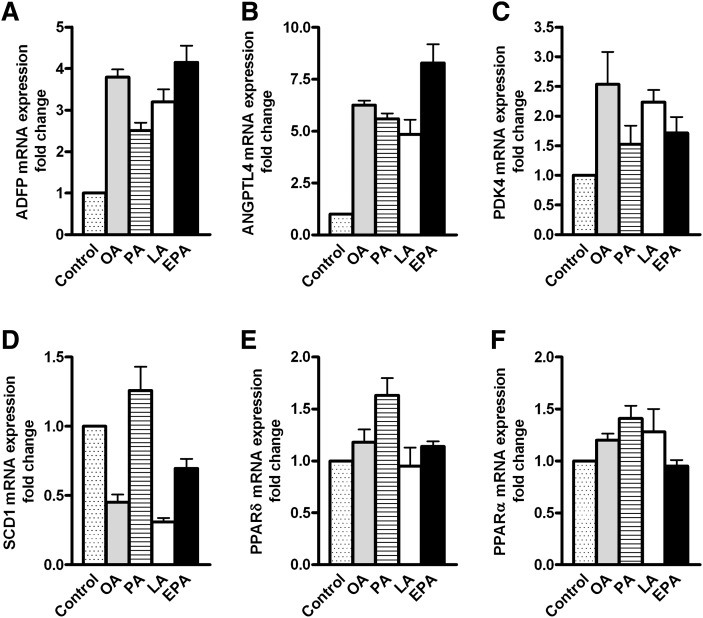
To examine whether the effects of fatty acid treatment could be explained by changes in gene expression, microarray analysis was performed. Treatment with EPA regulated most genes (40 upregulated and 112 downregulated genes), thereafter LA and OA, and PA regulated the fewest genes (11 upregulated and 0 downregulated genes, Table 1). Only 8 genes were induced by all fatty acids, including the PPARδ target genes angiopoietin-like 4 (ANGPTL4) and pyruvate dehydrogenase kinase isozyme 4 (PDK4), as well as the PAT-protein adipose differentiation-related protein (ADFP/adipophilin, Table 2). No genes were commonly repressed by all fatty acids. Exposure to EPA induced 24 genes and repressed 98 genes that were not affected by any of the other FAs. EPA was the only FA that induced acetyl-CoA carboxylase β [ACCβ/ACACB, fold change (FC) 1.4], which catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, the rate-limiting step in de novo fatty acid synthesis, and regulates mitochondrial fatty acid oxidation. The myokine interleukin 6 (IL-6) was only upregulated by EPA (FC 1.5), whereas the cholesterol transport protein ATP-binding cassette, subfamily A, member 1 (ABCA1) was only repressed by EPA (FC −2.1). Stearoyl-CoA desaturase 1 (SCD1), the enzyme that catalyzes a rate-limiting step in the synthesis of unsaturated fatty acids, was downregulated by OA and LA (FC −1.6 and −1.7, respectively). The fatty acid transporter CD36 was only upregulated by PA (FC 1.5).
TABLE 1.
Number of genes regulated by fatty acid treatment compared with control in myotubes identified by microarray analysis
| Treatment | Upregulated Number of Genes | Downregulated Number of Genes |
|---|---|---|
| OA | 16 | 20 |
| PA | 11 | 0 |
| LA | 20 | 19 |
| EPA | 40 | 112 |
Myotubes were incubated with fatty acids (100 µM) or BSA (40 µM, control) for 24 h and harvested for RNA isolation. Gene expression was measured by Affymetrix human NuGO Genechip arrays and fold change (FC) was calculated as log2-transformed expression level after fatty acid treatment divided by log2-transformed expression level after BSA treatment (control) for each individual donor. Mean fold changes were calculated based on the individual log2-transformed fold changes. Differentially expressed probe sets were identified using paired Intensity-based moderated t-statistics (IBMT, q-value) as explained in “Materials and Methods.” Genes with q < 0.2 was considered significantly regulated. Abbreviations: EPA, eicosapentaenoic acid; LA, linoleic acid; OA, oleic acid; PA, palmitic acid.
TABLE 2.
Genes upregulated by all fatty acids
| Gene Symbol | EPA | LA | OA | PA |
|---|---|---|---|---|
| PDK4 | 4.58 | 4.41 | 5.97 | 4.93 |
| ANGPTL4 | 3.45 | 2.42 | 2.72 | 2.42 |
| ADFP | 2.68 | 2.07 | 2.27 | 1.90 |
| CAT | 1.59 | 1.51 | 1.53 | 1.60 |
| ECH1 | 1.51 | 1.57 | 1.63 | 1.69 |
| IMPA2 | 1.48 | 1.55 | 1.48 | 1.37 |
| ACADVL | 1.44 | 1.34 | 1.42 | 1.40 |
| SLC25A20 | 1.39 | 1.30 | 1.30 | 1.34 |
Myotubes were incubated with fatty acids (100 µM) or BSA (40 µM, control) for 24 h and harvested for RNA isolation. Gene expression was measured by Affymetrix human NuGO Genechip arrays and fold change (FC) was calculated as log2-transformed expression level after fatty acid treatment divided by log2-transformed expression level after BSA treatment (control) for each individual donor. Mean fold changes were calculated based on the individual log2-transformed fold changes. Differentially expressed probe sets were identified using paired Intensity-based moderated t-statistics (IBMT, q-value) as explained in Materials and Methods. Genes with q < 0.2 was considered significantly regulated. The table shows FC for the genes upregulated by all fatty acids tested. Abbreviations: ACADVL, acyl-CoA dehydrogenase, very long chain; ADFP, adipose differentiation-related protein; ANGPTL4, angiopoietin-like 4; CAT, catalase; ECH1, enoyl CoA hydratase 1, peroxisomal; EPA, eicosapentaenoic acid; IMPA2, inositol(myo)-1(or 4)-monophosphatase 2; LA, linoleic acid; OA, oleic acid; PA, palmitic acid; PDK4, pyruvate dehydrogenase kinase, isozyme 4; SLC25A20, solute carrier family 25 (carnitine/acylcarnitine translocase), member 20.
To verify the results from microarray analysis, real-time RT-PCR on selected genes was performed. This confirmed that ADFP, ANGPTL4, and PDK4 were induced by all FAs tested, whereas SCD1 was repressed only by OA and LA (Fig. 9). PPARα and PPARδ were not regulated by any of the FAs used in the present study (Fig. 9). Furthermore, to obtain some indication of gene regulation by ALA and DHA, we investigated the expression level of ABCA1, one of the genes observed to be regulated only by EPA, after DHA and ALA pretreatment. Real-time RT-PCR confirmed that EPA pretreatment reduced the mRNA level of ABCA1 and showed that ABCA1 was downregulated to a similar extent also by ALA and DHA exposure (data not shown).
Fig. 9.
Gene regulation by fatty acids. Myotubes were incubated with fatty acids (100 µM) or BSA (40 µM, control) for 24 h and harvested for RNA isolation. Real-time PCR was performed with platinum Taq polymerase and SYBR green on an iCycler PCR machine as described in “Materials and Methods” on selected genes to confirm the microarray data. ADFP, adipose differentiation-related protein; ANGPTL4, angiopoietin-like 4; EPA, eicosapentaenoic acid; LA, linoleic acid; OA, oleic acid; PA, palmitic acid; PDK4, pyruvate dehydrogenase kinase isozyme 4; PPAR, peroxisome proliferator-activated receptor; SCD1, stearoyl-CoA desaturase 1.
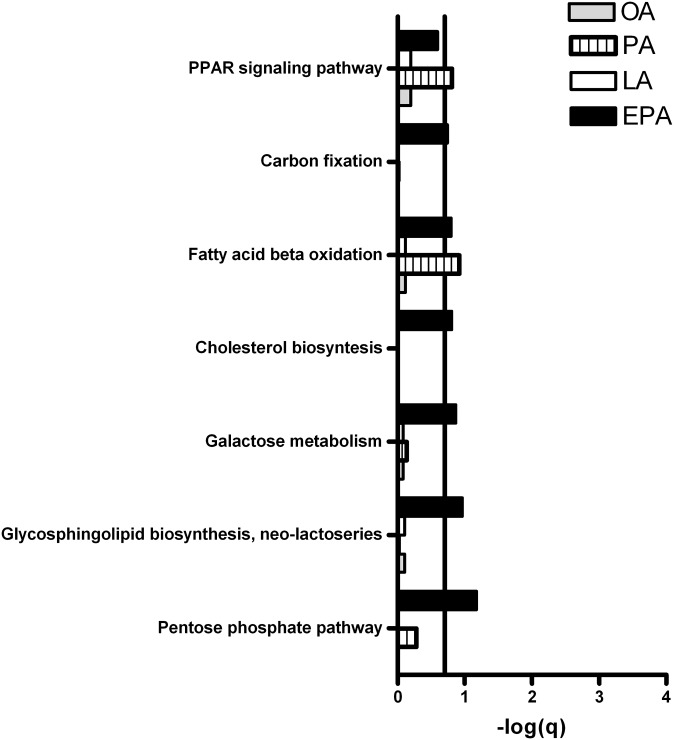
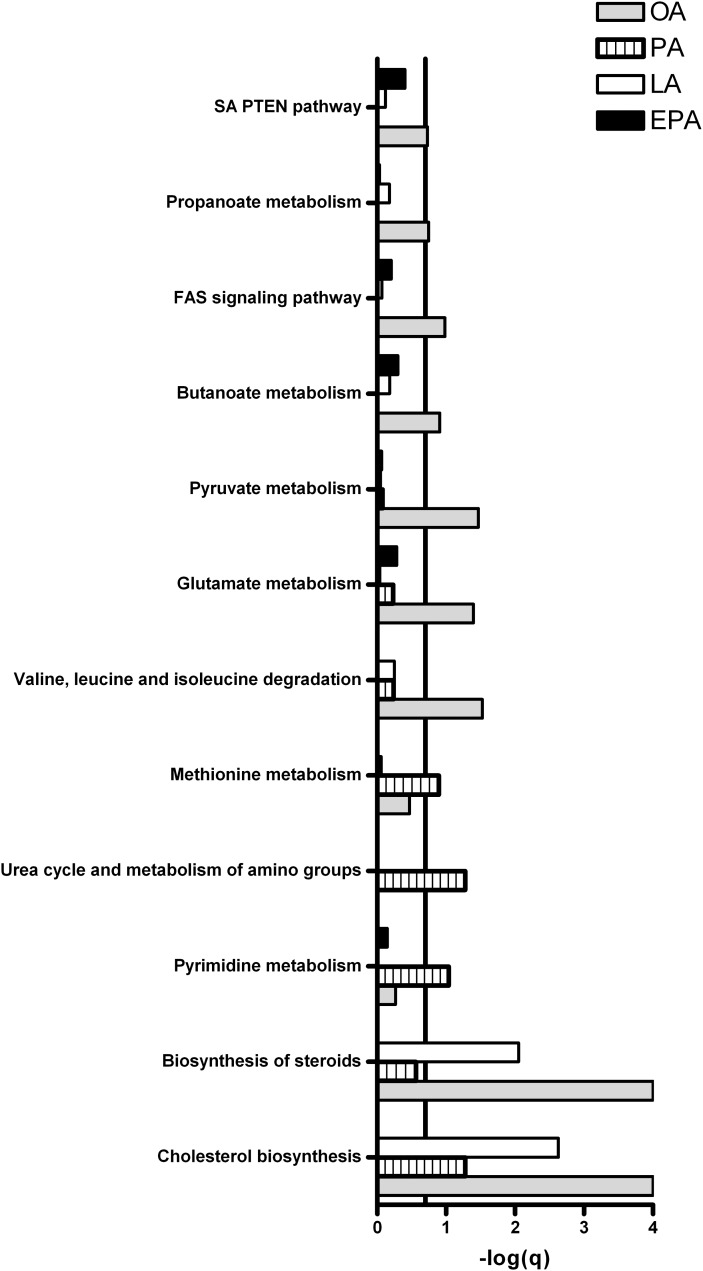
GSEA was performed for functional analysis of changes in gene expression. Pathways with FDR (q-value) < 0.2 (that is, −log q > 0.7) were considered significantly regulated. Figures 10 and 11 show that pathways involved in metabolic processes were significantly regulated in myotubes after treatment with OA, PA, LA, or EPA compared with control. Treatment with OA, LA, and PA downregulated cholesterol synthesis, while EPA upregulated this pathway. Fatty acid β-oxidation was induced by EPA and PA, but not by OA and LA. Furthermore, pathways involved in carbohydrate metabolism, such as the pentose phosphate pathway, galactose metabolism, and glycosphingolipid biosynthesis, were induced in myotubes after exposure to EPA, but not by the other fatty acids.
Fig. 10.
Pathways involved in metabolic processes upregulated by fatty acids in myotubes. Myotubes were incubated with fatty acids (100 µM) or BSA (40 µM, control) for 24 h and harvested for RNA isolation. Gene expression was measured by Affymetrix human NuGO Genechip arrays and GSEA was performed to identify pathways regulated by OA, PA, LA, and EPA compared with control as explained in “Materials and Methods.” Pathways with FDR (q-value) < 0.2 (that is, −log(q) > 0.7) were considered significantly regulated. Line marks −log(q) = 0.7, the cut-off for significance for the GSEA. In short, GSEA identifies pathways in which more genes are found to be regulated by microarray analysis than one would expect on the basis of chance. EPA, eicosapentaenoic acid; GSEA, gene set enrichment analysis; LA, linoleic acid; OA, oleic acid; PA, palmitic acid.
Fig. 11.
Pathways involved in metabolic processes downregulated by fatty acids in myotubes. Myotubes were incubated with fatty acids (100 µM) or BSA (40 µM, control) for 24 h and harvested for RNA isolation. Gene expression was measured by Affymetrix human NuGO Genechip arrays, and GSEA was performed to identify pathways regulated by OA, PA, LA, and EPA compared with control as explained in Fig. 10. Pathways with FDR (q-value) < 0.2 (that is, −log(q) > 0.7) were considered significantly regulated. Line marks −log(q) = 0.7, the cut-off for significance for the GSEA. EPA, eicosapentaenoic acid; GSEA, gene set enrichment analysis; LA, linoleic acid; OA, oleic acid; PA, palmitic acid.
DISCUSSION
In human myotubes, 24 h pretreatment with 100 µM of the n-3 FA EPA increased the suppressibility caused by acute exposure to glucose on fatty acid metabolism compared with pretreatment with OA, and EPA increased the substrate-regulated flexibility of the cells compared with control and OA pretreatment. Furthermore, pretreatment with ALA and DHA increased substrate-regulated flexibility to the same extent as EPA, indicating that the observed effects of EPA could be due to a general quality of n-3 FAs. These results suggest a possible favorable effect of n-3 FAs on skeletal muscle substrate handling and metabolic switching. EPA, LA, and PA significantly increased the cellular adaptability toward an increased acute exposure to fatty acids compared with control and OA pretreatment. This finding indicates that the increase in FA adaptability, in contrast to suppressibility and substrate-regulated flexibility, is due to a more general effect of fatty acids on skeletal muscle lipid metabolism, although this does not apply to OA. These data also fit with data from the microarray analysis, where all fatty acids were found to upregulate certain genes involved in fatty acid β-oxidation: enoyl CoA hydratase 1 (ECH1), acyl-CoA dehydrogenase very long chain (ACADVL), and solute carrier family 25 (carnitine/acylcarnitine translocase) member 20 (SLC25A20). Furthermore, pathway analysis showed that fatty acid β-oxidation was upregulated by EPA and PA. The pathway analysis also showed that exposure to EPA, in contrast to the other fatty acids, affected carbohydrate metabolism, supporting the functional data showing the specific effect of n-3 FAs on suppressibility and substrate-regulated flexibility compared with the other FAs. Real-time accumulation studies of fatty acids showed that EPA per se accumulated significantly less and was more oxidized to ASM than the other fatty acids. LXR-activation with T0901317 increased OA uptake and oxidation, as well as glucose uptake and oxidation. Furthermore, T0901317 counteracted the effect of EPA on suppressibility and adaptability, but not on substrate-regulated flexibility of the cells.
In the present study we observed eight genes commonly upregulated by EPA, LA, OA, and PA, indicating a general fatty acid effect, probably due to PPAR activation. Four of these genes, ANGPTL4, PDK4, ADFP, and IMPA2, were also found to be induced by both PA and LA in myotubes by Staiger et al. (47). In addition we showed that catalase (CAT) and a few genes involved in fatty acid β-oxidation (ACADVL, SLC25A20, ECH1) were upregulated by all FAs examined. The changes in adaptability cannot solely be explained by regulation of gene expression because no genes were found to be commonly regulated by EPA, LA, and PA, but not by OA.
Gene set enrichment analysis showed that EPA upregulated specific pathways involved in carbohydrate metabolism, indicating that the effects of n-3 FAs on metabolic switching are partially due to enhanced glucose utilization. In the present study we did not observe increased glucose oxidation after pretreatment with EPA compared with control, in contrast to previous reports (23, 24). This finding might be due to different experimental conditions, such as incubation time and concentration of fatty acids used. However, pretreatment with EPA increased acute fatty acid uptake, but not oxidation, in accordance with previous findings (23, 24). Furthermore, EPA was found to induce gene expression of ACACB, which controls fatty acid β-oxidation through production of malonyl-CoA and inhibition of carnitine palmitoyltransferase 1 (CPT1). This upregulation might reflect an increased capacity to control fatty acid oxidation rate. The present study also showed that pretreatment with EPA increased the expression level of IL-6. It has been shown that exposure to IL-6 increases phosphorylation and activation of AMP-activated protein kinase (AMPK) in skeletal muscle and thereby regulates muscle substrate utilization (48, 49). Therefore, increased phosphorylation of AMPK might contribute to the effect of n-3 FAs on metabolic switching in the myotubes.
LXRs, important regulators of cholesterol, lipid, and glucose metabolism (50–54), might be involved in regulation of metabolic switching. Recently, Stenson et al. showed that LXR regulated the switch between glucose and FA oxidation in adipocytes (55). In the present study, pretreatment with T0901317 increased acute uptake and oxidation of OA and glucose in myotubes. This result is in accordance with previous findings showing that exposure to T0901317 increased acute PA and glucose uptake and oxidation in the same cell model (56, 57). Stenson et al. showed that the LXR agonist GW3965 decreased glucose oxidation, but it increased fatty acid oxidation in human and murine adipocytes (55). This finding indicates that glucose oxidation is regulated by LXR in a tissue-specific manner. Treatment with T0901317 did not affect the adaptability or suppressibility itself, but it counteracted the effect of EPA on these parameters. Exposure to T0901317 showed no effect on substrate-regulated flexibility. The blunted increase in suppressibility and adaptability after treatment with EPA in combination with T0901317 could be explained by suppressive effects of LXR on PPAR target genes (58, 59). However, the fact that PPARδ activation did not influence adaptability or suppressibility does not support this explanation.
The counteractive effects of T0901317 on EPA-induced increase in adaptability and suppressibility could also indicate that EPA's effects on metabolic switching are due to LXR antagonism, as PUFAs are shown to act as LXR antagonists (25, 26). Although T0901317 counteracted EPA's effect on suppressibility and adaptability, the T0901317-mediated augmentation in glucose uptake and oxidation was increased after exposure to EPA, indicating that not all of the effects of T0901317 and PUFAs are counteractive. PPARs and LXRs are shown to perform reciprocal induction of each other (60–62), and these results further underline the complex interplay between these nuclear receptors.
The mechanisms by which n-3 FAs affect metabolic switching may not only involve gene regulation but may also be linked to different membrane incorporation of fatty acids (63–65) and, thereby, increased plasma membrane fluidity, as well as altered structure or dynamics of the mitochondrial membranes (65–67). According to the “membrane pacemaker theory,” in which n-3 FA content is correlated with metabolic rate, an increased proportion of PUFAs in membranes is proposed to increase the activity of membrane-associated proteins and thereby increase cellular metabolic activity and membrane leak-pump cycles (65–68). In the present study, mitochondrial mass of myotubes was unaffected by all pretreatments examined, suggesting that the metabolic effects of EPA is not due to increased mitochondrial biogenesis. However, mitochondrial dynamics and function might be altered.
The ability of fatty acids to differently change metabolic switching of myotubes might also be due to different accumulations of fatty acids, changes in the level of intracellular lipid species, such as acyl-CoA, diacylglycerol (DAG), and triacylglycerol (TAG), and lipid utilization. In this study, myotubes accumulated less labeled EPA per se compared with OA, PA, and LA. EPA was more oxidized to ASM than the other FAs, which might explain the reduced accumulation. However, pretreatment with EPA, as well as OA and LA, did increase the number of LDs per nucleus, whereas the volume and intensity of LDs were unaffected. The amount of neutral lipids after different FA pretreatments followed the same pattern as the number of LDs, but the distribution into different lipid classes was not changed by distinct FAs. Pretreatment with T0901317 also increased the number of LDs. The increased number of LDs in T0901317-treated myotubes might be due to the ability of T0901317 to stimulate lipogenesis, uptake of fatty acids, and TAG formation (56, 69). EPA, on the other hand, has been shown to decrease lipogenesis in several tissues, such as liver, intestine, and adipose (70–72). In accordance with our data, previous studies have shown that pretreatment with EPA increased acute fatty acid uptake, promoted accumulation of TAG, and reduced the level of total acyl-CoA in human myotubes (23, 24). Increased storage of TAG may protect against cell damage due to a reduction in lipotoxic intermediates (24, 73, 74) and might contribute to the favorable effects of n-3 FAs on metabolic switching. The impact of LDs and the regulation of LDs by different fatty acids are currently under further investigation.
Metabolic inflexibility might be caused by lifestyle factors and their interaction with intrinsic characteristics of skeletal muscle. In vitro adaptability of myotubes has been shown to be positively correlated with in vivo metabolic flexibility and insulin sensitivity (8), and it might reflect the ability to adapt to a high-fat diet and protect against obesity. In the present study, pretreatment with EPA, LA, and PA increased in vitro adaptability of myotubes. Unexpectedly, Ukropcova et al. found that in vitro suppressibility was inversely correlated with in vivo metabolic flexibility and insulin sensitivity (8). These observations might be inappropriate to compare, as the in vivo experiments were done in the presence of insulin, whereas the in vitro experiments were done in the absence of insulin. However, in our study, treatment with EPA was found to increase both in vitro suppressibility and substrate-regulated flexibility. Suppressibility reflects the ability of glucose to suppress FA oxidation, possibly reflecting the reverse Randle cycle (8, 31, 32). Metabolic inflexible muscle is characterized by an impaired switch from lipid to glucose oxidation after a meal, which might be reflected by decreased suppressibility. Improvement of adaptability, suppressibility, and substrate-regulated flexibility through changes in diet, weight, and physical activity might reflect a therapeutic beneficial effect for individuals with obesity and T2D.
CONCLUSION
Our study suggests a positive role for n-3 FAs compared with other FAs in improving overall energy metabolism and metabolic switching in skeletal muscle that might contribute to the beneficial effects of dietary intake of n-3 FAs. We also suggest the use of three parameters, suppressibility, adaptability, and substrate-regulated flexibility, in functional studies of fuel selection and energy metabolism in cell cultures.
Acknowledgments
The authors are thankful for the use of the NORMIC-UiO a FUGE/EMBIO-supported imaging platform at the Department of Molecular Biosciences, University of Oslo, as well as for the excellent technical assistance of Camilla Stensrud and Mari-Ann Baltzersen.
Footnotes
Abbreviations:
- ALA
- α-linolenic acid
- ASM
- acid-soluble metabolite
- CE
- cholesteryl ester
- DAG
- diacylglycerol
- DHA
- docosahexaenoic acid
- EPA
- eicosapentaenoic acid
- GSEA
- gene set enrichment analysis
- IMCL
- intramyocellular lipid
- LA
- linoleic acid
- LD
- lipid droplet
- LXR
- liver X receptor
- OA
- oleic acid
- PA
- palmitic acid
- PPAR
- peroxisome proliferator-activated receptor
- PUFA
- polyunsaturated fatty acid
- RXR
- retinoid X receptor
- T2D
- type 2 diabetes
- TAG
- triacylglycerol
This work was funded by the University of Oslo, the European Nutrigenomics Organisation (NuGO), the Norwegian Diabetes Foundation, the Freia Chocolade Fabriks Medical Foundation, and the Anders Jahre's Foundation.
REFERENCES
- 1.Kelley D. E., Reilly J. P., Veneman T., Mandarino L. J. 1990. Effects of insulin on skeletal muscle glucose storage, oxidation, and glycolysis in humans. Am. J. Physiol. 258: E923–E929. [DOI] [PubMed] [Google Scholar]
- 2.Andres R., Cader G., Zierler K. L. 1956. The quantitatively minor role of carbohydrate in oxidative metabolism by skeletal muscle in intact man in the basal state; measurements of oxygen and glucose uptake and carbon dioxide and lactate production in the forearm. J. Clin. Invest. 35: 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelley D. E., Mandarino L. J. 2000. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes. 49: 677–683. [DOI] [PubMed] [Google Scholar]
- 4.Corpeleijn E., Mensink M., Kooi M. E., Roekaerts P. M. H. J., Saris W. H. M., Blaak E. E. 2008. Impaired skeletal muscle substrate oxidation in glucose-intolerant men improves after weight loss. Obesity (Silver Spring). 16: 1025–1032. [DOI] [PubMed] [Google Scholar]
- 5.Kelley D. E., Goodpaster B., Wing R. R., Simoneau J. A. 1999. Skeletal muscle fatty acid metabolism in association with insulin resistance, obesity, and weight loss. Am. J. Physiol. 277: E1130–E1141. [DOI] [PubMed] [Google Scholar]
- 6.Blaak E. E., van Aggel-Leijssen D. P., Wagenmakers A. J., Saris W. H., van Baak M. A. 2000. Impaired oxidation of plasma- derived fatty acids in type 2 diabetic subjects during moderate- intensity exercise. Diabetes. 49: 2102–2107. [DOI] [PubMed] [Google Scholar]
- 7.Mensink M., Blaak E. E., van Baak M. A., Wagenmakers A. J., Saris W. H. 2001. Plasma free fatty acid uptake and oxidation are already diminished in subjects at high risk for developing type 2 diabetes. Diabetes. 50: 2548–2554. [DOI] [PubMed] [Google Scholar]
- 8.Ukropcova B., McNeil M., Sereda O., de Jonge L., Xie H., Bray G. A., Smith S. R. 2005. Dynamic changes in fat oxidation in human primary myocytes mirror metabolic characteristics of the donor. [comment] J. Clin. Invest. 115: 1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodpaster B. H., Katsiaras A., Kelley D. E. 2003. Enhanced fat oxidation through physical activity is associated with improvements in insulin sensitivity in obesity. Diabetes. 52: 2191–2197. [DOI] [PubMed] [Google Scholar]
- 10.Corpeleijn E., Saris W. H. M., Blaak E. E. 2009. Metabolic flexibility in the development of insulin resistance and type 2 diabetes: effects of lifestyle. Obes. Rev. 10: 178–193. [DOI] [PubMed] [Google Scholar]
- 11.Sial S., Coggan A. R., Hickner R. C., Klein S. 1998. Training-induced alterations in fat and carbohydrate metabolism during exercise in elderly subjects. Am. J. Physiol. 274: E785–E790. [DOI] [PubMed] [Google Scholar]
- 12.Gan S. K., Kriketos A. D., Ellis B. A., Thompson C. H., Kraegen E. W., Chisholm D. J. 2003. Changes in aerobic capacity and visceral fat but not myocyte lipid levels predict increased insulin action after exercise in overweight and obese men. Diabetes Care. 26: 1706–1713. [DOI] [PubMed] [Google Scholar]
- 13.Lee J. S., Pinnamaneni S. K., Eo S. J., Cho I. H., Pyo J. H., Kim C. K., Sinclair A. J., Febbraio M. A., Watt M. J. 2006. Saturated, but not n-6 polyunsaturated, fatty acids induce insulin resistance: role of intramuscular accumulation of lipid metabolites. J. Appl. Physiol. 100: 1467–1474. [DOI] [PubMed] [Google Scholar]
- 14.Storlien L. H., Kraegen E. W., Chisholm D. J., Ford G. L., Bruce D. G., Pascoe W. S. 1987. Fish oil prevents insulin resistance induced by high-fat feeding in rats. Science. 237: 885–888. [DOI] [PubMed] [Google Scholar]
- 15.Ramel A., Martinez A., Kiely M., Morais G., Bandarra N. M., Thorsdottir I. 2008. Beneficial effects of long-chain n-3 fatty acids included in an energy-restricted diet on insulin resistance in overweight and obese European young adults. Diabetologia. 51: 1261–1268. [DOI] [PubMed] [Google Scholar]
- 16.Kabir M., Skurnik G., Naour N., Pechtner V., Meugnier E., Rome S., Quignard-Boulange A., Vidal H., Slama G., Clement K., et al. 2007. Treatment for 2 mo with n 3 polyunsaturated fatty acids reduces adiposity and some atherogenic factors but does not improve insulin sensitivity in women with type 2 diabetes: a randomized controlled study. Am. J. Clin. Nutr. 86: 1670–1679. [DOI] [PubMed] [Google Scholar]
- 17.Browning L. M., Krebs J. D., Moore C. S., Mishra G. D., O'Connell M. A., Jebb S. A. 2007. The impact of long chain n-3 polyunsaturated fatty acid supplementation on inflammation, insulin sensitivity and CVD risk in a group of overweight women with an inflammatory phenotype. Diabetes Obes. Metab. 9: 70–80. [DOI] [PubMed] [Google Scholar]
- 18.Krebs J. D., Browning L. M., McLean N. K., Rothwell J. L., Mishra G. D., Moore C. S., Jebb S. A. 2006. Additive benefits of long-chain n-3 polyunsaturated fatty acids and weight-loss in the management of cardiovascular disease risk in overweight hyperinsulinaemic women. Int. J. Obes. 30: 1535–1544. [DOI] [PubMed] [Google Scholar]
- 19.Fedor D., Kelley D. S. 2009. Prevention of insulin resistance by n-3 polyunsaturated fatty acids. Curr. Opin. Clin. Nutr. Metab. Care. 12: 138–146. [DOI] [PubMed] [Google Scholar]
- 20.Haugaard S. B., Madsbad S., Hoy C-E., Vaag A. 2006. Dietary intervention increases n-3 long-chain polyunsaturated fatty acids in skeletal muscle membrane phospholipids of obese subjects. Implications for insulin sensitivity. Clin. Endocrinol. (Oxf.). 64: 169–178. [DOI] [PubMed] [Google Scholar]
- 21.Pickersgill L., Litherland G. J., Greenberg A. S., Walker M., Yeaman S. J. 2007. Key role for ceramides in mediating insulin resistance in human muscle cells. J. Biol. Chem. 282: 12583–12589. [DOI] [PubMed] [Google Scholar]
- 22.Coll T., Eyre E., Rodriguez-Calvo R., Palomer X., Sanchez R. M., Merlos M., Laguna J. C., Vazquez-Carrera M. 2008. Oleate reverses palmitate-induced insulin resistance and inflammation in skeletal muscle cells. J. Biol. Chem. 283: 11107–11116. [DOI] [PubMed] [Google Scholar]
- 23.Aas V., Rokling-Andersen M. H., Kase E. T., Thoresen G. H., Rustan A. C. 2006. Eicosapentaenoic acid (20:5 n-3) increases fatty acid and glucose uptake in cultured human skeletal muscle cells. J. Lipid Res. 47: 366–374. [DOI] [PubMed] [Google Scholar]
- 24.Wensaas A. J., Rustan A. C., Just M., Berge R. K., Drevon C. A., Gaster M. 2009. Fatty acid incubation of myotubes from humans with type 2 diabetes leads to enhanced release of beta-oxidation products because of impaired fatty acid oxidation: effects of tetradecylthioacetic acid and eicosapentaenoic acid. Diabetes. 58: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ou J., Tu H., Shan B., Luk A., DeBose-Boyd R. A., Bashmakov Y., Goldstein J. L., Brown M. S. 2001. Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing ligand-dependent activation of the LXR. Proc. Natl. Acad. Sci. USA. 98: 6027–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshikawa T., Shimano H., Yahagi N., Ide T., Amemiya-Kudo M., Matsuzaka T., Nakakuki M., Tomita S., Okazaki H., Tamura Y., et al. 2002. Polyunsaturated fatty acids suppress sterol regulatory element-binding protein 1c promoter activity by inhibition of liver X receptor (LXR) binding to LXR response elements. J. Biol. Chem. 277: 1705–1711. [DOI] [PubMed] [Google Scholar]
- 27.Gottlicher M., Widmark E., Li Q., Gustafsson J. A. 1992. Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA. 89: 4653–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottlicher M., Demoz A., Svensson D., Tollet P., Berge R. K., Gustafsson J. A. 1993. Structural and metabolic requirements for activators of the peroxisome proliferator-activated receptor. Biochem. Pharmacol. 46: 2177–2184. [DOI] [PubMed] [Google Scholar]
- 29.Sampath H., Ntambi J. M. 2005. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu. Rev. Nutr. 25: 317–340. [DOI] [PubMed] [Google Scholar]
- 30.Randle P. J., Garland P. B., Hales C. N., Newsholme E. A. 1963. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1: 785–789. [DOI] [PubMed] [Google Scholar]
- 31.Sidossis L. S., Stuart C. A., Shulman G. I., Lopaschuk G. D., Wolfe R. R. 1996. Glucose plus insulin regulate fat oxidation by controlling the rate of fatty acid entry into the mitochondria. J. Clin. Invest. 98: 2244–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidossis L. S., Wolfe R. R. 1996. Glucose and insulin-induced inhibition of fatty acid oxidation: the glucose-fatty acid cycle reversed. Am. J. Physiol. 270: E733–E738. [DOI] [PubMed] [Google Scholar]
- 33.Gaster M., Kristensen S. R., Beck-Nielsen H., Schroder H. D. 2001. A cellular model system of differentiated human myotubes. APMIS. 109: 735–744. [DOI] [PubMed] [Google Scholar]
- 34.Wensaas A. J., Rustan A. C., Lovstedt K., Kull B., Wikstrom S., Drevon C. A., Hallen S. 2007. Cell-based multiwell assays for the detection of substrate accumulation and oxidation. J. Lipid Res. 48: 961–967. [DOI] [PubMed] [Google Scholar]
- 35.Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 36.Skrede S., Bremer J., Berge R., Rustan A. 1994. Stimulation of fatty acid oxidation by a 3-thia fatty acid reduces triacylglycerol secretion in cultured rat hepatocytes. J. Lipid Res. 35: 1395–1404. [PubMed] [Google Scholar]
- 37.Gaster M., Rustan A. C., Aas V., Beck-Nielsen H. 2004. Reduced lipid oxidation in skeletal muscle from type 2 diabetic subjects may be of genetic origin: evidence from cultured myotubes. Diabetes. 53: 542–548. [DOI] [PubMed] [Google Scholar]
- 38.Dai M., Wang P., Boyd A. D., Kostov G., Athey B., Jones E. G., Bunney W. E., Myers R. M., Speed T. P., Akil H., et al. 2005. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 19: 185–193. [DOI] [PubMed] [Google Scholar]
- 40.Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sartor M. A., Tomlinson C. R., Wesselkamper S. C., Sivaganesan S., Leikauf G. D., Medvedovic M. 2006. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 7: 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storey J. D., Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. [comment] Proc. Natl. Acad. Sci. USA. 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahlquist K. D., Salomonis N., Vranizan K., Lawlor S. C., Conklin B. R. 2002. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 31: 19–20. [DOI] [PubMed] [Google Scholar]
- 45.Kanehisa M., Goto S. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy S., Martin S., Parton R. G. 2009. Lipid droplet-organelle interactions; sharing the fats. Biochim. Biophys. Acta. 1791: 441–447. [DOI] [PubMed] [Google Scholar]
- 47.Staiger H., Haas C., Machann J., Werner R., Weisser M., Schick F., Machicao F., Stefan N., Fritsche A., Haring H-U. 2009. Muscle-derived angiopoietin-like protein 4 is induced by fatty acids via peroxisome proliferator-activated receptor (PPAR)-delta and is of metabolic relevance in humans. Diabetes. 58: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Khalili L., Bouzakri K., Glund S., Lonnqvist F., Koistinen H. A., Krook A. 2006. Signaling specificity of interleukin-6 action on glucose and lipid metabolism in skeletal muscle. Mol. Endocrinol. 20: 3364–3375. [DOI] [PubMed] [Google Scholar]
- 49.Ruderman N. B., Keller C., Richard A-M., Saha A. K., Luo Z., Xiang X., Giralt M., Ritov V. B., Menshikova E. V., Kelley D. E., et al. 2006. Interleukin-6 regulation of AMP-activated protein kinase. Potential role in the systemic response to exercise and prevention of the metabolic syndrome. Diabetes. 55(Suppl. 2): S48–S54. [DOI] [PubMed] [Google Scholar]
- 50.Alberti S., Schuster G., Parini P., Feltkamp D., Diczfalusy U., Rudling M., Angelin B., Bjorkhem I., Pettersson S., Gustafsson J. A. 2001. Hepatic cholesterol metabolism and resistance to dietary cholesterol in LXRbeta-deficient mice. J. Clin. Invest. 107: 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peet D. J., Turley S. D., Ma W., Janowski B. A., Lobaccaro J. M., Hammer R. E., Mangelsdorf D. J. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 93: 693–704. [DOI] [PubMed] [Google Scholar]
- 52.Cao G., Liang Y., Broderick C. L., Oldham B. A., Beyer T. P., Schmidt R. J., Zhang Y., Stayrook K. R., Suen C., Otto K. A., et al. 2003. Antidiabetic action of a liver x receptor agonist mediated by inhibition of hepatic gluconeogenesis. J. Biol. Chem. 278: 1131–1136. [DOI] [PubMed] [Google Scholar]
- 53.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D. J., et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14: 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laffitte B. A., Chao L. C., Li J., Walczak R., Hummasti S., Joseph S. B., Castrillo A., Wilpitz D. C., Mangelsdorf D. J., Collins J. L., et al. 2003. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc. Natl. Acad. Sci. USA. 100: 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stenson B. M., Ryden M., Steffensen K. R., Wahlen K., Pettersson A. T., Jocken J. W., Arner P., Laurencikiene J. 2009. Activation of liver x receptor regulates substrate oxidation in white adipocytes. Endocrinology. 150: 4104–4113. [DOI] [PubMed] [Google Scholar]
- 56.Kase E. T., Wensaas A. J., Aas V., Hojlund K., Levin K., Thoresen G. H., Beck-Nielsen H., Rustan A. C., Gaster M. 2005. Skeletal muscle lipid accumulation in type 2 diabetes may involve the liver X receptor pathway. Diabetes. 54: 1108–1115. [DOI] [PubMed] [Google Scholar]
- 57.Kase E. T., Thoresen G. H., Westerlund S., Hojlund K., Rustan A. C., Gaster M. 2007. Liver X receptor antagonist reduces lipid formation and increases glucose metabolism in myotubes from lean, obese and type 2 diabetic individuals. Diabetologia. 50: 2171–2180. [DOI] [PubMed] [Google Scholar]
- 58.Ide T., Shimano H., Yoshikawa T., Yahagi N., Amemiya-Kudo M., Matsuzaka T., Nakakuki M., Yatoh S., Iizuka Y., Tomita S., et al. 2003. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. II. LXRs suppress lipid degradation gene promoters through inhibition of PPAR signaling. [comment] Mol. Endocrinol. 17: 1255–1267. [DOI] [PubMed] [Google Scholar]
- 59.Yoshikawa T., Ide T., Shimano H., Yahagi N., Amemiya-Kudo M., Matsuzaka T., Yatoh S., Kitamine T., Okazaki H., Tamura Y., et al. 2003. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. I. PPARs suppress sterol regulatory element binding protein-1c promoter through inhibition of LXR signaling. [comment] Mol. Endocrinol. 17: 1240–1254. [DOI] [PubMed] [Google Scholar]
- 60.Tobin K. A., Steineger H. H., Alberti S., Spydevold O., Auwerx J., Gustafsson J. A., Nebb H. I. 2000. Cross-talk between fatty acid and cholesterol metabolism mediated by liver X receptor-alpha. Mol. Endocrinol. 14: 741–752. [DOI] [PubMed] [Google Scholar]
- 61.Colin S., Bourguignon E., Boullay A-B., Tousaint J-J., Huet S., Caira F., Staels B., Lestavel S., Lobaccaro J-M. A., Delerive P. 2008. Intestine-specific regulation of PPARalpha gene transcription by liver X receptors. Endocrinology. 149: 5128–5135. [DOI] [PubMed] [Google Scholar]
- 62.Chinetti G., Lestavel S., Bocher V., Remaley A. T., Neve B., Torra I. P., Teissier E., Minnich A., Jaye M., Duverger N., et al. 2001. PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. [comment] Nat. Med. 7: 53–58. [DOI] [PubMed] [Google Scholar]
- 63.Chapkin R. S., McMurray D. N., Davidson L. A., Patil B. S., Fan Y-Y., Lupton J. R. 2008. Bioactive dietary long-chain fatty acids: emerging mechanisms of action. Br. J. Nutr. 100: 1152–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma D. W. L., Seo J., Switzer K. C., Fan Y-Y., McMurray D. N., Lupton J. R., Chapkin R. S. 2004. n-3 PUFA and membrane microdomains: a new frontier in bioactive lipid research. J. Nutr. Biochem. 15: 700–706. [DOI] [PubMed] [Google Scholar]
- 65.Hulbert A. J., Turner N., Storlien L. H., Else P. L. 2005. Dietary fats and membrane function: implications for metabolism and disease. Biol. Rev. Camb. Philos. Soc. 80: 155–169. [DOI] [PubMed] [Google Scholar]
- 66.Hulbert A. J., Else P. L. 1999. Membranes as possible pacemakers of metabolism. J. Theor. Biol. 199: 257–274. [DOI] [PubMed] [Google Scholar]
- 67.Hulbert A. J., Else P. L. 2000. Mechanisms underlying the cost of living in animals. Annu. Rev. Physiol. 62: 207–235. [DOI] [PubMed] [Google Scholar]
- 68.Hulbert A. J. 2003. Life, death and membrane bilayers. J. Exp. Biol. 206: 2303–2311. [DOI] [PubMed] [Google Scholar]
- 69.Cozzone D., Debard C., Dif N., Ricard N., Disse E., Vouillarmet J., Rabasa-Lhoret R., Laville M., Pruneau D., Rieusset J., et al. 2006. Activation of liver X receptors promotes lipid accumulation but does not alter insulin action in human skeletal muscle cells. Diabetologia. 49: 990–999. [DOI] [PubMed] [Google Scholar]
- 70.Kim H. J., Takahashi M., Ezaki O. 1999. Fish oil feeding decreases mature sterol regulatory element-binding protein 1 (SREBP-1) by down-regulation of SREBP-1c mRNA in mouse liver. A possible mechanism for down-regulation of lipogenic enzyme mRNAs. J. Biol. Chem. 274: 25892–25898. [DOI] [PubMed] [Google Scholar]
- 71.Howell G. I., Deng X., Yellaturu C., Park E. A., Wilcox H. G., Raghow R., Elam M. B. 2009. N-3 polyunsaturated fatty acids suppress insulin-induced SREBP-1c transcription via reduced trans-activating capacity of LXRα. Biochim. Biophys. Acta. 1791: 1190–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kopecky J., Rossmeisl M., Flachs P., Kuda O., Brauner P., Jilkova Z., Stankova B., Tvrzicka E., Bryhn M. 2009. Symposium on “Frontiers in adipose tisssue biology” n-3 PUFA: bioavailability and modulation of adipose tissue function. Proc. Nutr. Soc.. [DOI] [PubMed] [Google Scholar]
- 73.Liu L., Zhang Y., Chen N., Shi X., Tsang B., Yu Y-H. 2007. Upregulation of myocellular DGAT1 augments triglyceride synthesis in skeletal muscle and protects against fat-induced insulin resistance. J. Clin. Invest. 117: 1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Listenberger L. L., Han X., Lewis S. E., Cases S., Farese R. V., Jr., Ory D. S., Schaffer J. E. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA. 100: 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]