Abstract
Radioisotope-based and mass spectrometry coupled to chromatographic techniques are the conventional methods for monitoring HMG-CoA reductase (HMGR) activity. Irrespective of offering adequate sensitivity, these methods are often cumbersome and time-consuming, requiring the handling of radiolabeled chemicals or elaborate ad-hoc derivatizing procedures. We propose a rapid and versatile reverse phase-HPLC method for assaying HMGR activity capable of monitoring the levels of both substrates (HMG-CoA and NADPH) and products (CoA, mevalonate, and NADP+) in a single 20 min run with no pretreatment required. The linear dynamic range was 10–26 pmol for HMG-CoA, 7–27 nmol for NADPH, 0.5–40 pmol for CoA and mevalonate, and 2–27 nmol for NADP+, and limit of detection values were 2.67 pmol, 2.77 nmol, 0.27 pmol, and 1.3 nmol, respectively.
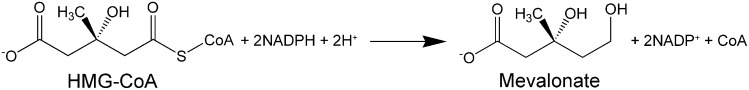
HMG-CoA reductase (HMGR) is the enzyme that catalyze the four-electron reductive deacylation of HMG-CoA to CoA and mevalonate (Fig. 1) (1). This reaction is the controlling step in the biosynthesis of sterols and isoprenoids (2, 3); hence, a large number of studies on the modulation of HMGR activity are continuously performed in the effort of developing new drugs in the treatment of hypercholesterolemic disorders (1).
Fig. 1.
Schematic representation of HMGR enzymatic reaction.
HMGR activity is conventionally assayed using elaborate radiochemical assay (4–9), chromatographic techniques coupled with mass spectrometry (10–15), or spectrophotometrically by monitoring the decrease in the absorbance of cofactor NADPH at 340 nm (16).
Herein, as an alternative for laboratories with no access to the expensive LC/MS equipment, we propose a rapid and adequately sensitive HPLC-based method capable of monitoring both the levels of all the species involved in the equilibrium in a single analysis and the kinetics of HMGR-catalyzed reactions.
MATERIALS AND METHODS
Reagents
HMGR, HMG-CoA, NADPH, NADP+, CoA, potassium phosphate, sodium phosphate, magnesium sulfate, phenyl-methane-sulfonyl-fluoride, tosyl-phenyl-alanyl-chloromethyl-ketone, EDTA, DTT, and DMSO were all purchased from Sigma-Aldrich. HPLC grade methanol was obtained from JT Baker. All solvents and reagents were of the highest purity available.
HPLC analysis
A HPLC system Gold (Beckman Coulter Inc.) equipped with a UV/VIS detector and HPLC column heater (Alltech) was used for the analysis. Reaction mixture consisting of HMGR (0.4 µM), NADPH (2.68 mM), and HMG-CoA (1.55 µM) diluted in the activity buffer was incubated at 37°C, and aliquots were withdrawn at indicated times and separated by HPLC. Each species (both isolated analytes and incubation mixtures) were injected and separated on a reverse phase Phenomenex Luna C18 column (5 μm particle size, 250 × 4.6 mm equipped with a 5 mm guard column of the same phase), and thermostated at 26°C, with the following linear gradient of potassium phosphate 100 mM (solvent A) and methanol (solvent B): 10%–30% B up to 15 min and 30%–10% B for 5 min at flow rate of 0.8 ml/min, UV/VIS detector set at 260 nm. An injection volume of 10 μl was used throughout. After each chromatographic elution, column was regenerated with two column volumes of 60% methanol. HMG-CoA, NADPH, and NADP+ were directly monitored, whereas mevalonate was determined by monitoring CoA production (mevalonate/CoA 1:1 stoichiometric ratio; Fig. 1). Enzyme catalytic activity was performed at 37°C in a 100 mM sodium phosphate buffer containing 1 mM EDTA, 10 mM DTT, 2% DMSO, and 1 mM magnesium sulfate, pH 6.8, following the reaction for different times of incubation. No interfering signal was detected upon injection of the activity buffer. Each analysis was repeated in quadruplicate. Detection of the HMGR levels and activity in cell homogenate was carried out on human colon cancer cell line HCT116, previously shown to express HMGR (17). Cells were suspended in the activity buffer added with phenyl-methane-sulfonyl-fluoride and tosyl-phenyl-alanyl-chloromethyl-ketone as proteases inhibitors and lysated using a insulin syringe. Total cellular enzyme levels were quantified according to the standard addition method (18); different amounts of the isolated enzyme (30–130 ng per mg of total proteins) were added to the reaction mixture (NADPH, HMG-CoA, and cell lysate, dissolved in the activity buffer) and detecting the product of CoA after 60 min. This mixture was finally centrifuged at 10,000 g for 10 min. These quantities were normalized to the total cell lysate proteins detected using Bradford assay (19) with BSA as standard.
RESULTS AND DISCUSSION
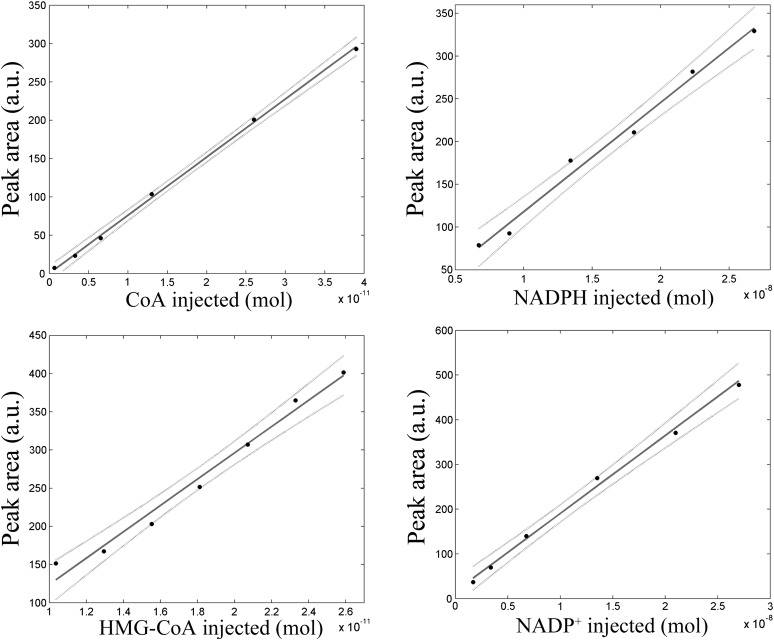
Calibration curves were established for each single analyte over adequate ranges of concentrations (Fig. 2). Dynamic ranges of linearity for the individual samples (determined by simple linear regression analysis), along with limits of detection (LODs) and limits of quantification (LOQs) (determined as ratio between 3 and 5 times the RMSE of the linear fit and the slope of the calibration curve, respectively) are reported in Table 1. The linearity of the standard curves was evaluated with a Chi-square χ2 goodness-of-fit test (P > 0.05).
Fig. 2.
Calibration curves of CoA, NADPH, HMG-CoA, and NADP+. Linear fits (solid lines) and 95% confidence bound (dotted lines) are reported. Each experimental point was the average of three replicates.
TABLE 1.
LOD and LOQ of the analytes tested with this reverse phase-HPLC method
| Analyte Tested | LOD | LOQ |
|---|---|---|
| NADPH | 2.77 nmol | 4.62 nmol |
| NADP+ | 1.33 nmol | 2.22 nmol |
| HMG-CoA | 2.67 pmol | 4.45 pmol |
| CoA | 0.27 pmol | 0.45 pmol |
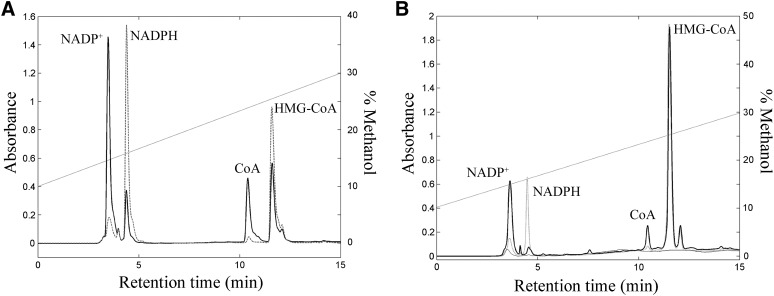
The substrates and the products of the enzymatic reaction were clearly and completely separated from each other, as shown in Fig. 3A.
Fig. 3.
A: Representative chromatographic profiles of reaction mixture in the absence (dashed line) and in the presence (after 60 min of incubation) of HMGR (solid line). B: Chromatographic profiles of cell lysate in the absence (gray line) and in the presence of reaction mixture after 0 and 60 min of incubation (dashed and solid line, respectively). Methanol gradient is reported (dotted line).
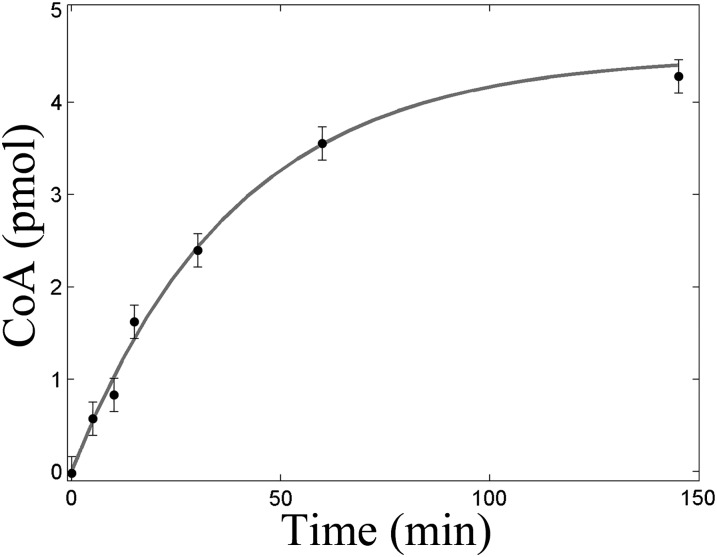
Time course of mevalonate/CoA production was analyzed according to a mono-exponential model (Fig. 4), and bi-exponential analysis did not significantly improve the quality of the fit, as assessed with F-Test (95% confidence).
Fig. 4.
Time course of CoA/mevalonate production fitted to mono-exponential model (solid line): CoA = Rmax×(1 − ek×time) with Rmax = 4.5 pmol (maximum equilibrium response at [HMG-CoA] = 1.55 µM) and k = 0.026min−1 (pseudo-first order rate constant). SD to fit are reported.
The assay reproducibility was evaluated by comparison of intra- and inter-day variability over 5 days by performing four replicates each day, with maximum variation coefficient associated to peak area CVintra = 5.48% and CVinter = 6.31%, and to retention time CVintra = 0.14% and CVinter = 0.23%.
The same method was successfully applied to a cellular lysate, reporting comparable chromatographic profiles without any significant interfering compounds (Fig. 3B).
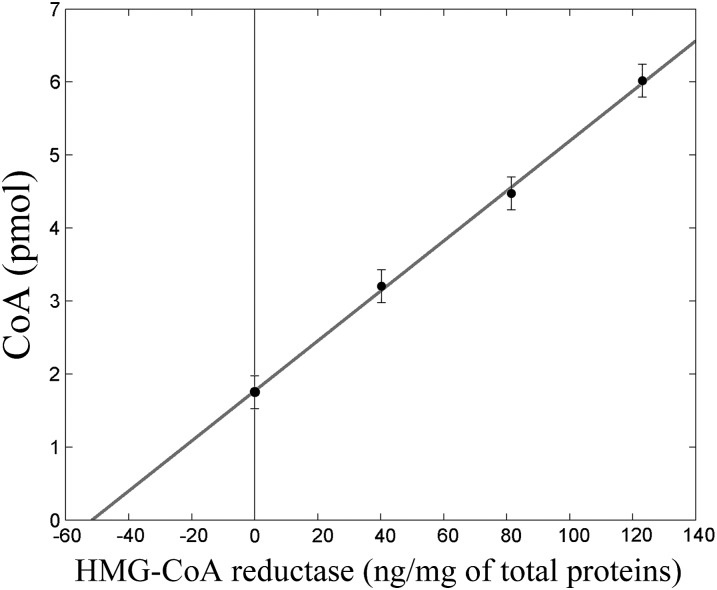
Multiple standard additions of the isolated enzyme to reaction mixture containing cell lysate showed a linear correlation between CoA/mevalonate produced and total enzyme. The measured concentration of the cellular active enzyme was 52 ± 2 ng/mg of total proteins (Fig. 5), corresponding to 15.58 ± 0.48 units/mg cell protein, consistent with previously studies on tumor cells (9).
Fig. 5.
Linear dependence between isolated enzyme added and CoA/mevalonate produced in a cell lysate reaction mixture. Quantity of the enzyme present in the cell lysate (normalized by the total quantity of proteins) is derived from the intercept on the X-axis.
Finally, the proposed method was validated by comparison with a commercially available spectrophotometric assay kit (Cod. CS1090, Sigma-Aldrich), which provided comparable levels of HMGR in HCT116 cell lysates (14.8 ± 0.4 units/mg of total cell proteins).
In conclusion, this method could represent a useful tool for both rapid and low-cost routine assays of HMGR activity, and quantitation of all the species involved in the equilibrium.
Footnotes
Abbreviations:
- HMGR
- HMG-CoA reductase
- LOD
- limit of detection
- LOQ
- limit of quantification
REFERENCES
- 1.Istvan E. S., Deisenhofer J. 2001. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 292: 1160–1164. [DOI] [PubMed] [Google Scholar]
- 2.Istvan E. S., Palnitkar M., Buchanan S. K., Deisenhofer J. 2000. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 19: 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tachibana A., Yano Y., Otani S., Taniguchi M. 1998. Non-radiochemical 3-hydroxy-3-methylglutaryl-coenzyme A synthase assay by reversed-phase HPLC without using ion-pair reagent. J. Ferment. Bioeng. 86: 523–526. [Google Scholar]
- 4.Caruso M. G., Notarnicola M., Santillo M., Cavallini A., Di Leo A. 1999. Enhanced 3-hydroxy-3-methyl-glutaryl coenzyme A reductase activity in human colorectal cancer not expressing low density lipoprotein receptor. Anticancer Res. 19: 451–454. [PubMed] [Google Scholar]
- 5.McWhinney V. J., Pond W. G., Mersmann H. J. 1996. Ontogeny and dietary modulation of 3-hydroxy-3-methylglutaryl-CoA reductase activities in neonatal pigs. J. Anim. Sci. 74: 2203–2210. [DOI] [PubMed] [Google Scholar]
- 6.Ong K. K., Khor H. T., Tan D. T. 1991. Assay of 3-hydroxy- 3-methylglutaryl CoA reductase activity using anionic-exchange column chromatography. Anal. Biochem. 196: 211–214. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen L. B., Shefer S., Salen G., Ness G. C., Tint G. S., Zaki F. G., Rani I. 1990. A molecular defect in hepatic cholesterol biosynthesis in sitosterolemia with xanthomatosis. J. Clin. Invest. 86: 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shefer S., Hauser S., Lapar V., Mosbach E. H. 1972. HMG CoA reductase of intestinal mucosa and liver of the rat. J. Lipid Res. 13: 402–412. [PubMed] [Google Scholar]
- 9.Bailey J. M., Wu J. D. 1977. Lipid metabolism in cultured cells. XVI. Lipoprotein binding and HMG CoA reductase levels in normal and tumor virus-transformed human fibroblasts. J. Lipid Res. 18: 512–516. [PubMed] [Google Scholar]
- 10.Jauhiainen M., Monkkonen H., Raikkonen J., Monkkonen J., Auriola S. 2009. Analysis of endogenous ATP analogs and mevalonate pathway metabolites in cancer cell cultures using liquid chromatography-electrospray ionization mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2967–2975. [DOI] [PubMed] [Google Scholar]
- 11.Liu A., Kushnir M. M., Roberts W. L., Pasquali M. 2004. Solid phase extraction procedure for urinary organic acid analysis by gas chromatography mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 806: 283–287. [DOI] [PubMed] [Google Scholar]
- 12.Saini G. S., Wani T. A., Gautam A., Varshney B., Ahmed T., Rajan K. S., Pillai K. K., Paliwal J. K. 2006. Validation of the LC-MS/MS method for the quantification of mevalonic acid in human plasma and determination of the matrix effect. J. Lipid Res. 47: 2340–2345. [DOI] [PubMed] [Google Scholar]
- 13.Honda A., Mizokami Y., Matsuzaki Y., Ikegami T., Doy M., Miyazaki H. 2007. Highly sensitive assay of HMG-CoA reductase activity by LC-ESI-MS/MS. J. Lipid Res. 48: 1212–1220. [DOI] [PubMed] [Google Scholar]
- 14.Ndong-Akoume M. Y., Mignault D., Perwaiz S., Plaa G. L., Yousef I. M. 2002. Simultaneous evaluation of HMG-CoA reductase and cholesterol 7alpha-hydroxylase activities by electrospray tandem MS. Lipids. 37: 1101–1107. [DOI] [PubMed] [Google Scholar]
- 15.Park E. J., Lee D., Shin Y. G., Lantvit D. D., van Breemen R. B., Kinghorn A. D., Pezzuto J. M. 2001. Analysis of 3-hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors using liquid chromatography-electrospray mass spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 754: 327–332. [DOI] [PubMed] [Google Scholar]
- 16.Edwards P. A., Lemongello D., Fogelman A. M. 1979. Improved methods for the solubilization and assay of hepatic 3-hydroxy- 3-methylglutaryl coenzyme A reductase. J. Lipid Res. 20: 40–46. [PubMed] [Google Scholar]
- 17.Yang Z., Xiao H., Jin H., Koo P. T., Tsang D. J., Yang C. S. 2010. Synergistic actions of atorvastatin with gamma-tocotrienol and celecoxib against human colon cancer HT29 and HCT116 cells. Int. J. Cancer. 126: 852–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxberg B. E., Kowalski B. R. 1979. Generalized Standard Addition Method. Anal. Chem. 51: 1031–1038. [Google Scholar]
- 19.Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]