Abstract
Cholesterol 7α-hydroxylase (CYP7A1) plays a key role in maintaining lipid and bile salt homeostasis as it is the rate-limiting enzyme converting cholesterol to bile acids. Deficiency of CYP7A1 leads to hyperlipidemia in man and mouse. Hyperlipidemia is often seen in patients when treated with high-dose retinoic acid (RA), but the molecular mechanisms remain elusive. Our present study revealed that CYP7A1 mRNA expression is greatly repressed by RA in both human hepatocytes and HepG2 cells where increased fibroblast growth factor 19 (FGF19) and small heterodimer partner (SHP) expressions were also observed, suggesting farnesoid X receptor (FXR) and retinoid X receptor (RXR) were activated. Promoter reporter assays demonstrate that all-trans RA (atRA) specifically activated FXR/RXR. However, detailed molecular analyses indicate that this activation is through RXR, whose ligand is 9-cis RA. Knocking down of FXR or RXRα by small interference RNA (siRNA) in human hepatocytes increased CYP7A1 basal expression, but the repressive effect of atRA persisted, suggesting there are also FXR/RXR-independent mechanisms mediating atRA repression of CYP7A1 expression. Chromatin immunoprecipitation (ChIP) assay and cell transfection results indicate that PGC-1α plays a role in the FXR/RXR-independent mechanism. Our findings may provide a potential explanation for hyperlipidemic side effects observed in some patients treated with high-dose RA.
Keywords: nuclear receptor, cholesterol, bile acid, hyperlipidemia
Mammals dispose of cholesterol mainly by converting cholesterol to bile acids where cholesterol 7α-hydroxylase (CYP7A1) is the rate-limiting enzyme (1). Genetic deficiency of CYP7A1 in humans leads to hyperlipidemia, including hypercholesterolemia and hypertriglyceridemia (2). Target deletion of Cyp7a1 in mice results in hypercholesterolemia as well, although this depends on their genetic background (3, 4). Hyperlipidemia is common in patients with chronic cholestasis where CYP7A1 expression is also reduced (5). Regulation of the expression of CYP7A1 is mainly transcriptional and tightly controlled by nuclear receptors (6). Hepatocyte nuclear factor 4α (HNF4α, NR2A1) is the major activator of transcription together with the peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α) (7, 8). The liver-related homolog-1 (LRH-1, NR5A2) is also a positive activator (8–10). While bile acids are the product of this enzymatic reaction, a feedback loop exists where bile acids can repress the expression of CYP7A1 (11), via the bile acid farnesoid X receptor (FXR, NR1H4) (8–10, 12). Bile acid activation of FXR upregulates the expression of the small heterodimeric partner (SHP, NR0B2) and fibroblast growth factor 19 (FGF19). The latter two in turn repress CYP7A1 expression (10, 13, 14). As a negative regulator, SHP inhibits the activation of HNF4α and LRH-1 in hepatocytes. Earlier studies indicated that CYP7A1 expression in hepatocytes was also inhibited by ileal secretion of FGF19 that circulates to the liver and binds to the FGF19 receptor FGFR4 on the plasma membrane of hepatocytes through the JNK signal transduction pathway (13, 15). However, activation of FXR can also increase FGF19 expression in the liver, suggesting that hepatic FGF19 may also repress CYP7A1 expression by an autocrine/paracrine mechanism in humans (14). Elevated FGF19, diminished CYP7A1, and unchanged levels of SHP mRNA expression have also been reported in the livers of patients with extrahepatic bile duct obstruction, findings which are reversed by biliary drainage, further supporting this mechanism (5). Together, these observations suggest that the FXR-FGF19 pathway may play a more important role than FXR-SHP in regulating CYP7A1 expression in vivo in human liver. The role of FXR in downregulation of CYP7A1 expression is further confirmed in Fxr−/− mice, where Cyp7a1 expression is increased (16).
FXR has been characterized as the bile acid nuclear receptor (17–19). It plays a pivotal role in maintaining bile salt and lipid homeostasis by regulating the expression of several key genes involved in bile acid synthesis, metabolism, and transport in the liver (16). In addition to CYP7A1, sterol 27-hydroxylase (CYP27A1) and Na+/taurocholate cotransporting polypeptide (NTCP, SLC10A1) are targets of FXR as well, mediated through SHP (20, 21). CYP27A1 initiates the acidic pathway of bile acid synthesis. FXR also directly regulates the expression of UDP glucuronosyltransferase 2B4 (UGT2B4), the bile salt export pump (BSEP, ABCB11), and the organic solute transporter α and β (OSTα-OSTβ) (22–25). Furthermore, FXR modulates the expression of genes involved in lipid and glucose metabolism, such as phospholipid transfer protein (PLTP), apolipoprotein C2 (ApoC2), ApoC3, and phosphoenolpyruvate carboxykinase (PEPCK) (26–28).
Chenodeoxycholic acid (CDCA) is considered to be the endogenous ligand for FXR (17). However, as a permissive nuclear receptor, FXR can also be activated through the ligand of its obligated heterodimeric partner, the retinoid X receptor (RXR, NR2B) (29, 30), where 9-cis retinoic acid (9cRA) is its specific ligand (31). However, Zavack et al. have reported that all-trans reti-noic acid (atRA) and TTNPB 4-[(E)-2-(5,6,7,8-Tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid activate mouse Fxr in a reporter assay and suggest that the metabolites of these two compounds may be the true ligands for FXR (32). While characterizing a primitive Fxr from the marine skate Leucoraja erinacea, we have also observed that atRA is a conserved activator of FXR in a reporter assay using cotransfection of skate Fxr and human RXRα expression constructs (33). These observations raise two questions: (1) Is atRA or its metabolite a true ligand for FXR? and (2) Does atRA modulate the expression of FXR targets? Intriguingly, high-dose RA (1.0–1.5 mg/kg/day) has been used to treat patients with leukemia, certain cancers, and some dermatological disorders, where retinoic acid receptors (RAR, NR1A) are the presumed target. However, numerous reports have documented that hyperlipidemia is often a major side effect, including hypercholesterolemia and hypertriglyceridemia (34–39). Increased total serum cholesterol levels were also seen in some gallstone patients treated with high-dose CDCA (40). The molecular mechanisms underlying these side effects of RA treatment remain to be determined.
In this report, we find that atRA represses CYP7A1 expression in both human primary hepatocytes and hepatoma HepG2 cells. We also demonstrate that this repression of CYP7A1 is mediated through both FXR/RXR-dependent and independent mechanisms. These findings may help to explain the hyperlipidemia observed in some patients, such as those with acute promyelocytic leukemia (APL) or acne, who are treated with high-dose RA.
MATERIAL AND METHODS
Materials
Chemicals were purchased from Sigma, except where otherwise specified. Cell culture media (DMEM), fetal bovine serum (FBS), penicillin/streptomycin, trypsin, and PBS were from Invitrogen (Carlsbad, CA). HMM medium is from Lonza (Walkersville, MD). Matrigel were purchased from BD Sciences (Bedford, MA). Luciferase assay kit was purchased from Promega (Madison, WI). [35S]L-Methionine and ECL reagents were purchased from Amersham (Piscataway, NJ). DNA oligos and sequencing were provided by the Keck Biotechnology Center at Yale University. UVI3001 and UVI3003 are gifts from Prof. Angel R. de Lera (Universidade de Vigo, Spain) (41).
Plasmid constructs
Human FXR (hFXR), RXRα (hRXRα) and I-BABP promoter reporter constructs are described in a previous article (33). Mouse HNF4α and PGC-1α expression plasmids were provided by Dr. Peter Edwards (University of California at Los Angeles, Los Angeles, CA). Human FXR W469A mutant (pCMX-hFXR-W469A) was a gift from Dr. David Mangelsdorf (University of Texas Southwestern Medical Center, Dallas, TX). Mouse Fxr-LBD and TK-gal4 reporter constructs were provided by Dr. David Moore (Baylor College of Medicine, Houston, TX). Constructs pCMX-hFXR-ΔH12, pCMX-hRXRα-ΔC19 were generated by mutating P463X in hFXR and D444X in hRXRα, respectively, using the QuickChange kit (Stratagene, La Jolla, CA). pPac0 and pPacRnLuc plasmids for gene expression in Drosophila S2 cells were provided by Dr. Robert Diasio (University of Alabama, Birmingham, AL). pPac-hFXR and pPac-hRXRα were made by inserting hFXR and hRXRα full-length coding regions into pPac0 vector. GST-FXR LBD and GST-FXR-LBDΔH1-5 expression constructs was made by inserting hFXR LBD or the C-terminal of hFXR LBD (AA341-472) into BamHI and XhoI sites of pGEX-5X-3 vector (Amersham). All mutants were confirmed by DNA sequencing. For the GST-hSRC-1 construct, a 680bp DNA fragment encoding the human steroid receptor coactivator 1 (hSRC-1) amino acid 561-782 was amplified from HepG2 cell cDNA library using PfuUltra DNA polymerase (Stratagene), confirmed by sequencing, and cloned into BamHI and XhoI sites of a pGEX-5X-3 vector. The primers are listed in Table 1.
TABLE 1.
DNA sequence of PCR primers and siRNA oligos
| Name | Sequence 5′→ 3′ |
|---|---|
| hRXRα-siRNA1 | TGACGGAGCTTGTGTCCAATT |
| hRXRα-siRNA2 | CAACAAGGACTGCCTGATTTT |
| hRXRα-siRNA3 | GCAAGGACCTGACCTACACTT |
| hRXRα-siRNA4 | GCAAGGACCGGAACGAGAATT |
| hFXR P463X | CAAGTTTACCTGACTTCTCTGTGAAATCTGG |
| hRXRα D444X | GCTCATCGGGTAGACACCCATTGACACCTTCCT |
| hFXR-F21 | GCTGGATCCCGTCTGGGCATTCTGAC |
| hFXR-F35 | GCTGGATCCCAACAAAGTCATGCAGGG |
| hFXR-R22 | CGCACTCGAGTCACTGCACGTCCCAGATT |
| hSRC-1-F | GCTGGATCCTCAGGCAGATGAGCTCAC |
| hSRC-1-R | CGCACTCGAGATCCATCTGTTCTTTCTTTTCC |
siRNA oligonucleotides
Four sets of double-strand, small interference RNA (siRNA) oligos that target human RXRα were chemically synthesized and annealed to duplex by Dharmacon (Chicago, IL) (Table 1). FXR siRNA and control RNA oligo, siCONTROL RISC-Free siRNA (sequence proprietary) were also purchased from Dharmacon.
Cell culture and transfection
Human hepatocytes were obtained from the Liver Tissue Procurement and Distribution System of the National Institutes of Health (Dr. Stephen Strom, University of Pittsburgh), and maintained on collagen-coated plates using HMM medium (containing dexamethasone and insulin, each 0.1 μM) and overlaid with Matrigel. Purified monoclonal anti-human FGF-19 antibody was purchased from R&D Systems (Minneapolis, MN). HepG2 and HEK293T cells were grown at 37°C in DMEM medium with 10% FBS in 5% CO2 atmosphere. Drosophila S2 cells (kindly provided by Dr. Tian Xu's lab at Yale University) were maintained in Schneider's Drosophila Medium (Invitrogen) supplemented with 10% FBS at 25°C incubation. Lipofectamine 2000 (Invitrogen) was used for transfection of human primary hepatocytes, HepG2, and HEK293T cells, whereas Effectene from Qiagen (Valencia, CA) was used for transfection of Drosophila S2 cells. All cells were transfected in serum-free medium for 16 h. After transfection, cells were treated with specified chemicals for an additional 24 h in serum-free or charcoal-stripped medium.
Quantitative real-time PCR analysis
Total RNA was extracted, purified, and converted to cDNA as template. Real-time PCR was performed in an ABI7500 Sequence Detection System. TaqMan primers/probe of GAPDH, SHP, CYP7A1, BSEP, OSTα, and OSTβ are as previously described (42), whereas primers/probe of FGF19, CYP27A1, and NTCP were purchased from ABI. GAPDH was used as reference for normalizing the data.
Western blot analyses
Protein was extracted from samples and analyzed as previously described (42). Polyclonal antibodies against FXR, RXRα, HNF4α, PGC-1α, and SH-PTP were from Santa Cruz and used at dilutions of 1:200, 1:500, 1:150, 1:150, and 1:1000, respectively. For Western blot analysis, total cell lysate (∼50 µg protein) were separated on a 10% SDS-polyacrylamide gel. After electrophoresis, proteins were transferred onto polyvinylidene fluoride (PVDF) microporous membranes (BioRad), the membranes were blocked with 5% nonfat milk, followed by primary antibody and HRP-conjugated secondary antibody incubation. The immune complexes were detected with the enhanced chemiluminescence reagent kit, and the signals were recorded on X-ray film.
Luciferase reporter assay
A dual-luciferase assay was applied for functional characterization by cotransfecting hFXR construct with the pCMX-hRXRα, pGL3-hI-BABP, and phRL-CMV plasmids (constitutively expressing Renilla luciferase) into HEK293T or HepG2 cells as previously described (33). The firefly luciferase activity was normalized by Renilla luciferase activity. Data are presented as mean ± SD of three or more independent experiments, and each transfection was performed in triplicate.
3H-atRA binding assay and coactivator recruitment assay
The GST, GST-hFXR LBD, GST-hFXR LBDΔH1-5, and GST-hSRC-1 (AA561-782) fusion proteins were expressed in E. coli DH5α and purified using glutathione-Sepharose 4B beads. Different amounts of 3H-atRA were directly incubated with proteins bound on Sepharose 4B beads and analyzed as described previously (43). 35S-labeled hFXR and hRARα were synthesized using the TNT T7 QuickCoupled transcription/translation kit (Promega) according to the manufacturer's instructions. The GST pulldown assay was performed as Lew et al. described (44).
ChIP assay
HepG2 cells were grown in 10-cm dishes and treated with 5 µM atRA or solvent (DMSO) for 24 h. The cells are fixed/cross-linked by directly adding formaldehyde to 1% (final concentration). The chromatin immunoprecipitation (ChIP) assay was performed using a kit from Millipore-Upstate (Temecula, CA). Antibodies against HNF4α or PGC-1α were purchased from Santa Cruz. PCR was performed as Li et al. described (45).
Statistical analysis
Data are expressed as means ± SD. Differences between experimental groups were assessed for significance using the two-tailed paired Student t-test. A P value of < 0.05 was considered statistically significant.
RESULTS
AtRA represses CYP7A1 expression in HepG2 cells
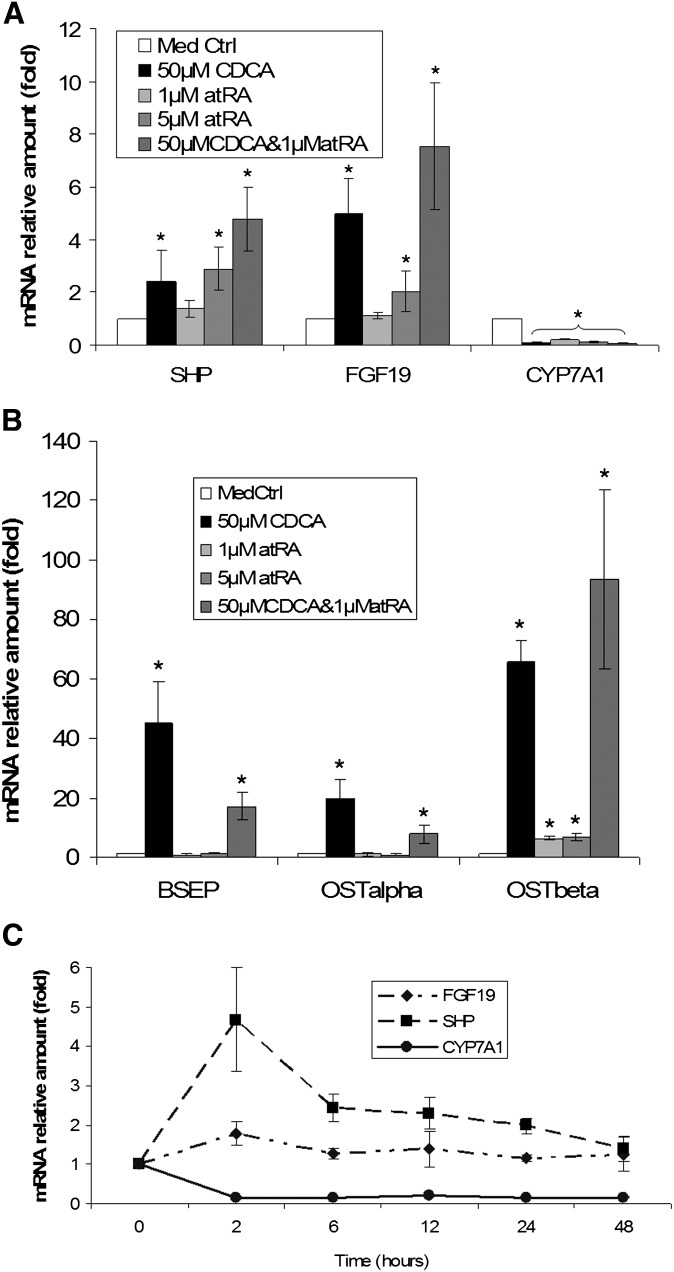
To assess whether atRA modulates the expression of FXR targets involved with bile acid synthesis, metabolism, and transport, as well as lipid catabolism, we treated HepG2 cells with atRA and/or CDCA for 24 h and measured the expression of several FXR targets in the liver using real-time quantitative PCR. Strikingly, 1 μM and 5 μM atRA reduced CYP7A1 mRNA levels to 21% and 13% of levels in control cells, respectively, whereas 50 μM CDCA reduced CYP7A1 expression to 8.3%. The combination of 1 μM atRA and 50 μM CDCA was synergistic and suppressed CYP7A1 mRNA expression further to 4.7% of control values. Consistent with this observation, expression levels of SHP and FGF19 mRNA in these cells were markedly increased (Fig. 1A). UGT2B4 and OSTβ mRNA expression levels were also significantly induced by atRA or CDCA treatment alone, with the combination demonstrating synergistic effects (Fig. 1B and data not shown). In contrast, atRA alone did not significantly increase BSEP or OSTα mRNA levels, whereas 50 μM CDCA stimulated their mRNA expression by 45- and 20-fold, respectively. Interestingly, 1 μM atRA partially antagonized CDCA's stimulation of both BSEP and OSTα (Fig. 1B). A time-course experiment showed that 5 μM atRA decreased CYP7A1 mRNA expression in association with increased SHP and FGF19 mRNA expression in HepG2 cells beginning 2 h after treatment, while CYP7A1 expression remained low throughout the time period (Fig. 1C).
Fig. 1.
atRA represses CYP7A1 mRNA expression in HepG2 cells. HepG2 cells were treated with indicated chemicals for 24 h. A: atRA and CDCA synergistically represses CYP7A1 expression accompanied by induction of SHP and FGF19. B: atRA induces OSTβ expression, but not BSEP or OSTα. C: Time course of the expression of CYP7A1, SHP, and FGF19 after 5 μM atRA treatment. *P < 0.05; n = 4. atRA, all-trans retinoic acid; CDCA, chenodeoxycholic acid; CYP7A1, cholesterol 7 alpha-hydroxylase; FGF19, fibroblast growth factor 19; OST, organic solute transporter; SHP, small heterodimer partner.
AtRA represses CYP7A1 expression in human hepatocytes
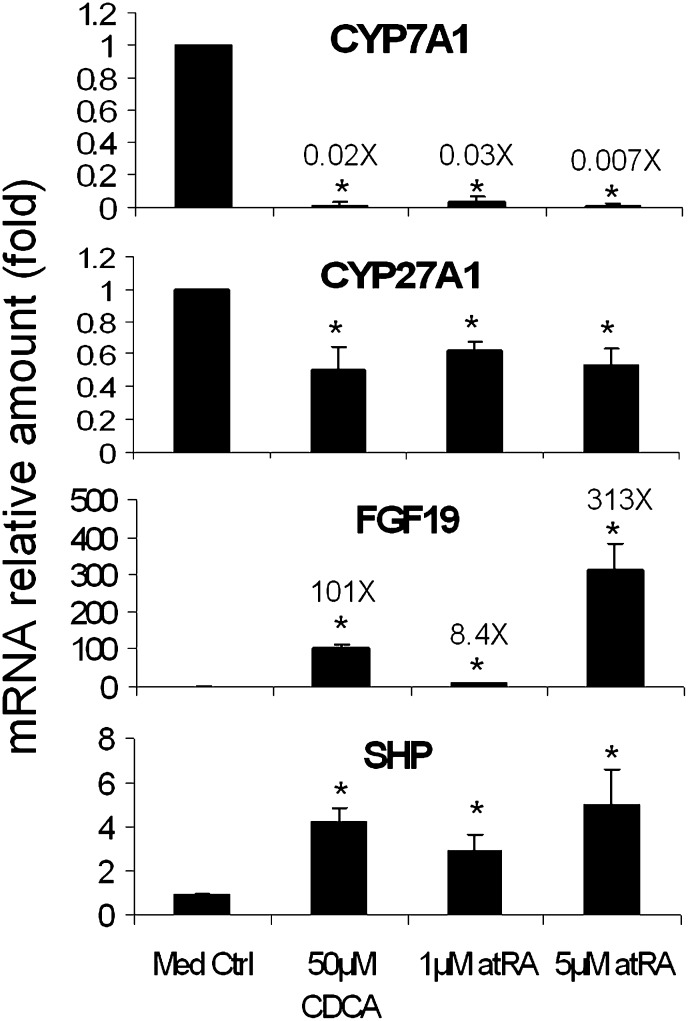
To determine whether atRA has the same effect on CYP7A1 expression in human primary hepatocytes as seen in HepG2 cells, we treated human hepatocytes with atRA for 24 h. In this experiment, both 1 μM and 5 μM atRA decreased CYP7A1 mRNA expression to 3% and <1% of the controls, respectively, a greater repression than in HepG2 cells. Consistent with this observation, a greater induction of FGF19 mRNA expression was detected in human hepatocytes than in HepG2 cells (8.4- and 313-fold, respectively) (Fig. 2). As a positive control, 50 μM CDCA reduced CYP7A1 to 2% and increased FGF19 by 101-fold. In addition, CYP27A1 and NTCP were significantly reduced to around 50% in CDCA- or atRA-treated cells, whereas SHP mRNA expression was increased (Fig. 2 and data not shown). Because marked increases in FGF19 expression and greater decreases of CYP7A1 were seen in atRA-treated human hepatocytes, we determined if antagonizing FGF19 could diminish atRA repression of CYP7A1. Purified FGF19 monoclonal antibody was added to human hepatocytes for 24 h. As expected, 10 μg/ml FGF19 antibodies reduced 5 μM atRA repression of CYP7A1 by nearly 4-fold (CYP7A1 mRNA increased from 0.67% to 2.6% of DMSO vector control). Of note, FGF19 antibody alone or control mouse IgG did not alter mRNA expression of CYP7A1, FGF19, or SHP. Interestingly, SHP mRNA was further increased by additions of FGF19 antibody to atRA-treated cells (from 4.4-fold by atRA alone to nearly 6-fold after FGF19 antibody was added). This result suggests that compromised FGF19 function may be compensated in part by increases in SHP expression as part of the mechanisms of CYP7A1 regulation. Taken together, atRA greatly repressed CYP7A1 expression, along with substantial increases in FGF19 and SHP expression in both HepG2 cells and primary human hepatocytes, suggesting that FXR/RXR activation may play a central role in regulation of these genes.
Fig. 2.
atRA demonstrates great repression of CYP7A1 mRNA expression (more than 95% repression) in primary human hepatocytes, where FGF19 was greatly induced. *P < 0.05; n = 4. atRA, all-trans retinoic acid; CYP7A1, cholesterol 7 alpha-hydroxylase; CYP27A1, sterol 27-hydroxylase; FGF19, fibroblast growth factor 19; SHP, small heterodimer partner.
AtRA specifically activates FXR/RXR in reporter assays
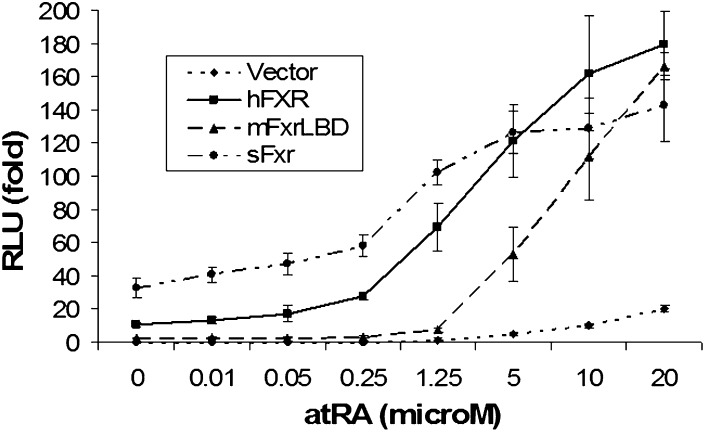
AtRA is known to activate FXR/Fxrs in reporter assays (32, 33). To confirm that this activation is specific for FXR, pGL3-hI-BABP and hRXRα expression constructs were cotransfected into HEK293T cells with expression plasmids of hFXR or human RARα (hRARα) or empty vector pcDNA3. After transfection, the cells were treated with 1 μM atRA for 24 h. Dual-luciferase assays indicate that significant activation was only detected in hFXR cotransfected cells, not in cells cotransfected with the empty vector or hRARα (data not shown), indicating that atRA stimulation of the FXR-response element (FXRE) reporter activity is FXR/RXR-dependent. To test the specificity of atRA in this activation, we next treated HEK293T cells with different doses of atRA after transfecting with hFXR, mouse Fxr LBD, and skate Fxr. As shown in Fig. 3, all three FXR/Fxrs were greatly activated by atRA in a dose-dependent manner with an EC50 ∼2.5 μM. Of note, the reporter construct for mouse Fxr LBD activates TK gal4 response elements but not an FXRE. In addition, when the same concentrations were used in these transfected cells for 24 h, we observed stronger activation of FXR/RXR by 9cRA and 13-cis RA than atRA, whereas 1 μM of all-trans retinol or all-trans retinal had no significant effects. Further, 2.5 μM atRA significantly stimulated the activity of the human BSEP minimal promoter by 3-fold in reporter assays when HEK293T cells were cotransfected with hFXR and hRXRα, in contrast to endogenous BSEP mRNA expression in HepG2 cells. Taken together, these findings suggest that RAs are conserved activators for FXR/RXR.
Fig. 3.
atRA activates human, mouse, and skate FXRs in reporter assay. The FXR response element (FXRE) in human I-BABP promoter and FXR, RXR, and Renilla luciferase expression constructs were transfected into HEK293T cells. After transfection, the cells were treated with indicated amount of atRA for additional 24 h (n = 4). Data normalized to Renilla luciferase activity and expressed as fold changes by setting pcDNA3 vector-transfected and medium-treated cells as 1. atRA, all-trans retinoic acid; FXR, farnesoid X receptor; RXR, retinoid X receptor.
AtRA is not a ligand for FXR
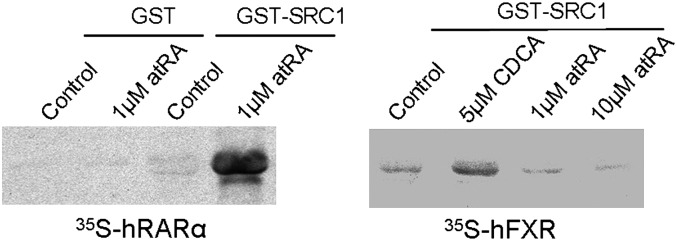
To examine whether atRA can bind to the FXR ligand binding domain, we generated expression constructs for a GST-hFXR LBD and a truncated hFXR LBD. The fusion proteins were purified and used for a direct binding assay with 3H-atRA. However, we did not observe any difference in binding between the GST-hFXR LBD fusion protein and the truncated form, to which no ligand is supposed to bind. This assay indicates that atRA does not specifically bind to the LBD of hFXR. To further confirm this negative finding, we performed a steroid receptor coactivator recruitment assay. In this experiment, we used GST-hSRC1 fusion protein to pull down in-vitro translated 35S-methionine-labeled hFXR as previously described (44). As Fig. 4 shows, 5 μM CDCA triggered hFXR binding to the hSRC1 fragment, whereas neither 1 μM nor 10 μM atRA did. In contrast, 1 μM atRA greatly induced hRARα binding to the hSRC1 fragment. These findings indicate that atRA is not a true ligand for FXR.
Fig. 4.
Coactivator recruit assay using recombinant human steroid receptor coactivator 1 (hSRC1) and 35S-methanoine labeled FXR and RARα. Purified GST-hSRC1 (fragment) fusion protein on glutathione beads were used to pull down 35S-FXR and 35S-RARα with or without addition of CDCA or atRA. atRA, all-trans retinoic acid; CDCA, chenodeoxycholic acid; FXR, farnesoid X receptor; GST, glutathione S-transferase; RARα, retinoic acid receptor α; SRC1, steroid receptor coactivator 1.
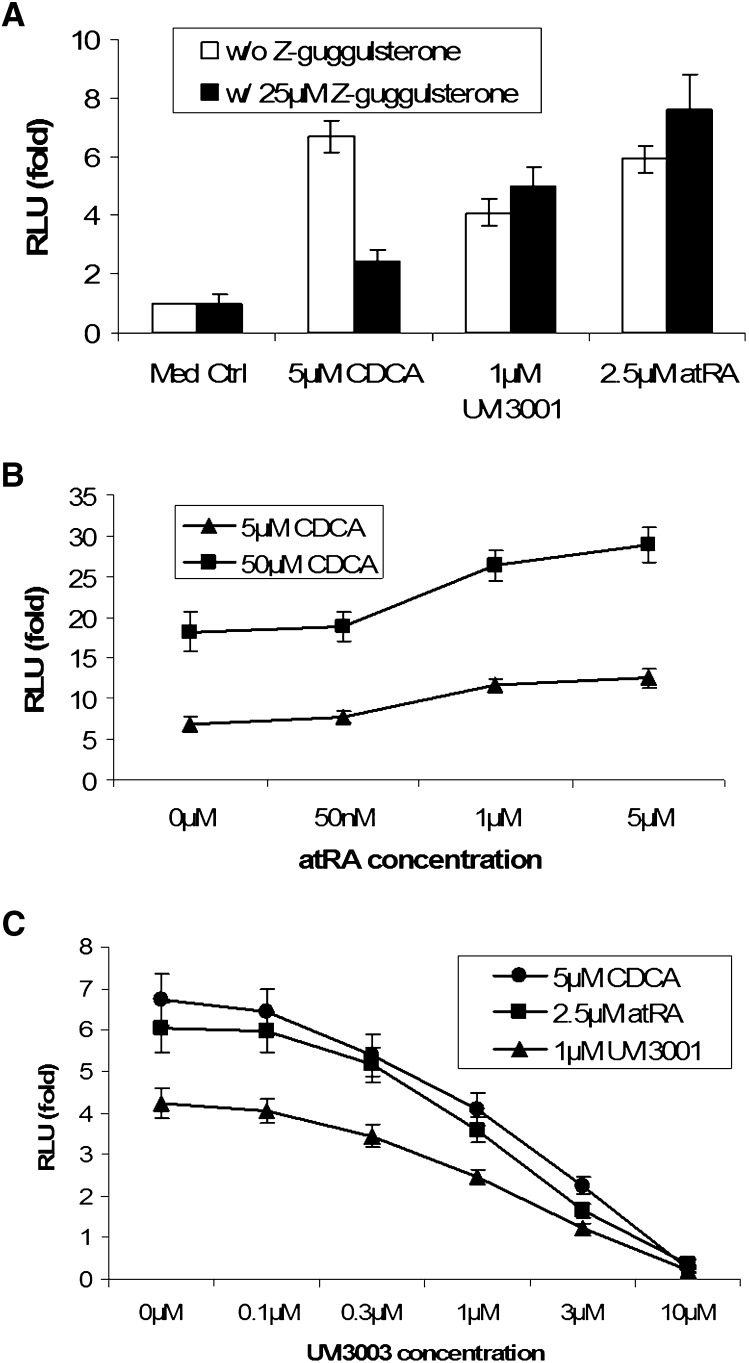
Guggulsterone antagonized CDCA but not atRA activation of FXR/RXR in reporter assays
To explore whether any metabolites of atRA might be ligands for FXR, we performed competition experiments with guggulsterone, a known antagonist for FXR (46), in the reporter assay. As Fig. 5A shows, 25 μM guggulsterone greatly repressed CDCA activation of FXR/RXR, in agreement with previous findings. In contrast, guggulsterone slightly increased atRA and UVI3001 (a specific RXR agonist) activation of FXR/RXR. This result rules out the possibility that an atRA metabolite might be a ligand for FXR and suggests that atRA or its metabolites may permissively stimulate FXR/RXR activity by activating RXR. If this is the case, one would expect a synergistic effect when the transfected cells are treated with CDCA and atRA. As predicted, CDCA and atRA synergistically induced FXR/RXR activity in the reporter assay in a dose-dependent manner (Fig. 5B). This synergistic effect was also seen when a minimal human BSEP promoter reporter construct was used in the assay. To further assess the role of RXR in this activation, we treated FXR/RXR transfected cells with RXR antagonist UVI3003 in combination with atRA, UVI3001, and CDCA. In this experiment, UVI3003 inhibited the activation of all three compounds on FXR/RXR in a dose-dependent manner (Fig. 5C), indicating a critical role for RXR in FXR/RXR activation.
Fig. 5.
Reporter assays indicate that atRA or its metabolites may not be ligands for FXR. Human I-BABP promoter luciferase construct was cotransfected with hFXR and RXRα expression vectors into HEK293T cells, and then treated with indicated chemicals for 24 h (n = 4). A: Guggulsterone, an antagonist of FXR, inhibits CDCA, but not UVI3001 (an RXR-specific agonist) or atRA on FXR activation. B: atRA and CDCA synergistically activate hFXR. C: RXR antagonist UVI3003 inhibited FXR activation, indicating a critical role of RXR in this activation. atRA, all-trans retinoic acid; CDCA, chenodeoxycholic acid; FXR, farnesoid X receptor; RXRα, retinoid X receptor α.
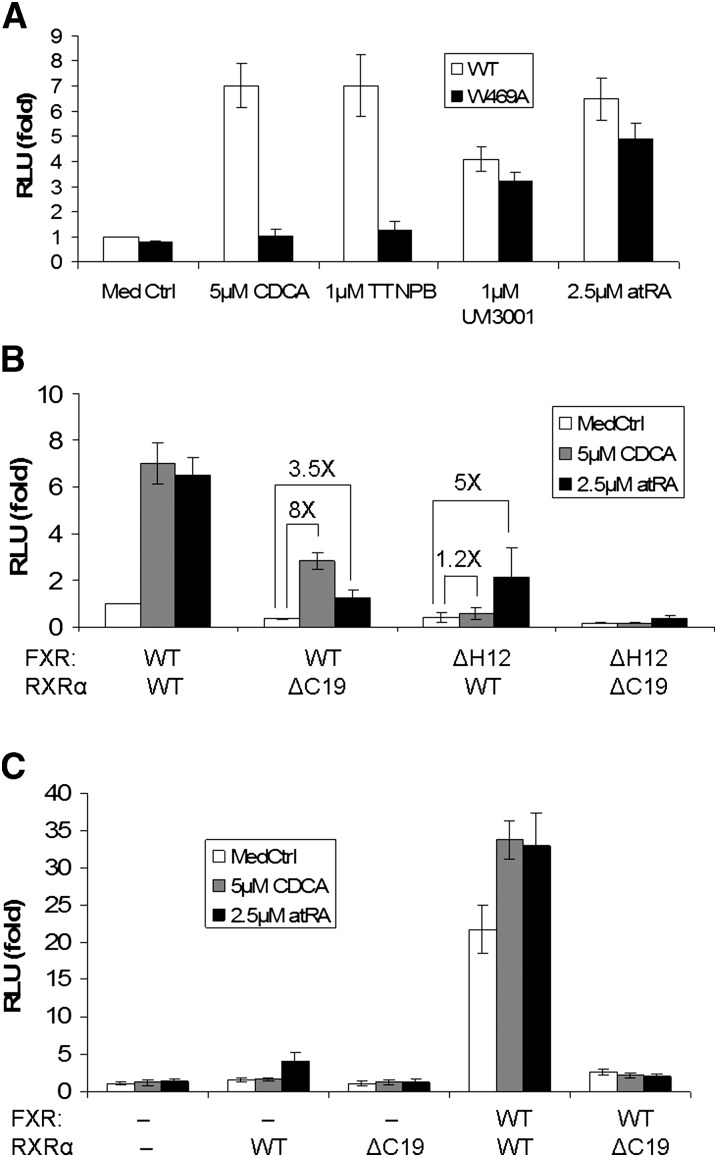
Functionally deficient mutations of RXR but not FXR abolished FXR/RXR activation by atRA
The Helix 12 (H12) in LBD, also called active function 2 (AF2), is a key motif in nuclear receptor ligand-dependent activation, including FXR and RXRα. It plays two roles: (1) binding to ligands and (2) interacting with coactivators, although these may occur at different residues. To further confirm that atRA stimulates FXR/RXR activity by activating RXR but not FXR, we tested three mutants: hFXR W469A, hFXR ΔH12, and hRXRα ΔC19. The W469A mutation only disables FXR-specific ligand binding, but the H12 is still capable of binding to the coactivators. However, truncation of H12 not only disables FXR ligand binding but also greatly compromises FXR/RXR interaction with coactivators. As expected, the hFXR W469A mutation completely abolished the stimulatory effects of two high-affinity FXR ligands, CDCA and TTNPB, in transfected HEK293T cells. In contrast, inductions of activity by atRA or UVI3001, while slightly reduced compared with the wild-type control, were essentially unaffected (Fig. 6A). Similar results were obtained with the hFXR ΔH12 mutation (Fig. 6B). However, when hRXRα was mutated, induction by atRA was greatly diminished (Fig. 6B). This observation was further confirmed in transfected Drosophila S2 cells where endogenous RXR is deficient (Fig. 6C). Together, these findings strongly indicate that RXR is the key component required for activation of FXR/RXR by atRA and that this permissive activation by atRA leads to repression of CYP7A1 expression in both human hepatocytes and HepG2 cells.
Fig. 6.
Reporter assays indicate that atRA activates FXR/RXR through RXR. Human I-BABP promoter luciferase construct was cotransfected with hFXR and RXRα expression vectors into HEK293T or Drosophila S2 cells, and then treated with indicated chemicals for 24 h (n = 4). A: Mutation in FXR (W469A) abolishes activation by CDCA and TTNPB (two FXR ligands) but not UVI3001 (an RXRα specific agonist) and atRA in transfected HEK293T cells. B: Mutant of RXRα (ΔC19) diminished atRA activation of FXR/RXR in transfected HEK293T cells. C: Mutation of RXR (ΔC19) abolishes FXR/RXR activation on human I-BABP promoter in S2 cells (deficient in RXR), again indicating that RXR is essential in FXR activation. atRA, all-trans retinoic acid; CDCA, chenodeoxycholic acid; FXR, farnesoid X receptor; RXRα, retinoid X receptor α.
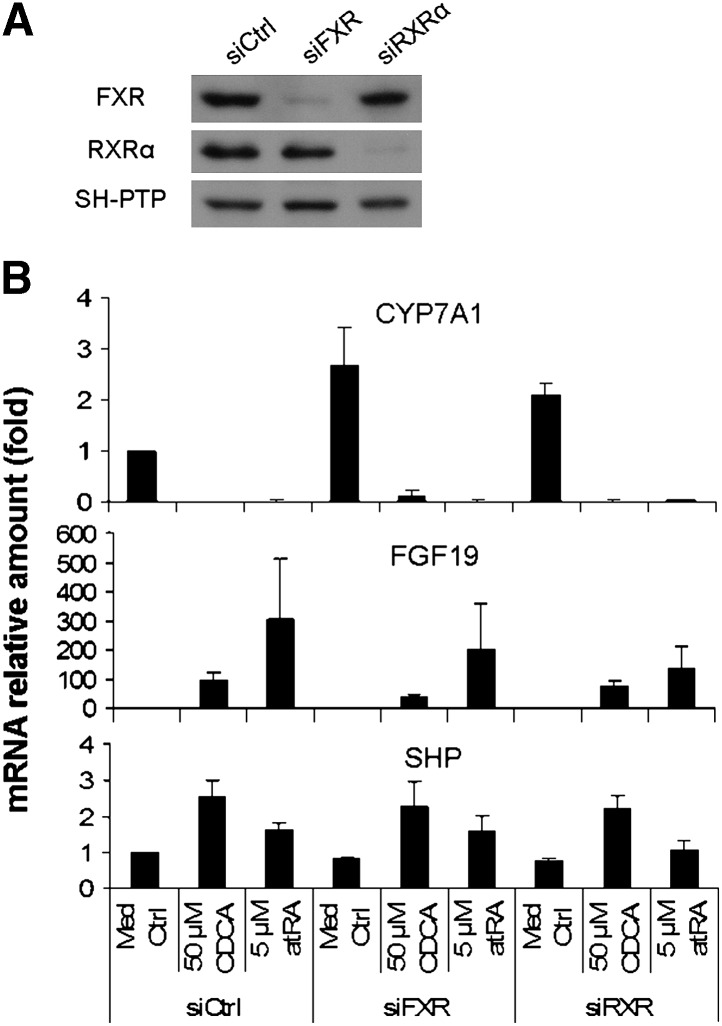
CYP7A1 mRNA expression was not abolished in FXR and RXRα knockdown human hepatocytes and HepG2 cells
To further determine the functional role of FXR/RXRα in repression of CYP7A1 expression by atRA, we knocked down the expression of FXR and RXRα in human hepatocytes using siRNA transfection. Western blot demonstrated that both FXR and RXRα protein expression were markedly reduced in human hepatocytes by these siRNAs (Fig. 7A). Interestingly, the basal mRNA expression of CYP7A1 increased by 2.7-fold in siFXR cells, while FGF19 mRNA expression decreased by 50%. In contrast, SHP mRNA levels remained essentially unchanged in all siRNA-transfected cells (Fig. 7B). These results suggest that FXR regulates CYP7A1 expression predominantly through FGF19 instead of SHP in human hepatocytes. However, when these cells were treated with 5 μM atRA for 24 h, substantial repression of CYP7A1 mRNA was seen in these cells, although reduced induction of FGF19 was observed in atRA-treated siFXR and siRXRα cells. The persistent repression of CYP7A1 by atRA suggests that there may be atRA-mediated FXR/RXR-independent mechanisms in this regulation.
Fig. 7.
Knockdown of FXR or RXRα diminished but not abolish atRA repression of CYP7A1 mRNA expression in siRNA transfected primary human hepatocytes. Human hepatocytes were transfected with siFXR, siRXRα, or siCtrl for 24 h, and then treated with atRA or CDCA for additional 24 h (n = 4). A: Western blot of FXR and RXRα from siRNA transfected human hepatocytes, where SH-PTP serves as loading control. B: CYP7A1, FGF19, and SHP mRNA expression in these cells, data normalized to GAPDH, n = 4. atRA, all-trans retinoic acid; CDCA, chenodeoxycholic acid; CYP7A1, cholesterol 7 alpha-hydroxylase; FGF19, fibroblast growth factor 19; FXR, farnesoid X receptor; RXR, retinoid X receptor; SHP, small heterodimer partner.
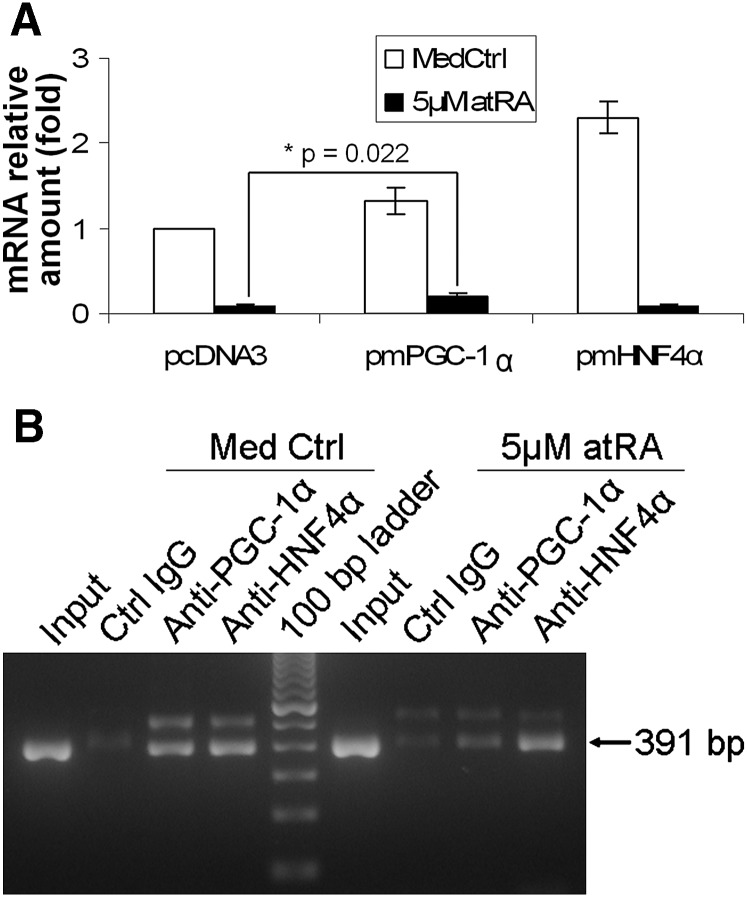
AtRA repression of CYP7A1 expression was diminished in PGC-1α transfected HepG2 cells
To determine whether other atRA-mediated transcriptional events might also play a role in repression of CYP7A1, we examined the effects of cotransfection with PGC-1α, a key coactivator of HNF4α-induced CYP7A1 expression. HepG2 cells were transfected with mouse PGC-1α, mouse HNF4α, or pcDNA3 empty vector for 16 h and then treated with atRA for an additional 24 h. As shown in Fig. 8A, transfection of mouse PGC-1α and HNF4α increased endogenous CYP7A1 mRNA by 30% and 130%, respectively, confirming that both PGC-1α and HNF4α are positive regulators in CYP7A1 expression. When these cells were treated with 5 μM atRA, CYP7A1 mRNA was decreased to 9.5% (± 2.5%) and 8.7% (± 2.8%) in the empty vector and HNF4α-transfected cells, respectively, of the basal expression with the empty vector (Fig. 8A). In contrast, atRA repression of CYP7A1 mRNA was significantly diminished in PGC-1α-transfected cells, which decreased to only 19.9% (± 5.2%) (P = 0.022, n = 3) of the basal expression with the empty vector. This result suggests that overexpression of PGC-1α can attenuate atRA repression of CYP7A1 expression. In other words, even though the total amount of PGC-1α may not change, depletion of PGC-1α levels in the CYP7A1 promoter may lead to decreased CYP7A1 mRNA expression. To further assess this possibility, we performed ChIP assays. Fig. 8B demonstrates that 5 μM atRA reduced PGC-1α but not HNF4α binding to the CYP7A1 promoter, whereas Western blot demonstrated that atRA did not change the level of nuclear expression of these two proteins (data not shown).
Fig. 8.
PGC-1α also plays an important role in CYP7A1 expression. A: Diminished repression of atRA in CYP7A1 mRNA expression in PGC-1α but not HNF4α or empty vector-transfected HepG2 cells. *P = 0.022; n = 3. B: ChIP assay detected reduced PGC-1α in CYP7A1 promoter after atRA treatment. atRA, all-trans retinoic acid; ChIP, chromatin immunoprecipitation; CYP7A1, cholesterol 7 alpha-hydroxylase; HNF4α, hepatocyte nuclear factor 4α; PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1 alpha.
DISCUSSION
High doses of RAs have been used to treat patients with APL, acne, and other dermatological disorders, often resulting in hyperlipidemia (34–39). To our knowledge, there is no report of liver RA concentrations in these patients, but we speculate that they should be at levels of no less than 5 μM. This is because (1) the liver is the major organ for storage and metabolism of RAs; (2) RA levels greater than 50 μM are toxic to hepatocytes (47); and (3) liver toxicity (elevated serum aminotransferases) are often seen in RA-treated patients (38). In this study we have examined potential mechanisms for these side effects. Here we demonstrate that a high dose (5 μM) of atRA represses CYP7A1 expression in both HepG2 cells and primary human hepatocytes (Figs. 1 and 2). Because CYP7A1 is the major enzyme that converts cholesterol into bile acids, inhibition of this metabolic pathway by RA could account for development of hypercholesterolemia in RA-treated patients.
Human CYP7A1 expression is tightly regulated by nuclear receptors. HNF4α and LRH-1 are positive regulators, whereas FXR, PXR, CAR, VDR, and PPARα are negative regulators. Our finding that atRA activated FXR/RXR in promoter reporter assays (Fig. 3) suggests that reduced CYP7A1 expression in atRA-treated cells may be mediated through the FXR/RXR signaling pathway. In support of this hypothesis, we found that atRA strongly induced the mRNA levels of FGF19 and SHP, two FXR targets that independently mediate FXR repression of CYP7A1 expression in both HepG2 cells and primary human hepatocytes (Figs. 1 and 2). Notably, 5 μM atRA showed greater induction of FGF19 and repression of CYP7A1 in primary human hepatocytes than in HepG2 cells (313-fold versus 2-fold for FGF19, and 0.7% versus 13% for CYP7A1, respectively). As Fgf15 (the mouse ortholog of human FGF19) plays a major role in Fxr-induced Cyp7a1 repression in vivo in mice and as Song et al. have recently demonstrated FXR-FGF19 autocrine signaling in regulating CYP7A1 expression in human hepatocytes (14), we propose that atRA repression of human CYP7A1 is mediated mainly through FGF19 in the FXR-dependent signal pathway. In line with this speculation, Schaap et al. have observed large increases in FGF19 but not SHP mRNA in the liver of cholestatic patients where substantially reduced CYP7A1 expression was also detected (5). FXR's role in modulating cholesterol homeostasis is further supported by the observation that serum cholesterol levels are increased in some gallstone patients treated with the FXR ligand CDCA while cholesterol levels are reduced in mice treated with the FXR antagonist guggulsterone (40, 46).
To elucidate how atRA activates FXR/RXR, we next confirmed that this activation was mediated via the FXRE using the human I-BABP gene and the human BSEP gene promoter reporter assays (Fig. 3), where RXRα alone or in combination with RARα did not show any stimulation in the same assay (data not shown). We then ruled out the possibility that atRA might be a ligand for FXR using both a direct binding assay and a coactivator recruitment assay (Fig. 4 and data not shown). Third, we explored the possibility that metabolites of atRA might be ligands for human FXR as this hypothesis has been proposed for mouse Fxr (32). However, our data indicate that this activation is mediated through RXR, the obligated heterodimeric partner of FXR. As Fig. 5A shows, atRA activates FXR/RXR in the same manner as UVI3001 (an RXRα selective agonist) but not like CDCA, the FXR agonist. In addition, CDCA and atRA demonstrated synergistic effect in this activation (Fig. 5B). If the metabolites of atRA are ligands for FXR, these metabolites should compete with CDCA for the FXR ligand binding pocket, and the ligand with higher affinity would have the dominant effect in this activation, where a synergistic effect would not be expected. Furthermore, mutations of RXRα but not FXR diminished atRA activation of FXR/RXR in reporter assays (Fig. 6). Altogether, these observations lead to the conclusion that atRA activation of FXR/RXR is mediated by RXRα most likely after conversion to 9cRA (48, 49), which is the specific ligand for RXRα (31). This permissive activation of FXR via its heterodimeric partner has also been observed with other synthetic RXR ligands (29, 30), where reduced Cyp7a1 mRNA expression was seen in vivo in mice. However, hypercholesterolemia was not seen in those mice after administration of a synthetic RXR agonist for 7 days (29).
To further assess the role of FXR/RXR in atRA repression of CYP7A1, we knocked down FXR or RXRα in human hepatocytes. In this experiment, increased basal CYP7A1 mRNA and diminished atRA repression were observed in the FXR knockdown cells, which were accompanied by reduced basal expression and induction of FGF19. This finding further supports a role for FXR/RXR in the regulation of CYP7A1 expression. Nevertheless, as atRA continues to repress the expression of CYP7A1 in these knockdown studies, atRA must also be activating via FXR/RXR-independent mechanisms as part of this regulation, although it is also possible that the remnant FXR and RXRα in the knockdown cells may still be capable of repressing CYP7A1 expression. To explore this possibility further, we found that overexpression of PGC-1α, a coactivator of HNF4α in the CYP7A1 promoter, diminished the ability of atRA to repress CYP7A1 expression in transfected HepG2 cells (Fig. 8A). In addition, ChIP assays also confirmed that atRA reduced the binding of PGC-1α to the CYP7A1 promoter (Fig. 8B). Together, these results indicate that PGC-1α is also a critical regulator in CYP7A1 expression. Thus, it is likely that when cells are treated with atRA, many additional nuclear receptors may be activated, including RARs and the permissive nuclear receptors PPARs and LXRs. Activation of these nuclear receptors may compete with HNF4α for PGC-1α and result in decreased CYP7A1 expression. However, we cannot rule out that other FXR/RXR-independent mechanisms might be also involved in mediating atRA repression of CYP7A1 expression. In fact, Marrapodi and Chiang have reported that activation of PPARα by its agonist can also inhibit human CYP7A1 expression in HepG2 cells (50). Therefore, it is also possible that atRA may repress CYP7A1 expression through PPARα signaling, as PPARα is a permissive nuclear receptor as well (29, 30). These possibilities remain to be examined.
FXR regulates the expression of its targets in a ligand- and promoter-selective fashion (44, 51), although details of the mechanisms remain elusive. In this report, we also found that atRA regulated FXR targets in a selective manner. For example, atRA alone stimulated the mRNA expression of SHP, FGF19, and OSTβ, but not BSEP or OSTα (Figs. 1 and 2). However, we cannot rule out the possibility that there may be as yet undescribed retinoic acid response elements in SHP, FGF19, and OSTβ genes. Further, atRA and CDCA demonstrated a synergistic effect in their mRNA expression. This synergistic effect on SHP expression and the attenuation of BSEP induction was also reported by others when HepG2 cells were treated with CDCA and 9cRA (52, 53). However, in those studies reduced FXR/RXR binding to the FXRE in the human BSEP promoter was observed in gel-shift and pulldown assays. Alternatively, as we suggest here, the supply of PGC-1α might also affect the expression of nuclear receptor targets by reducing the availability of this or other cofactors for specific gene expression. Finally, in contrast to previous reports (52, 53), we detected synergistic effects of atRA and CDCA in the minimal promoters of both human I-BABP and BSEP genes in reporter assays. Longer promoter regions (>1.5 kb) of the human BSEP gene were used in those reports and could account for these differences.
In addition to CYP7A1, atRA and CDCA also mildly but significantly reduced CYP27A1 and NTCP mRNA expression in human hepatocytes (Fig. 2 and data not shown). Previous reports have indicated that induction of SHP expression by CDCA in human cells accounts for the inhibitory effects on the expression of these two genes (20, 21). As increased SHP expression was also observed in atRA-treated human hepatocytes, we speculate that reduced expression of CYP27A1 and NTCP may be mediated through the same pathway.
SUMMARY
The present study demonstrates that atRA specifically represses human CYP7A1 expression. This suppression is mediated through both FXR/RXR-dependent and independent mechanisms. Neither atRA nor its metabolites are ligands for FXR. Rather, atRA and other RAs are capable of conversion to 9cRA, and then permissively activate FXR/RXR. We propose that these findings may provide a potential explanation for hyperlipidemia in patients treated with high doses of RA.
Acknowledgments
The authors thank Dr. John Chiang for critically reviewing the manuscript and comments. We greatly appreciate Dr. Peter Edwards, Dr. David Mangelsdorf, and Dr. David Moore for providing plasmid constructs. We thank Dr. Angel R. de Lera (Universidade de Vigo, Spain) for providing UVI3001 and UVI3003.
Footnotes
Abbreviations:
- APL
- acute promyelocytic leukemia
- atRA
- all-trans retinoic acid
- BSEP/Bsep
- bile salt export pump
- CDCA
- chenodeoxycholic acid
- ChIP
- chromatin immunoprecipitation
- CYP7A1
- cholesterol 7 alpha-hydroxylase
- FGF19
- fibroblast growth factor 19
- FXR/Fxr
- farnesoid X receptor
- HNF4α
- hepatocyte nuclear factor 4α
- LBD
- ligand binding domain
- OST
- organic solute transporter
- PGC-1α
- peroxisome proliferator-activated receptor gamma coactivator 1 alpha
- RA
- retinoic acid
- RARα
- retinoic acid receptor α
- RXR
- retinoid X receptor
- SHP
- small heterodimer partner
- siRNA
- small interference RNA
- SRC-1
- steroid receptor coactivator-1.
This work was supported by an American Liver Foundation PBC Fund for the Cure (S.Y.C.) and National Institutes of Health Grants DK-34989 and DK-25636 (J.L.B.). Normal human hepatocytes were obtained through the Liver Tissue Cell Distribution System, Pittsburgh, PA, which was funded by National Institutes of Health Contract N01-DK-7-0004/HHSN267200700004C. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Chiang J. Y. 2004. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J. Hepatol. 40: 539–551. [DOI] [PubMed] [Google Scholar]
- 2.Pullinger C. R., Eng C., Salen G., Shefer S., Batta A. K., Erickson S. K., Verhagen A., Rivera C. R., Mulvihill S. J., Malloy M. J., et al. 2002. Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J. Clin. Invest. 110: 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erickson S. K., Lear S. R., Deane S., Dubrac S., Huling S. L., Nguyen L., Bollineni J. S., Shefer S., Hyogo H., Cohen D. E., et al. 2003. Hypercholesterolemia and changes in lipid and bile acid metabolism in male and female cyp7A1-deficient mice. J. Lipid Res. 44: 1001–1009. [DOI] [PubMed] [Google Scholar]
- 4.Schwarz M., Lund E. G., Setchell K. D., Kayden H. J., Zerwekh J. E., Bjorkhem I., Herz J., Russell D. W. 1996. Disruption of cholesterol 7alpha-hydroxylase gene in mice. II. Bile acid deficiency is overcome by induction of oxysterol 7alpha-hydroxylase. J. Biol. Chem. 271: 18024–18031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaap F. G., van der Gaag N. A., Gouma D. J., Jansen P. L. 2009. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology. 49: 1228–1235. [DOI] [PubMed] [Google Scholar]
- 6.Chiang J. Y. 2009. Bile acids: regulation of synthesis. J.Lipid Res. 50: 1959–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin D. J., Campos J. A., Gil G., Osborne T. F. 2003. PGC-1alpha activates CYP7A1 and bile acid biosynthesis. J. Biol. Chem. 278: 50047–50052. [DOI] [PubMed] [Google Scholar]
- 8.Bhalla S., Ozalp C., Fang S., Xiang L., Kemper J. K. 2004. Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha. Functional implications in hepatic cholesterol and glucose metabolism. J. Biol. Chem. 279: 45139–45147. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6: 517–526. [DOI] [PubMed] [Google Scholar]
- 10.Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., Mangelsdorf D. J. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 6: 507–515. [DOI] [PubMed] [Google Scholar]
- 11.Stravitz R. T., Hylemon P. B., Heuman D. M., Hagey L. R., Schteingart C. D., Ton-Nu H. T., Hofmann A. F., Vlahcevic Z. R. 1993. Transcriptional regulation of cholesterol 7 alpha-hydroxylase mRNA by conjugated bile acids in primary cultures of rat hepatocytes. J. Biol. Chem. 268: 13987–13993. [PubMed] [Google Scholar]
- 12.Chiang J. Y., Kimmel R., Weinberger C., Stroup D. 2000. Farnesoid X receptor responds to bile acids and represses cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J. Biol. Chem. 275: 10918–10924. [DOI] [PubMed] [Google Scholar]
- 13.Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang Y., Mansfield T. A., Kliewer S. A., et al. 2003. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 17: 1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song K. H., Li T., Owsley E., Strom S., Chiang J. Y. 2009. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 49: 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim I., Ahn S. H., Inagaki T., Choi M., Ito S., Guo G. L., Kliewer S. A., Gonzalez F. J. 2007. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 48: 2664–2672. [DOI] [PubMed] [Google Scholar]
- 16.Sinal C. J., Tohkin M., Miyata M., Ward J. M., Lambert G., Gonzalez F. J. 2000. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 102: 731–744. [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 3: 543–553. [DOI] [PubMed] [Google Scholar]
- 18.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., et al. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284: 1365–1368. [DOI] [PubMed] [Google Scholar]
- 19.Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. 1999. Identification of a nuclear receptor for bile acids. Science. 284: 1362–1365. [DOI] [PubMed] [Google Scholar]
- 20.Chen W., Chiang J. Y. 2003. Regulation of human sterol 27-hydroxylase gene (CYP27A1) by bile acids and hepatocyte nuclear factor 4alpha (HNF4alpha). Gene. 313: 71–82. [DOI] [PubMed] [Google Scholar]
- 21.Eloranta J. J., Jung D., Kullak-Ublick G. A. 2006. The human Na+-taurocholate cotransporting polypeptide gene is activated by glucocorticoid receptor and peroxisome proliferator-activated receptor-gamma coactivator-1alpha, and suppressed by bile acids via a small heterodimer partner-dependent mechanism. Mol. Endocrinol. 20: 65–79. [DOI] [PubMed] [Google Scholar]
- 22.Barbier O., Torra I. P., Sirvent A., Claudel T., Blanquart C., Duran-Sandoval D., Kuipers F., Kosykh V., Fruchart J. C., Staels B. 2003. FXR induces the UGT2B4 enzyme in hepatocytes: a potential mechanism of negative feedback control of FXR activity. Gastroenterology. 124: 1926–1940. [DOI] [PubMed] [Google Scholar]
- 23.Denson L. A., Sturm E., Echevarria W., Zimmerman T. L., Makishima M., Mangelsdorf D. J., Karpen S. J. 2001. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology. 121: 140–147. [DOI] [PubMed] [Google Scholar]
- 24.Ananthanarayanan M., Balasubramanian N., Makishima M., Mangelsdorf D. J., Suchy F. J. 2001. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J. Biol. Chem. 276: 28857–28865. [DOI] [PubMed] [Google Scholar]
- 25.Landrier J. F., Eloranta J. J., Vavricka S. R., Kullak-Ublick G. A. 2006. The nuclear receptor for bile acids, FXR, transactivates human organic solute transporter-alpha and -beta genes. Am. J. Physiol. Gastrointest. Liver Physiol. 290: G476–G485. [DOI] [PubMed] [Google Scholar]
- 26.Kast H. R., Nguyen C. M., Sinal C. J., Jones S. A., Laffitte B. A., Reue K., Gonzalez F. J., Willson T. M., Edwards P. A. 2001. Farnesoid X-activated receptor induces apolipoprotein C–II transcription: a molecular mechanism linking plasma triglyceride levels to bile acids. Mol. Endocrinol. 15: 1720–1728. [DOI] [PubMed] [Google Scholar]
- 27.Claudel T., Inoue Y., Barbier O., Duran-Sandoval D., Kosykh V., Fruchart J., Fruchart J. C., Gonzalez F. J., Staels B. 2003. Farnesoid X receptor agonists suppress hepatic apolipoprotein CIII expression. Gastroenterology. 125: 544–555. [DOI] [PubMed] [Google Scholar]
- 28.Stayrook K. R., Bramlett K. S., Savkur R. S., Ficorilli J., Cook T., Christe M. E., Michael L. F., Burris T. P. 2005. Regulation of carbohydrate metabolism by the farnesoid X receptor. Endocrinology. 146: 984–991. [DOI] [PubMed] [Google Scholar]
- 29.Repa J. J., Turley S. D., Lobaccaro J. A., Medina J., Li L., Lustig K., Shan B., Heyman R. A., Dietschy J. M., Mangelsdorf D. J. 2000. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science. 289: 1524–1529. [DOI] [PubMed] [Google Scholar]
- 30.Shulman A. I., Larson C., Mangelsdorf D. J., Ranganathan R. 2004. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell. 116: 417–429. [DOI] [PubMed] [Google Scholar]
- 31.Heyman R. A., Mangelsdorf D. J., Dyck J. A., Stein R. B., Eichele G., Evans R. M., Thaller C. 1992. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 68: 397–406. [DOI] [PubMed] [Google Scholar]
- 32.Zavacki A. M., Lehmann J. M., Seol W., Willson T. M., Kliewer S. A., Moore D. D. 1997. Activation of the orphan receptor RIP14 by retinoids. Proc. Natl. Acad. Sci. USA. 94: 7909–7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai S. Y., Xiong L., Wray C. G., Ballatori N., Boyer J. L. 2007. The farnesoid X receptor FXRalpha/NR1H4 acquired ligand specificity for bile salts late in vertebrate evolution. Am. J. Physiol. Regul. Integr. Comp. Physiol. 293: R1400–R1409. [DOI] [PubMed] [Google Scholar]
- 34.Ellis C. N., Swanson N. A., Grekin R. C., Goldstein N. G., Bassett D. R., Anderson T. F., Voorhees J. J. 1982. Etretinate therapy causes increases in lipid levels in patients with psoriasis. Arch. Dermatol. 118: 559–562. [PubMed] [Google Scholar]
- 35.Bershad S., Rubinstein A., Paterniti J. R., Le N. A., Poliak S. C., Heller B., Ginsberg H. N., Fleischmajer R., Brown W. V. 1985. Changes in plasma lipids and lipoproteins during isotretinoin therapy for acne. N. Engl. J. Med. 313: 981–985. [DOI] [PubMed] [Google Scholar]
- 36.O'Leary T. J., Simo I. E., Kanigsberg N., Walker J., Goodall J. C., Ooi T. C. 1987. Changes in serum lipoproteins and high-density lipoprotein composition during isotretinoin therapy. Clin. Invest. Med. 10: 355–360. [PubMed] [Google Scholar]
- 37.Fex G. A., Aronsson A., Andersson A., Larsson K., Nilsson-Ehle P. 1996. In vivo effects of 13-cis retinoic acid treatment on the concentration of proteins and lipids in serum. Eur. J. Clin. Chem. Clin. Biochem. 34: 3–7. [DOI] [PubMed] [Google Scholar]
- 38.Zane L. T., Leyden W. A., Marqueling A. L., Manos M. M. 2006. A population-based analysis of laboratory abnormalities during isotretinoin therapy for acne vulgaris. Arch. Dermatol. 142: 1016–1022. [DOI] [PubMed] [Google Scholar]
- 39.Brelsford M., Beute T. C. 2008. Preventing and managing the side effects of isotretinoin. Semin. Cutan. Med. Surg. 27: 197–206. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Aguilar F., Breto M., Alegre B., Berenguer J. 1985. Increase in serum total cholesterol and low-density lipoprotein cholesterol by high-dose chenodeoxycholic acid in patients with radiolucent gallstones significantly reversed during preventive low dose after gallstone dissolution. Digestion. 31: 225–233. [DOI] [PubMed] [Google Scholar]
- 41.Nahoum V., Perez E., Germain P., Rodriguez-Barrios F., Manzo F., Kammerer S., Lemaire G., Hirsch O., Royer C. A., Gronemeyer H., et al. 2007. Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function. Proc. Natl. Acad. Sci. USA. 104: 17323–17328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai S. Y., Gautam S., Nguyen T., Soroka C. J., Rahner C., Boyer J. L. 2009. ATP8B1 deficiency disrupts the bile canalicular membrane bilayer structure in hepatocytes, but FXR expression and activity are maintained. Gastroenterology. 136: 1060–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang N., Schule R., Mangelsdorf D. J., Evans R. M. 1991. Characterization of DNA binding and retinoic acid binding properties of retinoic acid receptor. Proc. Natl. Acad. Sci. USA. 88: 3559–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lew J. L., Zhao A., Yu J., Huang L., De Pedro N., Pelaez F., Wright S. D., Cui J. 2004. The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. J. Biol. Chem. 279: 8856–8861. [DOI] [PubMed] [Google Scholar]
- 45.Li T., Jahan A., Chiang J. Y. 2006. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 43: 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urizar N. L., Liverman A. B., Dodds D. T., Silva F. V., Ordentlich P., Yan Y., Gonzalez F. J., Heyman R. A., Mangelsdorf D. J., Moore D. D. 2002. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science. 296: 1703–1706. [DOI] [PubMed] [Google Scholar]
- 47.Biagini C. P., Boissel E., Borde F., Bender V. E., Bouskila M., Blazy F., Nicaise L., Mignot A., Cassio D., Chevalier S. 2006. Investigation of the hepatotoxicity profile of chemical entities using Liverbeads and WIF-B9 in vitro models. Toxicol. In Vitro. 20: 1051–1059. [DOI] [PubMed] [Google Scholar]
- 48.Urbach J., Rando R. R. 1994. Isomerization of all-trans-retinoic acid to 9-cis-retinoic acid. Biochem. J. 299: 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shih T. W., Lin T. H., Shealy Y. F., Hill D. L. 1997. Non-enzymatic isomerization of 9-cis-retinoic acid catalyzed by sulfhydryl compounds. Drug Metab. Dispos. 25: 27–32. [PubMed] [Google Scholar]
- 50.Marrapodi M., Chiang J. Y. 2000. Peroxisome proliferator-activated receptor alpha (PPARalpha) and agonist inhibit cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J. Lipid Res. 41: 514–520. [PubMed] [Google Scholar]
- 51.Dussault I., Beard R., Lin M., Hollister K., Chen J., Xiao J. H., Chandraratna R., Forman B. M. 2003. Identification of gene-selective modulators of the bile acid receptor FXR. J. Biol. Chem. 278: 7027–7033. [DOI] [PubMed] [Google Scholar]
- 52.Hoeke M. O., Plass J. R., Heegsma J., Geuken M., van Rijsbergen D., Baller J. F., Kuipers F., Moshage H., Jansen P. L., Faber K. N. 2009. Low retinol levels differentially modulate bile salt-induced expression of human and mouse hepatic bile salt transporters. Hepatology. 49: 151–159. [DOI] [PubMed] [Google Scholar]
- 53.Kassam A., Miao B., Young P. R., Mukherjee R. 2003. Retinoid X receptor (RXR) agonist-induced antagonism of farnesoid X receptor (FXR) activity due to absence of coactivator recruitment and decreased DNA binding. J. Biol. Chem. 278: 10028–10032. [DOI] [PubMed] [Google Scholar]