Abstract
Genetic variability in the FADS1-FADS2 gene cluster [encoding delta-5 (D5D) and delta-6 (D6D) desaturases] has been associated with plasma long-chain PUFA (LCPUFA) and lipid levels in adults. To better understand these relationships, we further characterized the association between FADS1-FADS2 genetic variability and D5D and D6D activities in adolescents. Thirteen single nucleotide polymorphisms (SNPs) were genotyped in 1,144 European adolescents (mean ± SD age: 14.7 ± 1.4 y). Serum phospholipid fatty acid levels were analyzed using gas chromatography. D5D and D6D activities were estimated from the C20:4n-6/C20:3n-6 and C20:3n-6/C18:2n-6 ratios, respectively. Minor alleles of nine SNPs were associated with higher 18:2n-6 levels (1.9E-18 ≤ P ≤ 6.1E-5), lower C20:4n-6 levels (7.1E-69 ≤ P ≤ 1.2E-12), and lower D5D activity (7.2E-44 ≤ P ≤ 4.4E-5). All haplotypes carrying the rs174546 minor allele were associated with lower D5D activity, suggesting that this SNP is in linkage disequilibrium with a functional SNP within FADS1. In contrast, only the rs968567 minor allele was associated with higher D6D activity (P = 1.5E-6). This finding agrees with an earlier in vitro study showing that the minor allele of rs968567 is associated with a higher FADS2 promoter activity. These results suggest that rare alleles of several SNPs in the FADS gene cluster are associated with higher D6D activity and lower D5D activity in European adolescents.
Keywords: genetics, adolescents, lipoproteins, triglyceride, arachidonic acid
Long-chain PUFA (LCPUFA) have a key role in maintaining biological functions in humans; they are components of the cell membrane, serve as substrates for the synthesis of inflammatory eicosanoids (leukotrienes and prostaglandins), act as signaling molecules, and regulate gene expression (1, 2). The FA composition of plasma and body tissues has been associated with certain pathologies, such as obesity, metabolic syndrome, cardiovascular disease, diabetes, immune system diseases, and psychiatric disorders (3, 4). However, the mechanisms underlying these associations have not been clearly established.
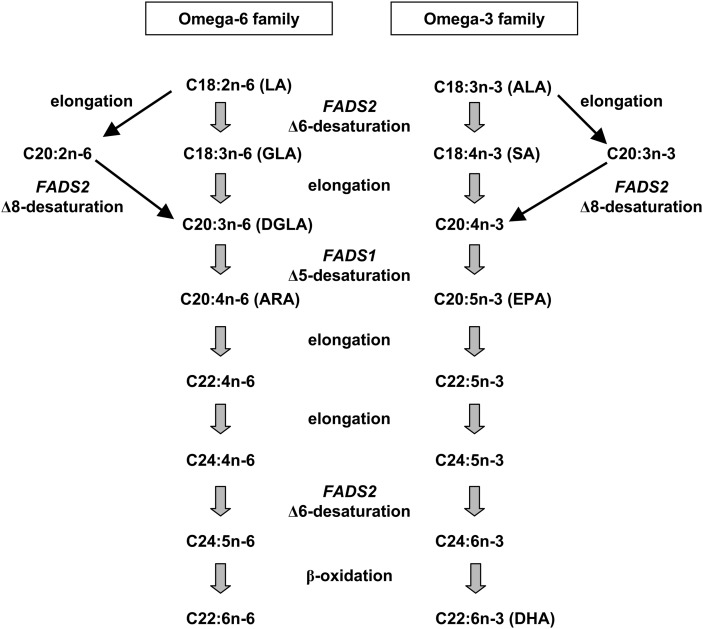
Tissue LCPUFA levels are determined by both dietary intake and endogenous synthesis via the successive elongation and desaturation of dietary FA precursors (Fig. 1). Delta-5 desaturase (D5D) and delta-6 desaturase (D6D), two enzymes required for the synthesis of LCPUFA in mammals (5), are respectively encoded by the FADS1 and FADS2 genes (located in a cluster on chromosome 11q12-13.1). Although both desaturases are expressed in most human tissues, the highest expression levels are observed in the liver (6). In mice, deletion of the FADS2 gene abolishes the initial step in the metabolic cascade and results in the absence of linoleic and α-linolenic acid derivatives and eicosanoid synthesis, inhibition of platelet aggregation and thrombus formation, and alterations in reproductive function (7).
Fig. 1.
Pathways for LCPUFA synthesis from n-6 and n-3 essential FAs. ALA: α-linoleic acid; ARA: arachidonic acid; DGLA: dihomo-γ-linoleic acid; DHA: docosahexaenoic acid; EPA: eicosapentaenoic acid; GLA: γ-linoleic acid; LA: linoleic acid; SA: stearidonic acid.
Many studies have reported associations between genetic variants in the FADS1-FADS2 gene cluster and plasma or erythrocyte LCPUFA levels (8–15). The observed associations between several single nucleotide polymorphisms (SNPs) on one hand and high levels of linoleic acid levels and low levels of arachidonic acid on the other suggested a defect in desaturase activity in adults. Other investigations have found a consistent association between FADS1-FADS2 gene cluster SNPs and low levels of LDL- and HDL-cholesterol and high levels of triglycerides (TG), suggesting that alterations in desaturase activities also affect plasma lipoprotein levels (9, 13, 16–19). At present, it is not known which desaturase (i.e., D5D, D6D, or both) is involved in these associations. To address this question, we further characterized the association between the genetic variability in this cluster and surrogate estimates of D5D and D6D activities separately in a sample of 1,144 European adolescents.
SUBJECTS AND METHODS
The HELENA study
The recruitment and phenotyping of the adolescents participating in the HELENA cross-sectional study (“Healthy Lifestyle in Europe by Nutrition in Adolescence,” www.helenastudy.com) have been described previously (20). Briefly, a total of 3,865 adolescents were recruited between 2006 and 2007. Data were collected in a total of 10 centers from 9 European countries. Subjects were randomly selected from schools by using a proportional cluster sampling methodology and taking age into account. One-third of the classes were randomly selected for blood collection; this resulted in a total of 1,155 blood samples. The body mass index (BMI) was available for 1,144 adolescents (i.e., the final sample in the present study).
Data were collected on a detailed case report form in accordance with standardized procedures. In each center, trained researchers carried out complete physical examinations, including weight, height, and blood pressure measurements. The protocol was approved by the appropriate investigational review board for each investigating center. Written, informed consent was obtained from each adolescent and both of his/her parents or legal representatives. Participation in the study was voluntary. The subjects’ clinical characteristics are presented in Table 1.
TABLE 1.
Characteristics of the sample
| HELENA Study | |
|---|---|
| Number of subjects | 1,144 |
| Boys/girls | 549/595 |
| Age (years) | 14.7(1.4) |
| Total cholesterol (mmol/l) | 4.16(0.71) |
| HDL (mmol/l) | 1.43 (0.28) |
| LDL (mmol/l) | 2.44 (0.63) |
| TG (mmol/l) | 0.78 (0.38) |
| BMI (kg/m2) | 21.3(3.8) |
| n-6 FAs* | |
| Linoleic acid (C18:2n-6) | 22.1 (2.5) |
| γ-Linolenic acid (C18:3n-6) | 0.088 (0.037) |
| Dihomo-γ-linolenic acid (C20:3n-6) | 2.98 (0.68) |
| Arachidonic acid (C20:4n-6) | 9.67 (1.63) |
| n-3 FAs* | |
| α-linolenic acid (C18:3n-3) | 0.14 (0.08) |
| Stearidonic acid (C18:4n-3) | 0.052 (0.08) |
| Eicosapentaenoic acid (C20:5n-3) | 0.49 (0.32) |
| Docosahexaenoic acid (C22:6n-3) | 2.95 (0.94) |
Data are mean (SD). * FA measurements are available for 1,034 subjects. Results for FAs are presented in percent of total FAs.
Venous blood samples were drawn after a 10 h overnight fast. Blood samples were sent to a central laboratory (the Analytical Laboratory at the University of Bonn, Bonn, Germany). Serum TGs, HDL, and LDL cholesterol and glucose levels were enzymatically assayed on the Dimension RxL clinical chemistry system (Dade Behring, Schwalbach, Germany). Blood for DNA extraction was collected in EDTA K3 tubes, stored at the Analytical Laboratory at the University of Bonn, and then sent to the Genomic Analysis Laboratory at the Institut Pasteur de Lille (Lille, France). DNA was extracted from white blood cells with the Puregene kit (QIAGEN, Courtaboeuf, France) and stored at −20°C.
After Folch extraction performed on serum samples, the phospholipid fraction was separated using thin-layer chromatography. The phospholipid band was scraped off and the FAs were converted into their methyl esters by transesterification with methanol/hydrochloric acid. The phospholipid fraction's FA methyl esters were analyzed using gas chromatography (Model 3900, Varian GmbH, Darmstadt, Germany) on a 30 m × 0.25 mm × 0.25 µm polyethylene glycol column (Zebron ZB-WAXplus, Phenomenex Ltd, Aschaffenburg, Germany). Peaks of interest were identified by comparison with authentic FA methyl ester standards (Sigma-Aldrich, Deisenhofen, Germany). FAs were expressed as a percentage area by integrating the area under the peak and dividing the results by the total area for all FAs. The coefficients of variations were <4.4% for all FA analyses.
Although C18:3 n-6 is the immediate product of D6D, it is rapidly elongated to C20:3n-6 (21) resulting in very low C18:3n-6 concentrations (mean = 0.088 ± 0.034%). Therefore, as previously described (22), (C20:3n-6):(C18:2n-6) and (C20:4n-6):(C20:3n-6) ratios were used as surrogate estimates of the D6D and D5D activities, respectively.
SNP selection and genotyping
With the criteria used in our SNP selection procedure [a minor allele frequency (MAF) over 0.1 and tag SNPs with an r2 value above 0.8], the November 2008 release 24 of the HapMap database described five haplotype blocks and five independent SNPs that span the whole FADS1-FADS2 gene cluster. In the present study, we selected one SNP from each of the haplotype blocks (block 1: rs174546; block 2: rs174589; block 3: rs2072114; block 4: rs174611; block 5: rs174616) and the five independent SNPs (rs174570, rs174602, rs498793, rs968567, and rs526126). The SNP rs965867 was of particular interest, because it was recently shown to be functional (23). To cover the full range of genetic variability, we also used the NCBI database to select three SNPs (rs174572, rs2072113 and rs174587) whose linkage disequilibrium status with the other SNPs was unknown. Subjects were genotyped on an Illumina system (using VeraCode technology for rs174589 and GoldenGate technology for the other SNPs).
Haplotype analysis
Haplotype analysis was performed using a two-step strategy similar to that described by Tregouet et al. (24). Briefly, haplotype frequencies derived from all the studied polymorphisms were first estimated independently of any phenotype. On the basis of the inferred haplotype structure and the 2K-1 possible combinations of 1 to K polymorphisms, we computed a minimal set of polymorphisms (called HtSNPs) that was sufficient for characterizing all haplotypes with a frequency ≥ 0.01. The HtSNPs were then used to test for associations between FADS1 and FADS2 gene haplotypes and LCPUFA levels and ratios. To reduce the haplotype dimension and select the most informative and parsimonious haplotype configuration when predicting phenotypic variability, we applied the maximum likelihood model to all possible 1 to k-loci combinations of polymorphisms that could be derived from the set of k HtSNPs. For each model (including one with no polymorphisms), an information criterion (AIC) was calculated (25). All AIC values were rescaled by subtracting the smallest AIC value obtained in the set of models. According to a rule derived by extensive Monte Carlo simulation, all models with a rescaled AIC ≤ 2 could be considered as “equivalent” to the model with the lowest minAIC. The most parsimonious of the latter (corresponding to the minimal haplotype configuration) was selected. Haplotype analyses were performed using the Thesias software package (http://ecgene.net/genecanvas) (26) and were adjusted for age, gender, BMI, and center.
Statistical methods
Statistical analyses were performed with SAS software (SAS Institute Inc., Cary, NC). Departure from Hardy-Weinberg equilibrium within the study groups was evaluated using a Chi-square test. Inter-locus linkage disequilibrium was assessed using Haploview software. The C18:3n-6, C18:3n-3, C18:4n-3, C20:5n-3, and C22:6n-3 values were log-transformed before analysis to achieve a normal distribution. The association between genotypes and quantitative variables was estimated with a general linear regression model (GLM procedure) assuming an allele-dose effect. The extent of inter-center heterogeneity was assessed using a genotype × center interaction term in the GLM procedure. All tests were adjusted for age, gender, BMI, and center. Bonferroni correction was applied to take multiple testing into account [P-value threshold = 0.0003 (i.e., 0.05/165 tests (11 SNPs × 15 traits)].
Power calculations were performed using Quanto v1.2.4 (27). On the basis of the effects reported in genome-wide association studies and with a MAF of 31% (such as for rs174546, for example), the statistical power values needed to detect a significant association (P < 0.0045; i.e., 0.05/11 SNPs) with serum HDL-cholesterol (β = −0.09 mmol/l) (17), LDL-cholesterol (β = −0.09 mmol/l) (19), and TG (β = +0.06 mmol/l) (17) levels in our study were 99%, 63%, and 69%, respectively.
RESULTS
Genetic data and selection of relevant SNPs for association studies
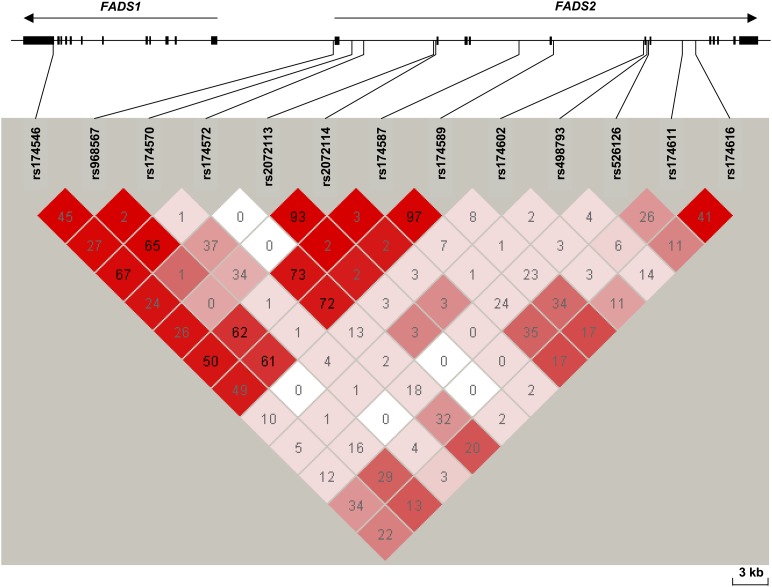
Thirteen SNPs in FADS1 and FADS2 were genotyped. The characteristics of the SNPs (including their position, genotype frequency, and MAF) are presented in Table 2. The mean genotyping success rate was 99.5%. All genotype distributions conformed to Hardy-Weinberg proportions and genotype distribution did not differ significantly across the different geographic regions (supplementary Table I). The minor allele frequencies ranged from 0.11 to 0.48. The SNPs’ linkage disequilibrium patterns were assessed by using both the D’ and r2 values (Fig. 2). Two SNPs from the NCBI database (rs174587 and rs2072113) were in strong linkage disequilibrium with two different SNPs reported in the HapMap database. The rs174587 SNP was in strong linkage disequilibrium with rs174589 (block 2) (r2 = 0.97) and rs2072113 was tagged by the rs2072114 SNP (block 3) (r2 = 0.93). Hence, we included the following 11 SNPs in our analyses: rs174546, rs968567, rs174570, rs174572, rs2072114, rs174589, rs174602, rs498793, rs526126, rs174611, and rs174616.
TABLE 2.
Main characteristics of the SNPs and the genotype distribution in the HELENA Study
| Position | Alleles (1/2) | GSR (%) | 11 | 12 | 22 | MAF | HW | |
|---|---|---|---|---|---|---|---|---|
| FADS1 | ||||||||
| rs174546 | 3′UTR | C/T | 99.8 | 555 (0.49) | 479 (0.42) | 108 (0.09) | 0.31 | 0.752 |
| FADS2 | ||||||||
| rs968567 | 5′UTR | C/T | 99.9 | 803 (0.70) | 303 (0.27) | 37 (0.03) | 0.16 | 0.204 |
| rs174570 | intron 1 | C/T | 100 | 910 (0.79) | 217 (0.19) | 17 (0.02) | 0.11 | 0.328 |
| rs174572 | intron 1 | C/T | 100 | 677 (0.59) | 407 (0.36) | 60 (0.05) | 0.23 | 0.908 |
| rs2072113 | intron 1 | C/T | 99.8 | 918 (0.80) | 207 (0.18) | 17 (0.02) | 0.11 | 0.179 |
| rs2072114 | intron 1 | A/G | 100 | 906 (0.79) | 220 (0.19) | 18 (0.02) | 0.11 | 0.274 |
| rs174587 | intron 4 | C/T | 100 | 743 (0.65) | 358 (0.31) | 43 (0.04) | 0.20 | 0.988 |
| rs174589 | intron 5 | C/G | 97.0 | 715 (0.64) | 352 (0.32) | 43 (0.04) | 0.20 | 0.969 |
| rs174602 | intron 5 | T/C | 98.6 | 693 (0.61) | 373 (0.33) | 62 (0.06) | 0.22 | 0.209 |
| rs498793 | intron 6 | T/C | 99.8 | 419 (0.37) | 532 (0.46) | 191 (0.17) | 0.40 | 0.317 |
| rs526126 | intron 6 | C/G | 99.2 | 735 (0.65) | 355 (0.31) | 45 (0.04) | 0.20 | 0.795 |
| rs174611 | intron 7 | T/C | 100 | 599 (0.52) | 464 (0.41) | 81 (0.07) | 0.27 | 0.490 |
| rs174616 | intron 7 | G/A | 99.8 | 311 (0.27) | 574 (0.50) | 257 (0.23) | 0.48 | 0.800 |
GSR: genotyping success rate; Major allele: 1; Minor allele: 2; HW: P-value for Hardy-Weinberg equilibrium. Data are n (frequency).
Fig. 2.
Structure of the FADS gene cluster and linkage disequilibrium patterns for the 13 investigated FADS SNPs. The red scale is for D’ values and numbers are for r2 values. Only the rs498793 SNP is in negative linkage disequilibrium (D’<0) with the others.
Association of FADS SNPs with LCPUFA, desaturase activities, serum lipid levels, and BMI
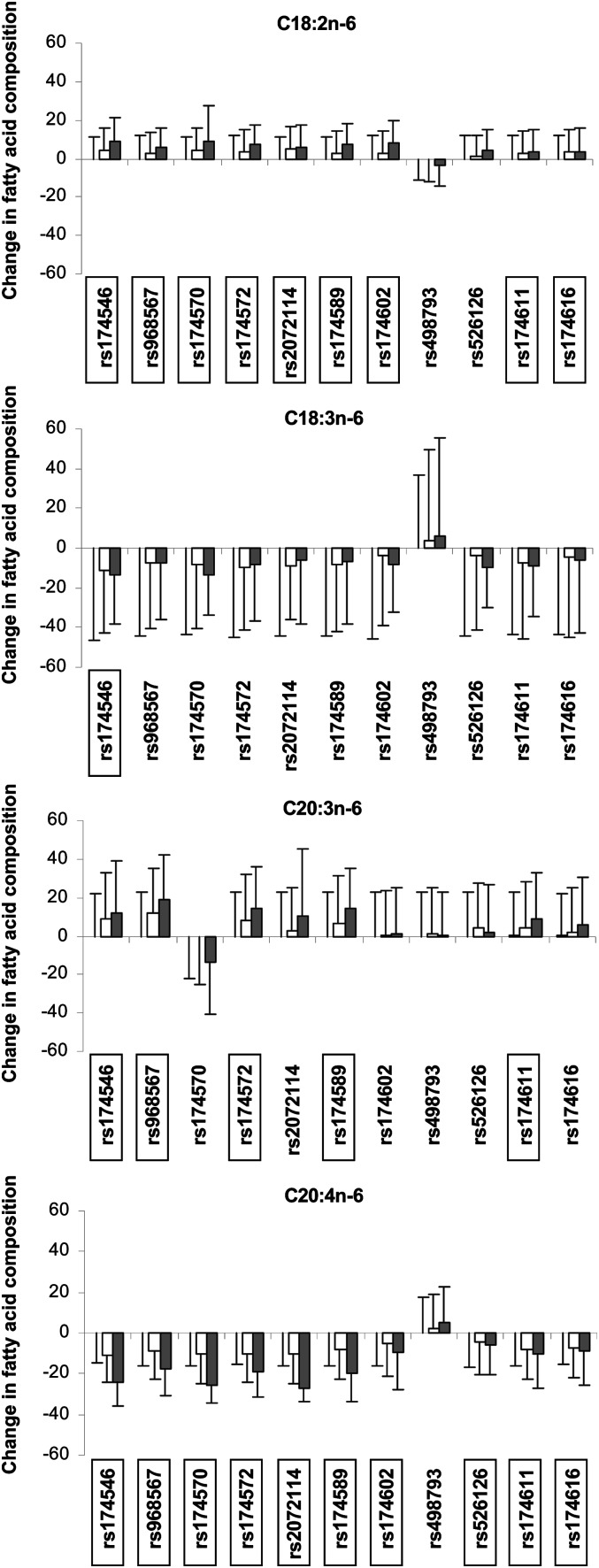
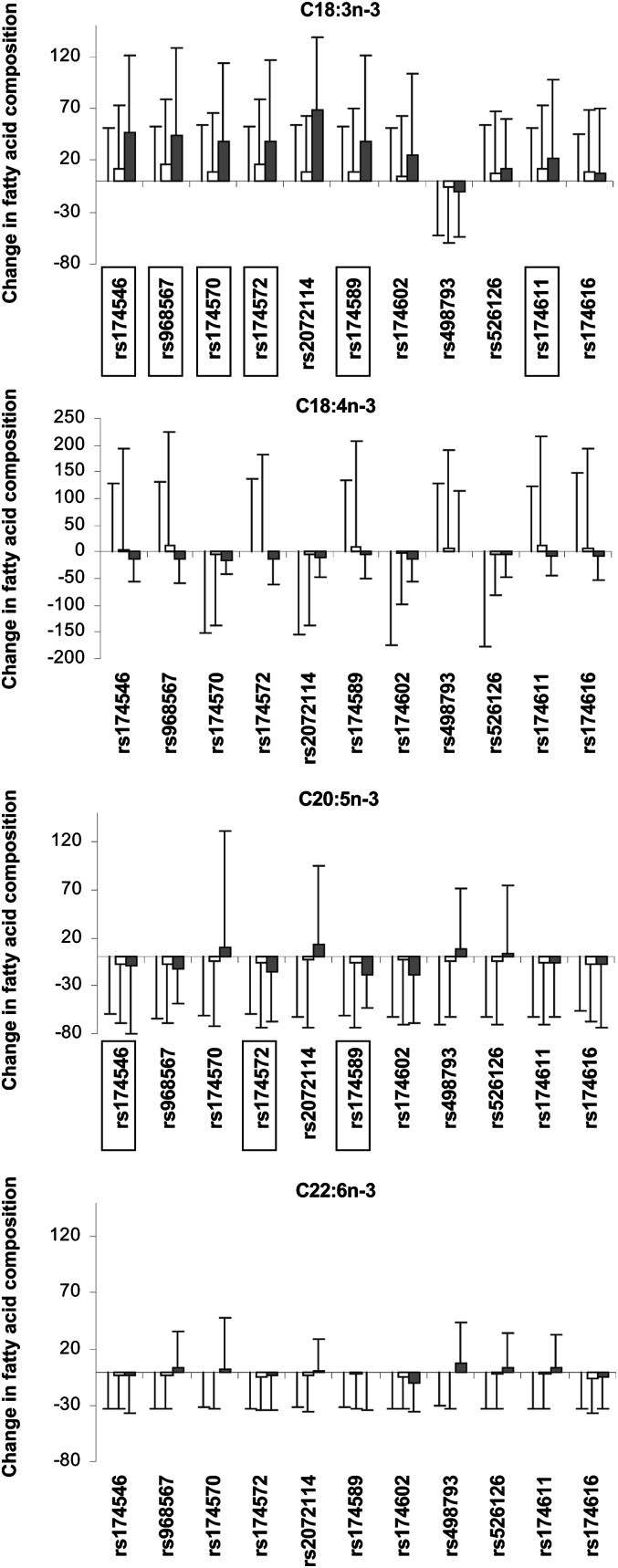
The minor alleles of all SNPs except rs498793 and rs526126 were significantly associated with higher levels of C18:2n-6 (1.9E-18 ≤ P ≤ 6.1E-5) (Fig. 3 and supplementary Table II). In contrast, only the rs174546 minor allele was significantly associated with lower levels of C18:3n-6 (P = 0.0001). The minor alleles of five SNPs (rs174546, rs968567, rs174572, rs174589, and rs174611) were significantly associated with higher levels of C20:3n-6 (3.1E-18 ≤≤ P ≤ 0.0002). The minor alleles of all SNPs except rs498793 were associated with lower levels of C20:4n-6 (7.1E-69 ≤ P ≤ 1.3E-5). In the n-3 FA family, the minor alleles of all SNPs except rs2072114, rs174602, rs498793, rs526126, and rs174616 were associated with significantly higher levels of C18:3n-3 (0.0002 ≤ P ≤ 2.0E-13) but none showed an association with the C18:4n-3 product (Fig. 4 and supplementary Table II). The rare alleles of three SNPs (rs174546, rs174572, and rs174589) were associated with significantly lower levels of C20:5n-3 (4.2E-6 ≤ P ≤ 0.0002), but none showed an association with C22:6n-3.
Fig. 3.
Association of FADS genotypes with variations in n-6 LCPUFA levels (%). In each graph and for each SNP, the first bar represents the reference value [the mean level (± SD) of the n-6 LCPUFA in homozygotes for the major allele]. The second and third bars indicate the n-6 LCPUFA level in heterozygote and homozygotes for the minor allele, respectively, calculated as a percentage of the reference value. White and black bars represent heterozygotes and homozygotes, respectively. All tests were adjusted for age, gender, BMI, and center. Significant associations (P < 0.0003) are indicated by boxes around the SNP.
Fig. 4.
Association of FADS genotypes with variations in n-3 LCPUFA levels (%). In each graph and for each SNP, the first bar represents the reference value [the mean level (±SD) of the n-3 LCPUFA in homozygotes for the major allele]. The second and third bars indicate the n-3 LCPUFA level in heterozygote and homozygotes for the minor allele, respectively, calculated as a percentage of the reference value. White and black bars represent heterozygotes and homozygotes, respectively. All tests were adjusted for age, gender, BMI, and center. Significant associations (P < 0.0003) are indicated by boxes around the SNP.
We calculated the (C20:3n-6):(C18:2n-6) and (C20:4n-6):(C20:3n-6) ratios as surrogate estimates of the D6D and D5D activities, respectively (see “Methods”). Only the rs968567 rare allele was significantly associated with higher D6D activity (P = 1.5E-6) (Table 3). In contrast; the minor alleles of all SNPs except rs498793 and rs526126 were associated with lower D5D desaturase activity (7.4E-44 ≤ P ≤ 4.4E-5) (Table 3). Because the plasma levels of C20:4n-3 were not available in the present study, the D6D and D5D activities could not be estimated from n-3 LCPUFA.
TABLE 3.
D6D and D5D activities according to genotypes
| 11 | 12 | 22 | P* | |
|---|---|---|---|---|
| D6D activity | ||||
| rs 174546 | 0.14 (0.04) | 0.14 (0.04) | 0.14 (0.04) | 0.04 |
| rs968567 | 0.13 (0.04) | 0.15 (0.04) | 0.15 (0.03) | 1.5E-06 |
| rs 174570 | 0.14 (0.04) | 0.13 (0.04) | 0.11 (0.05) | 0.01 |
| rs 174572 | 0.14 (0.04) | 0.14 (0.04) | 0.14 (0.03) | 0.01 |
| rs2072114 | 0.14 (0.04) | 0.14 (0.04) | 0.14 (0.06) | 0.55 |
| rs 174589 | 0.14 (0.04) | 0.14 (0.04) | 0.14 (0.03) | 0.01 |
| rs 174602 | 0.14 (0.04) | 0.14 (0.04) | 0.13 (0.04) | 0.03 |
| rs498793 | 0.14 (0.04) | 0.14 (0.04) | 0.14 (0.04) | 0.25 |
| rs526126 | 0.14 (0.04) | 0.14 (0.04) | 0.13 (0.04) | 0.76 |
| rs174611 | 0.14 (0.04) | 0.14 (0.04) | 0.14 (0.04) | 0.24 |
| rs174616 | 0.14 (0.04) | 0.14 (0.04) | 0.14 (0.04) | 0.98 |
| D5D activity | ||||
| rs 174546 | 3.82 (1.07) | 3.14 (0.85) | 2.61 (0.72) | 7.4E-44 |
| rs968567 | 3.64 (1.06) | 2.95 (0.77) | 2.51 (0.74) | 4.6E-27 |
| rs 174570 | 3.49 (1.06) | 3.16 (0.95) | 3.02 (0.63) | 1.2E-06 |
| rs 174572 | 3.70 (1.09) | 3.06 (0.81) | 2.60 (0.69) | 6.1E-31 |
| rs2072114 | 3.53 (1.06) | 3.06 (0.88) | 2.41 (0.69) | 3.0E-12 |
| rs 174589 | 3.63 (1.07) | 3.12 (0.87) | 2.50 (0.70) | 2.8E-21 |
| rs 174602 | 3.52 (1.09) | 3.29 (0.95) | 3.15 (0.95) | 4.4E-05 |
| rs498793 | 3.39 (1.08) | 3.40 (0.99) | 3.54 (1.09) | 0.23 |
| rs526126 | 3.53 (1.06) | 3.23 (0.97) | 3.29 (1.10) | 0.0003 |
| rs174611 | 3.64 (1.09) | 3.22 (0.94) | 2.99 (0.93) | 1.0E-12 |
| rs174616 | 3.70 (1.11) | 3.38 (0.97) | 3.21 (1.05) | 1.5E-08 |
Major allele: 1, minor allele: 2 . Data are mean (SD). (C20:3n-6):(C18:2n-6) and (C20:4n-6):(C20:3n-6) ratios were used as estimates of D6D and D5D desaturase activities, respectively (22). *P-values were adjusted for age, gender, BMI and center.
The mean serum total cholesterol, HDL-cholesterol, LDL-cholesterol, and TG levels and BMI are shown by genotype in supplementary Table III. We did not detect significant associations between the SNPs and these parameters.
Association of FADS haplotypes with LCPUFA and desaturase activities, lipid levels, and BMI
We performed haplotype analyses with the 11 SNPs to assess associations with LCPUFA and lipid levels, desaturase activities, and BMI. The 18 observed haplotypes (with a frequency ranging from 0.01 to 0.27) accounted for ≥80% of all possible haplotypes (supplementary Table IV, upper panel). This haplotype structure could be fully characterized by a subset of seven HtSNPs (rs174546, rs968567, rs174602, rs498793, rs526126, rs174611, and rs174616) (supplementary Table IV, lower panel). However, 14 of the 18 haplotypes have frequencies below 5%, precluding the possibility to analyze the associations with sufficient statistical power. Therefore, we further selected the most informative and parsimonious haplotype configurations with respect to the C18:2n-6 and C20:4n-6 levels as well as D5D and D6D activities, as described above (see “Methods”). The most informative haplotype configuration generated from this selection was the model including rs174546, rs968567, and rs174602 (see supplementary Table V).
We performed haplotype analysis using Thesias software (26) with this three-SNP combination (Table 4). The haplotype carrying the rs174602 minor allele (underlined) in combination with the frequent alleles of the rs174546 and rs968567 SNPs (CCC) was associated with neither LCPUFA levels nor D6D and D5D activities. In contrast, two of the four haplotypes carrying the minor allele of the rs174546 SNP (TCC and TTT) were associated with higher levels of the precursor C18:2n-6 (+15% and +8%, respectively). The two haplotypes bearing the minor alleles of the rs174546 and rs968567 SNPs (TTT and TTC) were associated with higher levels of the D6D product C20:3n-6 (+20% and +33%, respectively) but only TTC was consistently associated with higher D6D activity (+25%). All the haplotypes carrying the rs174546 minor allele (TCT, TCC, TTT, and TTC) were consistently associated with significantly lower levels of the D5D product C20:4n-6 (–26%, –25%, 22%, and –17%, respectively) and lower D5D activity (–33%, –24%, –38%, and –42%, respectively). Similar results were obtained when performing the haplotype analysis with rs174546 and rs968567 only (i.e., the best haplotype model with respect to D5D activity) (supplementary Table V). All the haplotypes carrying the rs174546 minor allele (TC and TT) were associated with higher levels of C18:2n-6 (precursor), lower levels of C20:4n-6 (product), and lower D5D activity (data not shown). Additionally, only the TT haplotype carrying the rs968567 minor allele was consistently associated with a higher D6D activity and higher levels of its product, C20:3n-6.
TABLE 4.
Haplotype frequencies and association with serum n-6 LCPUFAs, desaturase activities, lipids, and BMI
| Haplotype | CCT | CCC | TCT | TCC | TTT | TTC |
|---|---|---|---|---|---|---|
| Frequency | 0.605 | 0.091 | 0.061 | 0.077 | 0.113 | 0.052 |
| (n-6) LCPUFAs profile | ||||||
| C18:2n-6 (%) p* | 10.69 reference | 0.44 0.03 | 0.75 0.0009 | 1.64 <E-06 | 0.88 4.0E-06 | 0.59 0.05 |
| C18:3n-6 (%) p* | 0.047 reference | −0.003 0.83 | −0.007 0.05 | −0.009 0.03 | −0.008 0.04 | −0.006 0.37 |
| C20:3n-6 (%) p* | 1.43 reference | −0.06 0.31 | 0.20 0.0001 | −0.04 0.46 | 0.28 <E-06 | 0.47 <E-06 |
| C20:4n-6 (%) p* | 5.22 reference | −0.23 0.04 | −1.33 <E-06 | −1.31 <E-06 | −1.16 <E-06 | −0.90 <E-06 |
| Desaturase activities | ||||||
| D6D p* | 0.069 reference | −0.006 0.10 | 0.004 0.18 | −0.012 0.002 | 0.007 0.023 | 0.017 3.0E-05 |
| D5D p* | 1.91 reference | −0.04 0.64 | −0.63 <E-06 | −0.46 3.0E-05 | −0.72 <E-06 | −0.81 <E-06 |
| Lipids and BMI | ||||||
| Total cholesterol (mmol/l) p* | 2.08 reference | 0.09 0.12 | 0.02 0.82 | −0.07 0.14 | −0.06 0.18 | 0.03 0.67 |
| HDL-cholesterol (mmol/l) p* | 0.72 reference | 0.03 0.14 | −0.03 0.23 | −0.01 0.59 | −0.02 0.22 | 0.00 0.98 |
| LDL-cholesterol (mmol/l) p* | 1.21 reference | 0.06 0.24 | 0.06 0.37 | −0.06 0.29 | 0.00 0.93 | 0.06 0.36 |
| TG (mmol/l) p* | 0.39 reference | 0.01 0.76 | 0.11 0.0002 | −0.04 0.28 | −0.01 0.59 | 0.06 0.13 |
| BMI (kg/m2) p* | 10.68 reference | 0.32 0.29 | −0.45 0.24 | −0.07 0.79 | 0.07 0.77 | −0.38 0.38 |
Order of htSNPs used for the analysis: rs174546, rs968567, and rs174602. Minor alleles for each SNP are underlined. Values are the mean effects estimated for each haplotype by comparison to the reference haplotype combining the most frequent allele at each site. (C20:3n-6):(C18:2n-6) and (C20:4n-6):(C20:3n-6) ratios were used as estimates of D6D and D5D desaturase activities, respectively (22). *P-values were adjusted for age, gender, BMI, and center.
Regarding the n-3 LCPUFA (data not shown), two haplotypes carrying both the rs174546 and rs968567 minor alleles (TT T and TTC) were associated with higher precursor (C18:3 n-3) levels (+30%, P = 4.8E-5 and +71%, P = 6.0E-6, respectively). However, only the T CC haplotype, which was associated with a lower D5D activity, was consistently associated with lower C20:5n-3 and C22:6 n-3 (products) levels (−20%, P = 6.4E-5 and –19%, P = 5.1E-5, respectively).
Because significant associations between FADS haplotypes and LCPUFA levels were detected, the haplotype analysis was next extended to lipid levels and BMI (Table 4). None of the haplotypes was associated with total cholesterol, HDL-cholesterol, or LDL-cholesterol levels or BMI. In contrast, the haplotype carrying the minor allele of the rs174546 SNP (TCT) was significantly associated with higher TG levels (P = 1.8E-4).
The present study revealed the presence of strong associations between nine SNPs in the FADS cluster and D5D activity determined as the ratio of C20:4n-6 to C20:3n-6 in serum phospholipids. Haplotypes carrying the rs174546 minor allele were consistently associated with both lower C20:4n-6 levels and lower D5D activity, suggesting that rs174546 could be in linkage disequilibrium with a functional SNP that affects FADS1. Furthermore, we found that the minor allele of rs968567 was associated with higher levels of D6D activity determined as the ratio of C20:3n-6 to C18:2n-6 in serum phospholipids. These data suggest that the genetic variability in the FADS gene cluster that affects LCPUFA levels is explained by both higher D6D activity and lower D5D activity in European adolescents.
The present study showed that the minor alleles of the rs174546, rs968567, rs174570, rs174572, rs2072114, rs174589, rs174602, rs174611, and rs174616 SNPs were associated with higher levels of C18:2n-6 and lower levels of C20:4n-6. Previous studies have typically used the C20:4n-6/C18:2n-6 ratio as an index of overall desaturase activity without distinguishing between D6D and D5D activities (11, 14). Using the C20:3n-6/C18:2n-6 and C20:4n-6/C20:3n-6 ratios, we were able to estimate the D6D and D5D activities separately (22). We found that the minor alleles of all SNPs except rs498793 (which is in negative linkage disequilibrium with the others) and rs526126 were associated with lower D5D activity. Only rs968567 was associated with higher D6D activity.
Of the nine SNPs associated with D5D activity, the largest effects were observed for rs174546 and rs968567 (β = −0.63 and −0.62, respectively). All the SNPs are in partial linkage disequilibrium and are located within an 80 kbp haplotype block containing both FADS1 and FADS2. Hence, these associations very likely reflect the same signal. Haplotypes carrying the rs174546 minor allele were consistently associated with lower D5D activity. It is noteworthy that the latter SNP is the only one located within FADS1 (3′ untranslated region). The rs174546 SNP may be in linkage disequilibrium with a functional SNP affecting D5D activity. For example, rs174546 is in strong linkage disequilibrium (r2 > 0.80) with the rs3834458 [T/del] promoter SNP (8). In an in vitro study of a patient with D6D deficiency, the presence of the T nucleotide was associated with a ∼6-fold lower promoter activity and D6D activity and lower plasma C20:4n-6 levels (28). However, this finding was not replicated in a recent study (23), suggesting that the earlier finding could be spurious, as also suggested by Baylin et al. (13). The rs174546 SNP is also in complete linkage disequilibrium with rs174547 (r2 = 1, according to HapMap CEU), a SNP located within FADS1 and the minor allele of which has been associated with lower FADS1 transcript levels in the human liver (17).
In contrast, only the rs968567 minor allele was associated with higher D6D activity. This SNP is located 299 bp upstream of the FADS2 translation start site. Our findings are consistent with an earlier report of similar significant associations between the minor allele of rs968567 and higher C18:2n-6 and C20:3n-6 levels and lower C20:4n-6 levels but which lacked data on D6D activity (8). Our haplotype analysis indicated that one haplotype carrying the rs968567 minor allele was associated with higher D6D activity. This association agrees with the results of a functional study showing that the rs968567 minor allele was linked to a 2- to 3-fold higher FADS2 promoter activity in HeLa and HepG2 cells (23).
Strong associations between the FADS cluster and desaturase activities estimated by n-6 FA ratios were shown, raising the question of possible consequences on the n-3 FA levels. The associations between FADS SNPs and n-3 LCPUFA were consistent with those observed with n-6 LCPUFA. Indeed, the minor alleles of three SNPs (rs174546, rs174572, and rs174589) were individually associated with much higher levels of C18:3n-3 (precursor) and, to a lesser extent, with lower levels of C20:5n-3 (product). A possible interpretation of this difference relates to the impact of dietary n-6 FA. Given that C18:2n-6 and C18:3n-3 compete for D6D and D5D and that the dietary intake of C18:2n-6 is by far much higher than that of n-3, the enzymes are likely to be saturated with n-6 FA, and this situation would facilitate the expression of a genetic defect in the n-6 pathway. In contrast, the C18:3n-3 levels markedly accumulate when both the competing effect of dietary C18:2n-6 and the genetic defect interact.
The present study also agrees with the earlier data showing associations between several SNPs in the FADS cluster and plasma, erythrocyte, breast milk, and adipose tissue LCPUFA levels in adults (8–15) and extends these observations to European adolescents. Previous data (including those from genome-wide association studies) have also revealed associations between SNPs in the FADS1-2 gene cluster on one hand and lower levels of HDL- (17, 18) and LDL-cholesterol (9, 16–19) and higher TG levels on the other (13, 16–18). In the present study, a haplotype analysis revealed a significant association between the rs174546 minor allele and higher TG levels in agreement with the literature on adults (13, 16–18). In contrast, we found no evidence for such associations for LDL- and HDL-cholesterol in adolescents despite a statistical power of 99% and 63%, respectively (see Methods). These results suggest that the impact of FADS SNPs on cholesterol metabolism might be lower in this age group than in adults. However, it must be acknowledged that adolescence may not be the optimal life period to analyze the plasma lipid/lipoprotein profile, because significant changes in these parameters occur during the transition from late childhood through adolescence (29). Therefore, very large genetic association studies or meta-analyses are required to fully explore the impact of FADS1-2 SNPs on lipid/lipoprotein metabolism in children.
This first association study of FADS cluster in adolescents had several strengths and limitations. Because the influence of behavioral and exogenous factors is less marked in adolescents than in adults, SNPs have a proportionally greater influence on the phenotype; hence, our study was more likely to reliably reveal genetic associations. Moreover, we assessed the impact of 11 tag-SNPs in order to cover the cluster's whole, common, genetic variability. Nevertheless, the selection of tag-SNPs with a MAF > 0.10 may prevent the identification of rare alleles with potentially stronger influences. For the first time, this study separately reports the association of FADS cluster polymorphisms with D6D and D5D activities. However, we used the ratio between individual serum FAs as surrogate of desaturase activities, because direct measures of enzyme activities are not feasible in large epidemiological studies. Although the study was performed in 10 centers from 9 countries, we found no evidence for heterogeneity of the association between the SNPs and the biochemical and anthropometric variables (data not shown). Another potential limitation relates to the fact that the dietary FA status was not recorded in the HELENA study. However, in an earlier study (13), dietary FAs did not differ significantly according to the FADS genotype. Lastly, our results would benefit from replication with an independent sample of adolescents.
In conclusion, our results strongly suggest that the SNPs that contribute to inter-individual variability in serum LCPUFA levels may affect not only D6D but also D5D activity. In parallel with recent data that have shown an influence of rs968567 in FADS2 gene transcription, our results also provide a rationale for performing additional functional studies on the FADS1 gene.
Supplementary Material
Acknowledgments
The authors acknowledge Dr. Louisa Goumidi for her help with haplotype analyses.
Footnotes
Abbreviations:
- BMI
- body mass index
- D5D
- delta-5 desaturase
- D6D
- delta-6 desaturase
- LCPUFA
- long-chain PUFA
- SNP
- single nucleotide polymorphism
- TG
- triglyceride
The HELENA Study receives funding from the European Union's Sixth RTD Framework Program (contract FOOD-CT-2005-007034), the Spanish Ministry of Education (EX-2007-1124, AGL2007-29784-E/ALI; AP-2005-3827), Universidad Politécnica de Madrid (CH/018/2008), and Cognis GmbH (Germany). The writing group takes sole responsibility for the content of this article and the European Union is not liable for any use that may be made of the information contained herein. This work was supported by the Conseil Régional du Nord-Pas de Calais, ERDF (European Regional Development Fund) in the frame of the CPER Cardio-diabete (convention 09220016).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five tables.
REFERENCES
- 1.Sampath H., Ntambi J. M. 2005. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu. Rev. Nutr. 25: 317–340. [DOI] [PubMed] [Google Scholar]
- 2.Jump D. B., Clarke S. D., Thelen A., Liimatta M., Ren B., Badin M. V. 1997. Dietary fat, genes, and human health. Adv. Exp. Med. Biol. 422: 167–176. [DOI] [PubMed] [Google Scholar]
- 3.Germano M., Meleleo D., Montorfano G., Adorni L., Negroni M., Berra B., Rizzo A. M. 2007. Plasma, red blood cells phospholipids and clinical evaluation after long chain omega-3 supplementation in children with attention deficit hyperactivity disorder (ADHD). Nutr. Neurosci. 10: 1–9. [DOI] [PubMed] [Google Scholar]
- 4.Calder P. C. 2006. Polyunsaturated fatty acids and inflammation. Prostaglandins Leukot. Essent. Fatty Acids. 75: 197–202. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura M. T., Nara T. Y. 2004. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 24: 345–376. [DOI] [PubMed] [Google Scholar]
- 6.Cho H. P., Nakamura M., Clarke S. D. 1999. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J. Biol. Chem. 274: 37335–37339. [DOI] [PubMed] [Google Scholar]
- 7.Stoffel W., Holz B., Jenke B., Binczek E., Gunter R. H., Kiss C., Karakesisoglou I., Thevis M., Weber A. A., Arnhold S., et al. 2008. Delta6-Desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 27: 2281–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaeffer L., Gohlke H., Muller M., Heid I. M., Palmer L. J., Kompauer I., Demmelmair H., Illig T., Koletzko B., Heinrich J. 2006. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum. Mol. Genet. 15: 1745–1756. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T., Shen J., Abecasis G. R., Kisialiou A., Ordovas J. M., Guralnik J. M., Singleton A., Bandinelli S., Cherubini A., Arnett D., et al. 2009. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 5: e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rzehak P., Heinrich J., Klopp N., Schaeffer L., Hoff S., Wolfram G., Illig T., Linseisen J. 2009. Evidence for an association between genetic variants of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster and the fatty acid composition of erythrocyte membranes. Br. J. Nutr. 101: 20–26. [DOI] [PubMed] [Google Scholar]
- 11.Xie L., Innis S. M. 2008. Genetic variants of the FADS1 FADS2 gene cluster are associated with altered (n-6) and (n-3) essential fatty acids in plasma and erythrocyte phospholipids in women during pregnancy and in breast milk during lactation. J. Nutr. 138: 2222–2228. [DOI] [PubMed] [Google Scholar]
- 12.Truong H., DiBello J. R., Ruiz-Narvaez E., Kraft P., Campos H., Baylin A. 2009. Does genetic variation in the Delta6-desaturase promoter modify the association between alpha-linolenic acid and the prevalence of metabolic syndrome? Am. J. Clin. Nutr. 89: 920–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baylin A., Ruiz-Narvaez E., Kraft P., Campos H. 2007. alpha-Linolenic acid, Delta6-desaturase gene polymorphism, and the risk of nonfatal myocardial infarction. Am. J. Clin. Nutr. 85: 554–560. [DOI] [PubMed] [Google Scholar]
- 14.Martinelli N., Girelli D., Malerba G., Guarini P., Illig T., Trabetti E., Sandri M., Friso S., Pizzolo F., Schaeffer L., et al. 2008. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am. J. Clin. Nutr. 88: 941–949. [DOI] [PubMed] [Google Scholar]
- 15.Malerba G., Schaeffer L., Xumerle L., Klopp N., Trabetti E., Biscuola M., Cavallari U., Galavotti R., Martinelli N., Guarini P., et al. 2008. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 43: 289–299. [DOI] [PubMed] [Google Scholar]
- 16.Plaisier C. L., Horvath S., Huertas-Vazquez A., Cruz-Bautista I., Herrera M. F., Tusie-Luna T., Aguilar-Salinas C., Pajukanta P. 2009. A systems genetics approach implicates USF1, FADS3, and other causal candidate genes for familial combined hyperlipidemia. PLoS Genet. 5: e1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aulchenko Y. S., Ripatti S., Lindqvist I., Boomsma D., Heid I. M., Pramstaller P. P., Penninx B. W., Janssens A. C., Wilson J. F., Spector T., et al. 2009. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 41: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabatti C., Service S. K., Hartikainen A. L., Pouta A., Ripatti S., Brodsky J., Jones C. G., Zaitlen N. A., Varilo T., Kaakinen M., et al. 2009. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 41: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno L. A., De Henauw S., Gonzalez-Gross M., Kersting M., Molnar D., Gottrand F., Barrios L., Sjostrom M., Manios Y., Gilbert C. C., et al. 2008. Design and implementation of the Healthy Lifestyle in Europe by Nutrition in Adolescence Cross-Sectional Study. Int J Obes (Lond). 32(Suppl. 5): S4–S11. [DOI] [PubMed] [Google Scholar]
- 21.Fan Y. Y., Chapkin R. S. 1998. Importance of dietary gamma-linolenic acid in human health and nutrition. J. Nutr. 128: 1411– 1414. [DOI] [PubMed] [Google Scholar]
- 22.Warensjo E., Rosell M., Hellenius M. L., Vessby B., De Faire U., Riserus U. 2009. Associations between estimated fatty acid desaturase activities in serum lipids and adipose tissue in humans: links to obesity and insulin resistance. Lipids Health Dis. 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lattka E., Eggers S., Moeller G., Heim K., Weber M., Mehta D., Prokisch H., Illig T., Adamski J. 2010. A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J. Lipid Res. 51: 182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tregouet D. A., Ricard S., Nicaud V., Arnould I., Soubigou S., Rosier M., Duverger N., Poirier O., Mace S., Kee F., et al. 2004. In-depth haplotype analysis of ABCA1 gene polymorphisms in relation to plasma ApoA1 levels and myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 24: 775–781. [DOI] [PubMed] [Google Scholar]
- 25.Akaike H. 1974. A new look at the statistical model identification. IEEE Trans. Autom. Control. 19: 716–723. [Google Scholar]
- 26.Tregouet D. A., Escolano S., Tiret L., Mallet A., Golmard J. L. 2004. A new algorithm for haplotype-based association analysis: the Stochastic-EM algorithm. Ann. Hum. Genet. 68: 165–177. [DOI] [PubMed] [Google Scholar]
- 27.Gauderman W. J., Morrison J. M. 2006. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. Internet: http://hydra.usc.edu/gxe.
- 28.Nwankwo J. O., Spector A. A., Domann F. E. 2003. A nucleotide insertion in the transcriptional regulatory region of FADS2 gives rise to human fatty acid delta-6-desaturase deficiency. J. Lipid Res. 44: 2311–2319. [DOI] [PubMed] [Google Scholar]
- 29.Moran A., Jacobs D. R., Jr, Steinberger J., Steffen L. M., Pankow J. S., Hong C. P., Sinaiko A. R. 2008. Changes in insulin resistance and cardiovascular risk during adolescence: establishment of differential risk in males and females. Circulation. 117: 2361–2368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.