Abstract
Obese individuals are both insulin resistant and have high levels of circulating free fatty acids (FFAs). In cell culture, saturated but not unsaturated fatty acids induce endoplasmic reticulum (ER) stress. We hypothesized that chronic exposure to low dose fatty acids would significantly attenuate the acute stress response to a saturated fatty acid challenge and that unsaturated fatty acids (oleate) would be more protective than saturated fatty acids (palmitate). The ER stress response to palmitate was reduced after low dose fatty acid exposure in human hepatoma cells. Palmitate and oleate gave distinctive transcript responses, both acutely and after chronic low dose exposure. Differentially regulated pathways included lipid, cholesterol, fatty acid, and triglyceride metabolism, and IκB kinase and nuclear factor κB kinase inflammatory cascades. Oleate reduced palmitate-induced changes significantly more than low dose palmitate and completely blocked palmitate-induced phosphoinositide 3 kinase inhibitor (PIK3IP1) as well as induction of GADD45A and B. These changes are predicted to alter the PI3 kinase pathway and the pro-apoptotic p38 MAPK pathway. We recapitulated the oleate response by small interfering RNA-mediated block of PIK3IP1 stimulation with palmitate and significantly protected cells from palmitate-mediated ER stress. We show that transcriptional responses to oleate and palmitate are distinct, broad, and often discordant. We identified several potential candidates that may direct the transcriptional networks and demonstrate that PIK3IP1 partially accounts for the protective effects of oleate.
Keywords: fatty acid, transcription, microarray, enrichment analysis, endoplasmic reticulum stress
Obesity is the most significant risk factor for impaired glucose homeostasis and type 2 diabetes and has reached epidemic proportions worldwide (1). Impaired insulin action with increasing body mass index is thought to be the etiology of altered glucose homeostasis, but the reason for that impaired insulin action remains obscure. One possible explanation is the elevation in FFA, which is observed in obese and insulin-resistant individuals (2–5). Consistent with this model, experimentally increased FFA in animals and humans reduces peripheral insulin action (6, 7). In a high-fat-fed dog model, insulin resistance developed with increases in both basal and nocturnal FFA levels approaching 1 mM (8), a level that routinely induces cellular stress in vitro. This model suggests that both basal FFA elevations and intermittent increases in FFA from meals and the nocturnal FFA rise contribute to insulin resistance.
FFA may alter multiple cellular processes, including transcription, translation, enzymatic activities, cellular signaling, ion homeostasis, membrane fluidity, and cell survival (9). Several recent studies have demonstrated that FFA induce the endoplasmic reticulum (ER) stress response in tissue culture (10, 11) and in animal studies (12, 13). ER stress responses in liver and adipose have been proposed as the link between obesity and insulin resistance (13, 14). In cell culture-based studies, long-chain saturated fatty acids such as palmitate (C16:0) and stearate (C18:0) induce ER stress at concentrations comparable to total FFA concentrations observed in obese humans, whereas the equivalent monounsaturated fatty acids (palmitolate, C16:1 and oleate, C18:1) do not induce ER stress (15–17) and may be protective (18, 19). The mechanism of this differential effect is unknown (14). However, whereas human and animal exposure is characterized by chronically elevated fasting FFA punctuated by acute postprandial and nocturnal increases, published cell culture experiments were based on acute FFA exposure. Recent studies demonstrated that chronic exposure of cells to low levels of the classic ER stress inducers tunicamycin and thapsigargin were protective of ER stress response and apoptosis induced by subsequent acute challenge (20). A similar model of long-term FFA exposure followed by an acute challenge would more closely recapitulate the physiologic situation, but whether chronic FFA elevations are toxic or protective is, to our knowledge, unknown and untested.
Based on these considerations, we hypothesized that chronic exposure to low levels of FFA would similarly attenuate the acute response to FFA levels comparable to those observed physiologically in the postprandial state. Furthermore, we hypothesized that low levels of unsaturated FFA would be more protective than saturated FFA. Finally, we sought to define the molecular mechanisms behind the differential response to saturated and unsaturated FFA. To test these hypotheses, we developed an in vitro system based on the human HepG2 hepatoma cell line in which cells were adapted in the presence of low doses of either palmitate (C16:0) or oleate (C18:1). We then challenged cells acutely with high doses of either FFA. After demonstrating the differential induction of ER stress, we explored the molecular basis of the protection using genome-wide transcript profiling. We further used genome wide transcript profiles to determine whether simultaneous exposure to oleate in the presence of palmitate induced the same protective response observed in the adaptation experiments. Finally, we demonstrated that the protective response of oleate is mediated in part by downregulation of the phosphoinositide 3 kinase inhibitor (PIK3IP1) by recapitulating the oleate effect with small interfering RNA (siRNA)-mediated knock-down of the PIK3IP1 gene.
MATERIALS AND METHODS
Experimental reagents
Palmitic and oleic acids were conjugated to fatty acid-free BSA as described (10, 18) (supplementary Data and Methods).
Cell culture
Human hepatoma (HepG2; American Type Culture Collection, Manassas, VA) cells were cultured in 5.6 mM glucose under standard conditions. For adaptation experiments, cells were grown for two passages in the presence of 0.25 mM oleate, 0.25 mM palmitate, or BSA (control) with exchange of culture medium daily to maintain the FFA concentration. Because palmitate-treated cells grew more slowly, more cells were seeded to achieve similar confluence. At 80–90% confluence in the third passage (treatment day 14), cells were washed and transferred to serum-free medium containing either 0.25 mM or 1 mM FFA (BSA-conjugated oleate or palmitate) or BSA control (Fig. 1). Cells were harvested after 6 h. Our fatty acid adaptive stress model was based on the adaptive stress model in MEF cells using chemically induced ER stress (tunicamycin and thapsigargin) (20). We selected an acute FFA dose that significantly induced ER stress and resulted in JNK1 and c-JUN phosphorylation by 6 h based on prior studies in our laboratory, which were within the observed physiologic range of total FFA in obese humans. We selected 0.25 mM FFA as an adaptive dose, because it does not induce ER stress in HepG2 cells at this time point. In a separate experiment, we treated HepG2 cells with palmitate, oleate, BSA control, or a mixture of palmitate and oleate for 12 h.
Fig. 1.
Experimental design. T25 and T75, 25 ml and 75 ml cell culture flasks; 6W, 6-well cell culture plate.
Inhibitor treatment
The PI3 kinase inhibitor LY294002 was dissolved in DMSO and used in a final concentration of 25–50 μM. The LY294002 dose was based on published literature and vendor recommendations. Cells were pretreated with LY294002 or DMSO (vehicle control) for 1 h and then treated with palmitate (0.5 mM), a mixture of 0.5 mM palmitate and 0.25 mM oleate, or BSA (control) for 12 h in serum free condition.
siRNA targeting of PIK3IP1
HepG2 cells were transfected with 100 nM PIK3IP1 siRNA or a nontargeting siRNA pool (Dharmacon, Chicago, IL) using DharmaFECT 4 transfection reagent (Dharmacon) according to an optimized protocol modified based on the manufacturer's instructions. Further details of our transfection protocol are described in supplementary Methods. After 24 h, siRNA-transfected cells were treated with either palmitate (0.5 mM) or BSA (control) for 12 h under serum and antibiotic-free conditions. The cells were treated as indicated in the figure legends and processed for RNA isolation and quantitative real-time PCR to analyze transcript level expression of ER stress pathway genes.
RNA isolation
RNA was isolated as described previously (10) (supplementary Data and Methods).
Quantitative real-time PCR to analyze transcript level
Measurements were conducted in at least two independent experiments for each condition, each in turn with two biological replicates yielding between four and six measures for each condition. Real-time quantitative PCR was performed as described (10) (supplementary Methods).
Immunoblot analysis
Phosphorylation of eIF2α, JNK1, and C-Jun were analyzed by immunoblot (10, 21) normalized to β-actin (supplementary Methods) for at least three independent experiments, each with two technical replicates.
Genome wide expression analysis
Genome wide transcriptome analysis and initial array processing was performed by GenUs Biosystems (Northbrook, IL) using Human Whole Genome 4 × 44k arrays (Agilent Technologies, Santa Clara, CA). We included genes in subsequent analyses if they were both above background and differed by <40% between biological replicates. Fold change was compared with BSA control. Additional details are provided in supplementary Methods.
Statistical and bioinformatic analysis
Differences between responses for quantitative real-time PCR and immunoblot were evaluated by Student's t-test, with a P < 0.05 considered significant. We used PermutMatrix version 1.9.3EN software (22) for unsupervised hierarchical clustering. Differentially expressed genes were categorized and annotated using singular and modular enrichment analysis implemented in the DAVID package (23, 24) (http://david.abcc.ncifcrf.gov/), Gene Set Enrichment Analysis (GSEA) using GeneTrail software (25) (http://genetrail.bioinf.uni-sb.de/), and pathway interaction network analysis using Ingenuity software v 7.1 (https://analysis.ingenuity.com). Statistical methods are detailed in the supplementary Methods.
RESULTS
Short-term oleate and palmitate induce unique gene expression profiles
We showed previously that palmitate but not oleate at comparable levels induced genes of the ER stress pathway (10). In confirmation of our previous studies (10), treatment of HepG2 cells with 1 mM palmitate for 6 h elevated the transcript levels of genes downstream of the PERK (EIF2AK3)-eIF2α ER stress pathway, including ATF3 (P = 0.009), DDIT3 (CHOP, P = 0.006), and PPP1R15A (GADD34, P = 0.014). Activation of the IRE1α (ERN1)-XBP1 ER stress pathway was also observed, with induction of ERN1 (P = 0.027) and splicing of XBP1. Palmitate also induced expression of STC2 (P = 0.037) and NRF2 (NFE2L2, P = 0.012) but not the major ER stress chaperone, HSPA5 (BiP/GRP78). Phosphorylation of eIF2α, JNK1, and c-JUN was induced by palmitate. In contrast, oleate did not induce the ER stress genes.
To expand this work, we compared the global gene expression response in HepG2 cells to palmitate (1 mM), oleate (1 mM), or BSA control. Palmitate induced 513 genes at least a 1.5-fold and reduced expression of 263 genes by at least 1.5-fold (supplementary Data and Table I). The most strongly upregulated gene was ATF3 (10.3-fold increased), whereas oxysterol binding protein 2 was 2.5-fold decreased. In contrast, 1 mM oleate upregulated only 51 transcripts but downregulated 306 unique genes (supplementary Data and Table II). Of 19 genes with ≥1.5-fold change by both palmitate and oleate, 4 were concordantly upregulated, 10 were concordantly downregulated, and 5 were discordant including the key ER stress response gene, stanniocalcin (supplementary Data and Table III).
We considered all probes showing a 1.5-fold or greater change with either palmitate or oleate (1531 probes). Hierarchical clustering showed distinctive profiles (supplementary Data; Fig. 1A). Modular enrichment analysis (23, 24) showed that palmitate altered pathways of apoptosis, inflammatory and stress response, oxidative stress, NFκβ activation, JAK-STAT signaling, and MAPK signaling (Table 1), whereas these pathways were not involved in the oleate response (supplementary Data and Table IV). In contrast, oleate primarily altered pathways of rhodopsin-like receptor (G-protein coupled receptor) activity, oxidative phosphorylation, and sterol and lipid biosynthesis.
TABLE 1.
Modular enrichment analysis of genes differentially expressed by acute palmitate exposure
| Functional Group | Description | No. of Category | Pa | Pb | No. of Genes in Group |
|---|---|---|---|---|---|
| 4 | Apoptosis (programmed cell death) | 10 | 6.14 × 10−5 | 9.86 × 10−5 | 111 |
| 10 | Response to stress including inflammatory response | 5 | 8.13 × 10−3 | 5.49 × 10−3 | 82 |
| 16 | Oxidative stress-induced gene expression via Nrf2 and MafK | 3 | 1.02 × 10−2 | 1.74 × 10−2 | 6 |
| 18 | Activation of NFκB transcription factor | 3 | 1.47 × 10−2 | 2.66 × 10−2 | 8 |
| 19 | JAK-STAT cascade | 8 | 4.22 × 10−2 | 2.83 × 10−2 | 17 |
| 22 | SOCS protein and JAK-STAT cascade | 7 | 4.40 × 10−2 | 4.78 × 10−2 | 17 |
| 23 | MAP kinase phosphatase activity | 23 | 4.16 × 10−2 | 4.90 × 10−2 | 21 |
Data are for HepG2 cells treated with 1 mM palmitate for 6 h and compared with controls (BSA). We show selected DAVID functional annotation cluster analysis groups with a geometric mean P-value ≤ 0.05.
Median P-value of all categories in a functional group.
Geometric mean P-value of all categories in a functional group.
GSEA considers all expressed genes by rank without a fold-change threshold (25) and thus may identify altered pathways in which no individual members are altered by 1.5-fold. Indeed, GSEA identified additional palmitate-induced changes in pathways of oxidative phosphorylation, fatty acid and lipid synthesis, lipid binding, and lipid transport (Table 2; supplementary Data and Table V). Again, oleate and palmitate acted on distinct pathways, with only 4 of 25 gene ontology (GO) categories showing concordant responses (Table 2). Whereas GO categories involving oxidative phosphorylation, mitochondrial electron transport, and lipid biosynthesis were all downregulated by palmitate, they were upregulated by oleate (Table 2). Notably, oleate and palmitate had strikingly different profiles for genes involved in oxidative phosphorylation (supplementary Data and Fig. IB).
TABLE 2.
Comparison of gene categories by GSEA between acute palmitate and oleate exposure in HepG2
| Palmitate |
Oleate |
||||
|---|---|---|---|---|---|
| Category Description | Category ID | Observed | Pa | Observed | Pa |
| Concordant: upregulated ↑ | |||||
| Ubiquitin-dependent protein catabolic process | GO:0006511 | 182 | 1.38 × 10−2 | 180 | 3.28 × 10−2 |
| Discordant: palmitate↓ oleate↑ | |||||
| Ribonucleoside triphosphate metabolic process | GO:0009199 | 42 | 1.56 × 10−2 | 43 | 3.84 × 10−2 |
| Respiratory electron transport chain | GO:0022904 | 48 | 1.66 × 10−2 | 47 | 3.69 × 10−3 |
| Peroxidase activity | GO:0004601 | 21 | 2.73 × 10−2 | 19 | 1.39 × 10−2 |
| Oxidoreductase activity, acting on peroxide as acceptor | GO:0016684 | 21 | 2.73 × 10−2 | 19 | 1.39 × 10−2 |
| Oxidoreductase activity, acting on NADH or NADPH, quinine, or similar compound as acceptor | GO:0016655 | 45 | 3.80 × 10−2 | 44 | 7.46 × 10−3 |
| Oxidative phosphorylation | GO:0006119 | 65 | 1.74 × 10−4 | 65 | 1.38 × 10−3 |
| NADH dehydrogenase (ubiquinone) activity | GO:0008137 | 40 | 4.14 × 10−2 | 39 | 7.76 × 10−3 |
| Monovalent inorganic cation transmembrane transporter activity | GO:0015077 | 87 | 5.66 × 10−3 | 83 | 3.28 × 10−2 |
| Mitochondrial ATP synthesis coupled electron transport | GO:0042775 | 45 | 3.01 × 10−2 | 44 | 2.46 × 10−3 |
| Membrane lipid metabolic process | GO:0006643 | 175 | 3.71 × 10−2 | 173 | 3.83 × 10−2 |
| Lipid biosynthetic process | GO:0008610 | 225 | 9.27 × 10−3 | 220 | 2.69 × 10−2 |
| Ion channel activity | GO:0005216 | 164 | 1.24 × 10−2 | 147 | 4.40 × 10−2 |
| Inorganic cation transmembrane transporter activity | GO:0022890 | 120 | 2.23 × 10−3 | 109 | 4.89 × 10−2 |
| G-protein coupled receptor protein signaling pathway | GO:0007186 | 340 | 3.13 × 10−4 | 285 | 5.17 × 10−4 |
| ER part | GO:0044432 | 478 | 1.57 × 10−5 | 468 | 4.00 × 10−2 |
| Concordant: downregulated ↓ | |||||
| Regulation of small GTPase mediated signal transduction | GO:0051056 | 179 | 2.84 × 10−2 | 178 | 4.76 × 10−2 |
| Phosphoinositide phospholipase C activity | GO:0004435 | 13 | 8.16 × 10−3 | 11 | 4.00 × 10−2 |
| Inositol or phosphatidylinositol phosphodiesterase activity | GO:0004434 | 13 | 8.16 × 10−3 | 11 | 4.00 × 10−2 |
Selected enriched categories common between acute 1 mM palmitate and 1 mM oleate treatment for 6 h in HepG2 cells compared with controls.
Benjamini and Hochberg corrected P-value.
Low dose palmitate and oleate attenuate the ER stress response to high dose palmitate
Rutkowski et al. (20) demonstrated protective adaptation and reduced ER stress response after exposure to low dose tunicamycin and thapsigargin. We asked whether HepG2 cells adapt similarly to low levels of palmitate or oleate over several passages. Thus, cells were adapted to 0.25 mM oleate or palmitate followed by a 1 mM acute challenge (Fig. 1). Indeed, adaptation to either low dose palmitate or low dose oleate blunted the expected increase in ER stress transcripts and phosphorylation response (eIF2α, JNK1, c-Jun) in response to 1 mM palmitate (Fig. 2), with oleate providing more protection.
Fig. 2.
Fatty acid adaptation attenuates the ER stress response to palmitate in HepG2 cells. A, B: ER stress pathway response to palmitate with and without low dose palmitate preexposure. Data is shown as mean ± SD for 4–6 biological replicates. C: Reduction in XBP1 splicing with palmitate preexposure. D, E: ER stress response to palmitate with and without oleate preexposure. Data for transcript levels are shown as mean ± SD for 4–6 biological replicates. F: Oleate protects against increased XBP1 splicing induced by 1 mM palmitate. G: Representative Western blot showing stress-induced phosphorylation of eIF2α, JNK1, and cJUN with and without palmitate and oleate preexposure. H–J: Quantification of all data from experiments represented in the Western blot in G, expressed as ratio of phosphorylated to unphosphorylated proteins. H: eIF2α. I: JNK1. J: Phosphorylated cJun normalized to actin. Data for protein levels are shown as mean ± SD for three biological replicates. Differences between palmitate response in unadapted cells (lane 2) and cells adapted in 0.25 mM oleate (lane 8) were significant for all three graphs (H: P = 0.0003; I: P = 0.0003; J: P < 0.00001). Transcript expression is shown as the 18S normalized value. AU, arbitrary unit of protein expression. XBP1 transcripts amplified from cDNA by PCR (197 bp XBP1-unspliced and 171 bp XBP1-spliced) were examined by agarose gel electrophoresis.
Distinct adaptive responses to 0.25 mM oleate and palmitate
We compared next the global transcript response in HepG2 cells to chronic low dose (0.25 mM) palmitate or low dose oleate (0.25 mM). Whereas palmitate upregulated 724 genes and downregulated 1,035 genes at least 1.5-fold (supplementary Data and Table VI), oleate induced 1,669 genes and downregulated 1,535 genes (supplementary Data and Table VII). Surprisingly, these adaptive responses shared only 417 upregulated genes and 452 downregulated genes. Among the shared upregulated pathways were mitochondrial activity, electron transport, oxidative phosphorylation, oxidoreductase activity, and cell cycle regulation. Shared downregulated pathways included phospholipid binding, glycerol metabolism, cytokine binding and cytokine-cytokine receptor interaction, immune response, ER membrane genes, insulin receptor signaling pathway, and cell adhesion (Table 3). Among the discordantly regulated pathways were lipid metabolism (endothelial lipase, hydroxysteroid dehydrogenase like 2, steroidogenic acute regulatory protein, phospholipase A2 group IIA, apolipoprotein L3), the I-κB kinase/NF-κB cascades, fatty acid metabolism and long-chain fatty acid-CoA ligase activity, fatty acid biosynthesis, acute inflammatory response, and hypoxia response genes. As with the acute responses, the adaptive responses to 2 week exposure to 0.25 mM palmitate and oleate were distinct when the 5,953 differentially expressed probes were examined by hierarchical clustering (supplementary Data and Fig. II), despite the shared pathways.
TABLE 3.
GSEA of response to chronic palmitate and oleate in HepG2 cells
| Palmitate |
Oleate |
||||
|---|---|---|---|---|---|
| Category Description | Category Id | Observed | Pa | Observed | Pa |
| Concordant upregulated ↑ | |||||
| Mitochondrial ATP synthesis coupled electron transport | GO:0042775 | 43 | 1.30 × 10−5 | 45 | 9.54 × 10−3 |
| Oxidative phosphorylation | GO:0006119 | 63 | 2.05 × 10−5 | 66 | 1.28 × 10−2 |
| Oxidoreductase activity, | GO:0016655 | 43 | 6.09 × 10−6 | 45 | 3.68 × 10−3 |
| Plk1 cell cycle regulation | TRANSPATH:CH000001002 | 9 | 9.11 × 10−3 | 10 | 3.80 × 10−2 |
| Mitotic cell cycle checkpoint | GO:0007093 | 29 | 3.52 × 10−2 | 28 | 1.87 × 10−4 |
| M phase of mitotic cell cycle | GO:0000087 | 189 | <1.0 × 10−6 | 189 | <1.0 × 10−6 |
| Cellular response to stress | GO:0033554 | 292 | 1.74 × 10−6 | 295 | <1.0 × 10−6 |
| DNA topoisomerase activity | GO:0003916 | 6 | 3.90 × 10−2 | 6 | 2.74 × 10−2 |
| DNA replication initiation | GO:0006270 | 19 | 2.45 × 10−3 | 22 | 1.39 × 10−4 |
| DNA helicase activity | GO:0003678 | 34 | 1.84 × 10−2 | 31 | 5.10 × 10−4 |
| DNA damage response, signal transduction | GO:0042770 | 39 | 1.79 × 10−2 | 40 | 7.39 × 10−4 |
| Concordant downregulated ↓ | |||||
| Phospholipid binding | GO:0005543 | 105 | 2.76 × 10−2 | 104 | 3.42 × 10−2 |
| Cytokine-cytokine receptor interaction | KEGG:4060 | 74 | 3.02 × 10−2 | 71 | 2.64 × 10−4 |
| Cytokine binding | GO:0019955 | 39 | 3.17 × 10−3 | 38 | 2.03 × 10−2 |
| Immune response | GO:0006955 | 294 | 1.55 × 10−4 | 292 | 2.64 × 10−4 |
| ER membrane | GO:0005789 | 402 | 9.80 × 10−3 | 409 | <1.0 × 10−6 |
| Insulin signaling pathway | KEGG:4910 or GO:0008286 | 103 | 3.91 × 10−2 | 32 | 1.83 × 10−2 |
| Cell adhesion molecules (CAMs) | KEGG:4514 | 58 | 1.29 × 10−4 | 56 | 1.08 × 10−2 |
| Caspase activity | GO:0030693 | 15 | 4.28 × 10−2 | 14 | 1.41 × 10−2 |
| Carbohydrate phosphatase activity | GO:0019203 | 6 | 4.65 × 10−2 | 7 | 1.98 × 10−2 |
| Transmembrane receptor protein tyrosine kinase signaling pathway | GO:0007169 | 129 | 2.16 × 10−3 | 135 | 1.75 × 10−2 |
| G-protein coupled receptor activity | GO:0004930 | 188 | 2.07 × 10−2 | 158 | <1.0 × 10−6 |
| Discordant: palmitate ↑ and oleate↓ or ↔ | |||||
| Steroid biosynthetic process | GO:0006694 or KEGG:100 | 21 | 1.14 × 10−2 | 59 | 2.48 × 10−5 |
| Response to oxidative stress | GO:0006979 | 74 | 4.39 × 10−2 | NE | |
| Discordant: palmitate ↓ and oleate ↔ | |||||
| Regulation of IκB kinase/NFκB cascade | GO:0043122 | 81 | 6.06 × 10−3 | NE | |
| Phosphatidylinositol phosphate kinase activity | GO:0016307 | 9 | 1.55 × 10−2 | NE | |
| Discordant: palmitate ↔ and oleate ↓ | |||||
| Glycerolipid metabolic process | GO:0046486 | NE | 20 | 9.85 × 10−6 | |
| Lipid biosynthetic process | GO:0008610 | NE | 220 | 4.09 × 10−5 | |
| Fatty acid biosynthetic process | GO:0006633 | NE | 48 | 2.18 × 10−2 | |
| Long-chain-fatty-acid-CoA ligase activity | GO:0004467 | NE | 7 | 4.60 × 10−2 | |
| Cholesterol metabolic process | GO:0008203 | NE | 62 | 3.29 × 10−6 | |
| Triacylglycerol metabolic process | GO:0006641 | NE | 17 | 7.22 × 10−5 | |
| Acute inflammatory response | GO:0002526 | NE | 48 | 1.00 × 10−2 | |
| Response to hypoxia | GO:0001666 | NE | 44 | 4.92 × 10−2 | |
| Discordant: palmitate ↔ and oleate ↑ | |||||
| Protein folding | GO:0006457 | NE | 153 | 2.08 × 10−2 | |
Selected enriched categories after 14 day exposure to 0.25 mM palmitate or 0.25 mM oleate in HepG2 cells compared with controls.
Benjamini and Hochberg corrected P-value. NE, no enrichment (↔).
Responses to 1 mM palmitate challenge after oleate and palmitate adaptation are distinct
We selected 203 probes that showed at least a 2-fold change with 6 h, 1 mM palmitate exposure in unadpated cells (189 increased, 14 decreased). Despite the similar effects on the narrower class of ER stress response transcripts, hierarchical clustering again showed distinct profiles after palmitate or oleate adaptation. Whereas palmitate-induced changes were reduced by at least 50% in 82 probes after oleate adaptation, palmitate adaptation similarly reduced the response by 50% or more in only 62 probes. This difference was readily apparent by hierarchical cluster analysis (Fig. 3A). Among the individual genes showing striking differences was PIK3IP1, which was induced by 2.6-fold by palmitate in unadapted cells. This response was reduced by 32% after palmitate adaptation but completely abolished by oleate adaptation. Table 4 lists other genes showing marked differences in response following oleate or palmitate adaptation.
Fig. 3.
Hierarchical clustering and heat maps of transcript profile in responses to oleate and palmitate. A: Hierarchical clustering with heat map of 203 probes that showed at least 2-fold change with 1 mM palmitate challenge with and without palmitate or oleate preexposure. B: Hierarchical clustering with heat map of 405 probes that showed at least a 2-fold change in expression after 12 h palmitate exposure (0.5 mM) when exposed to variable palmitate and oleate ratios.
TABLE 4.
Genes showing discordant or >50% difference in response between oleate and palmitate adaptation in HepG2 cells
| % Amelioration |
|||||
|---|---|---|---|---|---|
| Gene Symbol | Entrez Gene ID | Gene Name | Fold Changea | Palmitate | Oleate |
| PIK3IP1 | 113791 | Phosphoinositide-3-kinase interacting protein 1 | 2.58 | 32 | 100 |
| DUSP8 | 1850 | Dual specificity phosphatase 8 | 3.86 | 28 | 98 |
| GPR109B | 8843 | G protein-coupled receptor 109B | 3.21 | 2 | 91 |
| ZNF295 | 49854 | Zinc finger protein 295 | 2.12 | 5 | 84 |
| SLC25A25 | 114789 | Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 | 2.36 | 6 | 84 |
| GADD45B | 4616 | Growth arrest and DNA-damage-inducible, β | 2.77 | 21 | 74 |
| VNN2 | 8875 | Vanin 2 | 2.00 | −41 | 70 |
| SERPINB8 | 5271 | Serpin peptidase inhibitor, clade B (ovalbumin), member 8 | 2.29 | 9 | 63 |
| EGR1 | 1958 | Early growth response 1 | 5.66 | 3 | 56 |
| TUBB2B | 347733 | Tubulin, β 2B | 2.03 | −8 | 56 |
| SERPINE1 | 5054 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 2.10 | −54 | 48 |
| C10orf10 | 11067 | Chromosome 10 open reading frame 10 | 2.40 | −63 | 40 |
| TANC1 | 85461 | Tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | 3.08 | 88 | 38 |
| GATA6 | 2627 | GATA binding protein 6 | 2.60 | 108 | 31 |
| AREG | 374 | Amphiregulin (schwannoma-derived growth factor) | 3.66 | 91 | 25 |
| KLF5 | 688 | Kruppel-like factor 5 (intestinal) | 2.18 | 81 | 4.33 |
| IKBKE | 9641 | Inhibitor of κ light polypeptide gene enhancer in B-cells, kinase ϵ | 2.40 | 57 | −1 |
| DUSP2 | 1844 | Dual specificity phosphatase 2 | 3.31 | 50 | −4 |
| CYR61 | 3491 | Cysteine-rich, angiogenic inducer, 61 | 2.11 | 50 | −12 |
| THBS1 | 7057 | Thrombospondin 1 | 2.84 | 62 | −71 |
| SLC6A14 | 11254 | Solute carrier family 6 (amino acid transporter), member 14 | 2.24 | 37 | −73 |
Only genes with at least 2-fold upregulation by 1 mM palmitate exposure (6 h) in HepG2 cells and discordance between oleate and palmitate adaptation are shown.
Fold change is taken from the acute 1 mM response to palmitate compared with control (BSA).
We used GSEA to identify the pathways that characterized these responses. Oleate adaptation resulted in an attenuated palmitate response in pathways of cholesterol and triglyceride metabolism (GO categories GO 0008203 and GO 0006641 with 61 and 17 genes, respectively; corrected P < 2 × 10−5 for both). Oleate but not palmitate adaptation completely blocked the palmitate induction of 3-hydroxy-3-methylglutaryl-CoA synthase, mevalonate kinase, and phosphoenolpyruvate carboxykinase 1. A somewhat different picture emerged when we instead selected 23 genes from the interaction network that was most significantly upregulated by 1 mM palmitate (Ingenuity; https://analysis.ingenuity.com). This network included ATF3, CEBPB, DUSP1/8, GADD45A/B, JUN/JUNB/JUND, KLF5, LDLR, and LIPG (significance score 34; supplementary Data and Fig. III A). Oleate adaptation reduced the palmitate response for the 31 probes covering this network by an average 54% (P = 0.00003), with the most marked effect on genes upstream of the JNK and p38 pathways (DUSP1 and DUSP8 at 72% and 97% reduced response, respectively; supplementary Data and Fig. III B). In marked contrast, palmitate adaptation had no significant effect on the genes in this interactive network (P = 0.22).
Oleate protects against the saturated fatty acid response when mixed with palmitate
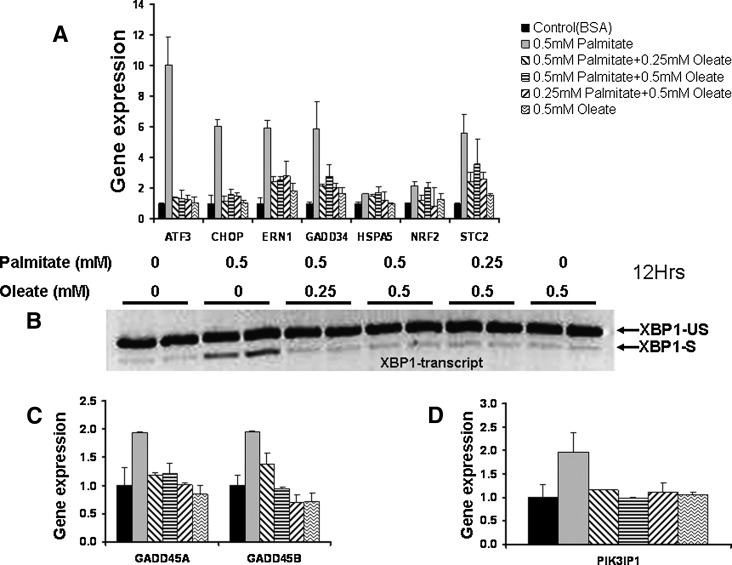
In contrast to the adaptation model, physiologic exposure to FFA involves a mixture of saturated and unsaturated fatty acids. Hence, we asked whether the preadaptation response was unique or whether unsaturated fatty acids could protect also against the saturated fatty acid stress response when mixed in an acute challenge. The ER stress response to 0.5 mM palmitate was nearly completely abolished by the addition of 0.25 mM oleate (Fig. 4). Additionally, oleate when added to palmitate attenuated the induction of GADD45A, GADD45B, and PI3KIP1. We considered the 405 transcripts that differed by at least 2-fold in response to 0.5 mM oleate and 0.5 mM palmitate (Fig. 3B). Oleate acted in a dose-dependent manner to attenuate the palmitate response, with 0.25 mM oleate reducing the response in 232/261 upregulated and 140/144 downregulated probes, and 0.5 mM oleate attenuating 260/261 upregulated and 143/144 downregulated transcript responses. The mean reduction in palmitate response when comparing 0.25 mM to 0.5 mM oleate was 54% versus 90% for upregulated and 29% versus 73% for downregulated transcripts (supplementary Data and Table VIII). Thus, oleate when mixed with palmitate provided similar protection to that observed when cells were adapted to the same oleate concentration for 14 days.
Fig. 4.
Effect of combined oleate and palmitate exposure on stress response in HepG2 cells. A: Changes in transcript profiles in response to 0.5 mM palmitate compared with oleate and mixed palmitate and oleate (see also supplementary Fig. V). B: XBP1 splicing response to palmitate and to mixed palmitate and oleate. C: Transcript level as in A for GADD45A and GADD45B. D: Transcript level of PIK3IP1, showing attenuated response in the presence of oleate. Gene expression is shown after normalization with 18S and considering control as 1. XBP-US, un-spliced XBP1 and XBP1-S, spliced XBP1. XBP1 transcripts amplified from cDNA by PCR (197 bp XBP1-unspliced and 171 bp XBP1-spliced) were examined by agarose gel electrophoresis. Data are shown as mean ± SD for four biological replicates.
PIK3IP1 mediates the palmitate-induced ER stress response
PIK3IP1 was induced by palmitate in unadapted cells. This induction was reduced by 32% after palmitate adaptation and completely abolished by oleate. PIK3IP1 was identified recently as a negative regulator of the p110 catalytic subunit of PI3 kinase (26), suggesting that downregulation of PIK3IP1 might mediate the protective effects of oleate. We first tested the effects of the pharmacologic PI3 kinase inhibitor LY294002 in cells treated with palmitate (0.5 mM) or palmitate and oleate (0.5 mM and 0.25 mM, respectively). As expected, LY294002 recapitulated the palmitate-induced ER stress and significantly reduced the oleate-mediated protection (supplementary Data and Fig. IV). In the presence of LY294002, proapoptotic factors CHOP (P = 0.004), ATF3 (P = 0.04), ATF4 (P = 0.02), and GADD34 (P = 0.04) were all increased in cells treated with both palmitate and oleate.
Based on evidence that PIK3IP1 may mediate the oleate response, we sought to recapitulate the oleate effect by siRNA-mediated silencing of the PIK3IP1 transcriptional response to palmitate. Palmitate-induced PIK3IP1 was significantly (P = 0.002) attenuated in HepG2 cells treated with PIK3IP1-siRNA compared with similar nontarget siRNA (Fig.5A and supplementary Fig. VI). Furthermore, siRNA-mediated reductions in PIK3IP1 significantly protected cells from palmitate-induced ER stress, with significant reductions in CHOP (P = 0.01), ATF3 (P = 0.01), and ATF4 (P = 0.04) (Fig. 5B). The palmitate-mediated XBP1 splicing was significantly attenuated (P = 0.001) by reduced PIK3IP1 (Fig. 5C). These results suggest that PIK3IP1 is an important mediator of the palmitate-induced ER stress response and part of the protective mechanism of oleate.
Fig. 5.
Silencing of the PIK3IP1 transcriptional response to palmitate protects cells against ER stress. A: Significant attenuation of palmitate-mediated induction of PIK3IP1 expression was achieved in HepG2 cells treated with PIK3IP1-siRNA compared with similar nontarget siRNA-treated cells. SiRNA-mediated knock-down of PIK3IP1 significantly attenuated palmitate-mediated ER stress response (B), including XBP1 splicing (C). XBP1 transcripts amplified from cDNA by PCR (197 bp XBP1-unspliced and 171 bp XBP1-spliced) were examined by agarose gel electrophoresis. Data are shown as mean ± SD for three biological replicates. NT-siRNA, nontargeting siRNA pool.
DISCUSSION
The principal aims of our study were to understand the differences in cellular response to saturated and unsaturated fatty acids and to determine whether the protective adaptation seen with preexposure to low levels of tunicamycin and thapsigargin could be replicated with FFAs. While the motivation for our studies was to mimic the exposure of liver to fatty acids in human obesity, we were necessarily limited to performing these experiments in human-derived cell lines and using nonphysiologic preparations of palmitate and oleate. Nonetheless, our results suggest important differences in oleate and palmitate responses. We have identified important pathways that are regulated in cells undergoing the adaptive response, and these pathways may reflect human physiology.
Previous studies have examined the cellular response to saturated and unsaturated fatty acids in HepG2 cells but at lower exposure levels. Vock et al. (27, 28) examined 0.2 mM oleate for 24 h and 50 μM palmitate. They identified only 14 genes altered by oleate and 11 genes altered by palmitate. Swagell et al. (29) found over 1.6-fold altered expression in 162 genes in the huh-7 human hepatocyte line after 48 h exposure to 150 μM palmitate. These experiments were similar in palmitate concentration and duration to the adaptive experiments performed in the present study, and in our hands, these concentrations did not induce markers of ER stress in HepG2 cells. In contrast, Li et al. (30) and Srivastava et al. (31) tested in HepG2 cells a 24 h exposure of 700 µM palmitate, oleate, and linoleate, a level that would induce ER stress and cytotoxicity with palmitate. Although these studies focused on the analytical approach, they reported fatty acid oxidation and electron transport as key markers of cytotoxicity and NADH dehydrogenase, mitogen activated protein kinases, extracellular signal regulated kinase, and JNK as potential regulators of cytotoxicity and lipid accumulation. Previous studies have not, however, evaluated the differences between chronic and acute exposure to saturated and unsaturated fatty acid or the adaptive response to prolonged, low level saturated or unsaturated fatty acid exposure and the effect on the response to a subsequent challenge with cytotoxic levels of saturated fatty acids.
We observed almost no overlap in the genes showing a 1.5-fold or greater response to palmitate when compared with oleate in HepG2 cells, thus suggesting distinct transcriptional responses. Notably, the transcript profiles we observed could be the result of altered gene transcription or altered mRNA stability; these mechanisms cannot be distinguished in steady-state measures of transcript levels and may both influence our observations. In the current manuscript, we have not attempted to distinguish the mechanism for differential transcript levels. Interestingly, GSEA analysis, which uses information from all expressed genes, showed differential expression of the same gene sets by both oleate and palmitate, but in distinctly opposite directions. Genes in electron transport chain and oxidative phosphorylation were upregulated by acute oleate exposure, whereas acute palmitate exposure downregulated these same processes. Given that protein folding in the ER is highly energy dependent, the downregulation of electron transport chain and oxidative phosphorylation by palmitate would be expected to reduce cellular ATP production and induce ER stress. Indeed, this model is supported by recent studies showing induction of ER stress response by chemicals that impair mitochondrial function by activating the JNK pathway (32); JNK inhibitors both ameliorated ER stress and restored mitochondrial function (32).
Palmitate exposure in HepG2 cells induced the PERK-eIF2α -ATF4 and IRE1-XBP1 arms of the ER stress pathways. Genes downstream of these arms were also upregulated, as were genes in other pathways (Table 1; supplementary Table V). Because of significant cross-talk between pathways, many differentially expressed genes overlap between pathways. This complex network of palmitate response is evident in our Ingenuity Pathway Analysis, which showed significant enrichment of 41 different canonical pathways, each in turn with 5–21 gene members (supplementary Data and Table IX). These 41 pathways together included only 92 differentially expressed genes, with 12 genes present in over 20 different pathways. The NFKB1 gene was a member of 37 of the 41 pathways. Taken together, these data suggest that a model based on a linear cascade of transcriptional regulation fails to capture the true complexity of the palmitate response. Instead, the response to saturated fatty acids including the ER stress response must be viewed in the context of the global transcriptional response network.
Rutkowski et al. (20) showed previously that activation of all three proximal activators of ER stress were attenuated by prior low dose exposure to thapsigargin or tunicamycin. They concluded that adaptation resulted from upregulation of the protein folding capacity, which in turn suppressed the unfolded protein response. Rutkowski et al. (20) used low doses of tunicamycin and thapsigargin that induced mild ER stress, challenged with the same dose, and showed that either tunicamycin or thapsigargin adaptation provided protection against ER stress induction by the other chemical. In contrast, we used a much lower concentration of oleate, which in itself does not induce ER stress, and palmitate, which does not induce ER stress at the adaptive doses used in this study, and we challenged with large doses of palmitate that induce a full ER stress response. In contrast to thapsigargin and tunicamycin, we did not observe the induction of ATF6 expression or of protein folding chaperones. Although fatty acid response appears to be distinct from the ER stress induced by tunicamycin and thapsigargin, we cannot exclude differences in the tissues studied (nonimmortalized MEF cells vs. immortalized HepG2 cells).
The mechanism by which monounsaturated, long-chain fatty acids protect against the saturated fatty acid-induced stress response is unknown, with limited previous data (11, 18). In the current study, oleate adaptation completely abolished the PIK3IP1 transcript response to palmitate in HepG2 cells. PIK3IP1 was recently identified by Zhu et al. (26) as a protein that shows homology to the regulatory p85 subunit of PI3K. PIK3IP1 binds to and negatively regulates the p110 subunit of PI3K, thus acting in place of the p85 regulatory subunit. PIK3IP1 overexpression reduced PI3K activity and thus Akt phosphorylation, whereas a reduction of PIK3IP1 increased PI3K activity and Akt phosphorylation (26). Inhibition of PI3K, including by increased PIK3IP1, may increase apoptosis in conditions of cellular stress (26). Hence, the silencing of the PIK3IP1 transcriptional response to palmitate by oleate pretreatment or simultaneous oleate/palmitate treatment may protect cells against saturated fatty acid-induced cellular stress and apoptosis. Several studies support this model. Beeharry et al. (33) also found that the protection afforded by lineoleic acid in RINm5F cells against palmitate-induced apoptosis was lost with PI3K inhibition. Gao et al. (34) showed that the beneficial effects of oleate on palmitate-induced insulin resistance in L6 myotubes was dependent on activation of PI3-kinase, likewise suggesting that oleate functions in part by maintaining insulin signaling through the PI3-kinase pathway. The siRNA-mediated knock-down of PIK3IP1 in the current study significantly protected HepG2 cells from palmitate-mediated ER stress. With our data, these studies strongly suggest that PIK3IP1 is one of the mediators of the palmitate-induced stress response and that adaptation acts in part to suppress this activation.
We also found evidence that a highly upregulated interaction network comprising 23 genes was involved in the adaptive response. This network includes GADD45A and GADD45B, which in turn bind to and activate MTK1 kinase, which is upstream of the JNK and p38 MAPK pathway. Thus, palmitate may act through GADD45A and GADD45B to induce apoptosis (35). Like PIK3IP1, palmitate-induced GADD45A and GADD45B transcription was reduced in HepG2 cells adapted first to oleate, thus providing another mechanism by which oleate protected against cellular stress responses and apoptosis. We made similar observations when palmitate and oleate were present simultaneously as a mixture.
Recently, palmitate conversion to lysophosphatidylcholine (LPC) but not ceramide was shown to cause apoptosis in hepatocytes (36). These authors suggested that oleate was protective by diverting palmitate from LPC to triglyceride formation. Other studies have shown that fatty acids that cannot be metabolized do not induce ER stress (37, 38). In the current study, palmitate upregulated genes involved in cholesterol and triglyceride metabolism, including HMGSC1, MVK, and PCK1. This upregulation was abolished by oleate pretreatment. Furthermore, the LPC acyltransferase 1 (AYTL2) gene was downregulated by palmitate (supplementary Data and Fig. V). Putting these observations together with the evidence that LPC may induce cellular stress, we propose another mechanism by which oleate may protect against palmitate-induced cellular stress. Oleate appears to alter saturated triglyceride metabolism, cholesterol metabolism, and LPC synthesis, which in turn may reduce the remodeling of the ER and mitochondrial membranes.
We have identified multiple important genes and pathways that are altered in cells undergoing an adaptive response to unsaturated fatty acids. These pathways may reflect the physiological response in humans and may provide targets to prevent lipotoxicity in the liver and β-cell. Our in vitro study validates the role of PIK3IP1 and the PI3 kinase pathway as mediators that partially account for the protective effects of oleate against palmitate-induced ER stress. Functional analyses of other novel pathways that have emerged from our study are required to further test the hypotheses generated by the transcriptional profiling reported here.
Supplementary Material
Acknowledgments
The authors thank Dr. Michael Falduto, Dr. Scott Magnuson, and Karen Torres of GenUs Biosystems Inc., Northbrook, IL, for their extensive help in microarray data acquisition and analysis. The authors also thank Winston Chu for helping in cell culture and maintenance.
Footnotes
Abbreviations:
- ER
- endoplasmic reticulum
- GO
- gene ontology
- GSEA
- Gene Set Enrichment Analysis
- LPC
- lysophosphatidylcholine
- PIK3IP1
- phosphoinositide 3 kinase inhibitor
- siRNA
- small interfering RNA
This work was supported by National Institutes of Health Grants DK-039311 and DK-071100). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. We also acknowledge the Research Service of the Department of Veteran's Affairs (Central Arkansas Veterans Healthcare System) and Research Enhancement Award Program (VA-REAP) for work completed at the John L. McClellan Veterans Hospital.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of data, methods, nine tables, and six figures.
REFERENCES
- 1.Ogden C. L., Carroll M. D., Curtin L. R., McDowell M. A., Tabak C. J., Flegal K. M. 2006. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 295: 1549–1555. [DOI] [PubMed] [Google Scholar]
- 2.Clore J. N., Allred J., White D., Li J., Stillman J. 2002. The role of plasma fatty acid composition in endogenous glucose production in patients with type 2 diabetes mellitus. Metabolism. 51: 1471–1477. [DOI] [PubMed] [Google Scholar]
- 3.Haus J. M., Kashyap S. R., Kasumov T., Zhang R., Kelly K. R., Defronzo R. A., Kirwan J. P. 2009. Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes. 58: 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kankaanpaa M., Lehto H. R., Parkka J. P., Komu M., Viljanen A., Ferrannini E., Knuuti J., Nuutila P., Parkkola R., Iozzo P. 2006. Myocardial triglyceride content and epicardial fat mass in human obesity: relationship to left ventricular function and serum free fatty acid levels. J. Clin. Endocrinol. Metab. 91: 4689–4695. [DOI] [PubMed] [Google Scholar]
- 5.Vessby B., Aro A., Skarfors E., Berglund L., Salminen I., Lithell H. 1994. The risk to develop NIDDM is related to the fatty acid composition of the serum cholesterol esters. Diabetes. 43: 1353–1357. [DOI] [PubMed] [Google Scholar]
- 6.Guilherme A., Virbasius J. V., Puri V., Czech M. P. 2008. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muoio D. M., Newgard C. B. 2008. Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9: 193–205. [DOI] [PubMed] [Google Scholar]
- 8.Kim S. P., Catalano K. J., Hsu I. R., Chiu J. D., Richey J. M., Bergman R. N. 2007. Nocturnal free fatty acids are uniquely elevated in the longitudinal development of diet-induced insulin resistance and hyperinsulinemia. Am. J. Physiol. Endocrinol. Metab. 292: E1590–E1598. [DOI] [PubMed] [Google Scholar]
- 9.Pegorier J. P., Le May C., Girard J. 2004. Control of gene expression by fatty acids. J. Nutr. 134: 2444S–2449S. [DOI] [PubMed] [Google Scholar]
- 10.Das S. K., Chu W. S., Mondal A. K., Sharma N. K., Kern P. A., Rasouli N., Elbein S. C. 2008. Effect of pioglitazone treatment on endoplasmic reticulum stress response in human adipose and in palmitate-induced stress in human liver and adipose cell lines. Am. J. Physiol. Endocrinol. Metab. 295: E393–E400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diakogiannaki E., Morgan N. G. 2008. Differential regulation of the ER stress response by long-chain fatty acids in the pancreatic beta-cell. Biochem. Soc. Trans. 36: 959–962. [DOI] [PubMed] [Google Scholar]
- 12.Ota T., Gayet C., Ginsberg H. N. 2008. Inhibition of apolipoprotein B100 secretion by lipid-induced hepatic endoplasmic reticulum stress in rodents. J. Clin. Invest. 118: 316–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozcan U., Cao Q., Yilmaz E., Lee A. H., Iwakoshi N. N., Ozdelen E., Tuncman G., Gorgun C., Glimcher L. H., Hotamisligil G. S. 2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 306: 457–461. [DOI] [PubMed] [Google Scholar]
- 14.Hotamisligil G. S., Erbay E. 2008. Nutrient sensing and inflammation in metabolic diseases. Nat. Rev. Immunol. 8: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo W., Wong S., Xie W., Lei T., Luo Z. 2007. Palmitate modulates intracellular signaling, induces endoplasmic reticulum stress, and causes apoptosis in mouse 3T3–L1 and rat primary preadipocytes. Am. J. Physiol. Endocrinol. Metab. 293: E576–E586. [DOI] [PubMed] [Google Scholar]
- 16.Karaskov E., Scott C., Zhang L., Teodoro T., Ravazzola M., Volchuk A. 2006. Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology. 147: 3398–3407. [DOI] [PubMed] [Google Scholar]
- 17.Lai E., Bikopoulos G., Wheeler M. B., Rozakis-Adcock M., Volchuk A. 2008. Differential activation of ER stress and apoptosis in response to chronically elevated free fatty acids in pancreatic beta-cells. Am. J. Physiol. Endocrinol. Metab. 294: E540–E550. [DOI] [PubMed] [Google Scholar]
- 18.Diakogiannaki E., Welters H. J., Morgan N. G. 2008. Differential regulation of the endoplasmic reticulum stress response in pancreatic beta-cells exposed to long-chain saturated and monounsaturated fatty acids. J. Endocrinol. 197: 553–563. [DOI] [PubMed] [Google Scholar]
- 19.Wei Y., Wang D., Topczewski F., Pagliassotti M. J. 2006. Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am. J. Physiol. Endocrinol. Metab. 291: E275–E281. [DOI] [PubMed] [Google Scholar]
- 20.Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. 2006. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4: e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma N. K., Das S. K., Mondal A. K., Hackney O. G., Chu W. S., Kern P. A., Rasouli N., Spencer H. J., Yao-Borengasser A., Elbein S. C. 2008. Endoplasmic reticulum stress markers are associated with obesity in nondiabetic subjects. J. Clin. Endocrinol. Metab. 93: 4532–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caraux G., Pinloche S. 2005. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 21: 1280–1281. [DOI] [PubMed] [Google Scholar]
- 23.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4: P3. [PubMed] [Google Scholar]
- 24.Huang W. da, Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 25.Backes C., Keller A., Kuentzer J., Kneissl B., Comtesse N., Elnakady Y. A., Muller R., Meese E., Lenhof H. P. 2007. GeneTrail–advanced gene set enrichment analysis. Nucleic Acids Res. 35: W186–W192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Z., He X., Johnson C., Stoops J., Eaker A. E., Stoffer D. S., Bell A., Zarnegar R., DeFrances M. C. 2007. PI3K is negatively regulated by PIK3IP1, a novel p110 interacting protein. Biochem. Biophys. Res. Commun. 358: 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vock C., Gleissner M., Klapper M., Doring F. 2008. Oleate regulates genes controlled by signaling pathways of mitogen-activated protein kinase, insulin, and hypoxia. Nutr. Res. 28: 681–689. [DOI] [PubMed] [Google Scholar]
- 28.Vock C., Gleissner M., Klapper M., Doring F. 2007. Identification of palmitate-regulated genes in HepG2 cells by applying microarray analysis. Biochim. Biophys. Acta. 1770: 1283–1288. [DOI] [PubMed] [Google Scholar]
- 29.Swagell C. D., Henly D. C., Morris C. P. 2005. Expression analysis of a human hepatic cell line in response to palmitate. Biochem. Biophys. Res. Commun. 328: 432–441. [DOI] [PubMed] [Google Scholar]
- 30.Li Z., Srivastava S., Yang X., Mittal S., Norton P., Resau J., Haab B., Chan C. 2007. A hierarchical approach employing metabolic and gene expression profiles to identify the pathways that confer cytotoxicity in HepG2 cells. BMC. Syst. Biol. 1: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava S., Li Z., Yang X., Yedwabnick M., Shaw S., Chan C. 2007. Identification of genes that regulate multiple cellular processes/responses in the context of lipotoxicity to hepatoma cells. BMC. Genomics. 8: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koh E. H., Park J. Y., Park H. S., Jeon M. J., Ryu J. W., Kim M., Kim S. Y., Kim M. S., Kim S. W., Park I. S., et al. 2007. Essential role of mitochondrial function in adiponectin synthesis in adipocytes. Diabetes. 56: 2973–2981. [DOI] [PubMed] [Google Scholar]
- 33.Beeharry N., Chambers J. A., Green I. C. 2004. Fatty acid protection from palmitic acid-induced apoptosis is lost following PI3-kinase inhibition. Apoptosis. 9: 599–607. [DOI] [PubMed] [Google Scholar]
- 34.Gao D., Griffiths H. R., Bailey C. J. 2009. Oleate protects against palmitate-induced insulin resistance in L6 myotubes. Br. J. Nutr. 102: 1557–1563. [DOI] [PubMed] [Google Scholar]
- 35.Miyake Z., Takekawa M., Ge Q., Saito H. 2007. Activation of MTK1/MEKK4 by GADD45 through induced N-C dissociation and dimerization-mediated trans autophosphorylation of the MTK1 kinase domain. Mol. Cell. Biol. 27: 2765–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han M. S., Park S. Y., Shinzawa K., Kim S., Chung K. W., Lee J. H., Kwon C. H., Lee K. W., Lee J. H., Park C. K., et al. 2008. Lysophosphatidylcholine as a death effector in the lipoapoptosis of hepatocytes. J. Lipid Res. 49: 84–97. [DOI] [PubMed] [Google Scholar]
- 37.Borradaile N. M., Han X., Harp J. D., Gale S. E., Ory D. S., Schaffer J. E. 2006. Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J. Lipid Res. 47: 2726–2737. [DOI] [PubMed] [Google Scholar]
- 38.Cunha D. A., Hekerman P., Ladriere L., Bazarra-Castro A., Ortis F., Wakeham M. C., Moore F., Rasschaert J., Cardozo A. K., Bellomo E., et al. 2008. Initiation and execution of lipotoxic ER stress in pancreatic beta-cells. J. Cell Sci. 121: 2308–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.