Abstract
Phospholipids and triacylglycerols (TAGs) are important classes of lipids in biological systems. Rapid methods have been developed for their characterization in crude samples, including MALDI time-of-flight MS. For mixtures, MALDI often selectively shows only some components. For example, phosphatidylcholine (PC) suppresses detection of other lipids. Most rapid MS methods detect either TAGs or phospholipids but not both. Herein, we demonstrate a simple approach to rapidly screen mixtures containing multiple lipid classes. To validate this approach, reference lipids [PC, tripalmitin (PPP), and phosphatidyl-ethanolamine (PE)] and real samples (beef, egg yolk) were used. In a binary mixture with a strong suppressor (PC), PPP was greatly suppressed. After a simple separation, suppression was virtually eliminated. A mixture of nominally nonsuppressing lipids (PE and PPP) was not adversely affected by separation. Ground beef and egg yolk were used to demonstrate detection of known lipid compositions where other methods have missed one or more lipids or lipid classes. Separation was performed using solid phase extraction with a PrepSep florisil column. A 10 min separation allows rapid screening for lipids and changes in lipids. It is sufficient to clearly detect all lipids and overcome suppression effects in complex lipid mixtures.
Keywords: lipids, suppression-effects, solid phase extraction
Lipids play important roles in biological systems. Analysis allows probing of both their biological roles and as use in foodstuffs or other products. Examples of applications where it is important to detect multiple lipid classes [i.e., phospholipids and triacylglycerols (TAGs)] include food analysis (1–3), cell biology (4), health effects (5), taxonomy (6), and other fields (7–11). However, because of different chemistries, it is often difficult to characterize one class of lipids in the presence of another, especially using rapid methods. Elegant methods have been developed for quantitative analysis in complex mixtures, but they are typically difficult to implement and are time consuming (8, 12). There has been much recent interest in rapid MALDI-MS analysis of lipids due to its speed of analysis and high sensitivity (5, 13, 14). Sample preparation is quick because derivatization is not required (i.e., no silylation), HPLC separation is not needed, limited buffer or salt contamination is tolerated (9, 11, 13), and spectra are easy to interpret (13).
MALDI-MS has functioned as both a qualitative and semiquantitative approach to measure and track effected biology-related phenomena. For example, Lay et al. (15) demonstrated a rapid method for the analysis of edible oils by MALDI-MS that allowed determination of the relative abundances of TAGs with sufficient accuracy to correctly identify blind-coded samples of various oils. Gidden et al. (16) have also reported using MALDI-MS to rapidly differentiate Escherichia coli and Bacillus subtilis based on the phospholipid profile and monitor changes in lipid content during the growth phases of the bacteria. However, this rapid MALDI-MS approach cannot currently be applied to experiments requiring the detection of multiple lipid classes in an unresolved mixture. Attempts to use direct MALDI-MS on such complex mixtures invariably results in entire classes of lipids being missed because of suppression effects.
Suppression in MALDI-MS is well known and has been demonstrated among lipid classes as well as within members of a single class. For example, phospholipids containing quaternary amines can suppress detection of other phospholipids in the sample (7, 11, 13, 17–20). The classical solution to this problem is chromatographic resolution of mixture components using HPLC. Fuchs et al. (18) have reported alternative methods to HPLC by coupling TLC with MALDI-MS. TLC/MALDI-MS worked well for phospholipids, but TAGs that were expected in the samples were not reported. Likewise, MALDI-MS of brain lipid extracts, using a silica gel cation exchanger for separation of components, have also been reported for the detection of phospholipids, but the detection of TAGs was again not reported (19).
Because some biological phenomena cause significant changes in the ratios of specific lipids within a class (14–16), a simple and rapid MALDI-based measurement technique is needed to monitor such changes. A rather lengthy LC-MS approach has been used for the separation of lipids, including phospholipids and TAGs (12). Although this approach is appropriate for quantitative analysis, it is not suitable for MALDI-MS or rapid screening. It is also not needed for measurements of large changes in relative lipid composition. The approach we report is the application of simple solid phase extraction (SPE) cartridges (rather than TLC or ion exchange) to produce a few fractions from which lipids and classes of lipids can be characterized by rapid and direct MALDI-MS analysis. SPE approaches have been used for separation of different lipid classes in foods (21, 22), oils (23), and biological tissues (23–25), most often after fatty acid methyl esterification to facilitate GC separation. In this case, we report MALDI-MS analysis of the intact lipids. Using a simple SPE separation, we report detection of both TAGs and phospholipids in a mixture.
MATERIALS AND METHODS
Herein, we demonstrate our approach using specific lipid standards and complex mixtures (beef and egg yolk) similar to those used in prior studies. A positive control for suppression included a mixture of phosphatidylcholine (PC) (strong suppressor) and tripalmitin (PPP) (reference TAG). A negative control involved a mixture of phosphatidylethanolamine (PE) (not a strong suppressor) and PPP. Comparison of the positive control spectra before and after SPE separation illustrates the extent of suppression, whereas spectra from the negative control tests for potential losses during separation or analysis.
Materials
PPP, l-α-phosphatidylcholine, distearoyl (PC), and l-α-phosphatidylethanolamine, dioleoyl (PE) were purchased from Sigma Aldrich. PrepSep florisil extraction columns (14 ml volume capacity, 1 g prepacked florisil) were purchased from Fisher Scientific. All chemicals [hexane, 2-propanol, 2,5-dihydroxybenzoic acid (DHB), and methanol] were analytical grade or better. Ground beef (80% lean, 20% fat) and hen eggs were purchased locally and used as obtained.
Lipid controls
Lipid standards were measured to the nearest 0.001 mg. Each lipid (1.500 mg) was placed in separate centrifuge tubes with 5 ml hexane/2-propanol (1:1, v/v) and vortexed several minutes to dissolve the solids. Mass spectra were then collected of the individual lipids. Afterwards, the PC solution was evaporated to approximately 1 ml under N2 and added to the PPP sample. A mass spectrum was subsequently collected of this lipid mixture. (Volume adjustment was used to ensure the sample volumes were approximately the same as when the lipids were analyzed individually). PC was selected as the positive control phospholipid in this experiment because of its prevalence in samples (11, 18) and because of its reported suppression effects on other phospholipids (7, 11, 13, 17–20). A sample of PE and PPP was also analyzed. In this case, suppression was not expected. This pair (negative control) of compounds was used to test for loss of signal from sample manipulation or during separation.
Beef and egg yolk lipids
The beef sample (0.25 g) was vortexed with 5 ml hexane/ 2-propanol (1:1, v/v) for 10 min. Insoluble tissue was removed, and a mass spectrum was obtained from this crude beef lipid extract. The egg white and yolk were separated from a single hen egg. The yolk (0.05 g) was vortexed with 5 ml hexane/2-propanol (1:1, v/v) for 10 min. The extract was separated from the insoluble solid matter using a pipet. A mass spectrum was obtained of this crude egg yolk lipid extract.
SPE separation
Lipid classes (standards or extracts) were separated from each other using disposable PrepSep florisil extraction columns. To prevent overloading the column, no more than 50 mg of lipids by weight (given 1 g florisil packing) was added to any given cartridge (26). Columns were preconditioned by washing with 5 ml hexane/2-propanol (1:1, v/v) with no collection of the filtrate. The lipid samples were added to the column, and the filtrate (which should contain TAGs) was collected by vacuum filtration into an Erlenmeyer flask. The column was then washed with 2.5 ml aliquots of hexane to remove any traces of undesirable compounds. This wash filtrate was collected and discarded. Lastly, the phospholipids were eluted with 5 ml 2-propanol (70%) and collected. Phospholipid and TAG fractions were analyzed by MALDI-MS as described below.
MS analysis
A 1 M matrix solution was prepared by dissolving DHB in 90% methanol. Samples and matrix (1 µL each) were mixed directly together in a ratio of 1:1, and 1 µL of this mixture was applied to a stainless steel MTP Multiprobe Adaptor MALDI target. A Bruker Ultraflex II (Bruker Daltonic GmbH, Bremen, Germany) MALDI- time of flight (TOF) was operated in the positive-ion reflectron mode. Spectra were acquired from m/z 400 to 1,650 with adjustment of the sample position and laser power to produce intense ions. The laser power was then kept constant, and 1,500 laser shots were obtained for each sample. Mass spectra were plotted to the same vertical scale.
RESULTS
Lipid standards
Intense ions are observed for PC as protonated [PC+H] and sodiated [PC+Na] adducts at m/z values of 790.6 and 812.6 (data not shown). Smaller peaks are also observed for PC as the protonated [2(PC)+H] and sodiated [2(PC)+Na] dimers. Gas-phase dimers are often observed using soft ionization MS and have been reported for MALDI-MS analysis of lipids (27). Typically, they arise from electrostatic attraction resulting in the sharing of a single ionizing cation between two analytes. PPP ions are observed (not shown) as sodiated adducts [PPP+Na] at m/z 829.7. TAGs, in general, are exclusively observed as salt-cationized ions rather than protonated molecules by MALDI-MS, because the protonated molecule is unstable and decomposes by rapid unimoleculear decay (28).
Using approximately 300 ng of each lipid, mass spectra were readily obtained. The ratio of the measured absolute peak intensities (PC:PPP) for the sodiated adduct ions was approximately the same for optimized spectra obtained using the same number of laser shots and identical conditions. However, when the PC and PPP were mixed in approximately equal proportions, a ratio of approximately 5:1 was observed for these two ions (not shown). This represents a significant suppression of the TAG in the mixture. Based on a report by Lou et al. (20), suppression of PPP (or any TAG) by PC is expected because of a more favorable competition for charge by its quaternary ammonium group. A simple SPE separation was used to recover the lipids for reanalysis, in two fractions, presumably without analyte-induced suppression. After rapid SPE separation of PC and PPP, the MALDI-TOF mass spectra again gave an approximately 1:1 ratio for the intensities of the sodium-adduct ions in the two spectra. Suppression effects were also seen using smaller amounts of PC (1:4 ratio), and likewise signal intensities were also recovered after SPE separation. In summary, the two analytes were readily detected with the expected relative abundances before and after separation but not in the mixture. For other pairs of lipids in which PC was present, the same sort of suppression would be expected (7, 11, 13, 17–20).
Some lipid mixtures do not show significant suppression in MALDI-MS spectra. The mass spectrum of a mixture of PE and PPP, e.g., had the expected peak ratios of each lipid class. Although the SPE extraction would not be needed for this specific mixture, the extra step did not result in significant sample loss. Spectra of individual components were similar both before and after separation, and the mixture spectrum was well represented by simple addition of the two individual component spectra.
Beef lipids
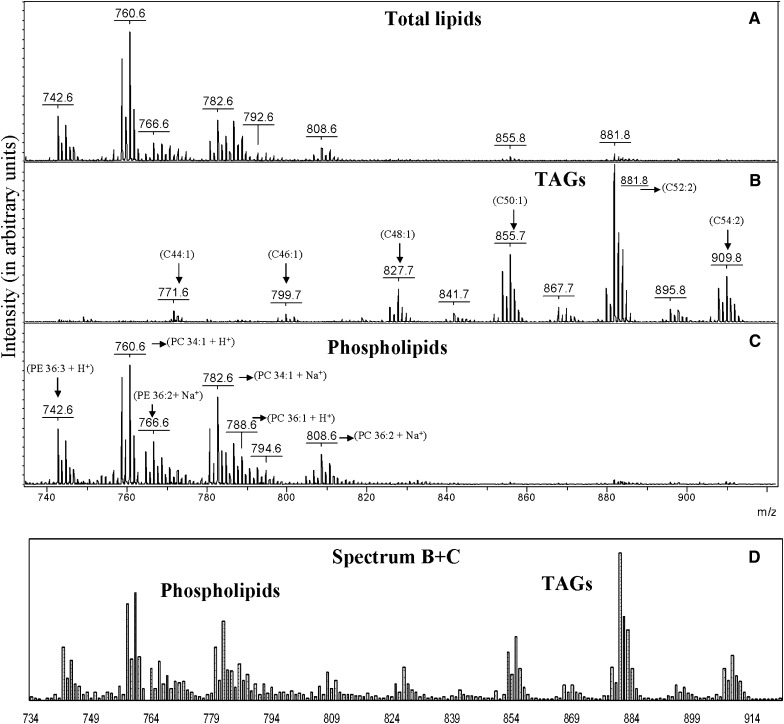
Ground beef has previously been shown to contain both phospholipids and TAGs, with the latter comprising 75–90% of the total lipid content (29). Of the different phospholipid classes, PC is the most abundant, comprising approximately 50% of the total phospholipids (30). A crude lipids fraction from “80:20” ground beef was analyzed by MALDI-TOF MS. The mass spectrum in Fig. 1A shows the lipids extracted from the beef sample before SPE separation. As expected because of the abundant PC, phospholipids are detected in the m/z range of 740–810, whereas TAGs, which should be detected between m/z 800 and 1,000, are almost entirely absent even though known to be present in much greater abundance. The only TAGs that were detected give small peaks at m/z 855.8 and 881.8. Interpretation of this mass spectrum, without knowledge of the effects of ion suppression, might lead to the conclusion that TAGs are present only in ground beef lipids in low abundance. Using this protocol, it might be possible to monitor changes in the phospholipid ratios but not changes in TAGs. For some applications (and for lipidomicis studies), measurement of the relative changes of TAG as well as phospholipids is important (14–16).
Fig. 1.
Partial (m/z 740–920) MALDI-TOF mass spectra of a ground beef lipids mixture (A), the TAGs (B), and phospholipids (C) fraction collected from the SPE cartridge, and a simulated spectrum (D) from the summation of the spectra (B, C) from the two lipid fractions showing the expected peak intensities for total lipids in the absence of suppression effects. Numbers in parenthesis indicate the number of carbon atoms and double bonds, respectively, in the fatty acid side chains.
Mass spectra of the two SPE-separated fractions are shown in Fig. 1B, C. In Fig. 1B, a complex set of TAGs is clearly detected. After this simple separation, changes in the TAGs composition, if any, could be easily determined from peak ratios in this spectrum. Assignment of peaks in the spectrum is in good agreement with the known fatty acids in ground beef (31). The carbon and double bond numbers from the TAG spectrum (Fig. 1B) range from C44:1 to 54:2, corresponding to fatty acids with carbon chains of C14:0 to C18:1, which are approximately 90% of fatty acids in ground beef (31). PE and PC, which account for approximately 80% of the phospholipids in ground beef (30), are also observed in great abundance in the phospholipid spectrum (Fig. 1C) for the same fatty acids. The most abundant TAGs and phospholipids are denoted in the spectra for the separated fractions (Fig. 1B, C).
Figure 1D is a calculated summation of the peak intensities in Fig. 1B, C. Considering that the TAGs are expected to be present in significantly greater abundance than the phospholipids in this sample, the expected intensities for TAG ions in Fig. 1D should probably be larger. In other words, the relative abundances of the TAGs remain less than expected based on reported values, even after separation. Most likely, this can be explained by different ionization efficiencies in the matrix. In the absence of suppression, lower ionization efficiencies are nevertheless expected for less polar compounds compared with more polar ones or preformed ions.
Generally, TAGs and phospholipids are observed in different m/z regions of the mass spectrum. However, there is some overlap that can make interpretation of spectra more difficult, especially for ions of different abundances and because of isotopic peaks. For example, if a sample contained the TAG 14:0/16:0/18:0 + Na+ (m/z 829.7) and PC 18:1/20:4 + Na+ (m/z 830.6) overlap would exist between the carbon isotope peak of the TAG and the monoisotopic peak from PC. In the beef lipid extract, two known TAGs (m/z 771.6 and m/z 799.7) (29) are observed overlapping the mass region normally associated with phospholipids. Even if there were no suppression effects, the overlap of members of these two classes could make it difficult to detect both. Simple SPE separation clearly resolves this issue. Moreover, knowledge of the lipid class often allows unambiguous assignment of the lipids based on molecular weight and known fatty acid composition. Thus, in addition to the reduction of suppression, separation minimizes problems that may arise from overlap in the mass regions for members of the two lipid classes. By minimizing this overlap, our approach greatly simplifies interpretation of the spectra.
Egg yolk lipids
TLC/MALDI-MS of egg yolk lipids was recently reported to demonstrate minimization of suppression and a separation method well suited for MALDI-MS analysis (18). Egg yolks contain approximately 30% lipids by weight (32), with phospholipids (31%) and neutral lipids (65%) being the most abundant species. Neutral lipids in this case include TAGs, diacylglycerols, monoacyglycerols, fatty acids, carotenoids, and sterols (33). Of the different phospholipid classes, PC, which frequently causes problems with suppression, is the most abundant, comprising approximately 26% of the total phospholipids (32). In the TLC/MALDI-MS report, reduction of suppression by phospholipids was achieved, but the abundant TAGs were not reported (18). Herein, we demonstrate the ability to detect both phospholipids and TAGs using a SPE approach rather than TLC.
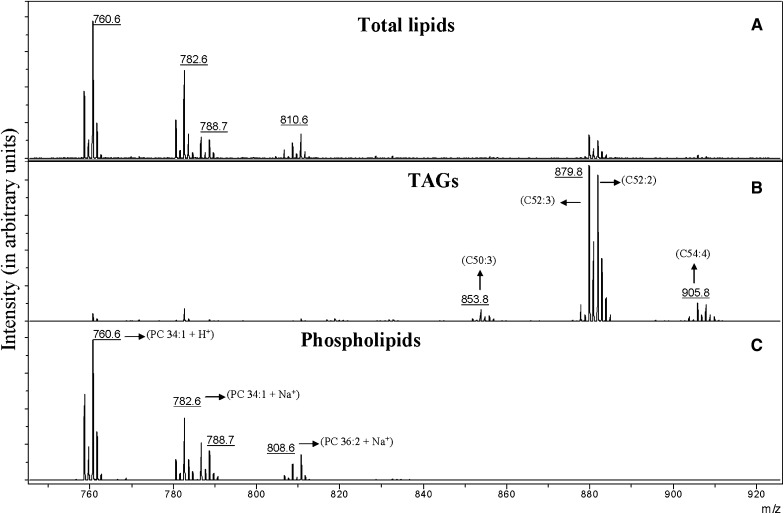
Figure 2A shows the mass spectrum of the lipids extracted from the egg yolk before application of SPE separation. Phospholipids are detected in the m/z range of 758–810, whereas TAGs, which should be detected between m/z 840 and 920, are almost entirely absent even though present in a greater abundance. The only TAGs that were detected give small peaks at m/z 879.8 and 881.8. As with the ground beef lipids, interpretation of this mass spectrum might lead to the conclusion that TAGs were either present in low abundance or absent.
Fig. 2.
Partial (m/z 740–920) MALDI-TOF mass spectra of an egg yolk lipids mixture (A), the TAGs (B), and phospholipids (C) fraction collected from the SPE cartridge. Numbers in parenthesis indicate the number of carbon atoms and double bonds, respectively, in the fatty acid side chains.
Mass spectra of the two separated fractions from a 15 mg-equivalent sample are shown in Fig. 2B, C. In Fig. 2B, the TAGs are clearly detected, and two additional groups of TAGs (C50:3-50:1 and C54:5-54:2) that were not previously detected in Fig. 2A are observed. The fatty acids in the separated TAG fraction range from C50:3 to 54:2, corresponding to fatty acids with carbon chains of C16:0–C18:2, which represent approximately 95% of fatty acids in egg yolk (31). The most abundant TAGs are labeled in the spectrum from this fraction.
Figure 2C shows the fraction containing the phospholipids. Little difference is observed in this spectrum compared with the same m/z range in the total egg yolk sample (Fig. 2A). Biologically important changes in the phospholipid composition could probably be detected in the crude mixture, but separated fractions would be needed to detect any changes in the TAG composition. The most abundant phospholipids are labeled in the spectrum for the separated fraction (Fig. 2C).
Fuchs et al. (18) reported analysis of egg yolk lipids by MS analysis of the extracts before and after TLC separation using MALDI and MALDI imaging, respectively. Our observed phospholipids (Fig. 2A, C) are in good agreement with those observed in their direct MALDI analysis of a total lipids fraction. It should be noted that in this previous study by Fuchs, no TAGs were reported in the samples (by MALDI or TLC imaging) despite their careful analysis of PC suppression effects on phospholipids from a total lipids extract. Their analysis using direct MALDI and MALDI TLC imaging only detected phospholipids and completely missed the TAGs that were more abundant in the sample. It is likely that the TAGs were completely suppressed by direct MALDI (without separation) and ran off of the TLC plate when analyzed by TLC/MALDI imaging. The advantage observed using TLC/MALDI was that some additional phospholipids were detected when the phospholipids themselves were separated by TLC. This highlights a limitation of the simple two-step SPE technique. It readily detects abundant members of each fraction but does not have sufficient chromatographic resolution to prevent all suppression within a SPE fraction. This can, perhaps, be resolved using different solvents and multiple elution steps if more detailed information about either phospholipids or other lipid classes is needed. Clearly, TLC and SPE separations are complementary. The SPE approach provides rapid separation into classes, both of which can be screened by MALDI-MS, whereas the TLC approach provides more resolution but may miss components (classes) with very different polarity. Future analysis will focus on development of more complex SPE techniques of this sort.
DISCUSSION
Analysis of a sample containing both phospholipid and TAG components by rapid MALDI-TOF MS resulted in mass spectra dominated by the phospholipids (in particular PC). For a lipid extract of beef and egg yolk, most TAGs, while present in greater abundance than the phospholipids, were not detected from an unresolved lipid/sample mixture. Whereas relative changes in phospholipid composition might be detectable by rapid MALDI on the mixture, variations in the TAGs could not. With simple SPE separation, mixture resolution was sufficient to allow detection of both TAGs and phospholipids from a complex mixture known to contain ion-suppressing components, such as PC. Although the SPE approach does not have the resolving power of the comparable rapid TLC approach, it provides broader coverage for mixtures containing analytes with disparate polarities. With either approach spectra are easier to interpret, because mass overlap of lipids/classes is minimized by the separation. Separation using the disposable, prepacked columns is fast and easy. This step required acquisition of two MALDI spectra rather than one, but the separation itself took only about 10 min.
Future studies will focus on additional lipid class separations in different samples, suppression by other phospholipids, a solvent system or additional SPE cartridge to separate the phospholipid classes, and a ZipTip® (Millipore) procedure that allows fractionation of the lipids during MALDI plate spotting rather than by using an SPE cartridge.
Footnotes
Abbreviations:
- DHB
- 2,5-dihydroxybenzoic acid
- PC
- phosphatidylcholine
- PE
- phosphatidyl-ethanolamine
- PPP
- tripalmitin
- SPE
- solid phase extraction
- TAG
- triacylglycerol
- TOF
- time-of-flight
This research was supported by National Institutes of Health NCRR Grant 5P20RR015569. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Carrasco-Pancorbo A., Navas-Iglesias N., Cuadros-Rodriguez L. 2009. From lipid analysis towards lipidomics, a new challenge for the analytical chemistry of the 21st century. Part I: modern lipid analysis. TrAC. Trends Analyt. Chem. 28: 263–278. [Google Scholar]
- 2.Yoshida H., Saiki M., Yoshida N., Tomiyama Y., Mizushina Y. 2009. Fatty acid distribution in triacylglycerols and phospholipids of broad beans (Vicia faba). Food Chem. 112: 924–928. [Google Scholar]
- 3.Harrabi S., Boukhchina S., Kallel H., Mayer P. M. 2009. Glycerophospholipid and triacylglycerol distribution in corn kernals (Zea Mays L.). J. Cereal Sci. In press. [Google Scholar]
- 4.Han X., Abendschein D. R., Kelley J. G., Gross R. W. 2000. Diabetes-induced changes in specific lipid molecular species in rat myocardium. Biochem. J. 352: 79–89. [PMC free article] [PubMed] [Google Scholar]
- 5.Schiller J., Suss R., Fuchs B., Muller M., Zschornig O., Arnold K. 2007. MALDI-TOF MS in lipidomics. Front. Biosci. 12: 2568–2579. [DOI] [PubMed] [Google Scholar]
- 5.Schiller J., Zschornig O., Petkovic M., Muller M., Arnhold J., Arnold K. 2001. Lipid analysis of human HDL and LDL by MALDI-TOF mass spectrometry and 31P-NMR. J. Lipid Res. 42: 1501–1508. [PubMed] [Google Scholar]
- 6.Ishida Y., Nakanishi O., Hirao S., Tsuge S., Urabe J., Sekino T., Nakanishi M., Kimoto T., Ohtani H. 2003. Direct analysis of lipids in single zooplankter individuals by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 75: 4514–4518. [DOI] [PubMed] [Google Scholar]
- 7.Schiller J., Suss R., Fuchs B., Muller M., Zschornig O., Arnold K. 2007. MALDI-TOF MS in lipidomics. Front. Biosci. 12: 2568–2579. [DOI] [PubMed] [Google Scholar]
- 8.Peterson B. L., Cummings B. S. 2006. A review of chromatographic methods for the assessment of phospholipids in biological samples. Biomed. Chromatogr. 20: 227–243. [DOI] [PubMed] [Google Scholar]
- 9.Schiller J., Suess R., Arnhold J., Fuchs B., Lessig J., Mueller M., Petkovic M., Spalteholz H., Zschoernig O., Arnold K. 2004. Matrix-assisted laser desorption and ionization time-of-flight (MALDI-TOF) mass spectrometry in lipid and phospholipid research. Prog. Lipid Res. 43: 449–488. [DOI] [PubMed] [Google Scholar]
- 10.Watson A. D. 2006. Lipidomics: a global approach to lipid analysis in biological systems. J. Lipid Res. 47: 2101–2111. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs B., Schiller J., Suess R., Zscharnack M., Bader A., Mueller P., Schuerenberg M., Becker M., Suckau D. 2008. Analysis of stem cell lipids by offline HPTLC-MALDI-TOF MS. Anal. Bioanal. Chem. 392: 849–860. [DOI] [PubMed] [Google Scholar]
- 12.Sommer U., Herscovitz H., Welty F. K., Costello C. E. 2006. LC-MS-based method for the qualitative and quantitative analysis of complex lipid mixtures. J. Lipid Res. 47: 804–814. [DOI] [PubMed] [Google Scholar]
- 13.Petkovic M., Schiller J., Muller M., Benard S., Reichl S., Arnold K., Arnhold J. 2001. Detection of individual phospholipids in lipid mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: Phosphatidylcholine prevents the detection of further species. Anal. Biochem. 289: 202–216. [DOI] [PubMed] [Google Scholar]
- 14.Batoy S. M. A. B., Borgmann S., Flick K., Griffith J., Jones J. J., Saraswathi V., Hasty A. H., Kaiser P., Wilkins C. L. 2009. Lipid and phospholipid profiling of biological samples using MALDI fourier transform mass spectrometry. Lipids. 44: 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lay J. O., Jr., Liyanage R., Durham B., Brooks J. 2006. Rapid characterization of edible oils by direct matrix-assisted laser desorption ionization time-of-flight mass spectrometry analysis of triacylglycerols. Rapid Commun. Mass Spectrom. 20: 952–958. [DOI] [PubMed] [Google Scholar]
- 16.Gidden J., Denson J., Liyanage R., Ivey D. M., Lay J. O., Jr 2009. Lipid compositions in Esherichia coli and Bacillus subtilis during growth as determined by MALDI-TOF and TOF/TOF mass spectrometry. Int. J. Mass Spectrom. 283: 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs B., Suess R., Nimptsch A., Schiller J. 2009. MALDI-TOF-MS directly combined with TLC: a review of the current state. Chromatographia. 69: S95–S105. [Google Scholar]
- 18.Fuchs B., Schiller J., Suess R., Schuerenberg M., Suckau D. 2007. A direct and simple method of coupling matrix-assisted laser desorption and ionization time-of-flight mass spectrometry (MALDI-TOF MS) to thin-layer chromatography (TLC) for the analysis of phospholipids from egg yolk. Anal. Bioanal. Chem. 389: 827–834. [DOI] [PubMed] [Google Scholar]
- 19.Johanson R. A., Buccafusca R., Quong J. N., Shaw M. A., Berry G. T. 2007. Phosphatidylcholine removal from brain lipid extracts expands lipid detection and enhances phosphoinositide quantification by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Anal. Biochem. 362: 155–167. [DOI] [PubMed] [Google Scholar]
- 20.Lou X., van Dongen J. L. J., Vekemans J. A. J. M., Meijer E. W. 2009. Matrix suppression and analyte suppression effects of quaternary ammonium salts in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: an investigation of suppression mechanism. Rapid Commun. Mass Spectrom. 23: 3077–3082. [DOI] [PubMed] [Google Scholar]
- 21.Prieto J. A., Ebri A., Collar C. 1992. Optimized separation of nonpolar and polar lipid classes from wheat flour by solid-phase extraction. J. Am. Oil Chem. Soc. 69: 387–391. [Google Scholar]
- 22.Garcia Regueiro J. A., Gibert J., Diaz I. 1994. Determination of neutral lipids from subcutaneous fat of cured ham by capillary gas chromatography and liquid chromatography. J. Chromatogr. A. 667: 225–233. [DOI] [PubMed] [Google Scholar]
- 23.Giacometti J., Milosevic A., Milin C. 2002. Gas chromatographic determination of fatty acids contained in different lipid classes after their separation by solid-phase extraction. J. Chromatogr. A. 976: 47–54. [DOI] [PubMed] [Google Scholar]
- 24.Bateman H. G., II, Jenkins T. C. 1997. Method for extraction and separation by solid phase extraction of neutral lipid, free fatty acids, and polar lipid from mixed microbial cultures. J. Agric. Food Chem. 45: 132–134. [Google Scholar]
- 25.Pernet F., Pelletier C. J., Milley J. 2006. Comparison of three solid-phase extraction methods for fatty acid analysis of lipid fractions in tissues of marine bivalves. J. Chromatogr. A. 1137: 127–137. [DOI] [PubMed] [Google Scholar]
- 26.Supelco Bulletin 910. Guide to solid phase extraction. 1997. accessed January 2010 at http://www.sigmaaldrich.com/Graphics/Supelco/objects/4600/4538.pdf.
- 27.Jackson S. N., Wang H. J., Woods A. S., Ugarov M., Egan T., Schultz J. A. 2005. Direct tissue analysis of phospholipids in rat brain using MALDI-TOFMS and MALDI-ion mobility-TOFMS. J. Am. Soc. Mass Spectrom. 16: 133–138. [DOI] [PubMed] [Google Scholar]
- 28.Gidden J., Liyanage R., Durham B., Lay J. O., Jr. 2007. Reducing fragmentation observed in the matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of triacylglycerols in vegetable oils. Rapid Commun. Mass Spectrom. 21: 1951–1957. [DOI] [PubMed] [Google Scholar]
- 29.Tanamati A., Oliveira C. C., Visentainer J. V., Matsushita M., de Souza N. E. 2005. Comparative study of total lipids in beef using chlorinated solvent and low-toxicity solvent methods. J. Am. Oil Chem. Soc. 82: 393–397. [Google Scholar]
- 30.Keller J. D., Kinsella J. E. 1974. Phospholipid changes and lipid oxidation during cooking and frozen storage of raw ground beef. J. Food Sci. 38: 1200–1204. [Google Scholar]
- 31.Nutrient Data Products and Services, United States Department of Agriculture. 2009. Accessed August 2009 at http://www.nal.usda.gov/fnic/foodcomp/Data/SR21/reports/sr21fg13.pdf (NDB No. 23572).
- 32.Juneja L. R. 1997. Egg yolk lipids. Hen Eggs: Their Basic and Applied Science. Yamamoto T., Juneja L. R., Hatta H., Mujo K., CRC Press, MA: 1–12. [Google Scholar]
- 33.Li-Chan E. C. Y., Powrie W. D., Nakai S. 1995. The chemistry of eggs and egg products. Egg Science and Technology. Stadelman W. J., Cotterill O. J., The Haworth Press, Inc., New York: 105–176. [Google Scholar]