Abstract
Pioglitazone, a hypoglycemic agent, has been shown to increase plasma HDL cholesterol, but the mechanism is incompletely understood. We further investigated effects of pioglitazone on transcriptional regulation of apolipoprotein (apo)A-I gene and functional properties of pioglitazone-induced apoA-I-containing particles. Pioglitazone dose-dependently stimulated apoA-I promoter activities in HepG2 cells. A peroxisome proliferator-activated receptor (PPAR)-response element located in site A (−214 to −192 bp, upstream of the transcription start site) of the promoter is required for pioglitazone-induced apoA-I gene transcription. Deletion of site A (−214 to −192 bp), B (−169 to −146 bp), or C (−134 to −119 bp), which clusters a number of cis-acting elements for binding of different transcription factors, reduced the basal apoA-I promoter activities, and no additional pioglitazone-sensitive elements were found within this region. Overexpression or knock-down of liver receptor homolog-1, a newly identified nuclear factor with strong stimulatory effect on apoA-I transcription, did not alter pioglitazone-induced apoA-I transcription. Pioglitazone-induced apoA-I transcription is mainly mediated through PPARα but not PPARγ in hepatocytes. Pioglitazone induced production of HDL enriched in its subfraction containing apoA-I without apoA-II, which inhibited monocyte adhesion to endothelial cells in vitro. In conclusion, pioglitazone increases apoA-I production by directly enhancing PPAR-response element-dependent transcription, resulting in generation of apoA-I-containing HDL particles with increased anti-inflammatory property.
Keywords: HDL/metabolism, atherosclerosis, nuclear receptors, cholesterol
Plasma HDL levels are inversely related to the development of atherosclerotic coronary heart disease (1). The anti-atherogenic effects of HDL have been largely attributed to the ability of apolipoprotein (apo)A-I- containing HDL particles to mediate cellular cholesterol efflux and thereby facilitate the removal of excess cholesterol from peripheral tissues and its delivery to the liver for elimination through reverse cholesterol transport pathway (2, 3). In addition, HDL also enhances fibrinolysis (4) and inhibits platelet aggregation, LDL oxidation (5), endothelial transmigration of monocytes (6), and pro-inflammatory cytokine-mediated expression of endothelial cell adhesion molecules (4–7). These pleiotropic properties of HDL have been thought to contribute to the beneficial effects of HDL on atherosclerotic cardiovascular disease.
Glitazones (a class of thiazolidinediones agents such as pioglitazone and rosiglitazone) are anti-hyperglycemic drugs used in the treatment of type 2 diabetes. These oral anti-diabetic agents reduce blood glucose levels by sensitizing peripheral tissues to insulin (8). In addition to improving glycemic control, glitazones also influence plasma lipid and lipoprotein levels (9). Treatment of type 2 diabetic patients with pioglitazone consistently increased plasma HDL-cholesterol and decreased triglycerides with generally minimal to no effect on total and LDL-cholesterol (10–13). Clinically, pioglitazone's effect on HDL-cholesterol (about 10–15% elevation) is greater than that observed with statins and several fibrates and just below that of niacin (3).
Initially, glitazones were designed as agonistic ligands of the nuclear receptor peroxisome proliferator-activated receptor (PPAR)-γ to enhance insulin action mainly in skeletal muscle and adipose tissues to promote glucose utilization and suppress intracellular lipolysis. PPARs (PPARα, γ, and δ) are members of the nuclear receptor family, and by ligand activating and hetero- dimerization of PPAR with retinoid X receptor and binding to the DNA sequence in the promoter region, modulate transcription of various target genes involved in glucose and lipid metabolism (14). The effects of pioglitazone to reduce triglycerides are mainly shown to be mediated by increased lipoprotein lipase expression through the activation of PPARγ and decreased plasma apoC-III, an inhibitor for lipoprotein lipase-mediated lipolysis (15). However, mechanisms of the effect of pioglitazone to raise HDL and apoA-I (the major protein of HDL) are incompletely understood. In initial studies, we have shown that pioglitazone did not change uptake of HDL protein and cholesterol ester but increased apoA-I and A-II de novo synthesis, secretion, and mRNA in human hepatoblastoma cells (HepG2) (16). These studies suggest that pioglitazone, by increasing hepatic apoA-I mRNA expression, increases apoA-I-containing HDL particles without influencing the HDL catabolic pathway in hepatocytes.
ApoA-I is mainly synthesized in the liver and intestine, and its expression levels are highly regulated by a number of transcription factors in response to nutrients, hormones, growth factors, cytokines, and drugs such as fibrates. These trans-acting factors bind to the specific DNA sequence elements in the promoter region to negatively or positively modulate the transcription of apoA-I gene (17, 18). Functional analysis of apoA-I gene promoter by mutagenesis identified that roughly three sections, A, B, and C, clustering cis-acting elements located between nucleotides −222 and −110 upstream from the transcription start site, are essential and sufficient for liver-specific expression of apoA-I gene in hepatocytes [reviewed in (19)]. These cis-acting elements in site A (−214 to −192) bind thyroid receptor, hepatic nuclear factor (HNF)-4, PPARα, RARα/β, retinoid X receptor-α, C/EBP, ARP-1, Rev-erbα, tumor necrosis factor-α, and interleukin-1; in site B (−169∼−146) bind glucocorticoids, estradiol, and HNF3b; and in site C (−134∼−119) bind HNF-4 and ARP-1. More recently, a new nuclear factor, liver receptor homolog-1 (LRH-1), with a strong stimulatory effect on apoA-I gene transcription, has been identified to be located in site C (20). Site C also has a direct repeat-1 sequence for binding a liver X receptor agonist TO901317 (21). Moreover, the regulation of the hepatic expression of apoA-I gene is controlled by synergistic interaction between these transcription factors (18). The present study was designed to further investigate the effects of pioglitazone on transcriptional regulation of apoA-I gene and properties of pioglitazone-induced apoA-I-containing HDL particles.
MATERIALS AND METHODS
Materials
Tissue culture materials, media, and FBS were obtained from Sigma Chemical Co. Human hepatoblastoma cell line (HepG2 cells) and human monocytic THP-1 cell line were obtained from American Type Culture Collection. Human aortic endothelial cells (HAEC) were purchased from Lonza Biologics. The polyclonal antibodies for human apoAI and apoAII were purchased from Calbiochem. All other chemicals used were of analytical grade.
Cell culture
HepG2 cells were maintained in DMEM containing 10% FBS supplemented with 100 units/ml penicillin G and 100 μg/ml streptomycin sulfate at 37°C in a humidified atmosphere of 95% air, 5% CO2. HepG2 cells were incubated in the presence or absence of pioglitazone as indicated. Human monocyte THP-1 cells and HAEC were grown in RPMI1640 (Invitrogen) and EMB2 (Cambrex Bio ScienceWalkersville, Inc.) medium, respectively.
Immunoprecipitation and Western blotting
HepG2 cells were grown in 6-well plates until they attained 75–80% confluence. HepG2 cells were incubated with various amount of pioglitazone (0–25 μM) at 37°C for 24–48 h. At the termination of the incubation, culture medium was collected. A 1 ml sample of culture medium was used to measure apoAI secreted into the media by immunopreciptation and Western blotting using anti-human apoA-I or apoA-II antibodies.
Plasmids
The human apoAI promoter DNA sequence (−470∼+8bp, using the transcription start site as +1) was isolated by PCR amplification from human genomic DNA (ClonTech) and cloned into pGL3 (R2.1) basic vector harboring the luciferase reporter cDNA. The primers used were: 5′-TCT TAC GCG TGG ACT AAA GAA GAG CAC TGG TG-3′ (sense) and 5′-CGC AGA TCT CAG TCT CTA AGC AGC CAG CTC TT-3′ (anti-sense). Underlined are restriction sites for MulI and BglII, respectively. The 470 bp apoA-I promoter sequence contained A, B, and C clusters for the binding of a number of transcriptional factors responsible for trans-acting regulating its expression (18–20). Deletions or mutations of the apoA-I promoter were made by PCR or mutagenesis with Stratagene Quick Change Mutagenesis kit with the following primers: to mutate the PPARE site by changing A to T with primers 5′-CGCCCCCACTGATCCCTTGTCCCCTGCCCTGCAGCC, and 5′-GGCTGCAGGGCAGGGGACAAGGGATCAGTGGGGGCG; to delete site A sequence GAACCCTTGA (−214∼−192) with primers, 5′-ACTCCCCTCCCGCCCCCACTCCCCTGCCCTGCAGCCCCCG and 5′-CGGGGGCTGCAGGGCAGGGGAGTGGGGGCGGGAGGGGAGT; to delete site B sequence, GTTTGCCCACTCTATTTGCCC (−169∼−146) with primers 5′-GCAGCCCCCGCAGCTTGCTGCCCAGGGACAGAGCTGATCC and 5′-GGA TCAGCTCTGTCCCTGGGCAGCAAGCTGCGGGGGCTGC; to delete site C sequence GCTGATCCTTGAACT (−134∼−119) with primers 5′-GCCC AGCCCCAGGGACAGAGCTTAAGTTCCACATTGCCAG and 5′-CTGGCAATGT GGAACTTAAG CTCTGT CCCTGGGGCT GGGC. All constructs were verified by DNA sequencing. Expression plasmids for PPARα, PPARγ, and LRH-1 were prepared by RT-PCR amplification and cloning into pcDNA3.1 mammalian expression vector (Invitrogen).
RT and quantitative real-time PCR
Total RNA extracted from cells was reverse transcribed with 100 units of SuperScript II reverse transcriptase (Invitrogen). Real-time PCR was performed by using the QuantiTect SYBG Green PCR reagents (Qiagen) in iCycler with primer sets: 5′-CAGACTGGCCGAGTACCACGCCAAGGCC and 5′-TCACTGGGTGTT GAGCTTCTTAGTGTAC for human apoA-1; and 5′-CGGCGACGACCCATTCGAAC, and 5′-GAATCGAAC CCTGATTCCCCG T for an endogenous reference 18S rRNA.
Transfection and dual luciferase assay
HepG2 cells were grown in 24-well or 48-well plates for 18–24 h. Cells were then transfected for 24 h with the pGL3-apoAI-Luc plasmid or pGL3 basic luciferase reporter vector as control, using 1.5 μl of FuGENE 6 transfection reagent (Roche). All cells were also cotransfected with 2 ng of phRL-TK (Int-) vector (Promega) containing wild-type Renilla luciferase (Rluc) as an internal control for transfection efficiency in each well. Twenty-four hour later, the transfected cells were washed with PBS two times, and then cultured for 48 h in DMEM containing 1 mg/ml BSA free of fatty acids and various amounts of pioglitazone as indicated. After washing with PBS, cells were lysed for luciferase assay using the Dual-Luciferase Reporter Assay System according to the manufacturer's protocol (Promega). Luminescence was measured on NOVOstar (BMG LABTECH GmbH, Offenburg, Germany). ApoA-I promoter activities are reflected by luciferase activities that are expressed as relative light units (RLU) to internal control Rluc. Data are expressed as mean ± SD of three experiments.
For small interfering RNA (siRNA) knock-down assay, 0.2 μg of pGL3-apoAI-Luc was cotransfected with 50 nM of siRNA (Ambion) in 24-well plates for 24 h and then treated with pioglitazone for 48 h. Luciferase assay was performed as described above.
Chromatin-immunoprecipitation assay
HepG2 cells (5 × 106), grown in 10 cm dishes or 75 cm2 flasks, were detached by trypsin-EDTA and resuspended in 9 ml DMEM medium. Cross-linking of proteins to DNA was performed by adding 270 μl of 37% formaldehyde (final 1%), and tubes were rocked gently at room temperature for 10 min. Cross-linking was quenched by adding 1 ml of 1.25 M glycine and gently rocking for 5 min at room temperature. Cells were washed three times with 10 ml ice-cold PBS. Cells were pelleted by centrifugation at 280 g for 5 min at 4°C and resuspended in 400 μl chromatin-immunoprecipitation (ChIP) buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 0.5% NP40, 1% Triton X-100) with protease inhibitors (complete tablet, Roche, Indianapolis, IN), and lysed on ice for 10 min. Nuclei were collected by centrifugation at 600 g for 5 min at 4°C and dissolved in 400 μl of ChIP buffer. Nuclei were resuspended 10 times by using a syringe with #26 needles before sonicating at an output of 40% for 5 s for a total 10 times in a Branson Sonifier 250 (Newtown, CT). Debris was cleared by centrifugation at 12,000 g for 15 min at 4°C, and the supernatant was split into 100 μl aliquots in 1.5 ml microcentrifuge tubes for immunoprecipitation (20 μl was saved for preparation of input DNA). Two micrograms of specific polyclonal antibodies against the human PPARα and PPARγ (SC-9000× and SC-7196×, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or normal rabbit IgG was added to each tube, and tubes were rotated overnight at 4°C. Fifteen microliters of protein A agarose beads (Roche, Inc., Indianapolis, IN) was added to 1.5 ml microcentrifuge tubes and rotated for 1 h at 4°C. Beads were collected by centrifugation at 2,000 g for 30 s at 4°C and then washed five times by removing the supernatant. Immunoprecipitates were eluted in 200 μl of 50 mM Tris-Cl (pH8.0), 1 mM EDTA, 1% SDS, and 50 mM NaHCO3 at 65°C for 10 min two times. Then 21 μl of 4 M NaCl was added into 400 μl sample (final 200 mM) to de-crosslink the protein-DNA complex at 65°C for 5 h. DNA was purified by phenol/chloroform extraction after digested with 20 μg/ml proteinase K at 55°C for 1 h. DNA samples were resuspended in 30 μl TE buffer. The 2 to 4 μl samples were used for PCR, which was performed at initial denature at 94°C for 2 min followed by 35 cycles at 94°C for 30s, 60°C for 45s, and 72°C for 1min, followed by 72°C for 7 min, with primers that amplified 264 bp of sequences containing the PPAR-response element (PPRE) site in the human apoA-1 promoter from −252 to +12 bp, 5′-CCGGGAGACCTGCAAGCCTGC and 5′-GCACCTCCTTCTCGCAGTCTC. PCR products were resolved on a 1.2% agarose gel.
Adhesion assay
THP-1 cells were labeled with fluorescence BCECF (Invitrogen) at 37°C for 30 min. HAEC (0.1–1 × 105 cells) were plated in 24- or 48-well plates and preincubated at 37°C for 24 h in the presence or absence of filter-concentrated medium (∼1.2 mg/ml protein) from cultured HepG2 cells that had been treated with pioglitazone or vehicle DMSO. The labeled THP-1 cells (2 × 105) and HAEC were co-incubated at 37°C for 10 min. Nonattached THP-1 cells were removed by gently washing three times with PBS containing 1% FBS. The cells were dissolved in PBS-0.1% SDS and the fluorescence was measured in NOVOstar (BMG LABTECH GmbH, Offenburg, Germany) with 485 nm excitation and 562 nm emission. Separate wells containing serial diluted labeled THP-1 cells were used for generating a standard curve. The ratio of fluorescence intensity of the attached cells to that of the total labeled cells applied to the well was calculated, and the adhesion activity of THP-1 monocytes to HAEC is expressed as percent of the number of attached monocytes of the total.
For immunodepletion of apoA-I, the conditioned medium was incubated overnight at 4°C in the presence of rabbit anti-apoA-I antibody (Calbiochem) at dilution of 1:20 or rabbit IgG control. The immunoprecipitated apoA-1 were absorbed on protein A-agarose beads. The supernatants were used in the adhesion assay.
Statistical analysis
Data presented are mean ± SD of three experiments. Student's t-test was utilized comparing to control or between two groups. One-way ANOVA analysis was used for the multiple and full set comparisons where appropriate. A P-value <0.05 was considered significant.
RESULTS
Pioglitazone stimulated apoA-I gene transcription
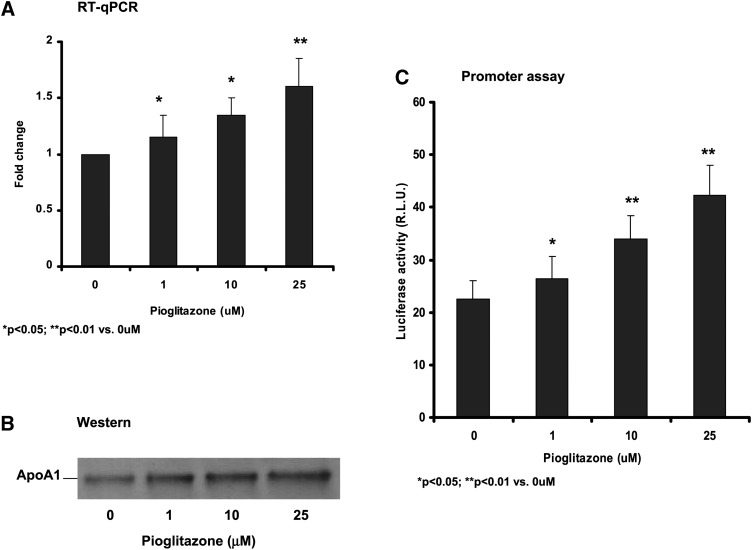
Pioglitazone (1–25 μM) increased apoA-I mRNA (Fig. 1A) and protein expression (Fig. 1B) in cultured human HepG2 hepatocytes. In transient transfection experiments with the reporter plasmids containing artificial PPAR elements, pioglitazone was shown to be a weak PPARα activator via a direct binding of pioglitazone to PPARα in nonhepatic cell system (22). To further test whether pioglitazone directly stimulates apoA-I transcription in hepatocytes, we constructed a luciferase reporter plasmid for the apoA-I promoter comprising the nucleotides −470 to +8 upstream of the apoA-I gene transcriptional stat site (+1) ,which is necessary and sufficient for expression of the apoAI gene in hepatocytes. Results of the luciferase assay showed that pioglitazone dose-dependently (1–25 μM) increased the apoA-I promoter activity in HepG2 cells (Fig. 1C).
Fig. 1.
Pioglitazone increased apoA-1 transcription and protein production. A: RT-qPCR analysis of apoA-1 mRNA expression in HepG2 cells. B: Western blotting of apoA-1 protein in the culture medium of HepG2 cells. C: Luciferase reporter assay of human apoA-1 gene promoter. Then 470 bp of the human apoA-1 promoter sequence was isolated by PCR amplification from human genomic DNA and cloned into the upstream of a Luciferase repoter in pGL3 (R2.1) basic vector. The pGL3-apoA-1-Luc plasmids were cotransfected into HepG2 cells with an internal control plasmid phRL-TK (Int-) Rluc. The cells were then treated with pioglitazone for 48 h as indicated. Luminescence in the transfected cell lysates was measured using the Dual-Luciferase Reporter Assay System. ApoAI promoter activities are reflected by luciferase activities that are expressed as RLU to internal control Rluc. Data are expressed as mean ± SD of three experiments.
PPRE is required for pioglitazone-induced apoA-I promoter activity
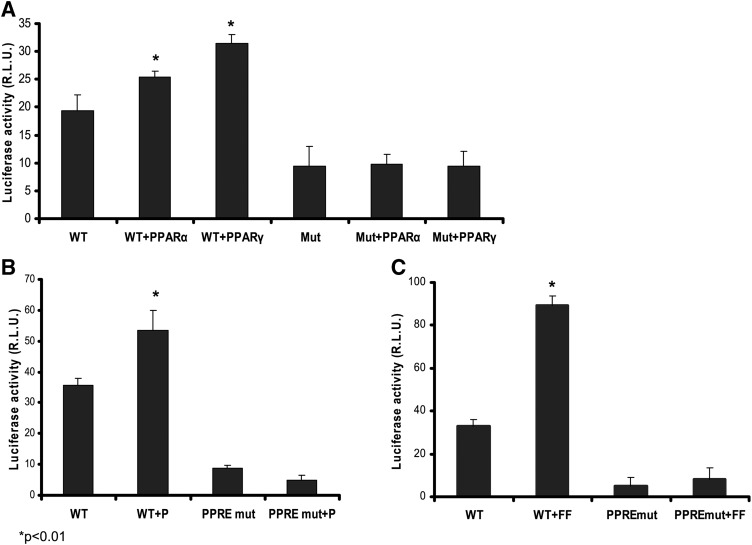
PPARα, but not PPARγ, is highly expressed in the liver. Agonists for PPARα, such as fenofibrate, have been shown to increase apoA-I gene expression (14). Although pioglitazone has been previously shown to weakly activate PPARα, the potential functional consequence has not been tested in hepatocytes. A PPARα binding sequence, the PPRE, is present in site A of the apoA-I promoter (TGAACCCTTGACCCC). Cotransfection with either PPARα or PPARγ increased the apoA-I promoter activities, suggesting that activation of both PPARα and PPARγ are able to stimulate apoAI transcription (Fig. 2A). Mutating the PPRE sequence by changing two nucleotides, A to T, dramatically reduced the promoter activity, indicating the importance of this binding element in the regulation of apoA-I expression in hepatocytes (Fig. 2). Transfecting HepG2 cells with PPARα and PPARγ did not reverse the mutated apoA-I promoter activity (Fig. 2A). Furthermore, these mutations abolished pioglitazone-induced apoA-I promoter activity, suggesting that the PPRE is required for the effect exerted by pioglitazone (Fig. 2B). Similarly, a PPARα ligand and activator, fenofibrate, significantly stimulates apoA-I transcription, but it did not reverse the apoA-I promoter activity when the PPRE binding site was mutated (Fig. 2C).
Fig. 2.
Mutated PPRE eliminated pioglitazone-induced apoA-1 gene transcription. The PPRE in site A of the human apoA-1 promoter region was mutated as described in “Materials and Methods.” A: Wild-type (TGAACCCTTGACCCC) or PPRE mutant (TGATCCCTTGTCCCC) pGL3-apoA-1-Luc was cotransfected with human PPARα or PPARγ expression plasmid. B: HepG2 cells were transfected with wild-type or PPRE mutant pGL3-apoA-1-Luc and then treated with 10 μM pioglitazone (+P) as indicated for 48 h. C: The transfected HepG2 cells were treated with 10 μM fenofibrate (+FF) for 48 h. Dual-luciferase assay was conducted, and the promoter activity is expressed as RLU to the internal control Rluc. Data are expressed as mean ± SD of three experiments.
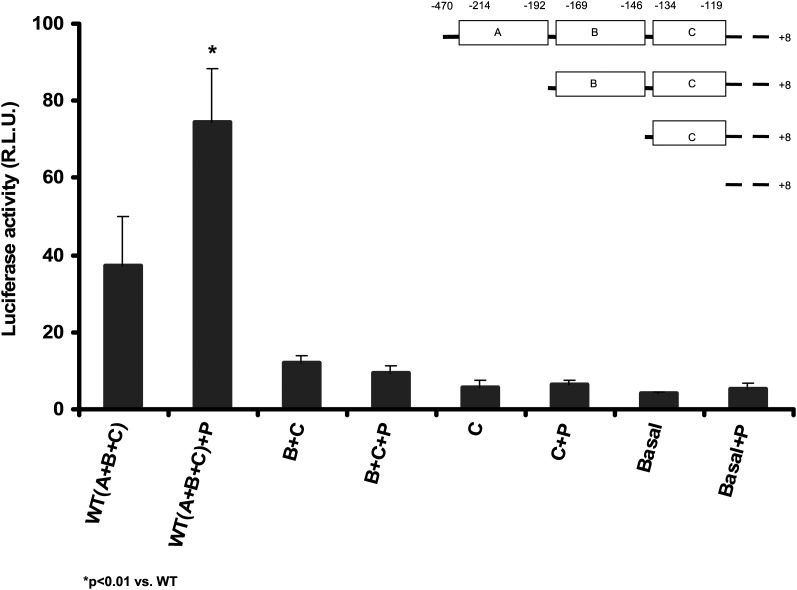
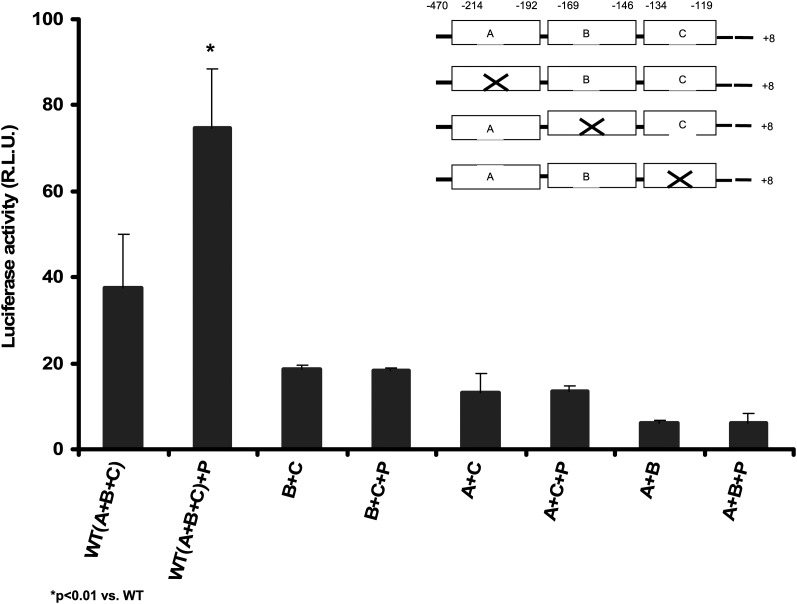
To examine whether there are additional potential pioglitazone-sensitive elements in the apoA-I promoter, we performed a deletion mapping assay. Deletion of site A (−214 to −192), site B (−169 to −146), and site C (−134 to −119) reduced the apoA-I promoter activity by approximately 3-, 6-, and 9-fold, respectively, compared with the full-length promoter, indicating that elements in these regions are important in regulation of its basal transcription activities (Fig. 3). When A, B, and C in each site was deleted individually, the basal promoter activities were also reduced (Fig. 4). No significant stimulatory effect by pioglitazone was found in these experiments. These data indicate that the integrity of all of A, B, and C sites in the apoA-I promoter is crucial for its full transcription activities and no other cis-elements responding to pioglitazone were found in these regions examined.
Fig. 3.
Deletion mapping potential on pioglitazone-sensitive elements in the human apoA-1 promoter. Three clusters, A, B, and C, containing the binding elements for a number of transcription factors in the apoA-1 promoter region were deleted sequentially by PCR, cloned into pGL3 (R2.1) basic vector, and transfected into HepG2 cells for luciferase assay. Then 10 μM pioglitazone (+P) was added as indicated and incubated for 48 h. Data are expressed as mean ± SD of three experiments. WT (A+B+C): pGL3-ApoA-1-Luc; B+C: deleted site A; C: deleted sites A and B; Basal: deleted sites A, B, and C.
Fig. 4.
Removal of sites A, B, or C reduced basal transcription of apoA-1 gene and abolished pioglitazone-induced apoA-1 gene transcription. Three clusters, A, B, and C, containing the binding elements for a number of transcription factors in the apoA-1 promoter region were deleted individually by mutagenesis and cloned into pGL3 (R2.1) basic vector and transfected into HepG2 cells for luciferase assay. Then 10 μM pioglitazone (+P) was added as indicated and incubated for 48 h. Data are expressed as mean ± SD of three experiments. WT: pGL3-ApoA-1-Luc; Del-A: deleted site A (−214∼−192: CCCACTGAACCCTTGACCCCT GC); Del-B: deleted site B (−169∼−146: GTTTGCCCACTCTATTTGCCC); Del-C: deleted site C (−134∼−119: GCTGATCCTTGAACT).
LRH-1 is not involved in pioglitazone-induced apoA-I transcription
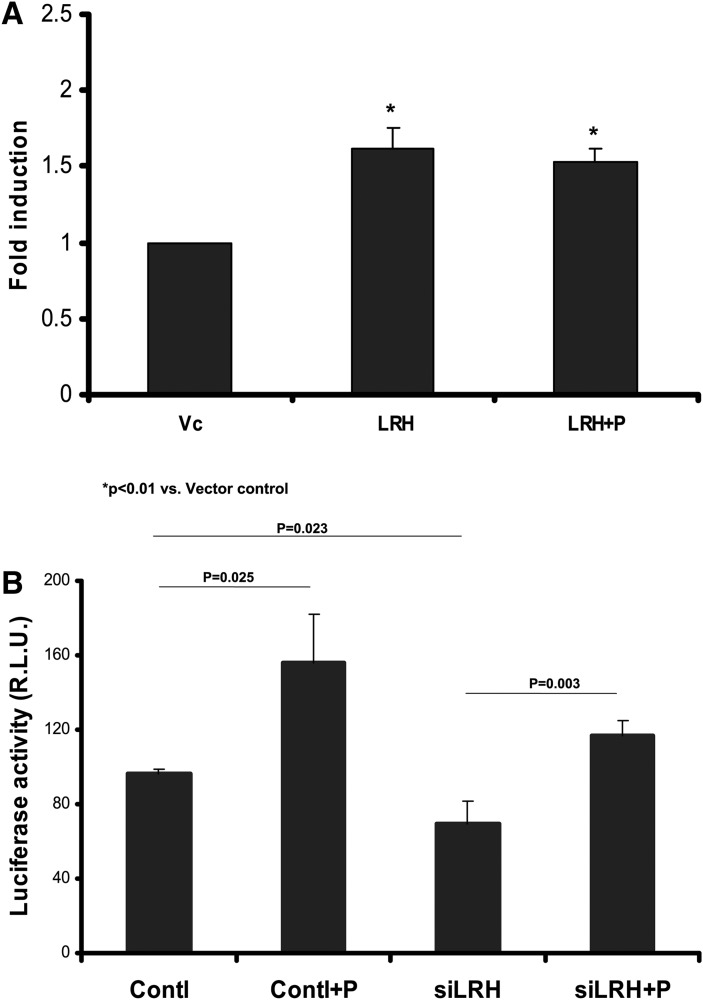
To further test our deletion mapping observation, we chose the LRH-1, a newly identified transcription factor located in site C (−133 to −117), which strongly stimulates apoA-1 expression in the liver (20). Overexpression of LRH-1 increased apoA-I promoter activity; addition of pioglitazone did not further increase the promoter activity (Fig. 5A). Furthermore, siRNA knocking-down LRH-1 reduced LRH-1 mRNA expression by 55% [RT and quantitative real-time PCR (RT-qPCR) fold change: 0.443 ± 0.145 vs. control 1.0 ± 0; P < 0.01) and decreased apoA-I transcription, but did not affect pioglitazone-induced activity, as pioglitazone similarly increased apoA-I transcription by 162% and 168% in control and siRNA-treated cells, respectively, suggesting that LRH-1, with its binding element in site C, is not involved in pioglitazone-induced apoA-I transcription (Fig. 5B).
Fig. 5.
LRH-1 is not involved in pioglitazone-induced apoA-1 gene transcription. A: pGL3-apoA-1-Luc was cotransfected with the LRH-1 expression plasmid (LRH) or empty vector control (Vc) and then treated with 10 μM of pioglitazone (LRH+P) as indicated for 48 h. Data are expressed as mean ± SD of three experiments for fold induction compared with control. B: Plasmid pGL3-apoA-1-Luc and 40 nM of siRNA nucleotides for the human LRH-1 (siLRH) were transfected into HepG2 cells and then treated with 10 μM of pioglitazone (+P) as indicated for 48 h. The scrambled nucleotides (Contl) were used as control. Data are expressed as mean ± SD of three experiments.
Pioglitazone-induced apoA-I transcription is mainly through PPARα but not PPARγ in HepG2 cells
RT and quantitative real-time PCR (RT-qPCR) indicated that PPARα expression is about 4-fold higher than PPARγ in HepG2 cells (ΔCt value normalized to reference 18S rRNA: 2.737 ± 0.664 vs. 0.986 ± 0.885; P < 0.01). However, pioglitazone did not significantly alter PPARα and PPARγ mRNA expression levels (data not shown).
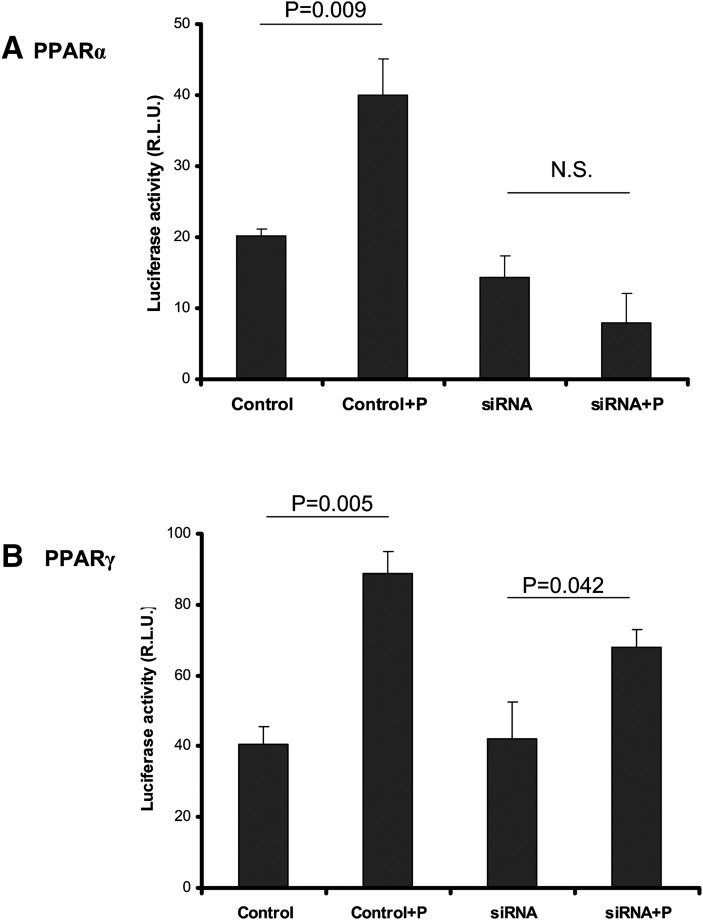
To investigate the role of PPARα and PPARγ in pioglitazone-induced apoA-I transcription, we first tested the effects of a PPARα agonist together with pioglitazone. No additive effects were observed when cells were treated with pioglitazone alone and PPARα agonist YW14643 plus pioglitazone, suggesting that both PPARγ ligand and PPARα agonist are able to bind the same PPRE in the apoA-I promoter (luciferase activity fold change: 1.39 ± 0.1 for 10 μM pioglitazone, 1.81 ± 0.39 for 10 μM WY14643, and 1.37 ± 0.22 for 10 μM pioglitazone plus 10 μM WY14643). We then performed siRNA knock-down assays. siRNA knocking-down reduced mRNA expression of PPARα by 44% (0.56 ± 0.087 vs. 1.0 ± 0; P < 0.01) and PPARγ by 40% (0.60 ± 0.048 vs. 1.0 ± 0; P < 0.01), respectively (RT-qPCR fold change). As a positive control for our silencing as says, knocking-down PPARα reduced fenofibrate-induced apoA-1 transcription by 74% (lucifease activity: 39.5 ± 3.1 vs. 14.4 ± 0.41; P < 0.01). Knock-down of PPARα eliminated pioglitazone-induced apoA-I transcription (Fig. 6A), whereas knock-down of PPARγ largely retained pioglitazone-induced apoA-I transcription (Fig. 6B), suggesting that PPARα but not PPARγ plays a major role in pioglitazone-induced apoA-I expression in hepatocytes.
Fig. 6.
siRNA knockdown assay. HepG2 cells in 24-well plates were cotransfected with 0.2 μg of pGL3-Luc-apoA-1 and 50 nM of siRNA (Ambion) for the human PPARα (A) and for the human PPARγ (B) for 24 h and then treated with pioglitazone (+P) for 48 h. Luciferase assay was performed as described in “Methods.” The scrambled nucleotides were used as control. Data are expressed as mean ± SD of three experiments. N.S.: not significant.
To further examine the role of PPARα and PPARγ in pioglitazone-induced apoA-I transcription, we performed a ChIP assay. The apoA-I promoter sequence from −252 to +12 bp containing the PPRE element was amplified by PCR in the DNA/chromatin fragments immunoisolated with anti-PPARα antibody but not with anti-PPARγ antibody (Fig. 7). The same apoA-I promoter region was amplified by PCR from the samples treated with Wy14643, a PPARα agonist that served as a positive control. These results clearly show that pioglitazone, by mainly interacting with PPARα but not PPARγ, increased apoA-I gene transcription in hepatocytes.
Fig. 7.
ChIP analysis of the binding of PPARα and PPARγ to the promoter of apoA-1 in HepG2 cells. HepG2 cells were grown to 80% confluence and then incubated in the presence of either vehicle (DMSO) or 10 uM of pioglitazone and Wy14643 for 48 h. ChIP were performed as described in “Materials and Methods.” Input: sheared nuclear lysates before subjecting to immuoprecipitation using: No Ab, rabbit IgG control, PPARα, anti-PPARα antibody, PPARγ, anti-PPARγ antibody.
Pioglitazone-induced apoA-I production is mainly in LP-A-I particles
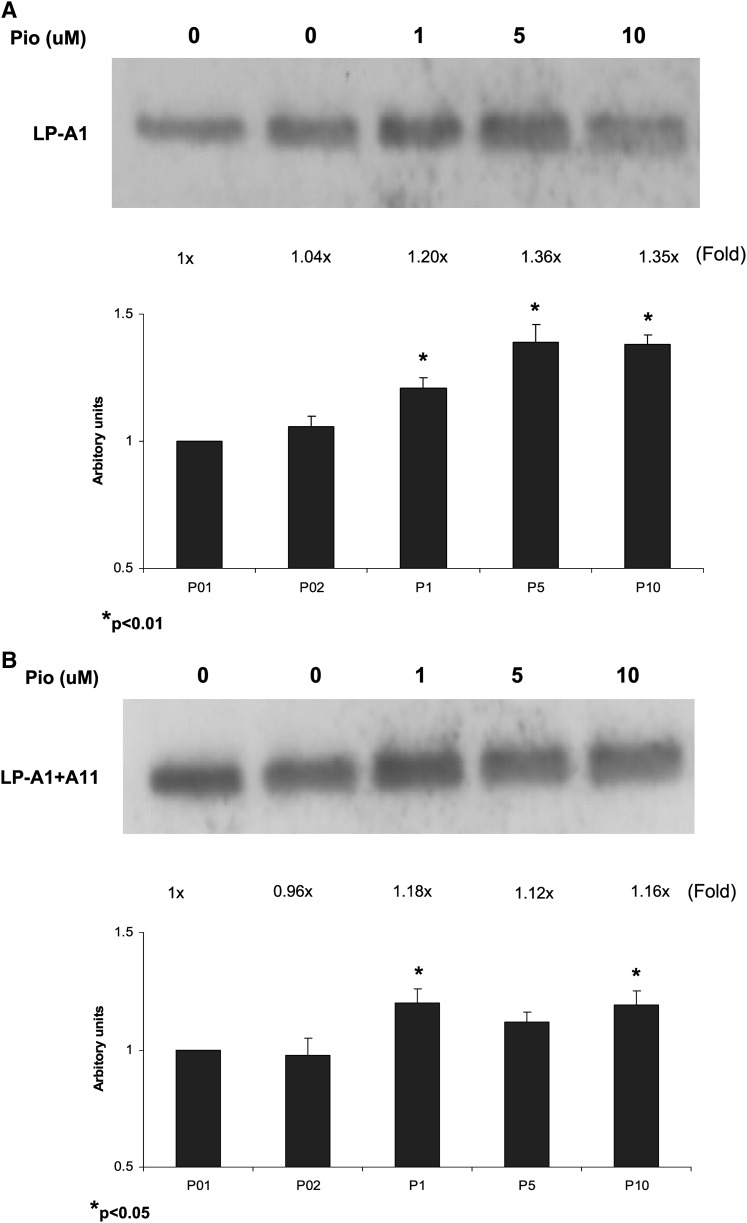
In HepG2 cells, we have previously shown that mainly two HDL particles are secreted, including HDL particles containing apoA-I only (LP-AI) or HDL particles containing both apoA-I and apoAII (LP-AI+AII) (16). To assess the distribution of pioglitazone-induced apo-A-I in these particles, we analyzed the secreted apoAI-containing particles by immunoprecipitation and Western blotting. As shown in Fig. 8, pioglitazone (1–10 μM) mainly increased apoA-I in LP-AI particles by 1.2- to 1.4-fold, and, to lesser degree, in LP-AI+AII particles by 1.1- to 1.2-fold (Fig. 8). These data indicate that the increased apoA-I by pioglitazone was mainly enriched in LP-AI particles.
Fig. 8.
Pioglitazone increased apoA-1 in LP-AI particles from HepG2 cells. HepG2 cells were incubated with pioglitazone for 48 h. LP-AI particles containing only apoA-1 (apoA-1 only HDL) were isolated from medium by immunoprecipitation first with apoAII antibody to remove apoAII-containing particles and then with apoAI antibody. LP-AI+AII particles (Total HDL) were isolated by immunoprecipitation with both apoAI and apoAII antibodies. Immunoprecipitates were then examined by Western blotting with apoA-1 antibody. Representative Western blots are shown with fold changes compared with control by densitometrical quantitation of three blots.
Conditioned media from pioglitazone-treated HepG2 cells increased the inhibition of monocyte adhesion to HAEC
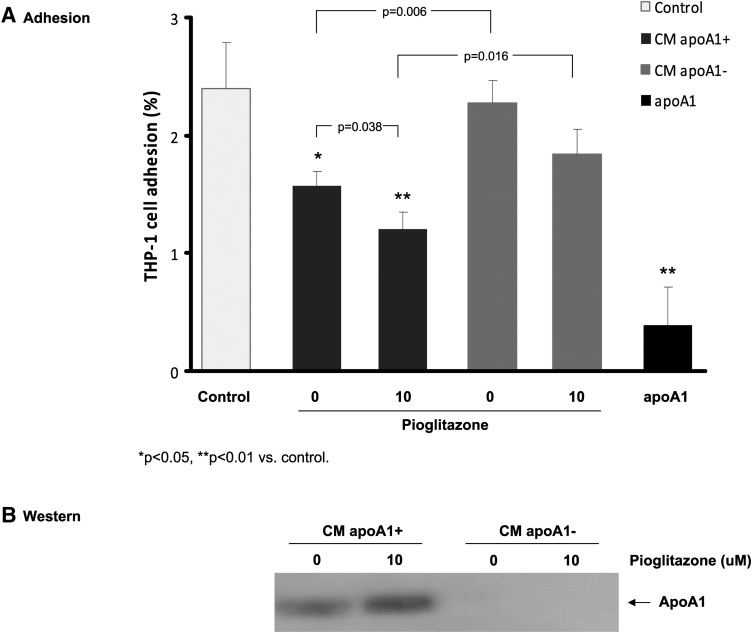
In previous studies, we have shown that pioglitazone-induced apoA-Icontaining particles were able to mediate cholesterol efflux from macrophages (16). To assess anti-inflammatory biological properties of HDL particles generated from pioglitazone-treated HepG2 cells, we tested the effect of apoA-I-containing HDL particles secreted by HepG2 cells on monocyte adhesion to HAEC, a critical initial inflammatory event involved in monocyte accumulation. As shown in Fig. 9A, the conditioned media (CM) from pioglitazone-treated (10 μM pioglitazone) HepG2 cells showed a stronger inhibitory effect on monocyte adhesion than the CM without pioglitazone treatment (0 μM pioglitazone, as treated with vehicle DMSO) (P = 0.038), suggesting an enhanced anti-inflammatory activity. During the adhesion assays, incubation of these CM for 24 h did not alter HAEC viability and/or proliferation as measured by CellTiter-Glo Luminescent Cell Viability kit (Promega; this assay is a highly sensitive method for assessing cell proliferation and cytotoxicity). The RLU for the control CM and pioglitazone-treated CM were 56,140 ± 482 and 60,717 ± 1,772, respectively.
Fig. 9.
Conditioned medium from pioglitazone-treated HepG2 cells inhibited monocyte adhesion to HAEC. A: HepG2 cells were incubated with DMSO vehicle (0 μM) or 10 μM pioglitazone for 48 h. The CM was filter concentrated and washed with PBS in an Amicon ultra centrifugal tube, Ultracel-50k (Amicon). The concentrated CM was then incubated 16–24 h at 4°C in the presence of rabbit anti-apoA-1 antibody (Calbiochem) at a dilution of 1:20 or in the presence of rabbit IgG as control for immunodepletion of apoA-1. The immunoprecipitated apoA-1 were removed by the binding to protein A-agarose beads. The supernatants were used in the adhesion assay of the fluorescence-labeled THP-1 monocytes to endothelial cells, as described in “Materials and Methods.” Control: The coincubation of labeled THP-1 cells and HAEC in assay (basal EMB2) medium; CM apoA-1+: the CM after immunoprecipitation with rabbit IgG control; CM apoA-1-: the conditioned medium after immunodepletion of apoA-1 with anti-apoAI. Then 1 μg purified apoAI was added as positive control in the adhesion assay. Data are expressed as mean ± SD of three experiments. One-way ANOVA analysis was used for the multiple comparisons between groups and control. B: Western blotting analysis of apoA-1 in the conditioned medium: CM apoA-1+ and CM apoA-1-.
To further test whether apoA-I in the CM is directly responsible for the reduced adhesion activity, we performed immunodepletion of apoA-I and compared the effects of CM containing apoA-I (CM apoA-1+) and CM without apoA-I (CM apoA-1-). As shown in Fig. 9, the CM apoA-1−, from either the 0 μM pioglitazone-treated or 10 μM pioglitazone-treated HepG2 cells, reduced their ability to inhibit the adhesion (P = 0.006 and P = 0.016). Comparisons between groups in CM apoA-1+ and control were significant (P for trend = 0.001) but not significant between groups in CM apoA-1- and control. These results suggest that apoA-1/apoA-1-containing particles significantly contribute to the inhibitory effects on the monocyte adhesion to the endothelial cells.
DISCUSSION
A number of clinical studies established that pioglitazone significantly increased serum HDL-cholesterol and decreased triglycerides, and pioglitazone produced more favorable lipid profiles than rosiglitazone in patients with type 2 diabetes (23–26). However, the effect of pioglitazone on the plasma apoA-I level has not been studied in detail. In a small number of nondiabetic patients with low HDL-cholesterol and metabolic syndrome, Szapary et al. (24) have recently shown that pioglitazone treatment for 6 weeks significantly increased mean apoA-I by 6.8%. Recently, Davidson et al. (26) also reported that pioglitazone treatment for 24 weeks increased serum apoA-I in patients with type 2 diabetes. In an attempt to understand the mechanisms of apoA-I increase by pioglitazone, using human hepatoblasoma cell line (HepG2 cells), we have previously shown that pioglitazone increased the de novo synthesis and secretion of apoA-I particles in the culture medium by increasing apoA-I mRNA levels without affecting HDL catabolic events in HepG2 cells (16).
Among the three synthetic glitazones with PPAR-agonist activities, pioglitazone has a much better effect on raising HDL and lowering triglycerides. The mechanism for this divergence has been suggested to be due to the ability of pioglitazone to weakly activate PPARα. In transient transfection experiments with the reporters containing four copies of the rat acyl-CoA oxidase PPRE, previous studies showed that pioglitazone appeared to be a weak PPARα activator via a direct binding of pioglitazone to the artificial PPRE in kidney-derived COS-1 cells (22). There is one PPRE in site A of the apoA-I promoter, and the PPARα expression level is much higher than PPARγ in the liver (14). We therefore hypothesized that a weak binding activity of pioglitazone to PPARα observed in a nonhepatocyte system would have the potential to initiate transcription of apoA-I gene in hepatocytes, resulting in an increased production of apoAI-containing HDL particles in the liver. In this study, we further assessed the effects of pioglitazone on apoAI transcriptional regulation in HepG2 cells. Using transfection studies in HepG2 cells with full-length apoA-I promoter, we showed that pioglitazone stimulated apoA-I transcription in HepG2 cells, a homogeneous background for its expression.
Furthermore, our studies using deletion mapping and site-directed mutagenesis have defined that a single PPARα binding element located in site A of the promoter region is required for pioglitazone-induced apoA-I expression in the hepatocyte system. In hepatocytes, we suggest that pioglitazone exerts its activating effects on apoA-I gene through a PPRE-dependent event. A modest stimulatory effect on apoA-I transcription reported in this study is consistent with a weaker binding activity of pioglitazone to PPARα observed in nonhepatic cells. In additional studies, we investigated the potential role of pioglitazone in the synergistic integrity of all three clustering cis-acting elements of A-, B-, and C-sites within the apoA-I gene in regulating apoA-I transcription in hepatocytes. Deletion of all sites A, B, or C individually markedly reduced basal apoA-I promoter activity, even when the PPRE in site A remained in the promoter. Our data suggest that the integrity of all three cis-acting element sites A, B, and C are essential and required for the full apoA-I transcription in HepG2 cells, similar to the previous reports (21). To further identify the possibility of additional cis-elements that may be operating in pioglitazone-induced apoA-I transcription, we assessed the effect of pioglitazone on apoA-I transcription with individually deleted regulatory sites A, B, and C in the promoter. In these studies, we were not able to identify the possibility of any other cis-elements in pioglitazone-mediated ApoA-I transcription in hepatocytes. Although our data suggest that PPRE is importantly linked to apoA-I transcription by pioglitazone, this study cannot completely exclude other factors that may participate in pioglitazone-induced apoA-I transcription. Because site B is mainly involved in hormonal regulated apoA-I transcription, it is unlikely that site B plays a major role in pioglitazone-mediated apoA-I transcription. Overexpression or knocking-down of a strong apoA-I activator LRH-1, a binding sequence located in site C (20), did not alter pioglitazone-induced apoA-I promoter activity. Furthermore, chromatin immunoprecipitation showed that pioglitazone increased the binding of PPARα to the apoA-1 promoter containing PPRE in vivo (Fig. 7). Our data indicate that the PPRE could be a major functional site for pioglitazone-induced apoA-I transcription in hepatocytes.
Previous studies have shown that pioglitazone is a PPARγ agonist but also a weaker PPARα activator; both PPARγ and PPARα can bind the PPRE in the promoter region of human apoA-I gene and pioglitazone can effectively compete for these bindings (22). Data from our cotransfection and siRNA knock-down studies and from other reports (16, 19, 22) indicate that pioglitazone, through activation of PPARα, increases apoA-I transcription and protein production, as PPARα is predominately expressed. Our ChIP assay data showed that pioglitazone-induced apoA-1 expression is mainly through the direct binding of PPARα but not PPARγ to PPRE element in apoA-I promoter in hepatocytes. In vivo studies would be helpful to further test whether pioglitazone will increase expression of apoA-I and plasma apoA-I levels as a compensatory mechanism through PPARγ when PPARα in the liver is suppressed or PPARγ expression is enhanced in the liver, such as in several murine models of obesity or diabetes (27).
To assess properties of HDL particles secreted from HepG2 cells that were treated with pioglitazone, we further analyzed apoAI-containing particles. We found that pioglitazone-induced apoA-I production is mainly enriched in LP-AI particles, and these apoAI-containing particles are more effective in inhibiting adhesion of THP-1 monocytes to HAEC ( These findings indicate that pioglitazone, by enhancing PPRE-dependent apoAI transcription, increased the de novo synthesis and secretion of apoAI-enriched LP-AI particles in the culture medium, which in turn participated in significantly inhibiting the adhesion of monocytes to aortical endothelial cells, a key initial inflammatory event associated with monocyte migration and accumulation during atherogenesis. Based on our previous and this study, we suggest that the pioglitazone-induced production of functionally active apoAI-containing HDL particles, by enhancing reverse cholesterol transport pathway and anti-inflammatory properties (two important beneficial functions of HDL), inhibit atherogenesis.
Although originally developed as an agent to control glycemia in diabetic patients, pioglitazone should also be considered as part of dyslipidemia treatment in diabetes in combination with lipid-regulating drugs including statins, niacin, and fibrates. Whereas fibrates and statins act via PPAR-activation, niacin mainly decreases apoA-I catabolism (28, 29). This information is important in combination therapy using drugs with complementary mechanisms of action to achieve aggressive HDL goals. Additional research is needed to assess the additive efficacy of such combinations not only on lipids, but also on reducing cardiovascular events in the high risk population of diabetics beyond monotherapy.
In summary, we suggest that pioglitazone, through PPRE-mediated events within cis-acting element site A in the apoA-I gene promoter, increases hepatic apoA-I mRNA expression, resulting in increased plasma levels of physiologically active apoA-I/HDL particles. We also suggest that the enhanced reverse cholesterol transport and anti- inflammatory properties of apoAI-enriched HDL particles generated by pioglitazone in hepatocytes may, at least in part, explain the cardio-protective actions of pioglitazone in inhibiting atherosclerosis in humans (26). It is also important to note that the effective concentrations of pioglitazone used in our studies (0.1–25 μmol/L) are comparable to the plasma levels observed in patients treated with pioglitazone (30), suggesting clinical relevance of our findings to the in vivo situation in humans. However, because the findings of this study were derived from in vitro studies using HepG2 cells, caution should be used in extrapolating in vitro findings of this study to in vivo conditions in humans. Further biochemical and functional characterization of apoAI-HDL particles in pioglitazone-treated patients may shed light on the cardio-protective properties of HDL particles and their impact on atherosclerotic events in humans. Additionally, our findings may be useful in forming the rationale for combination therapy using pioglitazone and other HDL-raising agents with different mechanisms to additively increase HDL levels in diabetic patients.
Footnotes
Abbreviations:
- Apo
- apolipoprotein
- ChIP
- chromatin-immunoprecipitation
- CM
- conditioned media
- HAEC
- human aortic endothelial cell
- HNF
- hepatic nuclear factor
- LP-AI
- HDL particles containing apoA-I only
- LP-AI+AII
- HDL particles containing both apoAI and apoAII
- LRH
- liver receptor homolog
- PPAR
- peroxisome proliferator-activated receptor
- PPRE
- PPAR-response element
- RLU
- relative light unit
- Rluc
- Renilla luciferase
- RT-qPCR
- reverse transcription and quantitative real-time PCR
- siRNA
- small interfering RNA
This study was supported, in part, by Takeda Pharmaceuticals North America Inc., and the Southern California Institute for Research and Education.
REFERENCES
- 1.Wong N. D., Malik S., Kashyap M. L. 2005. Dyslipidemia. Preventive Cardiology. Wong N. D., Black H. R., Gardin J. M., McGraw Hill Co., Inc, New York: 183–211. [Google Scholar]
- 2.Fielding C. J., Fielding P. E. 1995. Molecular physiology of reverse cholesterol transport. J. Lipid Res. 36: 211–228. [PubMed] [Google Scholar]
- 3.Meyers C. D., Kashyap M. L. 2005. Pharmacologic augmentation of high-density lipoproteins: mechanisms of currently available and emerging therapies. Curr. Opin. Cardiol. 20: 307–312. [DOI] [PubMed] [Google Scholar]
- 4.Saku K., Ahmed M., Glas-Greenwalt P., Kashyap M. L. 1985. Activation of fibrinolysis by apolipoproteins of high density lipoproteins in man. Thromb. Res. 39: 1–8. [DOI] [PubMed] [Google Scholar]
- 5.Banka C. L. 1996. High density lipoprotein and lipoprotein oxidation. Curr. Opin. Lipidol. 7: 139–142. [DOI] [PubMed] [Google Scholar]
- 6.Navab M., Imes S. S., Hama S. Y., Hough G. P., Ross L. A., Bork R. W., Valente A. J., Berliner J. A., Drinkwater D. C., Laks H. 1991. Monocyte transmigration induced by modification of low density lipoprotein in cocultures of human aortic wall cells is due to induction of monocyte chemotactic protein 1 synthesis and is abolished by high density lipoprotein. J. Clin. Invest. 88: 2039–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cockerill G. W., Rye K. A., Gamble J. R., Vadas M. A., Barter P. J. 1995. HDL inhibits cytokine-induced expression of endothelial cell adhesion molecules. Arterioscler. Thromb. Vasc. Biol. 15: 1987–1994. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann C. A., Colca J. R. 1992. New oral thiazolidinedione antidiabetic agents act as insulin sensitizers. Diabetes Care. 15: 1075–1078. [DOI] [PubMed] [Google Scholar]
- 9.Van Wijk J. P. H., de Koning E. J. P., Martens E. P., Rabelink T. J. 2003. Thiazolidinediones and blood lipids in type 2 diabetes. Arterioscler. Thromb. Vasc. Biol. 23: 1744–1749. [DOI] [PubMed] [Google Scholar]
- 10.Rosenblatt S., Miskin B., Glazer N. B., Prince M. J., Robertson K. E. 2001. The impact of pioglitazone on glycemic control and atherogenic dyslipidemia in patients with type 2 diabetes mellitus. Coron. Artery Dis. 12: 413–423. [DOI] [PubMed] [Google Scholar]
- 11.Gegick C. G., Altheimer M. D. 2001. Comparison of effects of thiazolidinediones on cardiovascular risk factors: observations from a clinical practice. Endocr. Pract. 7: 162–169. [DOI] [PubMed] [Google Scholar]
- 12.Kipnes M. S., Krosnick A., Rendell M. S., Egan J. W., Mathisen A. L., Schneider R. L. 2001. Pioglitazone hydrochloride in combination with sulfonylurea therapy improves glycemic control in patients with type 2 diabetes mellitus: a randomized, placebo-controlled study. Am. J. Med. 111: 10–17. [DOI] [PubMed] [Google Scholar]
- 13.Einhorn D., Rendell M., Rosenzweigh J., Egan J. W., Mathisen A. L., Schneider R. L. 2000. Pioglitazone hydrochloride in combination with metformin in the treatment of type 2 diabetes mellitus: a randomized, placebo-controlled study. The pioglitazone 027 Study Group. Clin. Ther. 22: 1395–1409. [DOI] [PubMed] [Google Scholar]
- 14.Barbier O., Torra I. P., Duguay Y., Blanquart C., Fruchart J. C., Glineur C., Staels B. 2002. Pleiotropic actions of peroxisome proliferator-activated receptors in lipid metabolism and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 22: 717–726. [DOI] [PubMed] [Google Scholar]
- 15.Nagashima K., Lopez C., Donovan D., Ngai C., Fontanez N., Bensadoun A., Fruchart-Najib J., Holleran S., Cohn J. S., Ramakrishnan R., et al. 2005. Effects of the PPARgamma agonist pioglitazone on lipoprotein metabolism in patients with type 2 diabetes mellitus. J. Clin. Invest. 115: 1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin S., Liu T., Kamanna V. S., Kashyap M. L. 2007. Pioglitazone stimulates apolipoprotein A-I production without affecting HDL removal in HepG2 cells: involvement of PPAR-alpha. Arterioscler. Thromb. Vasc. Biol. 27: 2428–2434. [DOI] [PubMed] [Google Scholar]
- 17.Higuchi K., Law S. W., Hoeg J. M., Schumacher U. K., Meglin N., Jr. Brewer H. B. 1988. Tissue-specific expression of apolipoprotein A-I (ApoA-I) is regulated by the 5′-flanking region of the human ApoA-I gene. J. Biol. Chem. 263: 18530–18536. [PubMed] [Google Scholar]
- 18.Widom R. L., Ladias J. A., Kouidou S., Karathanasis S. K. 1991. Synergistic interactions between transcription factors control expression of the apolipoprotein AI gene in liver cells. Mol. Cell. Biol. 11: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mooradian A. D., Haas M. J., Wong N. C. 2004. Transcriptional control of apolipoprotein A-I gene expression in diabetes. Diabetes. 53: 513–520. [DOI] [PubMed] [Google Scholar]
- 20.Delerive P., Galardi C. M., Bisi J. E., Nicodeme E., Goodwin B. 2004. Identification of liver receptor homolog-1 as a novel regulator of apolipoprotein AI gene transcription. Mol. Endocrinol. 18: 2378–2387. [DOI] [PubMed] [Google Scholar]
- 21.Huuskonen J., Vishnu M., Chau P., Fielding P. E., Fielding C. J. 2006. Liver X receptor inhibits the synthesis and secretion of apolipoprotein A1 by human liver-derived cells. Biochemistry. 45: 15068–15074. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto J., Kimura H., Moriyama S., Odaka H., Momose Y., Sugiyama Y., Sawada H. 2000. Activation of human peroxisome proliferator-activated receptor (PPAR) subtypes by pioglitazone. Biochem. Biophys. Res. Commun. 278: 704–711. [DOI] [PubMed] [Google Scholar]
- 23.Chiquette E., Ramirez G., Defronzo R. 2004. A meta-analysis comparing the effect of thiazolidinediones on cardiovascular risk factors. Arch. Intern. Med. 164: 2097–2104. [DOI] [PubMed] [Google Scholar]
- 24.Szapary P. O., Bloedon L. T., Samaha F. F., Duffy D., Wolfe M. L., Soffer D., Reilly M. P., Chittams J., Rader D. J. 2006. Effects of pioglitazone on lipoproteins, inflammatory markers, and adipokines in nondiabetic patients with metabolic syndrome. Arterioscler. Thromb. Vasc. Biol. 26: 182–188. [DOI] [PubMed] [Google Scholar]
- 25.Goldberg R. B., Kendall D. M., Deeg M. A., Buse J. B., Zagar A. J., Pinaire J. A., Tan M. H., Khan M. A., Perez A. T., Jacober S. J. 2005. A comparison of lipid and glycemic effects of pioglitazone and rosiglitazone in patients with type 2 diabetes and dyslipidemia. Diabetes Care. 28: 1547–1554. [DOI] [PubMed] [Google Scholar]
- 26.Davidson M., Meyer P. M., Haffner S., Feinstein S., Sr. D'Agostino R., Kondos G. T., Perez A., Chen Z., Mazzone T. 2008. Increased high-density lipoprotein cholesterol predicts the pioglitazone-mediated reduction of carotid intima-media thickness progression in patients with type 2 diabetes mellitus. Circulation. 117: 2123–2130. [DOI] [PubMed] [Google Scholar]
- 27.Matsusue K., Haluzik M., Lambert G., Yim S. H., Gavrilova O., Ward J. M., Brewer B., Jr., Reitman M. L., Gonzalez F. J. 2003. Liver-specific disruption of PPARgamma in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J. Clin. Invest. 111: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyers C. D., Kashyap M. L. 2004. Pharmacologic elevation of high-density lipoproteins: recent insights on mechanism of action and atherosclerosis protection. Curr. Opin. Cardiol. 19: 366–373. [DOI] [PubMed] [Google Scholar]
- 29.Jin F. Y., Kamanna V. S., Kashyap M. L. 1997. Niacin decreases removal of high density lipoprotein AI but not cholesterol ester by HepG2 cells. Arterioscler. Thromb. Vasc. Biol. 17: 2020–2028. [DOI] [PubMed] [Google Scholar]
- 30.Eckland D. A., Danhof M. 2000. Clinical pharmacokinetics of pioglitazone. Exp. Clin. Endocrinol. Diabetes. 108 (Suppl. 2): S234–S242. [Google Scholar]