Abstract
Exosomes are bioactive vesicles released from multivesicular bodies (MVB) by intact cells and participate in intercellular signaling. We investigated the presence of lipid-related proteins and bioactive lipids in RBL-2H3 exosomes. Besides a phospholipid scramblase and a fatty acid binding protein, the exosomes contained the whole set of phospholipases (A2, C, and D) together with interacting proteins such as aldolase A and Hsp 70. They also contained the phospholipase D (PLD) / phosphatidate phosphatase 1 (PAP1) pathway leading to the formation of diglycerides. RBL-2H3 exosomes also carried members of the three phospholipase A2 classes: the calcium-dependent cPLA2-IVA, the calcium-independent iPLA2-VIA, and the secreted sPLA2-IIA and V. Remarkably, almost all members of the Ras GTPase superfamily were present, and incubation of exosomes with GTPγS triggered activation of phospholipase A2 (PLA2)and PLD2. A large panel of free fatty acids, including arachidonic acid (AA) and derivatives such as prostaglandin E2 (PGE2) and 15-deoxy-Δ12,14-prostaglandinJ2 (15-d PGJ2), were detected. We observed that the exosomes were internalized by resting and activated RBL cells and that they accumulated in an endosomal compartment. Endosomal concentrations were in the micromolar range for prostaglandins; i.e., concentrations able to trigger prostaglandin-dependent biological responses. Therefore exosomes are carriers of GTP-activatable phospholipases and lipid mediators from cell to cell.
Keywords: exosome, phosphatidate phosphatase, arachidonic acid, docosahexaenoic acid, prostaglandin
Exosomes are nanovesicles (50–100 nm) released from viable cells, either constitutively or upon activation of cell secretion, but not from lysed or apoptotic cells (1). They are secreted from an intracellular compartment, the multivesicular bodies (MVB), or late endosomes (2). The “TfR/tetraspanin/Heat-Shock Protein”-containing exosomes originating from MVB differ from the “CD73 (5′-nucleotidase)/glycophorin/CD45”-containing microvesicles produced by plasma membrane shedding (3) and from “CD31/Annexin V”-containing apoptotic microparticles (4).
Exosomes were first characterized as a pathway for elimination of obsolete proteins during erythrocyte maturation, then as part of an essential process in the immune response, and recently as an enabler of the mechanism that modulates the translational activity of target cells by transferring selected micro RNA between cells (5). Whether exosomes participate dynamically in lipid metabolism is not known.
Exosomes appear to be involved in additional intercellular signaling beside soluble agonists. They interact with cell peripheral receptors, such as CD91 (6), a member of the LDL receptor-related proteins (LRP) receptors, and Tim4 (7), the phosphatidylserine receptor (8), a G protein coupled receptor (GPCR) member. Other nanovesicles similar to exosomes trigger the Notch signaling pathway (9).
The exosome biogenesis pathway can be “hijacked” by pathogens like the human immunodeficiency virus (HIV), by proteins like prions involved in Creutzfeld-Jacob disease (10), and by the amyloid precursor protein (APP) of Alzheimer's disease (11, 12).
Mast cell-derived exosomes trigger functional maturation of dendritic cells (DC) (6). The DC maturation process has been shown to involve lysophosphatidylcholine and secreted PLA2 (13), and prostaglandins (14). We previously reported that exosomes from RBL-2H3 cells contain a high amount of lysophosphatidylcholine (LPC) (15) and that phospholipase D was involved in exosome release (16).
We undertook a large analysis of RBL-2H3 exosome content by proteomic high-throughput analysis together with immunodetection and determination of lipolytic activities. We showed that exosomes can behave as “signalosomes” not only by transporting GTP-activatable phospholipases D2 (PLD2) and phospholipase A2 (PLA2), but also by carrying the whole set of prostaglandins, including prostaglandin E2 (PGE2) and the peroxysome proliferator activated receptor γ (PPARγ) agonist 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2). We observed that exosomes could traffic between resting or activated RBL-2H3 cells, thereby modulating RBL-2H3 cell activation by means of the lipid messengers they carry. In addition, exosomes could constitute a mechanism of entry for 15d-PGJ2, as the way this prostaglandin enters cells is as yet unknown (17–19).
EXPERIMENTAL PROCEDURES
Materials
For cell cultures, RPMI 1640, PBS, penicillin, streptomycin, L-glutamine, and FCS were purchased from Invitrogen. 4,4-Difluoro-4-bora-3a,4a-diaza-s-incadene (BODIPY)-PC as phospholipase substrate and BODIPY-ceramide for exosome labeling and uptake detection by immunofluorescence were obtained from Invitrogen Molecular Probes and stored in ethanol at −20°C after dilution. GTPγS was from Sigma. Methyl arachidonyl fluorophosphonate (MAFP) was from Calbiochem. Bromoenol lactone (BEL, or HaloEnol Lactone Suicide Substrate) was from Biomol International. Pyrrolidine-1 and Me-indoxam were generous gifts from Prof. M. H. Gelb (University of Washington, Seattle, WA). The cPLA2 monoclonal antibody (recognizing type IVA) and iPLA2 polyclonal antibody (recognizing type VIA) were from Santa Cruz Biotechnology. Mouse sPLA2-IIA and -V recombinant proteins and sPLA2 antibodies raised against type IIA and V sPLA2 were produced as described (20). Polyclonal antibodies against cyclooxygenase (COX)-1 and COX-2 were from Santa Cruz Biotechnology. Rabbit polyclonal anti-PLD antibody (N-PLD4) was from Johnson Pharmaceutical Research Institute (Raritan, NJ) and was kindly supplied by Dr. D. Uhlinger. The HA.11 monoclonal mouse anti-HA antibody (clone 16B12) was from Eurogentec. Anti-CD63 antibody was from Santa Cruz Biotechnology. The anti-LBPA antibody (6C4) was kindly supplied by Dr. T. Kobayashi, Riken Institute, Tokyo, Japan (21). Secondary antibodies labeled with horseradish peroxidase were from Santa Cruz Biotechnology and PhycoErythrine-labeled antibodies from BD Bioscience. FITC-labeled cholera toxin subunit was from Sigma. Isotype antibodies for flow cytometry were from Santa Cruz Biotechnology. Rhodamine-phosphatidylethanolamine (Rh-PE) was from Avanti Polar Lipids (Birmingham, AL). Protease inhibitor cocktail (P8340) was provided by Sigma. Chemical solvents were purchased from Sigma-Aldrich or from Merck for HPLC grade.
Methods
Cell lines.
RBL-2H3 (also referred to as RBLwt for RBL wild-type cells) were grown in RPMI 1640 supplemented with 10% (v/v) FCS, 4 mM L-glutamine, 140 units/ml penicillin, and 140 µg/ml streptomycin in a 5% CO2 humidified atmosphere at 37°C.
Cells overexpressing the human HA-tagged PLD2 (also referred to as RBLpld2 cells) were obtained by electroporation (250 V, 500 µF) of RBL-2H3 cells with linearized pcDNA3.1 vector containing the HA-tagged cDNA of human PLD2. PLD2 overexpressing cells were selected with G418 (500 µg/ml). Clones grown within one week were recovered in PBS-EDTA, mixed, expanded, and stored in liquid nitrogen. The characteristics of the RBLpld2 cell line are reported in the supplemental data.
RBL cell degranulation.
Cell secretion was monitored by the amount of 14C-serotonin released from the MVB compartment. Adherent cells were loaded overnight with 14C-serotonin, washed, and incubated for 4 h with saturating concentrations of IgE directed against dinitrophenol conjugated to serum albumin (DNP-HSA, Sigma). Cell activation was triggered by FcϵRI cross-linking with DNP-has, and the radioactivity released was measured by scintillation counting.
Exosome preparation.
For exosome preparation 1.5 × 107 adherent cells were harvested with PBS-EDTA and added into 250 ml complete RPMI medium in a spinner bottle for cell culture in suspension. Culture volume was doubled every day in the spinner bottles to maintain a cell density of around 0.25 × 106 cells/ml for good cell viability until about 109 cells were produced overall.
The cells were spun down, washed with DMEM medium, and concentrated to 108 cells in 10 ml of DMEM without FCS to avoid contamination by any microvesicles that might be present in the fetal calf serum. Exosomes were recovered following 20 min cell stimulation by ionomycin (1 µM final) and purified by differential centrifugations as reported previously (16). Correct exosome preparation required viable cells, which were checked by trypan blue exclusion. Briefly, viable activated cells were eliminated by centrifugation at 300 g for 5 min. To get rid of possible cell debris, the supernatant underwent two consecutive centrifugations at 2000 g for 20 min at 4°C and 10,000 g for 30 min at 4°C. Exosomes were isolated from the 10,000 g supernatant by ultracentrifugation at 110,000 g for 70 min at 4°C. The pellet was resuspended in PBS and centrifuged again at 110,000 g for 70 min at 4°C. The final pellet referred to as exosomes was resuspended in PBS for analysis. The quality of the preparations was checked by D2O/sucrose discontinuous gradient (1) and by electron microscopy (performed by D. Lankar, Institut Curie Paris; B. Payré, CMEAB, UPS Toulouse III, France). We also checked the size homogeneity of vesicles obtained using a Zetasizer Nano ZS90 (see below). Protein concentration was determined by the Lowry method (22) in the presence of 0.1% w/v SDS final.
Size distribution and zeta potential analysis of RBL-2H3-derived exosomes.
The Zetasizer Nano ZS 90 (Malvern Instruments, Orsay, France), allowed the analysis of particles with sizes ranging from 1 nm to 3 µm. Exosomes (50 µg from two pooled preparations) derived from RBLwt or RBLpld2 cells were diluted in 1 ml PBS, and parameters such as zeta potential (electronegativity) and size distribution were analyzed at 37°C according to the manufac- turer's instructions (see supplemental Fig. II).
Quantification of exosome vesicles.
The correlation between exosome protein content and the number of vesicles was established by FACS analysis on the basis of the method used to quantify the number of circulating microparticles (4). Exosomes were diluted in PBS-EDTA and the number of vesicles was taken as the number of events in the SSC/FSC quadrant.
Quantification of exosome internalization.
Exosomes were labeled with the fluorescent lipid probe BODIPY-ceramide so that fluorescence monitored the amount of vesicles directly (16). Fluorescent exosomes (25 µg proteins) were incubated with 106 adherent cells. At appropriate times, the excess of added exosomes removed, the cells washed, and cell-associated fluorescence monitoring internalized exosomes were extracted with butanol and quantified. The fluorescence was converted into µg exosome protein using a calibration curve as previously reported (16).
Confocal microscopy.
Internalization of fluorescent exosomes was monitored under a Zeiss LSM 510 confocal microscope on live cells using LSM 510 software. Cells (3 × 104 in RPMI medium buffered with 25 mM Hepes) were seeded in LabTek chambers and kept overnight in an incubator. Then medium was removed, and 0.5 ml of the same fresh medium was added. The LabTek chambers were placed into a microscope chamber adaptor warmed to 37°C and with CO2 flow. Exosomes (20 μg), previously made fluorescent by a 1 h incubation at 37°C with 1.2 μM BODIPY-ceramide (23) and washed, were added in a small volume (20 μl) into the cell medium and data acquisition started.
The compartment of exosome internalization in target cells was characterized by antibodies directed against late endosome markers. 2 × 105 cells were seeded on coverglass in 1 ml RPMI culture medium and incubated for 24 h with 75 µl anti-LBPA antibody (hybridoma supernatant) or 50 µl (10 µg) anti-CD63 antibody. Cells were washed with PBS, then overlaid with 0.5 ml culture medium, and 10 µg fluorescent (BODIPY-ceramide labeled) exosomes were added. Incubation proceeded for 4 h at 37°C. Cells were washed with PBS and fixed with 3.7% PFA for 20 min and washed again. The remaining PFA was quenched with 50 mM NH4Cl for 10 min. The cells were washed with PBS, then maintained for 30 min in PBS 3% BSA. Permeabilization was performed with 0.05% saponin in PBS 3% BSA for 10 min. The cells were washed and incubated 30 min with appropriate secondary antibodies (anti-mouse PE for LBPA and anti-goat FITC for CD63). Coverslips were mounted with Mowiol, and samples were examined under a LSM 510 confocal microscope.
To label the late endosome compartment with Rhodamine-PE, cells were incubated in suspension at 4°C with 3 µM final of the probe, washed with PBS 3% BSA, and incubated for an additional 3 h at 37°C. Cells were seeded on coverslips and pulsed for 4 h with fluorescent exosomes. After washing, cells were fixed with PFA and examined with the LSM 510.
Plasma membrane labeling was first performed on live RBLpld2 cells with fluorescent cholera toxin added in PBS containing 10% BSA at 4°C for 30 min. The cells were washed with PBS, fixed with 3% PFA for 20 min at 4°C, and permeabilized for 15 min at room temperature with 0.05% saponin in PBS-BSA. HA-PLD2 location was then detected by incubation with anti- HA antibody diluted at 1/50, followed by incubation with a se- condary antibody (45 min each antibody) at room temperature. Acquisition was performed with a Zeiss LSM 510 confocal microscope.
Measurement of phospholipase activities.
Identification of the various phospholipase activities in exosomes was performed by HPLC using a fluorescent phosphatidylcholine (BODIPY-PC) as substrate. Intact or sonicated (2 × 10 s output 4-5 Micro Tip, Branson Sonifier) exosomes (50 µg protein) were preincubated 10 min at room temperature in a total volume of 500 µl PBS containing 2 mM Ca2+/Mg2+ with 5 µl protease inhibitor cocktail, and as required 50 µM PLA2 inhibitors (MAFP for cPLA > iPLA2; pyrrolidine for cPLA2; Me-indoxam for sPLA2; BEL for iPLA2). When calcium was not required, Ca2+/Mg2+ free-PBS was used. Con- centrations of inhibitors and their specificity have already been documented (24, 25). When GTP dependency was checked, the nonhydrolysable analog of GTP (GTPγS) was added 10 min before monitoring phospholipase activity. Substrate (1 µl BODIPY-PC, 2.34 µM final) was supplied in ethanol (0.2–2%v/v final). The reaction was performed for 1 h at 37°C. Fluorescent lipids were extracted with 2 × 500 µl 1-n-butanol and were resolved by HPLC (see below).
Measurement of PA phosphatase activity.
Fluorescent phosphatidic acid (BODIPY-PA) was prepared from BODIPY-PC by in vitro hydrolysis with commercial phospholipase D. For enzymatic PAP activity measurement, 50 µg of exosomes were preincubated for 10 min at room temperature in a total volume of 500 µl of PBS with 2 mM Ca2+/Mg2+, with or without 100 µM GTPγS, in the presence of 5 µl protease inhibitor cocktail. The reaction was started by addition of 1 µl BODIPY-PA (1 µM final) supplied in ethanol and incubation proceeded at 37°C. Fluorescent lipids were extracted with 2 × 500 µl 1-n-butanol and resolved by HPLC.
HPLC analysis.
HPLC separation and quantification of BODIPY-PC-derived products were performed as already reported (26) on a silica diol column with a solvent flow rate of 0.4 ml/min. Fluorescent standards of lysophosphatidylcholine (LPC), phosphatidic acid (PA), diglycerides (DG) and phosphatidylethanol (PEt) were prepared by in vitro incubations of BODIPY-PC with appropriate lipolytic enzymes.
Calibration curves for quantification were plotted with BODIPY-PC as standard. HPLC peaks across chromatograms were identified by fluorescent standards injected in the middle of each series of samples to overcome variations in retention times.
Phospholipase immunodetection.
For phospholipases A2, cells and exosomes were lysed in Laemmli sample buffer at 95°C for 10 min and sonicated. 10 mM EDTA was added for phospholipase D detection. 40 µg of proteins were run on 7.5% SDS-PAGE and transferred onto PVDF membrane. The membranes were saturated with 5% nonfat milk in TBS 0.1% Tween 20 for 1 h at room temperature and blotted at 4°C overnight with mouse or rabbit primary antibodies supplied in blotting buffer. Membranes were then washed and incubated in TBS 0.1% Tween 20 with HRP-labeled anti-mouse IgG or anti-rabbit IgG secondary antibodies for 1 h at room temperature.
For sPLA2 detection, 40 µg of exosome proteins were separated on a 15% SDS-polyacrylamide gel, compared to 50 ng of group IIa and V recombinant sPLA2 proteins as standards, and transferred onto PVDF membrane. Membranes were saturated in NETG buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris pH 7.4, 0.05% Triton X-100, 0.25% gelatin), washed in PBS 0.05% Tween 20, and incubated in NETG buffer with HRP-labeled anti-rabbit IgG secondary antibodies.
In all cases, the signal was detected by the enhanced chemiluminescence system from GE Healthcare/Amersham.
Cyclooxygenase detection by flow cytometry.
Exosomes (5 µg proteins) were bound on 4 µm beads (5 µl aldehyde sulfate latex beads; Invitrogen) for 1 h at room temperature under gentle shaking. Unoccupied sites were saturated with vesicle-free FCS for 1 h at room temperature. Beads were spun down, washed with PBS, and resuspended in FACS buffer. Bound exosomes were incubated with anti-COX-1 or anti-COX-2 primary antibodies, or control isotype for 1 h at room temperature and spun down, then labeled for 30 min with secondary FITC-labeled antibody. COX expression was analyzed by flow cytometry on the FITC channel (FL-1) of a FACSCalibur analyzer (Becton-Dickinson) using settings previously reported (16).
Protein analysis.
High-throughput protein analysis was performed on 100 µg protein of purified exosome, first separated by one-dimensional SDS-PAGE. The protein gel lane was cut into 16 pieces, which were digested by trypsin. Tryptic peptides were analyzed by nanoLC-MS-MS with a Qstar XL spectrometer (Applied Biosystems). Data were searched against mouse entries in Sprot-Trembl with the Mascot software. Analyses were performed at the IPBS, CNRS, Toulouse, France (27).
Lipid analysis.
Determination of free fatty acids in exosomes was performed at the lipidomics facility of IFR-BMT (Institut Fédératif de Recherche Bio-Medicale de Toulouse), Toulouse, France. RBL-2H3 derived exosomes (100 µg) were incubated in PBS with 2 mM Ca2+/Mg2+ at 37°C for 4 h. The lipids were extracted by the Bligh and Dyer method (28) in the presence of EGTA in water. Fatty acids were methylated and further analyzed by gas chromatography with an HP5890 instrument (29).
Prostaglandins were quantified by GC-MS at the lipidomics facility of IMBL/INSA-Lyon, Villeurbanne, France. Lipids from RBL-2H3 derived exosomes (70 µg) were extracted with ethylacetate, derivatized into pentafluorobenzyl esters, purified by silica gel TLC using chloroform/ethanol (93:7, v/v), and then modified into trimethylsilyl ethers before analysis by GC-MS. Samples were spiked with 10 ng of deuterated prostaglandin standards (Cayman) and GC-MS was carried out with a Hewlett Packard quadrupole mass spectrometer interfaced with a Hewlett Packard gas chromatograph (30).
Data presentation.
HPLC profiles and phospholipase determinations representative of at least two independent experiments were plotted. Pooled exosome preparations from two to three experiments were used for protein and lipid analysis, which fulfilled the quality control stipulations of the respective facilities (IPBS and IFR-BMT, Toulouse, France; IMBL/INSA-Lyon, Villeurbanne, France). Confocal pictures were representative of at least two experiments performed by distinct operators. Errors bars corresponded to SEM from three determinations.
RESULTS
Lipid-related proteins in exosomes
To determine which of the diverse lipid-related proteins were present on the exosomes, we first performed an exhaustive protein analysis of the vesicles. The proteins found in the present study are reported in Table 1. Typical exosome markers, such as the transferring receptor, tetraspanins (CD63, CD81, CD82), and heat shock proteins, were detected (Table 1A), assessing the quality of the preparation. Exosomes were also characterized by their size and their electronegativity (supplemental Fig. II).
TABLE 1.
Partial protein content of RBL-2H3wt exosomes
| Protein Category | Accession Number | Protein Type | Observations |
|---|---|---|---|
| A) Exosome markers | Q62351 | Transferrin receptor | |
| P41731 | CD63 antigen | Tetraspanin | |
| P40237 | CD82 antigen | Tetraspanin | |
| P35762 | CD81 antigen | Tetraspanin | |
| P11499 | Heat-shock protein 90-β | ||
| P63017 | Heat-shock cognate 70 kDa (Hsc70) | Interacts with phospholipase A2 (iPLA2) | |
| B) Lipid-related proteins | Q8K4S1 | Phosphoinositide-specific phospholipase C ϵ | Involved in phosphoinositide signaling |
| Q9JJ00 | Phospholipid scramblase | Mix phospholipids between membrane leaflets | |
| Q05816 | Fatty acid binding protein (E-FABP) | Free fatty acid transporter | |
| Q5SRA8 | Prostaglandin F2 receptor negative regulator | Interacts with CD81 | |
| C) Phospholipase partners | P05064 | Fructose- bisphosphate aldolase | Interacts with phospholipase D (PLD2) |
| P63017 | Heat Shock protein 70 kDa | Interacts with phospholipase A2 (iPLA2) | |
| P67871 | Casein kinase II β subunit | Interacts with phospholipase A2 (sPLA2) | |
| D) GTP binding proteins | Heterotrimeric | ||
| P08752 | G(i)α-2 subunit | ||
| P62874 | G(i)G(s) β subunit 1 | ||
| P62880 | G(i)G(s) β subunit 2 | ||
| Monomeric GTPases | |||
| P35278 | Ras-related prorein Rab | ||
| Q9QUI0 | Transforming protein RhoA | Interacts and activates phospholipase D (PLD2) | |
| P63835 | Ras-related protein Rap-1A | ||
| Q61411 | Transforming protein P21/H-Ras-1 (c-H-Ras) | ||
| Q61820 | GTP-binding nuclear protein Ran | ||
| P62331 | ADP-ribosylation factor 6 | Interacts and activates phospholipase D (PLD2) | |
| Q8BGX0 | GTP-binding protein ARD-1 | ||
| (ADP-ribosylation factor domain protein 1) |
Only proteins related to exosome markers, lipid metabolism, and G proteins are reported. The overall analysis identified 382 different proteins. Observations are detailed in the text.
Regarding lipid-related proteins, we found a phospholipid scramblase, a protein that transports phospholipids between the two membrane leaflets, in both directions. The presence of this protein was consistent with the lack of membrane phospholipid asymmetry we reported earlier in RBL-derived exosomes (16). Also a member of the fatty acid binding proteins (E-FABP) was detected. FABPs constitute a multigene family of structurally homologous cytosolic proteins that bind and transport polyunsaturated fatty acids, such as arachidonic acid (AA) (31). Another type of protein was a prostaglandin F2 receptor negative regulator, also called FPRP (32). FPRP associates with the PGF2α receptor, thereby reducing ligand binding (33). However, the PGF2 receptor was not found in exosomes, and the presence of the FPRP protein might be better related to its ability to form a tight complex with the tetra- span molecule CD81 (32).
Among the phospholipases, only phospholipase Cϵ hydrolyzing phosphoinositides was detected (Table 1B). Note that proteins known to interact with phospholipases D and A2 were present (Table 1C). Fructose bisphosphate aldolase interacts directly with phospholipase D isoform PLD2 and inhibits its activity (34). The exosomes contained casein kinase II (cK2) that can phosphorylate PLD2 (35) and can also interact with sPLA2-IIA (36), precisely one of the sPLA2 isoforms we detected in the present work. Hsp 70, one of the typical exosome markers (Table 1A), has also been shown to interact with iPLA2 (37).
Phospholipase Cϵ has been shown to be regulated by G proteins, either the subunits of heterotrimeric G proteins or monomeric GTPases (18), both being recovered in the exosomes (Table 1D). This prompted us to consider that GTPases could participate in the regulation of exosome lipolytic enzymes. Note that exosomes contained almost all members of the Ras superfamily GTPases [ARF, Rho, Rap, Rab, p21Ras, and Ran (Table 1D)] except Cdc42 (38). Possible pathways connecting the Ras superfamily GTPases and phospholipases have been reported. The GTPases RhoA and Arf 6 (Table 1D) are direct activators of PLD2 (39, 40) from rat or human origin (41).
Exosomes contain the PLD/PAP pathway
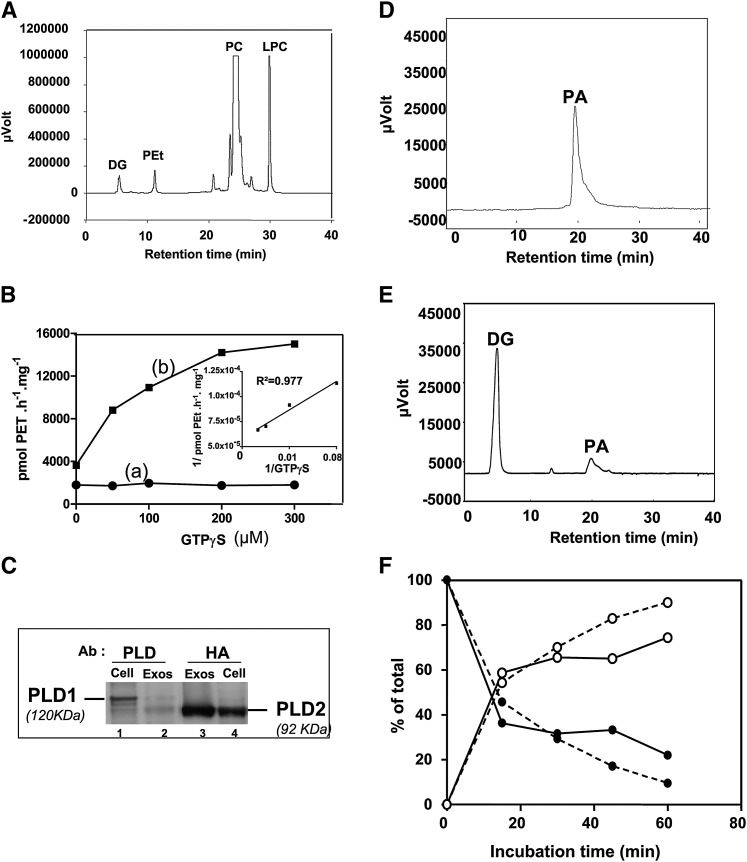
Fig. 1A reports the presence of DG, PEt, and LPC when RBLwt exosomes were incubated with the fluorescent and membrane-diffusible phosphatidylcholine. We investigated whether an autonomous regulation of the lipolytic enzymes involved in the production of these lipid mediators could occur in exosomes. Addition of GTPγS up to 300 µM in RBLwt exosomes had no effect on the PLD activity (Fig. 1B, curve a). The activity was not increased by exosome sonication. Immunodetection showed the selective sorting of the PLD2 isoform in exosomes (Fig. 1C, lane 2) compared with the parental cells, containing mainly PLD1 (Fig. 1C, lane 1). PLD2 activation in RBLwt exosomes could be repressed by aldolase A, which was reported by the protein analysis (Table 1 [P05064] fructose bis-phosphate aldolase) and has been established as a direct inhibitor of PLD2 by acting on its PH domain (34). Therefore, the occupation of the PH domain might prevent activation of the phospholipase.
Fig. 1.
Exosomes contain the phospholipase D/ phosphatidate phosphatase pathway. A: Presence of phospholipase-mediated second messengers on exosomes. Exosomes from RBLwt cells were incubated for 1 h at 37°C with BODIPY-phosphatidylcholine (BODIPY-PC) as substrate. DG, diglycerides; PEt, phosphatidylethanol; PC, phosphatidylcholine; LPC, lysophosphatidylcholine. DG indicates the presence of a DG producing enzyme (phospholipase C or phosphatidate phosphatase); PEt was indicative of a phospholipase D activity, whereas LPC accounted for phospholipase A2 activity. B: Comparative regulation by GTPγs of PLD activity in RBLwt or RBLpld2 exosomes. Exosomes prepared from (a) RBLwt cells (•) or from (b) RBLpld2 cells (▪)were incubated with BODIPY-PC for 1 h at 37°C in the presence of increasing concentrations of GTPγS. Activity of PLD is expressed as pmol PET(phosphatidylethanol)/h/mg protein. Inset: Double-reciprocal plot: 1/(activity) versus 1/[GTPγS]. C: The overexpressed human HA-PLD2 is targeted to exosomes. Cell lysate and RBLpld2 exosome samples were blotted either with the anti-PLD antibody (N-PLD4; see “Materials”), recognizing both PLD1 and PLD2 isoforms, or with the anti-HA antibody. D: Typical HPLC profile of BODIPY-labeled phosphatidic acid (BODIPY-PA) prepared from in vitro assay with phospholipase D (see “Methods”).The presence of positively charged triethylamine in the solvents delayed the elution of PA, leading to a characteristic asymmetrical peak. E: PA processing to yield diglyceride by RBLwt exosomes. Incubation of BODIPY-PA with exosomes proceeded for up to 1 h at 37°C, and then the products were separated as in Fig. 1D. F: Kinetics of PA hydrolysis. RBLwt exosomes were incubated with or without 100 µM GTPγS for various periods of time. The results are expressed as a percentage of the total products DG (○) + PA (•), with (solid lines) or without (dashed lines) GTPγS.
We expected to modify the natural stoichiometry between the putative inhibitor aldolase A and PLD2 by overexpressing the human HA-PLD2 in RBL cells (supplemental Fig. II), the hHA-PLD2 being targeted to exosomes (Fig. 1C, lanes 3, 4). Indeed, the basal PLD activity in RBLpld2 exosomes was twice as high as that of RBLwt exosomes (Fig. 1B; GTPγS = 0). When increasing amounts of GTPγS were added, a clear GTP dependency of the PLD activity in RBLpld2 exosomes was then observed (Fig. 1B, curve b). Phospholipase D activity generates the transphosphatidylation product PEt in the presence of ethanol (Fig. 1A), which competes with the water required to form PA. However, even in the absence of ethanol, PA was not detected across the chromatograms, suggesting the presence of a phosphatidate phosphatase (PAP1) on exosomes. When purified BODIPY-PA was injected into the HPLC system, the resulting peak exhibited a typical asymmetrical shape (Fig. 1D), which was not observed in any chromatograms obtained from exosome incubations with BODIPY-PC. Indeed, upon incubation of BODIPY-PA with exosomes, a strong conversion into diglycerides was observed (Fig. 1E). Kinetics analysis demonstrated that 60% of the PA was hydrolyzed into diglycerides within 15 min (Fig. 1F), indicating the presence of a very active PAP1 in intact exosomes. The kinetics of PA hydrolysis were similar in the presence or absence of GTPγS (Fig. 1F).
Exosomes from RBL cells carry GTP-activatable PLA2 and contain the three classes of PLA2
Fig. 1A reported the presence of PLA2 activity as evidenced by the high LPC content. During the course of the studies on PLD activation (Fig. 1) we noticed a GTP-dependent enhancement of the LPC peak, both on RBLwt and RBLpld2 exosomes.
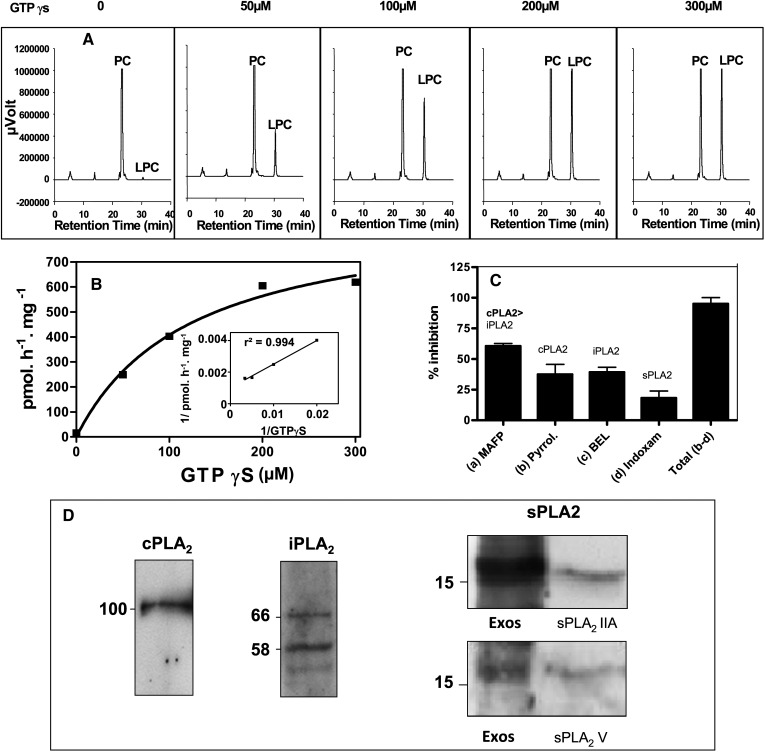
We then investigated whether a dynamic regulation of PLA2 activity might occur in RBLwt exosomes (Fig. 2). GTPγS was able to reveal PLA2 activity on intact exosomes incubated in calcium-free PBS and in the presence of the inhibitor MAFP (Fig. 2A). GTPγS dose-dependent PLA2 activation (Fig. 2B) fits a hyperbolic curve as shown by the linearity of the double-reciprocal plot (Fig. 2B, insert).
Fig. 2.
Exosomes are carriers of GTPγS-activatable phospholipases A2. A: HPLC profiles of exosome phospholipase A2 activity in the presence of GTPγS. Intact RBLwt exosomes were preincubated with MAFP for 10 min at room temperature. The reaction was started by adding BODIPY-PC in the presence of GTPγS at the indicated concentrations. Incubations were for 1 h at 37°C in Ca2+/Mg2+ free-PBS. B: Concentration-dependent effect of GTPγS on exosome MAFP-insensitive PLA2. Activity is expressed in pmol/h/mg of protein from HPLC profiles obtained in Fig. 2A. Inset: Double reciprocal plot: 1/(activity) versus 1/[GTPγS]. C: Effect of class-specific PLA2 inhibitors on total exosome PLA2 activity. Sonicated exosomes from RBLwt cells were preincubated for 10 min at room temperature with 50 µM of various class inhibitors—MAFP (cPLA2 > iPLA2 inhibitor), pyrrolidine (cPLA2 inhibitor), BEL (iPLA2 inhibitor), and Me-indoxam (sPLA2 inhibitor)—in the presence of 100 µM GTPγS. The reaction was started by addition of BODIPY-PC. The respective inhibitions are expressed as percentages of the total activity measured without inhibitor. The sum of inhibitions triggered by pyrolidine, BEL, and indoxam is indicated by the right bar (Total b-d). Results are means of four independent experiments +/−SEM for pyrolidine, BEL and indoxam treatments, and six independent experiments +/−SEM for MAFP treatment. E: Immunodetection of PLA2 class members in exosomes. The calcium-dependent cPLA2−IVA, calcium-independent iPLA2-VIA, and secreted sPLA2-IIA and V were detected in RBLwt-derived exosomes by Western blotting. Exos = exosomes; sPLA2 IIA = recombinant sPLA2-IIA; sPLA2 V = recombinant sPLA2-V.
Exosomes exhibited a higher PLA2 activity following sonication, indicating that the PLA2 were partly located in the exosome lumen. The relative parts played by each PLA2 class (cytosolic calcium-dependent cPLA2, cytosolic calcium-independent iPLA2, and secreted sPLA2) were next investigated in the presence of GTPγS on sonicated exosomes. We observed that MAFP decreased the total PLA2 activity by 60% (Fig. 2C). MAFP inhibits both cPLA2 and, to a lesser extent, iPLA2 (25). We next checked specific inhibitors. The specific cPLA2 inhibitor (pyrrolidine-1) reduced total PLA2 activity by 37% (Fig. 2C), whereas bromoenolactone (BEL), the specific iPLA2 inhibitor, abolished 39% of overall PLA2 activity (Fig. 2C). The concentration of inhibitors was 50 µM; i.e., above that used to inhibit the various PLA2 in cells (24, 25). The sum of the inhibitions triggered by pyrrolidine-1 and BEL in exosomes (Fig. 2C) led to the reduction of global PLA2 activity by 76%, indicating that another type of PLA2 activity was present. Me-indoxam, a specific inhibitor of secreted phospholipases (42) was checked, and it decreased total PLA2 activity by 19% (Fig. 2C). Together, the cumulative effect of the three inhibitors diminished total PLA2 activity by 95 ± 6.5%. The residual 5% activity might be related to PLA2 insensitive to the inhibitors, such as some secreted sPLA2 (43). Therefore, the three classes of PLA2 contributed to the global PLA2 activity detected in the exosomes.
We next assessed the presence of members of the three PLA2 classes by using specific antibodies (Fig. 2D). cPLA2-IVA was detected as a single band, whereas iPLA2-VIA was present as processed forms (44) The iPLA2-VIA can be cleaved at three different sites by caspase 3, which generates various processed forms depending upon the combination of the sites effectively cleaved (45). Fragmented forms of iPLA2, similar to those we reported in Fig. 2D, were observed in erythrocyte-derived exosomes (44). Proteolytic processing has been shown to enhance the iPLA2-VIA activity by removing part or complete ankyrin repeats suggested to function as a negative regulator (45). The form of 66 kDa we observed in Fig. 2D could correspond to the residual protein after caspase 3-mediated cleavage at the DVTD site of the iPLA2, leading to the release of the first ankyrin repeat (45) and making likely that the iPLA2-VIA is highly active in exosomes. Among the third class of PLA2, namely, secreted sPLA2, the presence of sPLA2-IIA and sPLA2-V groups was observed (Fig. 2D). Therefore, RBL exosomes concentrated members of each of the three classes of PLA2 and are thus a unique cell compartment.
Exosomes as carriers of bioactive lipids
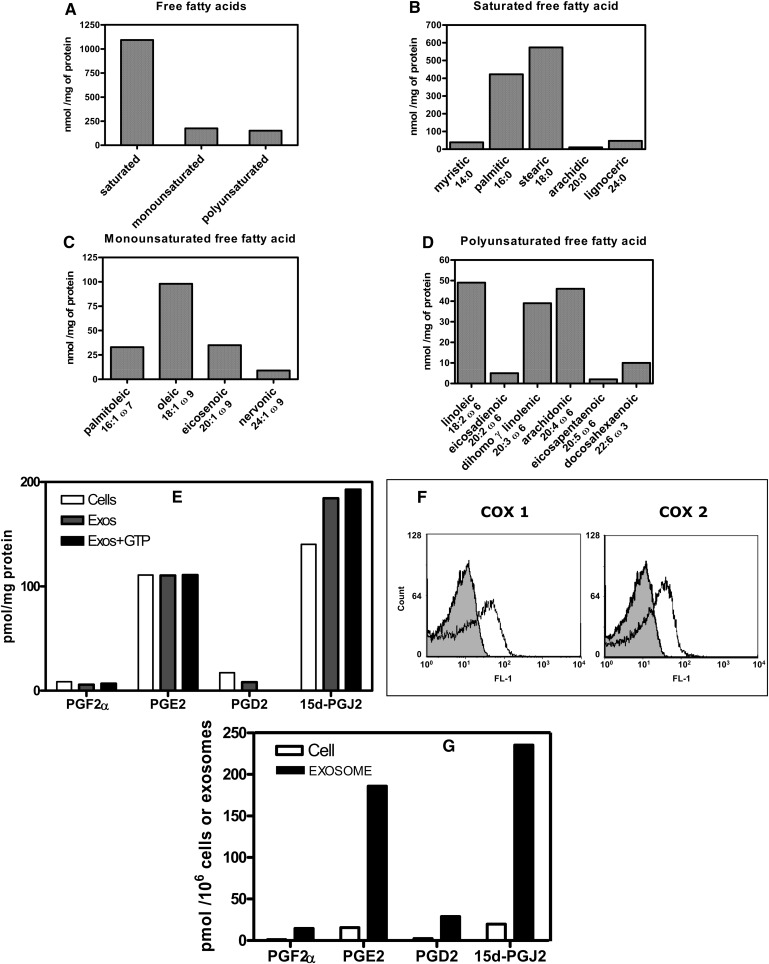
A large panel of free fatty acids was recovered from exosomes (Fig. 3A). Fatty acids could be carried from the parental cells or directly generated within the exosomes by the phospholipase A2 activities. We previously established that 1 mg of exosome protein contained 230 nmoles phospholipid [see Ref. (16)]. Therefore 230 nmoles free fatty acid could be potentially released by the respective PLA2 activities considering they displayed 100% efficiency, with an additional amount of free fatty acid originating from the lysophospholipase activity borne by cPLA2 and iPLA2. However, a total amount of 1,420 nmoles free fatty acid/mg exosome protein was measured (Fig. 3A), indicating that most of the exosome free fatty acid content was already present at the time of exosome membrane biogenesis. The chain length of the saturated fatty acids ranged from 14 to 24 carbons (Fig. 3B); the major ones were palmitic and stearic acids. Monounsaturated fatty acids were essentially from the omega-9 series (Fig. 3C); oleic acid was the most abundant. Polyunsaturated fatty acids almost exclusively contained members of the omega-6 series (Fig. 3D). Only one member of the omega-3 series, namely, docosahexaenoic acid (DHA), was detected. Interestingly, AA accounted for about 30% of the total polyunsaturated fatty acids.
Fig. 3.
Exosomes carry free fatty acids and arachidonic acid-derived bioactive lipids. A–D: Fatty acid distribution in exosomes. Details for analysis are reported in “Methods.” E: Prostaglandin content of exosomes and parent cells. Prostaglandins were quantified by GC-MS in untreated exosomes (Exos) or GTPγS-treated exosomes (Exos +GTP) and in parent cells (Cells) (RBLwt). GTPγS treatment was performed for 1 h at 37°C with 200 µM of the nucleotide. 15d-PGJ2 = 15-deoxy-Δ12,14-PGJ2. PGF2α, PGE2, PGD2 = prostaglandins F2α, E2, and D2 respectively. F: Exosomes contain cyclooxygenases 1 and 2. Analysis of exosome cyclooxygenase expression by flow cytometry, compared to control isotype (gray shaded curves). G: Comparative prostaglandin content of exosomes and parent cells. Prostaglandin content per mg protein plotted in Fig. 3E was converted into cell-equivalents or vesicle (exosome)-equivalents, and normalized to 106 cells or 106 exosome vesicles, respectively. 1 mg protein corresponded to (71.4 ± 0.54) × 105 cells and (5.96 ± 0.13) × 105 exosome vesicles.
We next investigated whether bioactive lipids derived from AA could be found in exosomes. Quantification of prostaglandins was performed by GC-MS and demonstrated the presence of mainly PGE2 and 15-deoxy-Δ12,14-PGJ2 [15d-PGJ2] (Fig. 3E). The 15d-PGJ2 was slightly enriched in the vesicles compared with the parent cells. The respective amounts of the various prostaglandins (PGF2α, PGE2, PGD2, and 15d-PGJ2) in exosomes was not enhanced by incubation with GTPγS (Fig. 3E), indicating that the prostaglandins originated either from the basal exosome PLA2 activities or were loaded in the exosome membrane at the time of their biogenesis in parent cells. Note that COX-1 and COX-2 involved in the early steps of prostaglandin biosynthesis were expressed in exosomes (Fig. 3F), indicating that exosomes could be autonomous biological structures for the biosynthesis of the various prostaglandins. In that respect, the AA concentration in exosome membrane (45 nmoles/mg protein; Fig. 3D) was in excessive compared with the total membrane prostaglandin concentration (0.31 nmoles/mg protein; Fig. 3E).
The number of exosome vesicles per unit protein was established and used to calculate the amount of prostaglandin associated with a defined number of vesicles. Compared with the same number of parental cells, exosomes carried from 12 to 15 times more prostaglandins (Fig. 3G). To our knowledge, this is the first report of vesicle-associated release of prostaglandins from cells.
To evaluate the potential of exosomes as vehicles of bioactive lipids, we investigated whether exosomes could traffic between RBL-2H3 cells.
Exosomes are internalized by resting and activated RBL-2H3 cells and concentrate into endosomes
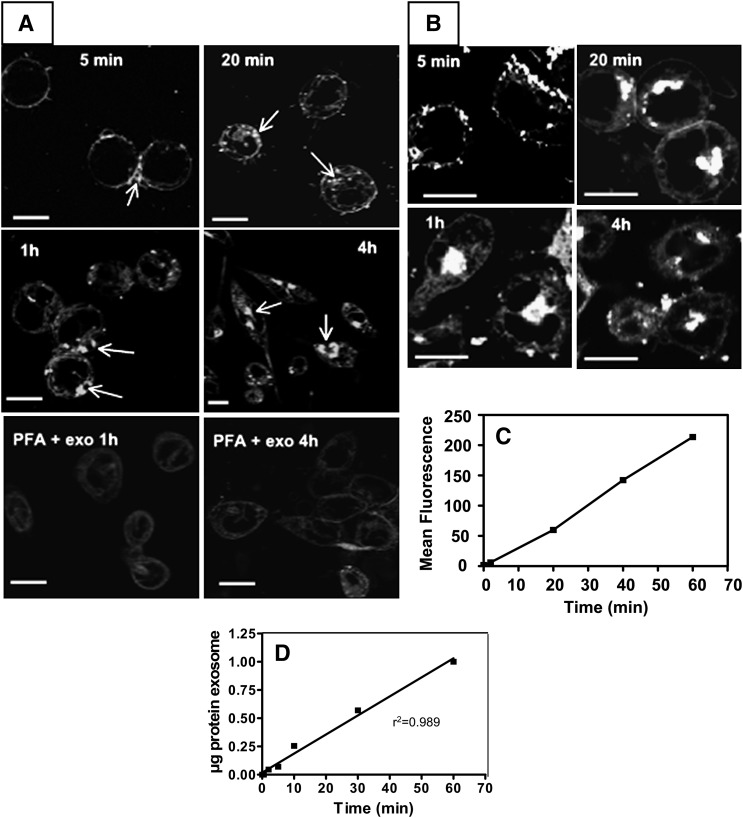
Confocal microscopy performed on living cells showed an accumulation of exosomes on the cell periphery detectable as soon as 5 min, with subsequent internalization leading to the formation of intracellular aggregates indicating storage in an endosomal compartment, as observed after 1 h and 4 h (Fig. 4A). Exosome uptake was an active process, as cross-linking of peripheral protein on target cells by paraformaldehyde impaired intracellular exosome accumulation (Fig. 4A, bottom panels). Only faint, diffuse cell labeling was observed in this case and was attributed to some exchange of the lipidic fluorescent probe between exosomes and the target cell during the step of exosome interaction with the peripheral cell membrane. We investigated whether activated cells, which release exosomes, were also able to internalize them. RBL-2H3 cells activated by Fcϵ-RI cross-linking appeared even more efficient at internalizing exosomes compared with resting ones (Fig. 4B).
Fig. 4.
Monitoring of exosome uptake by RBLwt cells A: Time-lapse monitoring of fluorescent (BODIPY-ceramide)-labeled exosome uptake by confocal microscopy. Uptake of exosomes labeled with BODIPY- ceramide (FITC-type probe) was monitored in a temperature-controlled CO2 chamber with Zeiss LSM 510 software, and pictures were taken at the indicated times. Controls were performed by treating cells with paraformaldehyde (PFA) then washing prior to the addition of exosomes. Uptake was then recorded for 1–4 h (bottom panels). Bars = 5 µm. B: Time-lapse monitoring of fluorescent (BODIPY-ceramide)-labeled exosome by activated RBLwt cells following Fcϵ cross-linking. The cells were incubated overnight with IgE directed against DNP-HSA and Fcϵ cross-linking was triggered by adding DNP-HSA at the same time as fluorescent exosomes. Uptake was monitored as in Fig. 4A. Bars = 5µm. C: Time course of exosome internalization recorded by flow cytometry. Mean fluorescence of cells incubated with BODIPY-ceramide-labeled exosomes and plotted versus time. After each incubation time, noninternalized exosomes were washed away prior to FACS analysis. D: Quantification of exosome internalization. Fluorescent exosomes were incubated as function of time with resting cells. After washing at appropriate times, cell-associated fluorescence monitoring internalized exosomes was extracted with butanol and converted into µg exosome protein. Average of two determinations.
Uptake was linear at least up to 1 h of incubation as shown by flow cytometry monitoring of exosome internalization (Fig. 4C). Similar data were obtained by measuring the BODIPY-ceramide content of target cells following organic extraction of the probe after exosome internalization. This procedure allowed us to quantify the amount of internalized exosomes. It was found that 1 μg of exosome was internalized in 1 h in 106 resting cells (Fig. 4D).
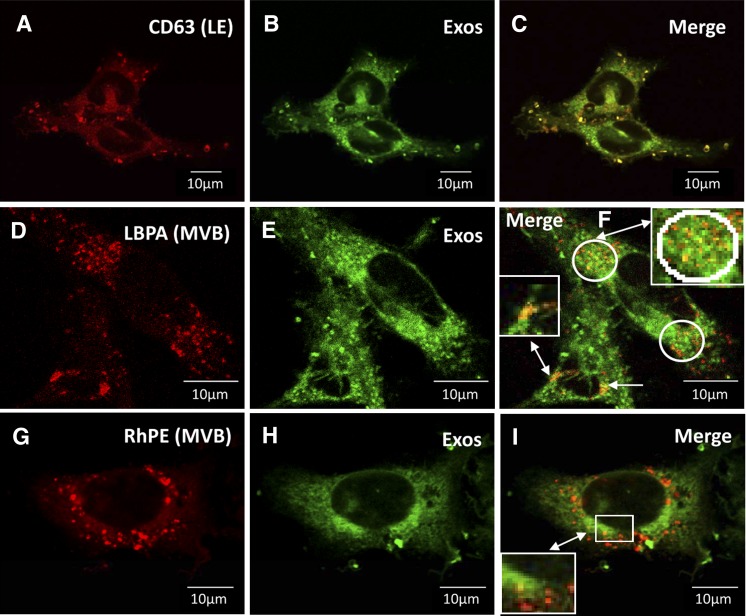
Late endosomes in RBL-2H3 cells feature a dual function: being able to release their contents upon stimulation and being recipients of an endocytosis activity (46). We investigated whether exosomes could be colocalized with endosome markers following their incubation with target cells. Three markers were checked: (i) CD63, a general marker of late endosomes in RBL cells (47); (ii) the lysolipid lysobisphospatidic acid (LBPA; also called BMP for bis[monoacylglycero]phosphate) that accumulates in MVB (21); i.e., late endosomes containing intralumenal vesicles (see supplemental Fig. IIa); and (iii) Rh-PE, a fluorescent lipidic probe that accumulates specifically in late endosomes. Fig. 5A–C shows that exogenously added exosomes were labeled inside the cells by anti-CD63 antibody internalized by fluid-phase endocytosis. Localization of exosomes inside the endocytic track was more precisely investigated by monitoring the MVB distribution with anti-LBPA labeling (Fig. 5D–F). Remarkably, colocalization of exosomes was observed inside MVBs located close to the nucleus (Fig. 5F, white circles and arrows). With Rh-PE as the MVB-specific probe, supplementary evidence was obtained that exogenous exosomes joined the MVB com- partment located close to the nucleus (Fig. 5G–I).
Fig. 5.
Characterization of the intracellular compartment accumulating exosomes. Antibodies against CD63 (late endosomes; LE) or LBPA (multivesicular bodies; MVB) and the MVB probe Rhodamine-PE (RhPE) were preinternalized in the cellular endocytic track. Cells were washed, and then fluorescent BODIPY-ceramide-labeled exosomes were pulsed for 4 h into cells. Excess exosomes were washed away and cells were fixed, permeabilized, and then CD63 or LBPA labeling was revealed by the appropriate secondary antibody. A–C: Endosome anti-CD63 labeling (A), exosome labeling (BODIPY-ceramide FITC) (B), and merge (C) showing the colocalization between exosomes and endosomes (orange and yellow spots represent different amounts of exosomes internalized). D–F: MVB anti-LBPA labeling (D), exosome labeling (BODIPY-ceramide FITC) (E), and merge (F) revealing various amounts of exosomes colocalized with MVBs (monitored by orange or yellow spots) occurring close to the nucleus (white circles and arrows) G–I: MVB Rhodamine-PE labeling (G), exosome labeling (BODIPY-ceramide FITC) (H), and merge (I) indicating different amounts of exosomes colocalizing with MVBs (monitored by orange or yellow spots) detected around the nucleus (white squares).
We estimated the concentration that could be reached by exosome-transported lipid mediators accumulated inside the endosomal compartment of a target cell. Considering an average diameter of 600 nm for an RBL-2H3 cell endosome (48), with an average number of 30 endosomes per RBL-2H3 cell (49), the resulting volume of the entire endosomal compartment is about 500 times lower than the total cell volume (Table 2). Therefore exosome-transported lipid mediators accumulated inside endosomes (Figs. 4, 5) were 500 times more concentrated than if they were diluted in the whole cell volume (Table 2). As a consequence, the resulting PGJ2 endosomal concentration reached 52 µM (Table 2). Other endosomal concentrations were 33µM, 2.4 µM, and 1.7 µM for PGE2, PGD2, and PGF2α respectively, whereas fatty acids, such as AA or DHA, reached millimolar concentrations (4 mM and 0.9 mM, respectively). Thus, target cell endosomes behave as a “concentrator compartment” of lipid mediators transported by exosomes, allowing micromolar concentrations of prostaglandins to be reached (i.e., concentrations able to trigger further biological responses, such as PGJ2-mediated PPARγ activation). Note that the extent of exosome internalization by cells (1 µg exosomes/106 cells/hr; Fig. 4D) was similar to the amount of exosomes released by 106 cells upon stimulation (1.44 ± 0.47 µg), suggesting an efficient cell-to-cell communication process.
TABLE 2.
Estimated concentration of exosome-transported PGJ2 in endosomes of target cells
| A |
B |
C |
D |
E |
F |
|
|---|---|---|---|---|---|---|
| Mean diameter | Mean volume of 106 cells or their endosomes | Exosomes internalized in 106 cells in 1 h | PGJ2 transported by 1 µg exosomes | Final PGJ2 concentrati on in cells or endosomes | Concentration ratio | |
| ml | µg | pmoles | µM | |||
| 1) Exosomes | 1 | 0.193 | ||||
| 2) Cell | 15 µm | 1.76 × 10−3 | 0.11 | 1 | ||
| 3) Endosomes | 600 nm | |||||
| 4) Total endosome compartment | 3.38 × 10−6 | 57.1 | 519 |
The amount of PGJ2 (column D) transported by the amount of exosomes plotted in column C was converted into µM concentrations (column E) either by considering the total volume of the recipient cells (column E line 2), or the total volume of late endosomes present in cells (column E line 4). The resulting values indicated that PGJ2 was about 500 times more concentrated in the total endosome compartment than in the total cell volume (column F). Column A: Values were obtained from literature data (see “Results”) and correspond to RBL-2H3 cells. Column B: The total volume of 106 cells (line 2) and the corresponding volume of their endosome compartment (line 4) were calculated from data in column A (1 ml = 1012 µm3). Column C: Net amount of exosome internalized in 106 cells in 1 h (from Fig. 4D). Column D: Amount of PGJ2 transported by the amount of exosome in column C; calculated from data in Fig. 3E [average of 193 pmol/mg protein in exosomes (Fig. 3E, right bars)]. Column E: PGJ2 concentrations obtained by dividing the amount of PGJ2 (column D) by the volume of cells (column B, line 2) or the volume of their total endosome compartment, considering an average of 30 endosomes per cell (column B, line 4). Column F: Ratio between PGJ2 concentrations in endosomal compartment versus total cell. The same calculation procedure was applied to the other prostaglandins and two typical fatty acids [arachidonic acid (AA) and docosahexaenoic acid (DHA)] from data in Fig. 3 and reported in “Results.”
30 endosomes per cell.
DISCUSSION
Exosomes are nanovesicles released from intact viable cells. They participate in cell-to-cell communication in various physiological and pathological situations, such as the immune response (50), inflammation (51), or atherogenesis (7). Mast cell-derived exosomes trigger functional maturation of dendritic cells (6). This maturation process involves secreted PLA2 (13) and prostaglandins (14). Therefore, we investigated the presence of lipid-related proteins and lipid mediators on exosomes derived from the mast cell line RBL-2H3.
High-throughput protein analysis reported the presence of only four proteins related to lipid metabolism. However, we revealed the presence of other lipolytic proteins by their activity and by immunodetection. The presence of a high content in monomeric G proteins led us to hypothesize specific regulation of these phospholipases in exosomes. The subfamilies of Ras GTPases reported in Table 1 are cytosolic proteins likely to be located inside the exosomes. Therefore, GTP must cross the exosome membrane to activate GTPases and, subsequently, phospholipases. GTP transporters might be present in exosomes, as they have been reported in synaptic vesicles (52), a type of vesicle very similar to exosomes.
The difference in activation of the PLD by GTP between RBLpld2 and RBLwt exosomes appeared related to the stoichiometry between PLD2 and aldolase A. Among the proteins recovered by protein analysis and reported in Table 1, aldolase (P05064) exhibited one of the highest expression scores, whereas PLDs were not even detected by high-throughput analysis. Purified aldolase dosedependently inhibits the PLD2 activity (34); the inhibitor interaction occurs at the PH domain of PLD2. This interaction might impair GTPases, such as RhoA or Arf6, to activate PLD2, or it might prevent another domain of PLD2, the phox homology domain (PX) that exhibits GAP activity, from activating GTPases (53).
Interaction domains on Arf 6 and RhoA with PLD2 have been mapped (39, 40); however, no direct interaction between any of the small G proteins reported in the present study and PLA2 have been reported so far. In whole cells, the cPLA2 and the sPLA2-IIA can be activated downstream of RhoA GTPases with subsequent effects on PGE2 formation (54). Similarly, iPLA2 activation downstream of RhoA has been suggested (55). However, in a cell-free system, such as exosomes, the signaling network between RhoA and the phospholipases might be different compared with whole cells, and exosomes might reveal a specific regulation of PLA2 activities.
Regarding the functional role of exosome phospolipases A2, the calcium-independent iPLA2 has been shown to allow the elimination of erythrocyte-derived exosomes by apoptotic cells (44). Concerning sPLA2, exosomes transporting sPLA2 IIA and V (Fig. 3) might account for the transcellular activity of these phospholipases reported to occur from activated RBL-2H3 cells (56). The group sPLA2-V was reported to be secreted from RBL-2H3 cells and to trigger eicosanoid biosynthesis in neighboring target granulocytic cells (56). The sPLA2-V activity appears related to IgE-dependent PGD2 formation and to enhanced exocytosis in RBL-2H3 cells (57). Vesicular secretion of cPLA2 has not been reported so far.
The set of prostaglandins transported by exosomes (Fig. 3) are derived from PGH2, and metabolic conversion of PGH2 has been shown to occur through a transcellular mechanism between two different types of cells, containing either COX-1 and COX-2 or the terminal prostaglandin synthases (58, 59). Transporting from cell to cell metabolic precursors, such as PGD2 (Fig. 3G), and enzymes, such as COX-1 and COX-2 (Fig. 3F), for the early steps of prostaglandin biosynthesis, exosomes could account for transcellular metabolism of prostanoids reported to occur between normal and tumor cells (59). Arachidonic acid present in exosomes (Fig. 3D) would serve as transcellular biosynthetic precursor. Although eicosanoid transcellular metabolism has been reported to occur at inflammation sites between different cell types (60), one can conceive that RBL-derived exosomes are a mixed population bearing either the COX-1 and COX-2 or the terminal prostaglandin synthases; therefore, exosome exchange between RBL-2H3 cells would be required to complete the entire prostanoid biosynthesis pathway. In this respect, we showed earlier that RBL-2H3 cells release three distinct subpopulations of exosomes (23). The present work opens further investigations to understand the mechanisms underlying the transcellular metabolism of eicosanoids.
This transcellular metabolism requires exosome trafficking between cells. We have shown that exosomes added to target cells are rapidly internalized (Fig. 4) into the endocytic track and join the MVB network located close to the nucleus (Fig. 5). It is likely that the GTP-dependent activation of PLD and PLA2 we observed in exosomes could occur inside the endocytic track of target cells. Many GTPases are present within the endocytosis track, some of them maintained in an active state even in unstimulated cells (61). GTP-activated phospholipases could participate in exosome fusion with the limiting membrane of the endosome, a process called “back-fusion” (62, 63). This process allows the lumen content of the exosomes to be released into the cytosol. Back-fusion molecular mechanisms require the lipid LBPA, whose biosynthesis involves a cPLA2-type activity (64, 65), as well as a combination of PLA2 and PLD activities (66). A previous report describes GTP-dependent cPLA2-mediated fusion of secretory granules (67). Phosphatidic acid resulting from PLD activity is a fusogenic compound in presence of calcium (68). Di- glycerides generated by the PI-PLCϵ (Table 1) or the PLD/PA phosphatase pathway (Fig. 1) could participate in exosome-endosome fusion processes by lowering the surface pressure of the phospholipids (69). More DG can be expected in RBLpld2 exosomes and could account for the modification of the biophysical parameters (size and electronegativity) shown in supplemental Fig. II. In addition, phospholipid mixing between exosome and endosome membranes triggered by the scramblase we reported in Table 1 would facilitate membrane fusion.
We established in this work that exosomes transport prostaglandins from the parent cells. RBL-2H3 cells feature a mast cell phenotype, and eicosanoids play an essential role in mast cell physiology by regulating their function in host defense and disease (70). PGE2 can block FcϵRI-mediated exocytosis of mast cells (70). Exosomes, during at least the first 5–20 min (Fig. 5), provide a vehicle for PGE2 to interact with its respective GPCRs on the periphery of target cells. Thereafter, exosome internalization provides the first mechanism described for 15deoxyΔ12,14-PGJ2 to enter the cells and possibly reach its intracellular targets. Actually, no specific peripheral receptors or mechanisms of entry have been identified to-date for this prostaglandin (17, 19). The exosome as a vehicle would allow the plasma membrane to be bypassed and 15d-PGJ2 to accumulate in the endosomes of target cells, from where the prostaglandin would be released into the cytosol after fusion between exosome and endosome membranes. A possible mechanism is summarized in Fig. 6.
Fig. 6.
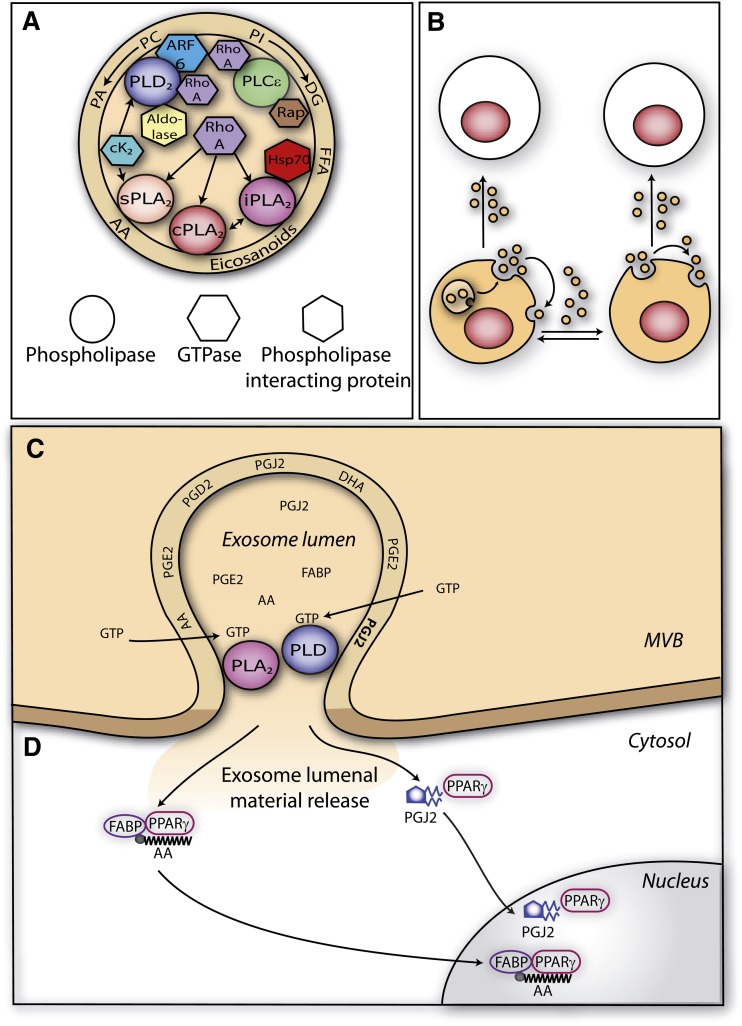
Exosomes as intercellular signalosomes carrying GTP-activatable phospholipases and prostaglandins A: Exosomes carry GTP-activatable phospholipases. Proteins detected in RBLwt-derived exosomes related to phospholipase activation and reported in Table 1 were represented together with the phospholipases detected in the present work. Arrows indicate possible activation pathways based on literature data (see “Discussion”). Note the presence of lipid mediators in the exosome membrane. B: Exosomes as intercellular “signalosomes.” Exosomes released upon cell activation can traffic between resting cells (white) and activated cells (color). Exosomes could trigger autocrine and paracrine-type signals. C and D: Possible mechanism of exosome-mediated bioactive lipid delivery from endosomes in target cells. Inside the intracellular compartment accumulating exosomes in target cells (endosomes, Fig. 5), phospholipases borne by exosomes could participate upon activation by GTP in the fusion between exosomal and endosomal membranes, allowing delivery of exosome content into the cytosol. Exosomes carry prostaglandins, such as the PPARγ agonist 15d-PGJ2 (Fig. 3) and a FABP (Table 1) which can bind arachidonic acid (Fig. 3) and interact with PPARγ (Fig. 6C). The 15d-PGJ2/PPARγ and FABP/arachidonic acid/PPARγ complexes would be further addressed to the nucleus (Fig. 6D).
Exosomes could also supply the 15d-PGJ2 already bound to its receptor, as a recent report indicates the presence of the PPARγ receptor among proteins found in exosomes isolated from human serum (71). Interestingly, the exosome FABP we report in Table1 could bind the AA present in exosomes (Fig. 3) and then interact directly with the PPARγ receptor, the resulting FABP-AA-PPARγ complex being subsequently addressed to the nucleus of target cells to regulate transcription (Fig. 6C) (72). In line with the possible modulation of nuclear receptors by exosome-carried mediators, note that PAP1, the diglyceride-generating enzyme we reported in Fig. 1, has recently been characterized as a transcriptional coactivator of the PPARα receptor (73).
Further experiments are required to support the functional role of exosomes in RBL-2H3 cells. As a first step, we evaluated whether exosomes could carry sufficient amounts of the prostaglandin 15d-PGJ2 to possibly trigger PPARγ activation in target cells. When added to cells, 15d-PGJ2 has been reported to trigger biological effects in the 10–40 µM range (74). Exosome accumulation in endosomes were allowed to reach values > 50 µM (Table 2); i.e., bioactive 15d-PGJ2 concentrations.
Because of the dynamic regulation of their phospholipases by GTP, exosomes appear to behave as “signalosomes” (Fig. 6A). The “signalosomes” would circulate between cells and might regulate their functions whether cells are resting or activated (Fig. 6B). Stimulated RBL-2H3 cells feature enhanced endocytosis (46) and could internalize exosomes they had just released. Preliminary data we obtained indicate that exosomes inhibited Fcϵ-mediated degranulation of RBL-2H3 cells. That this effect involves PGE2, which is known to inhibit FcϵRI-mediated exocytosis of mast cells (70), appears conceivable on the basis of data reported here. Also, by possibly providing 15d-PGJ2 to PPARγ of target cells, exosomes can repress the transcription of proinflammatory mRNAs (75). Circulating simultaneously with allergens that activate cells via FcϵRI receptors, exosomes appear as a signaling device able to modulate the FcϵRI-mediated mast cell response by means of phospholipases and lipid mediators that can be activated.
Supplementary Material
Acknowledgments
The authors thank Justine Bertrand-Michel (Toulouse) and Michel Guichardant (Lyon) for lipidomics analysis; Bruno Payré for performing the transmission electronic microscopy; P.Winterton for correcting the English manuscript; and Dr. Toshihide Kobayashi (Riken Institute, Tokyo, Japan) for supplying the anti-LBPA antibody.
Footnotes
Abbreviations:
- 15-d PGJ2
- 15-deoxy-Δ12,14-prostaglandinJ2
- AA
- arachidonic acid
- BEL
- bromo-eno-lactone
- DG
- diglycerides
- DHA
- docosahexaenoic acid
- FABP
- fatty acid binding protein
- LPC
- lysophosphatidylcholine
- MAFP
- methyl arachidonyl fluorophosphonate
- Me-indoxam
- methyl-indoxam
- MVB
- multivesicular bodies
- PA
- phosphatidic acid
- PAP
- phosphatidate phosphatase
- PC
- phosphatidylcholine
- PEt
- phosphatidylethanol
- PGE2
- prostaglandin E2
- PGF2α
- pros-taglandin F2α
- PLA2
- phospholipase A2
- PLC
- phospholipase C
- PLD2
- phospholipase D2
- PPAR
- peroxysome proliferator activated receptor
- Rh-PE
- rhodamine-phosphatidylethanolamine
This work was supported in part by funding from Agence Nationale pour la Recherche contre le SIDA (ANRS) (C.S) and by internal grants from INSERM.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Thery C., Amigorena S., Raposo G., Clayton A. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3: Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 2.Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., Geuze H. J. 1996. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183: 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hess C., Sadallah S., Hefti A., Landmann R., Schifferli J. A. 1999. Ectosomes released by human neutrophils are specialized functional units. J. Immunol. 163: 4564–4573. [PubMed] [Google Scholar]
- 4.Werner N., Wassmann S., Ahlers P., Kosiol S., Nickenig G. 2006. Circulating CD31+/annexin V+ apoptotic microparticles correlate with coronary endothelial function in patients with coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 26: 112–116. [DOI] [PubMed] [Google Scholar]
- 5.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J. J., Lotvall J. O. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9: 654–659. [DOI] [PubMed] [Google Scholar]
- 6.Skokos D., Botros H. G., Demeure C., Morin J., Peronet R., Birkenmeier G., Boudaly S., Mecheri S. 2003. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J. Immunol. 170: 3037–3045. [DOI] [PubMed] [Google Scholar]
- 7.Zakharova L., Svetlova M., Fomina A. F. 2007. T cell exosomes induce cholesterol accumulation in human monocytes via phosphatidylserine receptor. J. Cell. Physiol. 212: 174–181. [DOI] [PubMed] [Google Scholar]
- 8.Miyanishi M., Tada K., Koike M., Uchiyama Y., Kitamura T., Nagata S. 2007. Identification of Tim4 as a phosphatidylserine receptor. Nature. 450: 435–439. [DOI] [PubMed] [Google Scholar]
- 9.Ristorcelli E., Beraud E., Mathieu S., Lombardo D., Verine A. 2009. Essential role of Notch signaling in apoptosis of human pancreatic tumoral cells mediated by exosomal nanoparticles. Int. J. Cancer. 125: 1016–1026. [DOI] [PubMed] [Google Scholar]
- 10.Alais S., Simoes S., Baas D., Lehmann S., Raposo G., Darlix J. L., Leblanc P. 2008. Mouse neuroblastoma cells release prion infectivity associated with exosomal vesicles. Biol. Cell. 100: 603–615. [DOI] [PubMed] [Google Scholar]
- 11.Schorey J. S., Bhatnagar S. 2008. Exosome function: from tumor immunology to pathogen biology. Traffic. 9: 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharples R. A., Vella L. J., Nisbet R. M., Naylor R., Perez K., Barnham K. J., Masters C. L., Hill A. F. 2008. Inhibition of gamma-secretase causes increased secretion of amyloid precursor protein C-terminal fragments in association with exosomes. FASEB J. 22: 1469–1478. [DOI] [PubMed] [Google Scholar]
- 13.Perrin-Cocon L., Agaugue S., Coutant F., Masurel A., Bezzine S., Lambeau G., Andre P., Lotteau V. 2004. Secretory phospholipase A2 induces dendritic cell maturation. Eur. J. Immunol. 34: 2293–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thurnher M. 2007. Lipids in dendritic cell biology: messengers, effectors, and antigens. J. Leukoc. Biol. 81: 154–160. [DOI] [PubMed] [Google Scholar]
- 15.Laulagnier K., Motta C., Hamdi S., Roy S., Fauvelle F., Pageaux J. F., Kobayashi T., Salles J. P., Perret B., Bonnerot C., et al. 2004. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 380: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laulagnier K., Grand D., Dujardin A., Hamdi S., Vincent-Schneider H., Lankar D., Salles J. P., Bonnerot C., Perret B., Record M. 2004. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett. 572: 11–14. [DOI] [PubMed] [Google Scholar]
- 17.Scher J. U., Pillinger M. H. 2005. 15d-PGJ2: the anti-inflammatory prostaglandin? Clin. Immunol. 114: 100–109. [DOI] [PubMed] [Google Scholar]
- 18.Gandarillas N. L., Bunney T. D., Josephs M. B., Gierschik P., Katan M. 2009. In vitro reconstitution of activation of PLCepsilon by Ras and Rho GTPases. Methods Mol. Biol. 462: 379–389. [DOI] [PubMed] [Google Scholar]
- 19.Scher J. U., Pillinger M. H. 2009. The anti-inflammatory effects of prostaglandins. J. Investig. Med. 57: 703–708. [DOI] [PubMed] [Google Scholar]
- 20.Rouault M., Le Calvez C., Boilard E., Surrel F., Singer A., Ghomashchi F., Bezzine S., Scarzello S., Bollinger J., Gelb M. H., et al. 2007. Recombinant production and properties of binding of the full set of mouse secreted phospholipases A2 to the mouse M-type receptor. Biochemistry. 46: 1647–1662. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi T., Stang E., Fang K. S., de Moerloose P., Parton R. G., Gruenberg J. 1998. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 392: 193–197. [DOI] [PubMed] [Google Scholar]
- 22.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193: 265–275. [PubMed] [Google Scholar]
- 23.Laulagnier K., Vincent-Schneider H., Hamdi S., Subra C., Lankar D., Record M. 2005. Characterization of exosome subpopulations from RBL-2H3 cells using fluorescent lipids. Blood Cells Mol. Dis. 35: 116–121. [DOI] [PubMed] [Google Scholar]
- 24.Allal C., Buisson-Brenac C., Marion V., Claudel-Renard C., Faraut T., Dal Monte P., Streblow D., Record M., Davignon J. L. 2004. Human cytomegalovirus carries a cell-derived phospholipase A2 required for infectivity. J. Virol. 78: 7717–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gubern A., Casas J., Barcelo-Torns M., Barneda D., de la Rosa X., Masgrau R., Picatoste F., Balsinde J., Balboa M. A., Claro E. 2008. Group IVA phospholipase A2 is necessary for the biogenesis of lipid droplets. J. Biol. Chem. 283: 27369–27382. [DOI] [PubMed] [Google Scholar]
- 26.Gayral S., Deleris P., Laulagnier K., Laffargue M., Salles J. P., Perret B., Record M., Breton-Douillon M. 2006. Selective activation of nuclear phospholipase D-1 by g protein-coupled receptor agonists in vascular smooth muscle cells. Circ. Res. 99: 132–139. [DOI] [PubMed] [Google Scholar]
- 27.Bouyssie D., Gonzalez de Peredo A., Mouton E., Albigot R., Roussel L., Ortega N., Cayrol C., Burlet-Schiltz O., Girard J. P., Monsarrat B. 2007. Mascot file parsing and quantification (MFPaQ), a new software to parse, validate, and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: application to the proteomics study of membrane proteins from primary human endothelial cells. Mol. Cell. Proteomics. 6: 1621–1637. [DOI] [PubMed] [Google Scholar]
- 28.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 29.Payre B., de Medina P., Boubekeur N., Mhamdi L., Bertrand-Michel J., Terce F., Fourquaux I., Goudouneche D., Record M., Poirot M., et al. 2008. Microsomal antiestrogen-binding site ligands induce growth control and differentiation of human breast cancer cells through the modulation of cholesterol metabolism. Mol. Cancer Ther. 7: 3707–3718. [DOI] [PubMed] [Google Scholar]
- 30.Soares A. F., Nosjean O., Cozzone D., D'Orazio D., Becchi M., Guichardant M., Ferry G., Boutin J. A., Lagarde M., Geloen A. 2005. Covalent binding of 15-deoxy-delta12,14-prostaglandin J2 to PPARgamma. Biochem. Biophys. Res. Commun. 337: 521–525. [DOI] [PubMed] [Google Scholar]
- 31.Kitanaka N., Owada Y., Okuyama R., Sakagami H., Nourani M. R., Aiba S., Furukawa H., Watanabe M., Ono M., Ohteki T., et al. 2006. Epidermal-type fatty acid binding protein as a negative regulator of IL-12 production in dendritic cells. Biochem. Biophys. Res. Commun. 345: 459–466. [DOI] [PubMed] [Google Scholar]
- 32.Stipp C. S., Orlicky D., Hemler M. E. 2001. FPRP, a major, highly stoichiometric, highly specific CD81- and CD9-associated protein. J. Biol. Chem. 276: 4853–4862. [DOI] [PubMed] [Google Scholar]
- 33.Orlicky D. J., Berry R., Sikela J. M. 1996. Human chromosome 1 localization of the gene for a prostaglandin F2alpha receptor negative regulatory protein. Hum. Genet. 97: 655–658. [DOI] [PubMed] [Google Scholar]
- 34.Kim J. H., Lee S., Lee T. G., Hirata M., Suh P. G., Ryu S. H. 2002. Phospholipase D2 directly interacts with aldolase via Its PH domain. Biochemistry. 41: 3414–3421. [DOI] [PubMed] [Google Scholar]
- 35.Ganley I. G., Walker S. J., Manifava M., Li D., Brown H. A., Ktistakis N. T. 2001. Interaction of phospholipase D1 with a casein-kinase-2-like serine kinase. Biochem. J. 354: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimoyama Y., Sakamoto R., Akaboshi T., Tanaka M., Ohtsuki K. 2001. Characterization of secretory type IIA phospholipase A2 (sPLA2-IIA) as a glycyrrhizin (GL)-binding protein and the GL-induced inhibition of the CK-II-mediated stimulation of sPLA2-IIA activity in vitro. Biol. Pharm. Bull. 24: 1004–1008. [DOI] [PubMed] [Google Scholar]
- 37.Mancuso D. J., Jenkins C. M., Gross R. W. 2000. The genomic organization, complete mRNA sequence, cloning, and expression of a novel human intracellular membrane-associated calcium- independent phospholipase A(2). J. Biol. Chem. 275: 9937–9945. [DOI] [PubMed] [Google Scholar]
- 38.Konstantinopoulos P. A., Karamouzis M. V., Papavassiliou A. G. 2007. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat. Rev. Drug Discov. 6: 541–555. [DOI] [PubMed] [Google Scholar]
- 39.Bae C. D., Min D. S., Fleming I. N., Exton J. H. 1998. Determination of interaction sites on the small G protein RhoA for phospholipase D. J. Biol. Chem. 273: 11596–11604. [DOI] [PubMed] [Google Scholar]
- 40.Hiroyama M., Exton J. H. 2005. Localization and regulation of phospholipase D2 by ARF6. J. Cell. Biochem. 95: 149–164. [DOI] [PubMed] [Google Scholar]
- 41.Le Stunff H., Dokhac L., Bourgoin S., Bader M. F., Harbon S. 2000. Phospholipase D in rat myometrium: occurrence of a membrane-bound ARF6 (ADP-ribosylation factor 6)-regulated activity controlled by betagamma subunits of heterotrimeric G-proteins. Biochem. J. 352: 491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambeau G., Gelb M. H. 2008. Biochemistry and physiology of mammalian secreted phospholipases A2. Annu. Rev. Biochem. 77: 495–520. [DOI] [PubMed] [Google Scholar]
- 43.Singer A. G., Ghomashchi F., Le Calvez C., Bollinger J., Bezzine S., Rouault M., Sadilek M., Nguyen E., Lazdunski M., Lambeau G., et al. 2002. Interfacial kinetic and binding properties of the complete set of human and mouse groups I, II, V, X, and XII secreted phospholipases A2. J. Biol. Chem. 277: 48535–48549. [DOI] [PubMed] [Google Scholar]
- 44.Blanc L., Barres C., Bette-Bobillo P., Vidal M. 2007. Reticulocyte-secreted exosomes bind natural IgM antibodies: involvement of a ROS-activatable endosomal phospholipase iPLA2. Blood. 110: 3407–3416. [DOI] [PubMed] [Google Scholar]
- 45.Lauber K., Bohn E., Krober S. M., Xiao Y. J., Blumenthal S. G., Lindemann R. K., Marini P., Wiedig C., Zobywalski A., Baksh S., et al. 2003. Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell. 113: 717–730. [DOI] [PubMed] [Google Scholar]
- 46.Barbu A. E., Pecht I. 2005. Desensitization of mast cells' secretory response to an immuno-receptor stimulus. Immunol. Lett. 100: 78–87. [DOI] [PubMed] [Google Scholar]
- 47.Amano T., Furuno T., Hirashima N., Ohyama N., Nakanishi M. 2001. Dynamics of intracellular granules with CD63-GFP in rat basophilic leukemia cells. J Biochem. 129: 739–744. [DOI] [PubMed] [Google Scholar]
- 48.Grimberg E., Peng Z., Hammel I., Sagi-Eisenberg R. 2003. Synaptotagmin III is a critical factor for the formation of the perinuclear endocytic recycling compartment and determination of secretory granules size. J. Cell Sci. 116: 145–154. [DOI] [PubMed] [Google Scholar]
- 49.Tadokoro S., Kurimoto T., Nakanishi M., Hirashima N. 2007. Munc18-2 regulates exocytotic membrane fusion positively interacting with syntaxin-3 in RBL-2H3 cells. Mol. Immunol. 44: 3427–3433. [DOI] [PubMed] [Google Scholar]
- 50.Simons M., Raposo G. 2009. Exosomes—vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21: 575–581. [DOI] [PubMed] [Google Scholar]
- 51.Bhatnagar S., Shinagawa K., Castellino F. J., Schorey J. S. 2007. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 110: 3234–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santos T. G., Souza D. O., Tasca C. I. 2006. GTP uptake into rat brain synaptic vesicles. Brain Res. 1070: 71–76. [DOI] [PubMed] [Google Scholar]
- 53.Lee C. S., Kim I. S., Park J. B., Lee M. N., Lee H. Y., Suh P. G., Ryu S. H. 2006. The phox homology domain of phospholipase D activates dynamin GTPase activity and accelerates EGFR endocytosis. Nat. Cell Biol. 8: 477–484. [DOI] [PubMed] [Google Scholar]
- 54.Petry C., Fritz G., Pfeilschifter J., Huwiler A. 2004. Inhibition of Rho modulates cytokine-induced prostaglandin E2 formation in renal mesangial cells. Biochim. Biophys. Acta. 1636: 108–118. [DOI] [PubMed] [Google Scholar]
- 55.Maeda A., Ozaki Y., Sivakumaran S., Akiyama T., Urakubo H., Usami A., Sato M., Kaibuchi K., Kuroda S. 2006. Ca2+ -independent phospholipase A2-dependent sustained Rho-kinase activation exhibits all-or-none response. Genes Cells. 11: 1071–1083. [DOI] [PubMed] [Google Scholar]
- 56.Wijewickrama G. T., Kim J. H., Kim Y. J., Abraham A., Oh Y., Ananthanarayanan B., Kwatia M., Ackerman S. J., Cho W. 2006. Systematic evaluation of transcellular activities of secretory phospholipases A2. High activity of group V phospholipases A2 to induce eicosanoid biosynthesis in neighboring inflammatory cells. J. Biol. Chem. 281: 10935–10944. [DOI] [PubMed] [Google Scholar]
- 57.Sawada H., Murakami M., Enomoto A., Shimbara S., Kudo I. 1999. Regulation of type V phospholipase A2 expression and function by proinflammatory stimuli. Eur. J. Biochem. 263: 826–835. [DOI] [PubMed] [Google Scholar]
- 58.Salvado M. D., Alfranca A., Escolano A., Haeggstrom J. Z., Redondo J. M. 2009. COX-2 limits prostanoid production in activated HUVECs and is a source of PGH2 for transcellular metabolism to PGE2 by tumor cells. Arterioscler. Thromb. Vasc. Biol. 29: 1131–1137. [DOI] [PubMed] [Google Scholar]
- 59.Folco G., Murphy R. C. 2006. Eicosanoid transcellular biosynthesis: from cell-cell interactions to in vivo tissue responses. Pharmacol. Rev. 58: 375–388. [DOI] [PubMed] [Google Scholar]
- 60.Zarini S., Gijon M. A., Ransome A. E., Murphy R. C., Sala A. 2009. Transcellular biosynthesis of cysteinyl leukotrienes in vivo during mouse peritoneal inflammation. Proc. Natl. Acad. Sci. USA. 106: 8296–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kondo H., Shirakawa R., Higashi T., Kawato M., Fukuda M., Kita T., Horiuchi H. 2006. Constitutive GDP/GTP exchange and secretion-dependent GTP hydrolysis activity for Rab27 in platelets. J. Biol. Chem. 281: 28657–28665. [DOI] [PubMed] [Google Scholar]
- 62.van der Goot F. G., Gruenberg J. 2006. Intra-endosomal membrane traffic. Trends Cell Biol. 16: 514–521. [DOI] [PubMed] [Google Scholar]
- 63.Falguieres T., Luyet P. P., Gruenberg J. 2009. Molecular assemblies and membrane domains in multivesicular endosome dynamics. Exp. Cell Res. 315: 1567–1573. [DOI] [PubMed] [Google Scholar]
- 64.Shinozaki K., Waite M. 1999. A novel phosphatidylglycerol-selective phospholipase A2 from macrophages. Biochemistry. 38: 1669–1675. [DOI] [PubMed] [Google Scholar]
- 65.Hullin-Matsuda F., Kawasaki K., Delton-Vandenbroucke I., Xu Y., Nishijima M., Lagarde M., Schlame M., Kobayashi T. 2007. De novo biosynthesis of the late endosome lipid, bis(monoacylglycero) phosphate. J. Lipid Res. 48: 1997–2008. [DOI] [PubMed] [Google Scholar]
- 66.van Blitterswijk W. J., Hilkmann H. 1993. Rapid attenuation of receptor-induced diacylglycerol and phosphatidic acid by phospholipase D-mediated transphosphatidylation: formation of bisphosphatidic acid. EMBO J. 12: 2655–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sattar A. A., Haque R. 2007. Cytosolic PLA2 in zymogen granule fusion and amylase release: inhibition of GTP-induced fusion by arachidonyl trifluoromethyl ketone points to cPLA2 in G-protein-mediated secretory vesicle fusion. J Biochem. 141: 77–84. [DOI] [PubMed] [Google Scholar]
- 68.Blackwood R. A., Smolen J. E., Transue A., Hessler R. J., Harsh D. M., Brower R. C., French S. 1997. Phospholipase D activity facilitates Ca2+-induced aggregation and fusion of complex liposomes. Am. J. Physiol. 272: C1279–C1285. [DOI] [PubMed] [Google Scholar]
- 69.van Rossum D. B., Oberdick D., Rbaibi Y., Bhardwaj G., Barrow R. K., Nikolaidis N., Snyder S. H., Kiselyov K., Patterson R. L. 2008. TRP_2, a lipid/trafficking domain that mediates diacylglycerol-induced vesicle fusion. J. Biol. Chem. 283: 34384–34392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boyce J. A. 2007. Mast cells and eicosanoid mediators: a system of reciprocal paracrine and autocrine regulation. Immunol. Rev. 217: 168–185. [DOI] [PubMed] [Google Scholar]
- 71.Looze C., Yui D., Leung L., Ingham M., Kaler M., Yao X., Wu W. W., Shen R. F., Daniels M. P., Levine S. J. 2009. Proteomic profiling of human plasma exosomes identifies PPARgamma as an exosome-associated protein. Biochem. Biophys. Res. Commun. 378: 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan N. S., Shaw N. S., Vinckenbosch N., Liu P., Yasmin R., Desvergne B., Wahli W., Noy N. 2002. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol. Cell. Biol. 22: 5114–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reue K., Brindley D. N. 2008. Thematic Review Series: Glycerolipids. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 49: 2493–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cerbone A., Toaldo C., Laurora S., Briatore F., Pizzimenti S., Dianzani M. U., Ferretti C., Barrera G. 2007. 4-Hydroxynonenal and PPARgamma ligands affect proliferation, differentiation, and apoptosis in colon cancer cells. Free Radic. Biol. Med. 42: 1661–1670. [DOI] [PubMed] [Google Scholar]
- 75.Szanto A., Nagy L. 2008. The many faces of PPARgamma: anti-inflammatory by any means? Immunobiology. 213: 789–803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.