Abstract
Globins are heme-containing proteins that are best known for their roles in oxygen (O2) transport and storage. However, more diverse roles of globins in biology are being revealed, including gas and redox sensing. In the nematode Caenorhabditis elegans, 33 globin or globin-like genes have been recently identified, some of which are known to be expressed in the sensory neurons of the worm and linked to O2 sensing behavior. Here, we describe GLB-6, a novel globin-like protein expressed in the neurons of C. elegans. Recombinantly expressed full-length GLB-6 contains a heme site with spectral features that are similar to those of other bis-histidyl ligated globins, such as neuroglobin and cytoglobin. In contrast to these globins, however, ligands such as CO, NO, and CN− do not bind to the heme in GLB-6, demonstrating that the endogenous histidine ligands are likely very tightly coordinated. Additionally, GLB-6 exhibits rapid two-state autoxidation kinetics in the presence of physiological O2 levels as well as a low redox potential (−193 ± 2 mV). A high resolution (1.40 Å) crystal structure of the ferric form of the heme-domain of GLB-6 confirms both the putative globin-fold and bis-histidyl ligation and also demonstrates key structural features that can be correlated with the unusual ligand binding and redox properties exhibited by the full-length protein. Taken together, the biochemical properties of GLB-6 suggest that this neural protein would most likely serve as a physiological sensor for O2 in C. elegans via redox signaling and/or electron transfer.
Globins are hemoproteins of about 150 amino acids comprised of five to eight α-helices that adopt the classic globin fold. The heme b, or protophorphyrin IX, cofactor typically exists in either the reduced ferrous [Fe(II)] or oxidized ferric [Fe(III)] oxidation state. The heme iron center is coordinated equatorially by four pyrrole nitrogens from the porphyrin, and one of the two axial sites is invariably occupied by a histidine residue, often called the proximal histidine. The coordination at the remaining axial, or distal, site varies between different globins. In myoglobin (Mb) or hemoglobin (Hb), this distal site remains open in the ferrous state, allowing exogenous ligands, such as O2, CO and NO, to reversibly bind. Alternatively, the distal site in neuroglobin (Ngb) or cytoglobin (Cgb) is occupied by a second histidine, forming a bis-histidyl ligated heme site (1–4), which plays an important role in tuning heme ligand binding and redox potentials in these globin proteins (5, 6).
The globins have been mainly recognized as respiratory proteins involved in O2 storage and transport, based on our extensive knowledge of vertebrate Mb and Hb. However, genome libraries have revealed a plethora of globin and globin-like proteins varying in size, properties, and functions. Ngb and Cgb, which are found in nervous tissues (7) and non-neuronal cells (8), respectively, are two of the most recent additions to the family of vertebrate globins. The physiological function of these vertebrate globins is unclear, although a possible role for Ngb has been proposed in protecting neurons against hypoxia and ischemic injury (9–11). Non-vertebrate globins, on the other hand, are more diverse and have a wider variety of physiological functions (12, 13). These globins range from single domain globins to multi-domain and multi-subunit chimeric proteins containing both globin and non-globin domains. These include bacterial flavohemoglobins, which are proposed to detoxify NO through a reaction that forms nitrate (14–17), and globin-coupled sensor proteins, such as HemAT found in the archaeon Halobacterium salinarum and the gram-positive bacterium Bacillus subtilis(18), which are involved in aerotaxis transduction (19, 20).
In the nematode Caenorhabditis elegans, it was shown with reverse transcription polymerase chain reaction (RT-PCR) that 33 globin or globin-like genes are expressed (21–23). While little is still known about their function and biochemical properties, it has recently been shown that some of these globins may participate in a gas sensory signaling pathway in C. elegans. In particular, Bargmann and co-workers (24) and De Bono and co-workers (25) have independently shown that a neural globin, GLB-5, plays an important role in O2-dependent behavioral responses in wild-type strains of C. elegans. These studies have demonstrated that GLB-5 functions as an O2 sensor in the gas sensory neurons and modulates the neuronal and behavioral responses of the worm to changes in O2 concentration (26, 27). In addition, De Bono and co-workers have shown that recombinant GLB-5 exhibits absorption spectra that are very similar to those of Ngb and Cgb, indicative of a bis-histidyl ligated heme site (25). They further noted that GLB-5 oxidizes rapidly through reversible binding of O2, which would be an important feature for a signaling protein.
In this study, we present a detailed biochemical characterization of a C. elegans neural globin, GLB-6. Using spectroscopic, kinetic, and potentiometric methods, we demonstrate that this protein is a heme-binding protein with ligand binding and redox properties that are unusual for globins. Furthermore, we present a crystal structure of the heme domain of GLB-6 to provide a molecular level description of its novel globin-fold structure and a correlation to its biochemical properties. Taken together, we propose that GLB-6 could function as a redox sensor in C. elegans, playing an important role in O2 sensing in these worms.
MATERIALS AND METHODS
Construction of Expression Plasmids
Complementary DNA (cDNA) of the glb-6 gene from C. elegans strain N2 was provided by Prof. Cornelia Bargmann (The Rockefeller University, New York), which was obtained using RT-PCR as previously described (24). The cDNA was cloned into the PET-28b expression vector (Novagen) that was modified to include an N-terminal His6-tag followed by a tobacco etch virus (TEV) protease recognition site (ENLYFQG). As a result, the expression sequence of GLB-6 included an N-terminal sequence, MGSSHHHHHHSSGENLYFQGH, before the starting M1 residue.
Protein Expression
The expression vectors were transformed into E. coli Rosetta 2(DE3) cells (Novagen). Antibiotic selection was carried out with 35 µg/mL kanamycin and 34 µg/mL chloramphenicol. For expression, E. coli were grown in 4 L beveled flasks, each containing 1 L of modified Terrific Broth (12 g of casein enzymatic hydrolysate, 24 g of yeast extract, 4 mL of glycerol, 17 mM KH2PO4, and 72 mM K2HPO4). Cultures were grown at 37 °C to an OD600 of ~0.6. Upon induction with 1 mM isopropyl β-D-1-thiogalactopyranoside, 0.5 mM 5-aminolevulinic acid was also added to facilitate heme biosynthesis. Induction was allowed to occur at 25 °C for 17 hours before cells were collected by centrifugation and stored at −80 °C.
Protein Purification
Frozen cells were thawed on ice and resuspended in lysis buffer (50 mM sodium phosphate, pH. 8.0, 150 mM NaCl, 1 mM Pefabloc (Pentapharm), 1 mM benzamidine, 5 mM β-mercaptoethanol, and 5 % glycerol) with the addition of DNaseI and lysozyme (Sigma). Resuspended cells were lysed by homogenization with an Emulsiflex-C5 high-pressure homogenizer at 20000 psi (Avestin, Inc.). Lysed cells were centrifuged at 200000 g for 1 hour. The supernatant was collected and applied to a Ni-NTA column (Qiagen) equilibrated with buffer A (50 mM sodium phosphate, pH. 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol, 5% glycerol and 20 mM imidazole) at 4 °C. The column was washed with approximately ten column volumes of buffer A. GLB-6 was then eluted with buffer B (50 mM sodium phosphate, pH. 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol, 5% glycerol, and 200 mM imidazole). To cleave off the N-terminal His6-tag, TEV protease was added to the eluted protein sample at a ratio of 1:10 TEV versus GLB-6 based on A280, and the solution was dialyzed overnight at 4 °C against 2 L of buffer A. The dialyzed protein solution was then passed over a freshly prepared Ni-NTA column equilibrated with buffer A. The solution was eluted with buffer A and non-tagged GLB-6 was collected as flow-through. GLB-6 was then concentrated to approximately 5 mL and loaded onto a Superdex 75 HiLoad 26/60 gel filtration column (Pharmacia) that was pre-equilibrated with buffer C (50 mM HEPES, pH. 7.5, 150 mM NaCl, 2 mM DTT and 5% glycerol) at a flow rate of 0.5 mL/min. The aliquots containing the purified protein were collected, frozen in liquid N2, and stored at −80 °C. The yield was approximately 5 mg/L of E. coli culture. Purity was assessed to be > 95% by SDS-PAGE. Electrospray ion (ESI+) mass spectrometry confirmed the expected mass (43720.1 Da versus predicted 43720.9 Da; University of California at Berkeley, QB3 Mass Spectrometry Facility).
Characterization of the GLB-6 Heme Domain
To obtain a stable heme domain construct, several C-terminal constructs (residues 187–389, 190–389, 192–389, and 196–389) were initially generated based on sequence homology with other globins. Of these constructs, only the GLB-6 (187–389) construct bound heme and expressed well (the plasmid construction, protein expression and protein purification of the heme domain constructs were performed in the same manner as the full-length GLB-6 described above).
To further define the heme domain, limited proteolysis on GLB-6 (187–389) was performed by the addition of trypsin (Trypsin Gold, Promega) to the protein sample in a 1:1000 (w:w) ratio at 37 °C. A stable fragment at ~20 kDa was seen via SDS-PAGE. Mass fingerprinting using an Applied Biosystems 4800 matrix-assisted laser desorption/ionization time-of-flight spectrometer (MALDI-TOF, University of California at Berkeley, QB3 Mass Spectrometry Facility) was performed (Figure S1), which identified this fragment as residues M187-R355. Purified GLB-6 (187–355) was heme-bound and well expressed (protein expression and purification was performed in the same manner as the full-length GLB-6 described above) with a yield of 25–30 mg/L of E. coli culture. The mass was confirmed with ESI+ mass spectrometry (20010.1 Da versus predicted 20010.9 Da).
Crystallization of the Heme-Domain of GLB-6
GLB-6 (187–355) was exchanged into 20 mM MES (pH 5.5) with 200 mM NaCl and concentrated to 60 mg/mL (3.0 mM). Crystals were grown by sitting drop vapor diffusion by mixing 1 µL of the protein solution with 1 µL of the reservoir solution (1.0 M NaNO3 and 0.1 M Na(CH3COO) pH 4.5) and 1 µL of 10 mM praseodymium (III) acetate solution, which was equilibrated against 700 µL reservoir solution at 20 °C. Crystals began to appear within 6 hours. Cryoprotection was achieved by transferring the crystals stepwise into mother liquor solutions containing a final concentration of 35 % glycerol. Crystals were flash frozen and stored in liquid N2.
X-ray Data Collection, Phasing and Refinement
X-ray data were collected by using synchrotron radiation at beamlines 5.0.3 (λ = 0.9765 Å) and 8.3.1 (λ = 1.1158 Å) at the Advanced Light Source, Lawrence Berkeley National Laboratory. Diffraction images were collected at 100 K with 1 s exposure time and 1° oscillations per frame using inverse beam geometry. Data integration and scaling were performed using the HKL2000 suite with anomalous flags selected (28). Praseodymium containing crystals (4 Pr atoms per asymmetric unit) contained sufficient anomalous signal to 2.5 Å for experimental phase determination by single-wavelength anomalous dispersion (SAD). Substructure solution, phasing, density modification and preliminary model building were carried out using Autosol and Autobuild in PHENIX (29). Phases were extended to 1.40 Å by using rigid-body refinement of the initial model against the highest resolution data set. Iterative model building was performed using ARP/wARP (30) and manual model building was carried out by using the program Coot (31). Refinement was carried out using PHENIX (29) with TLS refinement parameters incorporated. Refinement statistics are given in Table 2. Stereochemical properties were assessed by MOLPROBITY (32) and PROCHECK (33). Coordinates are deposited in the RCSB Protein Data Bank under PDB accession code 3MVC.
Table 2.
Redox potentials of various heme proteins.
| Protein | Axial Ligation | E0’ (mV) | Reference |
|---|---|---|---|
| GLB-6 (1–389) | His/His | −193 ± 2 mV | this work |
| Truncated Hba | His/His | −195 | (64) |
| Rice Hbb | His/His | −143 | (64) |
| Ngbc | His/His | −115 | (64) |
| Cgbd | His/His | −28 | (64) |
| Cyt b5e | His/His | −2 | (64) |
| Mb H64V/V68Hf | His/His | −128 | (52) |
| Mb wt | (H2O)/His | +54 | (52) |
| CDH M65Hg | His/His | −53 | (49) |
| CDH wt | His/Met | +164 | (49) |
| Cyt c551 M61Hh | His/His | +55 | (50) |
| Cyt c551 wt | His/Met | +260 | (50) |
| Cyt c M80Hi | His/His | +41 | (51) |
| Cyt c wt | His/Met | +260 | (51) |
truncated hemoglobin from the cyanobacterium Synechocystis,
rice non-symbiotic hemoglobin,
human neuroglobin,
human cytoglobin,
bovine erythrocyte cytochrome b5,
human/pig myoglobin,
cellobiose dehydrogenase from white rot fungus Phanerochaete chrysosporium,
cytochrome c551 from bacterium Pseudomonas stutzeri,
cytochrome c from horse heart.
UV-vis Spectroscopy
Absorption spectra were recorded in an anaerobic cuvette on a Cary 3E or 300 spectrophotometer equipped with a temperature controller set at 10 °C. Spectra were recorded from proteins in a solution of buffer (20 mM HEPES pH 7.5 and 150 mM NaCl). Protein samples were prepared in an anaerobic chamber (Coy Laboratory Products) flushed with a 10:90 H2/N2 gas mixture (Praxair, Inc.). For both the full-length GLB-6 (1–389) and the heme-domain (187–355) construct, the ferric form was obtained upon purification. The ferrous form was prepared with the addition of sodium hydrosulfite (dithionite). Excess reductant was removed using a PD-10 desalting column (GE Healthcare) pre-equilibrated with degassed buffer in the anaerobic chamber. Ligand binding experiments with CO, NO and CN− were performed in the following manner: For CO binding, CO (Praxair, 99.99% purity) was added to the head space of a sealed Reacti-Vial (Pierce) containing ferrous GLB-6. For NO binding, an anaerobic solution of diethylamine NONOate (Cayman) in 10 mM NaOH was added to ferrous GLB-6 in the anaerobic chamber. For CN− binding, a solution of KCN was added directly to an aerobic solution of ferric GLB-6.
Resonance Raman Spectroscopy
Spectra were collected using 413.1 nm excitation from a Kr+ laser (Spectra-Physics model 2025). Raman scattering was detected with a cooled, back-illuminated CCD (LN/CCD-1100/PB; Roper Scientific) controlled by an ST-133 controller coupled to a subtractive dispersive double spectrograph. The laser power at the sample was 0.3–2 mW. A microspinning sample cell was used to minimize photo-induced degradation. The typical data acquisition time was 60 min. UV-Vis absorption spectra were obtained both before and after the Raman experiments to ensure that sample integrity was maintained. The Raman spectra were corrected for wavelength dependence of the spectrometer efficiency. The instrument was calibrated using cyclohexane, carbon tetrachloride, and toluene as standard solvents. The reported frequencies are accurate to ±1 cm−1, and the resolution of the spectra is 8 cm−1. For each Raman spectrum, the raw data were baseline corrected, and the buffer (20 mM HEPES pH 7.5 and 150 mM NaCl) background signal was subtracted.
Stopped-Flow Spectroscopy
Autoxidation rates were determined at 10 °C on a HiTech KinetAsyst stopped-flow instrument equipped with a diode array detector. Sample preparations were carried out in an anaerobic chamber as described above. GLB-6 was reduced with ~100 molar equivalent of dithionite for >10 minutes at 4 °C. The protein was then exchanged into a buffer for spectral measurements (50 mM HEPES, pH 7.5, 150 mM NaCl) using a PD-10 desalting column. Ferrous GLB-6 was loaded into a tonometer attached to a 3-way joint that was also connected to a 30 mL Luer-Lok syringe filled with anaerobic buffer. The joints were sealed with Parafilm. Prior to data acquisition, the stopped-flow sample syringe was made anaerobic through incubation with sodium hydrosulfite (~10 mM) for >15 minutes at 10 °C. The sodium hydrosulfite was subsequently flushed from the stopped-flow syringe with ~15 mL of anaerobic buffer delivered via the 3-way joint immediately before the anaerobic, ferrous protein was loaded for analysis. Saturated O2 solutions (1.28 mM) (34) were prepared by sparging 3–5 mL buffer in a septum-sealed vial with O2 gas (Praxair, Inc.) for >15 minutes at room temperature. Solutions of 74.2% O2 (950 µM) and 40.3% (516 µM) O2 were prepared by mixing saturated O2 solutions, delivered via a gas-tight syringe, with anaerobic buffer in a septum-sealed vial. Solutions of 21% (268 µM) O2 were prepared with air-saturated buffer. Solutions of 10.5% (134 µM) O2 were prepared by mixing air-saturated buffer, delivered via a gas-tight syringe, with anaerobic buffer in a septum-sealed vial. O2 stocks were made in duplicate or triplicate. Autoxidation reactions were initiated by equal mixing on the stopped-flow with the oxygenated buffer solutions. UV-visible spectra were acquired for 4.5 to 22.5 s (300 scans, 1.5 ms integration time) from 300–700 nm using the Kinetic Studio (TgK Scientific) software package. For each independently prepared O2 stock, 3–6 data sets were acquired. The spectral transition from 425 nm to 411 nm was fit to a two-state model using the SPECFIT Global Analysis System (Version 3.0.14).
Spectrochemical Redox Titration
Potentiometric titrations were performed as previously described (35) using an Oakton pH 1100 Series potentiometer. Titrations were carried out at 25 °C with stirring in the presence of redox mediator mix solution containing methyl viologen (−440 mV), anthraquinone-2,6-disulfonic acid (−184 mV), 2-hydroxy-1,4-naphthoquinone (−137 mV), 2,5-dihydroxy-1,4-benzoquinone (−60 mV), tetramethyl-p-benzochinon (Duroquinone) (+5 mV), 1,2-naphthoquinone (+157 mV) and ferricyanide (+356 mV). O2 was removed from the cuvette by flushing continuously with argon. Reductive titrations were performed by stepwise addition of dithionite solution and oxidative titrations were performed by stepwise addition of ferricyanide solution. The change in the heme oxidation state was monitored by the absorbance change in the α-band at 560 nm.
RESULTS AND DISCUSSION
Spectroscopic Characterization of GLB-6
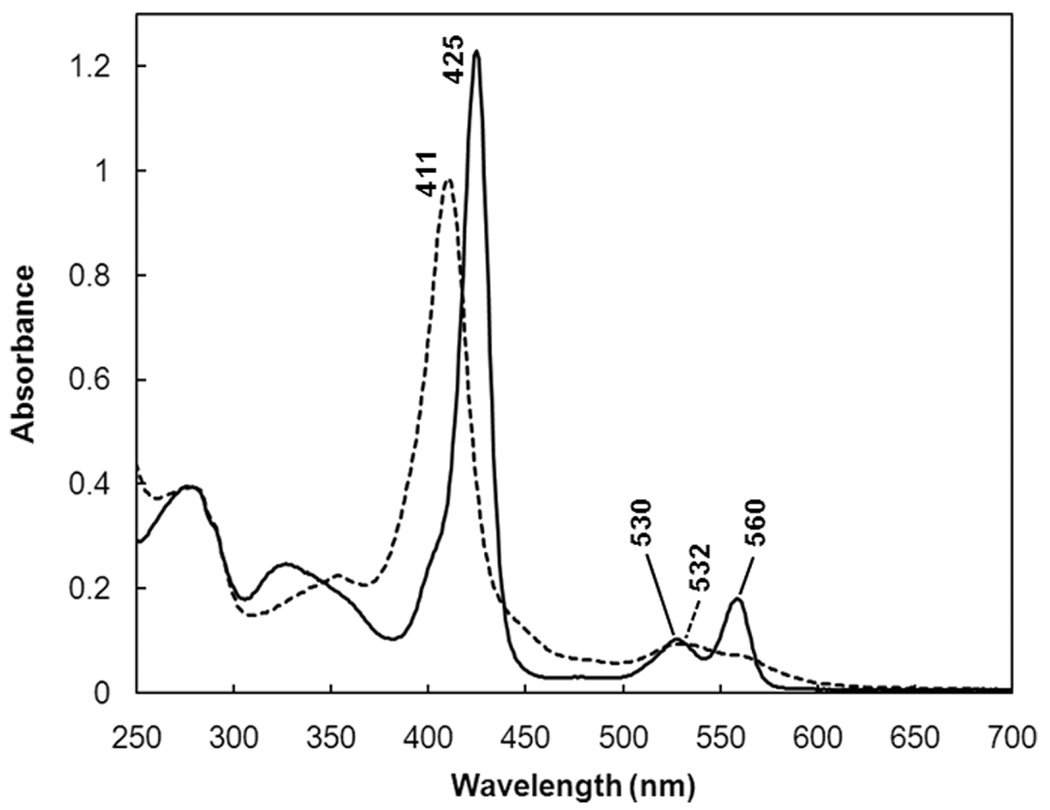
In order to characterize the spectral properties of GLB-6, the ferrous and ferric forms of the full-length protein were first investigated with UV-vis absorption spectroscopy (Figure 1). The spectrum of ferric GLB-6 exhibits a Soret band centered at 411 nm and a β band at 532 nm (dotted line, Figure 1). Ferrous GLB-6 exhibits a Soret band at 425 nm and α/β bands at 560 and 530 nm, with an α/β intensity ratio of approximately 2:1. These spectral features resemble those of cytochrome b5 (36) and globins such as neuroglobin (5, 7), cytoglobin (6) and nonsymbiotic plant hemoglobins (37, 38), suggestive of a low-spin (S = 1/2 for ferric and S = 0 for ferrous heme) bis-histidyl ligated heme site (39, 40). The heme domain construct of residues 187–355 also exhibits the same Soret and α/β band features as full-length GLB-6 in both the ferric and ferrous forms, indicating that the heme structures are very similar between the heme domain and the full-length constructs.
Figure 1.
UV-Vis absorption spectra of full-length ferrous GLB-6 (solid line) and ferric GLB-6 (dotted line). (Buffer = 20 mM HEPES, 150 mM NaCl, pH 7.5)
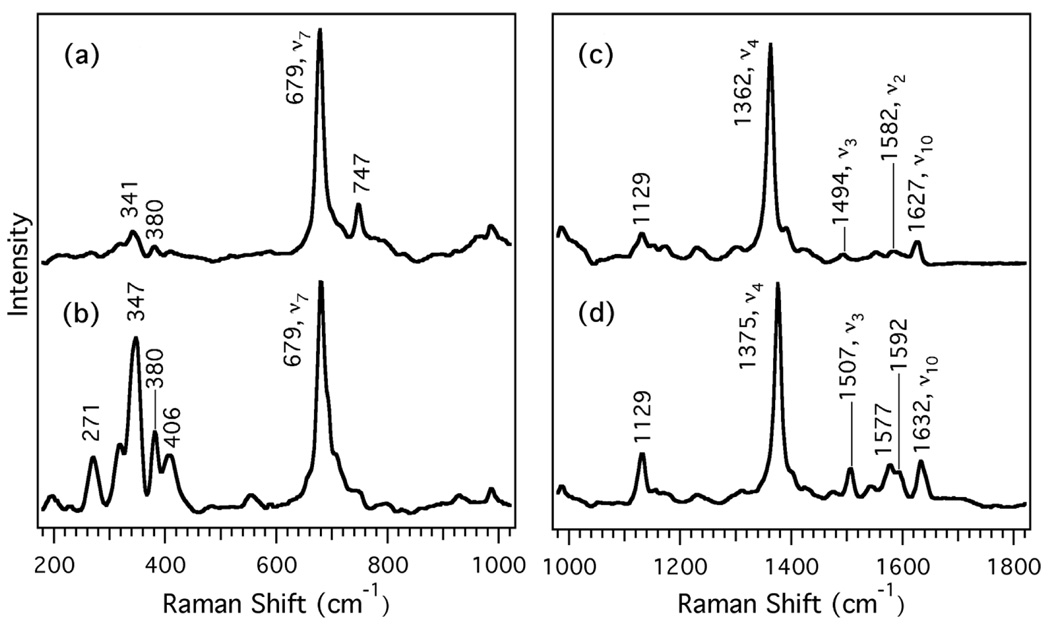
Resonance Raman spectra of the ferrous and ferric forms of GLB-6 are shown in Figure 2. The heme skeletal bands related to oxidation, coordination and spin states are found in the high frequency region. The most intense peak in this region, occurring between 1350–1370 cm−1, corresponds to the Cα-N stretching mode (ν4) and is sensitive to the oxidation state of the heme. In addition, ν3 is the coordination- and spin state-sensitive Cα-Cm stretching mode, and is typically detected in the 1470–1500 cm−1 range (39, 41). For GLB-6, ν4 and ν3 for the ferrous form are observed at 1362 cm−1 and 1494 cm−1, respectively (Figure 2c), while those of the ferric form are observed at 1375 cm−1 and 1507 cm−1, respectively (Figure 2d), consistent with a low-spin, bis-histidyl heme ligation in both the ferrous and ferric states. Due to the bis-histidyl state of ferrous GLB-6, no Fe-His stretching was observed in the low frequency region (at ~220 cm−1) (42, 43) (Figure 2a).
Figure 2.
Resonance Raman spectra of full-length ferrous GLB-6 (a, c) and ferric GLB-6 (b, d). The left panel (traces a and b) shows the lower frequency region and is normalized to ν7. The right panel (traces c and d) shows the high frequency region and is normalized to ν4. All spectra were acquired for 1 hour at room temperature with 413.1 nm excitation, room temperature, 2 mW.
Ligand Binding in GLB-6
Ligand binding experiments with CO, NO, and CN−, which are known to form highly stable complexes with ferrous (CO and NO) and ferric (CN−) heme sites, were performed to further investigate the biochemical properties of the protein (44). Interestingly, however, it was found that GLB-6 does not bind any of these ligands. No spectral change was observed when ferrous GLB-6 was treated with NO or when ferric GLB-6 was treated with CN−. For CO binding, a small blue-shift (~1 nm) of the Soret band (no change in the α and β band positions) and a slight decrease in the intensities of the Soret, α and β bands were observed when excess CO was added to ferrous GLB-6 anaerobically. These spectral changes are possibly due to some Fe-CO complex formation in the presence of excess CO. However, no sign of a Fe-CO or C–O stretch was observed in the resonance Raman spectrum, indicating that a very low amount of Fe-CO complex, if any, was present. Alternatively, exposing the ferrous-CO sample to air (in the presence of dithionite to prevent oxidation) resulted in regeneration of the fully reduced ferrous spectrum, likely due to the CO ligand being readily replaced by the endogenous histidine. Note that an O2 binding experiment could not be performed due to the rapid oxidation of GLB-6. However, the fact that the affinities of CO and NO to heme centers are typically much higher than O2 in other heme proteins (45) suggests that a stable O2 complex with the heme iron in GLB-6 is very unlikely.
The role of bis-histidyl ligation in slowing the on-rate of exogenous ligands has been previously demonstrated in Ngb using stopped-flow and flash photolysis experiments, in which the dissociation of the distal histidine (koff ~ 1 sec−1) was shown to be the rate limiting step in CO and O2 binding (5, 46, 47). However, with high intrinsic affinities for CO and O2 for the pentacoordinate heme, Ngb is able to form stable CO and O2 complexes. In GLB-6, the lack of ligand binding suggests that the Fe-His bonds in the GLB-6 heme site may be stronger than those of Ngb but also that the intrinsic affinities for exogenous ligands could be low even in the absence of the distal histidine, possibly due to a heme pocket that is unfavorable for stable heme-ligand complex formation.2
Autoxidation Rate of GLB-6
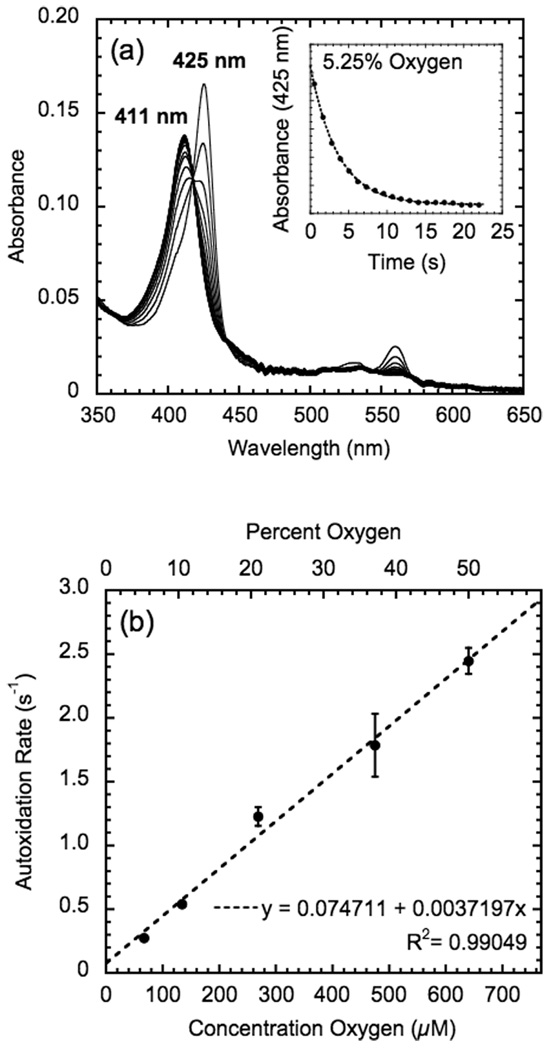
During the spectral characterization of GLB-6, it was found that the ferrous protein immediately oxidized to the ferric oxidation state when exposed to air. To quantify this rapid autoxidation of GLB-6, stopped-flow experiments were performed. A spectral transition from 425 nm to 411 nm was observed upon reaction of the ferrous form of full-length GLB-6 (1–389) with varying O2 concentrations (5.25, 10.5, 21.0, 37.1, and 50.0 % O2, which correspond to 67, 134, 268, 475, and 640 µM O2). In Figure 3a, spectral changes at 5.25 % O2 are shown as representative data with the trace of A425 given in the inset. Note that 5.25 % O2 is within the O2 concentration range preferred by C. elegans in the wild (5–12%) (26). The reactions at all O2 concentrations did not show any indication of the formation of an intermediate (isosbestic point at 418 nm, Figure 3A), such as a ferrous-oxy or ferric-superoxide complex. Therefore, the autoxidation rate was determined with a two-state, single-exponential model (i.e. ferrous → ferric), using a global fit analysis of the spectral window (375–700 nm). The observed first-order autoxidation rate constant kobs of full-length GLB-6 as a function of O2 concentration is shown in Figure 3b. No saturation of kobs was observed even at high O2 concentrations, and the data points were fit to a linear function with kobs = (3.72 × 103 M−1 s−1)[ O2] + 0.0747 s−1 (R2 = 0.990).3 Notably, the autoxidation rate of GLB-6 at ambient O2 levels (kobs = 1.23 s−1 at 21% O2) is orders of magnitude faster than other bis-histidyl ligated globins (Table 1). In these proteins, electron transfer occurs between the ferrous heme to the O2 ligand that is directly coordinated to the Fe center. Thus, autoxidation would require at least five sequential steps: (1) dissociation of the distal histidine from the heme, (2) coordination of O2 to the heme Fe(II), (3) electron transfer from the heme Fe(II) to O2 to generate a Fe(III)-superoxide complex, (4) dissociation of superoxide, leaving unligated Fe(III) heme, and (5) re-coordination of the distal histidine to the heme to reform a bis-histidyl Fe(III) heme site). GLB-6, on the other hand, lacks ligand binding as demonstrated above, and no intermediate was observed in the stopped-flow experiments at any O2 concentration. Thus, the autoxidation most likely occurs via direct outer-sphere electron transfer from the ferrous heme to the unbound O2 molecule (i.e. without direct coordination of O2 to the heme Fe center), generating ferric heme and superoxide in one chemical step.
Figure 3.
Autoxidation rates of full-length GLB-6 measured by stopped-flow spectroscopy. (a) Spectral changes with time for the oxidation of ferrous GLB-6. Experiments were performed at 10 °C with different O2 concentrations, 5.25, 10.5, 21.0, 37.1, and 50.0 % O2. The spectral changes at 5.25 % O2 are shown as representative data. The curve fitting of the change in A425 versus time using a single exponential model is shown in the inset. (b) The observed first-order autoxidation rate constant kobs of full-length GLB-6 is plotted as a function of O2 concentration. The fit to a linear function is shown as a dotted line.
Table 1.
Autoxidation rates of selected globins.
| Protein | Autoxidation Rate (hr−1) | Reference |
|---|---|---|
| GLB-6 (1–389) | 4,428 | this work |
| Cyt b5a | 20 | (58) |
| mNgbb | 19 | (5) |
| hNgbc | 5.4 | (5) |
| Lbd | 0.2 | (59) |
| rice Hbe | 0.08 | (60) |
| Sw Mbf | 0.055 | (61) |
| hHb (α chain)g | 0.032 | (62) |
| hHb (β chain)g | 0.0037 | (62) |
| Cgbh | stablei | (6, 63) |
trypsin-cleaved calf liver cytochrome b5,
mouse neuroglobin,
human neuroglobin,
soybean leghemoglobin,
rice non-symbiotic hemoglobin,
sperm whale myoglobin,
human hemoglobin,
human cytoglobin,
negligible autoxidation observed (>1000 s)
Redox Potential of GLB-6
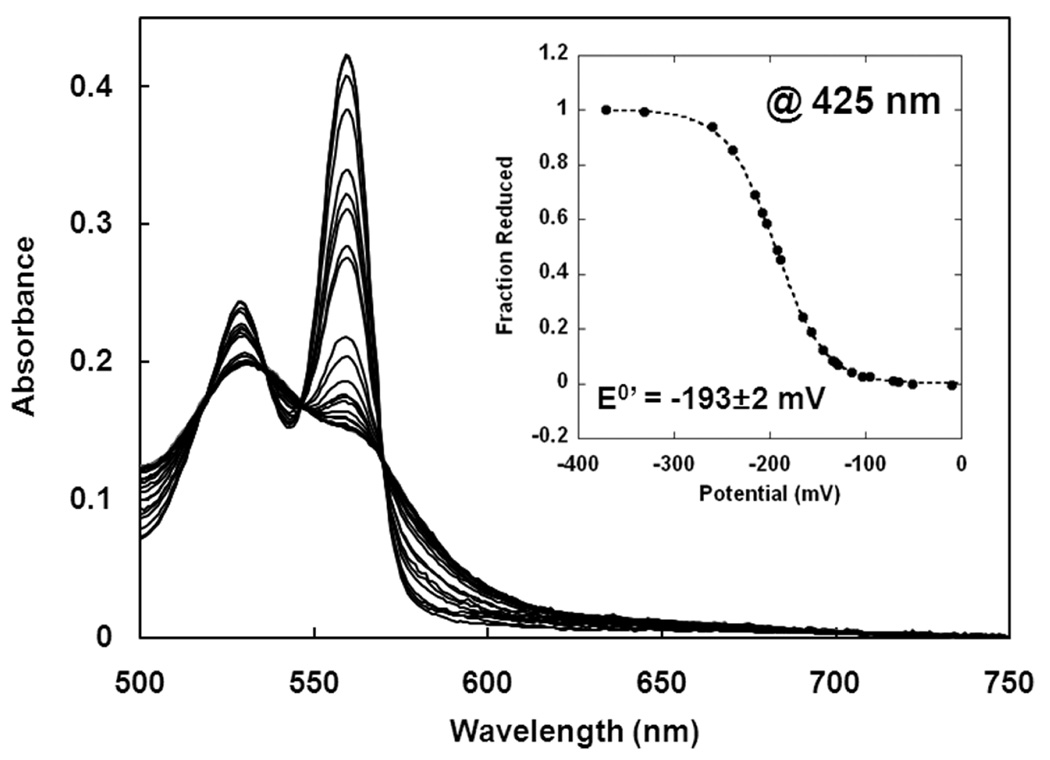
To better understand the thermodynamic properties of GLB-6, the redox potential of the full-length protein was measured by potentiometric redox titration (Figure 4). A560 of ferrous GLB-6 (i.e. the α-band maximum) minus A560 of ferric GLB-6 was normalized and plotted against ambient potential (Figure 4, inset) and the data were analyzed for the best fit to the Nernst equation with n = 1 (dotted lines in Figure 4, inset). The reduction midpoint potential (E0’) was −193 ± 2 mV versus the standard hydrogen electrode (SHE), which is comparable to those of other 6-coordinate b-type heme proteins (Table 2). This value is also lower than that of the O2/O2•− redox couple (−160 mV) (48), demonstrating that electron transfer from the ferrous heme in GLB-6 to O2 is a thermodynamically favorable process. Note that bis-histidyl ligated proteins generally exhibit lower redox potentials than those with different axial ligands, such as cytochromes c/c551, cellobiose dehydrogenase with Met/His (49–51) and Mb with H2O/His (52) (Table 2). Importantly, mutants with bis-histidyl ligation exhibit redox potentials that are ~200 mV lower than those of the corresponding wild type proteins. Thus, bis-histidyl ligation is a major factor in keeping the potential relatively low in GLB-6, which would be particularly critical if the protein is physiologically involved in electron transfer.
Figure 4.
Spectroelectrochemical potentiometric redox titrations of full-length GLB-6. The intensity of the α-band maxima, A560, of ferrous GLB-6 minus A560 of ferric GLB-6 is normalized and plotted versus the ambient potential (inset). The data were analyzed for the best fit to Nernst equations with n = 1 (inset, dotted line). The measured redox potential versus SHE, E0’, is −193 ± 2 mV, which is an average of three experiments. (Buffer: 100 mM potassium phosphate, 150 mM NaCl, pH 7.0, Temperature = 25 °C).
Crystal Structure of the Heme Domain of GLB-6
To gain structural insight into the biochemical properties of GLB-6, a high resolution crystal structure of the heme domain (residues 187–355) in the ferric form was solved to 1.40 Å by SAD analysis. The structure was refined to a final Rwork of 16.1 % (RFree of 20.1 %) (Table 3). The refined structure of ferric GLB-6 was built in a primitive monoclinic (P21) space group with two molecules in the asymmetric unit, each containing a b-type heme site. The final model was comprised of 156 and 153 amino acids in molecules A and B, respectively (no electron density was observed for residues 187–194 and 318–321 in molecule A and 187–194, 270–272 and 319–322 in molecule B), and it contained 139 water molecules and 4 praseodymium atoms. As shown in Figure 5, the tertiary structure of the GLB-6 heme domain consists of seven helices that correspond to the A, B, C, E, F, G and H helices, as anticipated of a globin-like domain.
Table 3.
Data collection, phasing, and refinement statistics.
| Data Collection and Refinement | Praseodymium | GLB-6 (187–355) | |
|---|---|---|---|
| Data Collection | Wavelength | 1.1158 | 0.9765 |
| Resolution (Å) | 50.0–1.69 (1.72–1.69) | 33.7–1.40 (1.42–1.40) | |
| Space Group | P21 | P21 | |
| Cell Dimensions | |||
| a, b, c (Å) | 37.55, 76.77, 62.68 | 37.58, 76.34, 62.69 | |
| β(°) | 91.6 | 91.7 | |
| Redundancy | 3.6 (3.3) | 4.2 (4.0) | |
| Completenessa (%) | 98.6 (82.8) | 97.0 (95.1) | |
| Rsym (%) | 5.4 (21.3) | 5.3 (29.4) | |
| I/σa | 19.9 (5.9) | 21.5 (3.6) | |
| Refinement | No. of Reflections | 127215 | |
| Rwork/Rfreeb (%) | 16.1/20.1 | ||
| No. Atoms | |||
| Protein | 2560 | ||
| Heme | 86 | ||
| Praseodymium | 4 | ||
| Solvent Molecules | 139 | ||
| B-factors | |||
| Protein | 25.7 | ||
| Water | 51.6 | ||
| Overall | 31.8 | ||
| Rms Deviation | |||
| Bond Lengths (Å) | 0.005 | ||
| Bond Angles (°) | 0.863 | ||
The values in parentheses relate to highest-resolution shells.
Rfree is calculated for a randomly chosen 5% of reflections.
Figure 5.
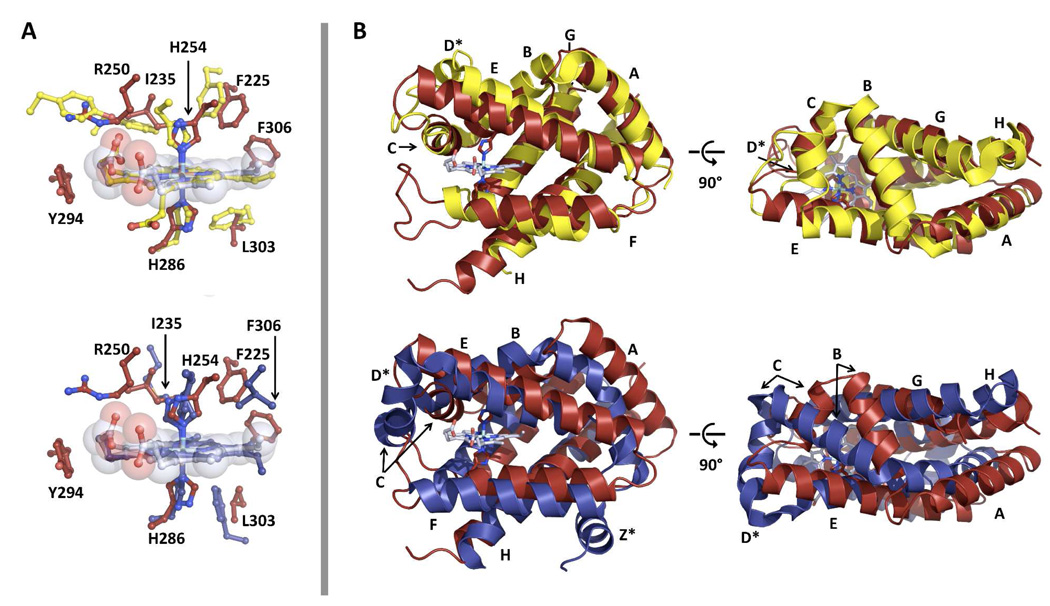
Structure of the heme domain of GLB-6. Ribbon structure of the GLB-6 heme domain (molecule A) is shown with helices labeled according to the canonical globin fold. His254 and His289, distal (above) and proximal (below) heme ligands, respectively, and the heme moiety are shown as stick representation. Praseodymium atoms are represented as orange spheres. Note that residues 318–321 could not be traced and are shown as black dots. Lower panel shows 2Fo-Fc composite simulated-annealing omit electron density map (1.0σ, gray mesh) calculated with heme moieties omitted.
The most conspicuous feature in the GLB-6 (187–355) structure is the bis-histidyl ligated heme site, which is consistent with our spectroscopic characterization (see above). The ferric iron center is ligated by H254 and H286 with Fe-N bond distances as 2.05 Å and 1.98 Å, respectively, reflecting strong Fe-N bonds. The bis-histidyl ligated heme structure is similar to those found in the crystal structures of human and murine Ngb (5, 7), human Cgb (6), nonsymbiotic rice Hb (37, 38) and most recently, a bacterial globin-coupled sensor from Geobacter sulfurreducens (53).
The GLB-6 heme domain structure exhibits several features that are distinct from those of other bis-histidyl ligated globins and help explain the unusual biochemical properties of the protein described above. Figure 6a and 6b (top) show a structural comparison between GLB-6 (red) and murine Ngb (yellow; PDB: 1Q1F), which is a good structural representative of other bis-histidyl ligated globins. The GLB-6 and Ngb structures superimpose to a relatively high degree overall (root mean square deviation over all Cα atoms = 2.211 Å). However, marked differences are found near the heme pocket where GLB-6 has an elongated E-helix and is missing a D-helix. The E-helix in GLB-6 has an extra helix loop (~4 amino acids) at the N-terminal side, and as a consequence, the distal His254 is positioned at the E11 position instead of E7 as in other globins (note that His254 is designated ‘distal’ based on sequence alignment with other globins). In addition, the D-helix is completely absent from the GLB-6 structure, and as a result, the C- and E-helices are directly connected. Importantly, these structural features could impose a tight restriction in the helical movement of the GLB-6 heme domain, partially inhibiting exogenous ligand binding.
Figure 6.
Heme (a) and overall (b) alignments of GLB-6 (red) with murine neuroglobin (yellow, PDB: 1Q1F) (top) and Geobacter sulfurreducens globin-coupled sensor (blue, PDB: 2W31) (bottom). Only residue assignments for GLB-6 are provided for clarity. Helices are labeled according to Figure 5. D-and Z-helices are marked with an asterisk to indicate that they do not exist in the GLB-6 structure.
Additionally, the F-helix in the GLB-6 structure is shorter than that of Ngb and is followed by a long F–G loop. Consequently, the proximal His286 is positioned at the end of the helix and is more exposed to the hydrophilic environment compared to Ngb in which the heme is imbedded in the hydrophobic pocket. Exposure to a more polar aqueous environment would thermodynamically favor the ferric state over the ferrous state of the heme site (54), consistent with the fact that GLB-6 exhibits a relatively low redox potential (GLB-6 = −193 mV vs. Ngb = −115 mV, Table 1) and undergoes spontaneous oxidation.
Figure 6a and 6b (bottom) show a structural comparison between GLB-6 (red) and a G. sulfurreducens globin-coupled sensor (blue; GsGCS, PDB: 2W31). GsGCS is a recently reported bis-histidyl ligated globin with an elongated E-helix (with an E11 distal histidine) and a missing D-helix, similar to GLB-6. Unlike GLB-6, however, GsGCS has been shown to bind various exogenous ligands (53). The difference in the ligand binding properties of the two globins may be explained by the low structural similarity at the A-, B-, and C-helix regions. This may be due to the presence of an extra helix (the Z-helix) in GsGCS at the N-terminus, which affects the A-, B- and C-helix orientations in the tertiary structure of GsGCS. Such structural differences could result in different movement and flexibility in the two protein structures, which would undoubtedly alter ligand binding properties.
Potential Role of GLB-6
The present study indicates that GLB-6 would most likely function as a redox sensor or electron transfer protein rather than a gas transporter or storage protein as with most other globins. This is supported by the fact that exogenous ligands have no detectable affinity for the heme and that GLB-6 has rapid redox kinetics (note that only oxidation kinetics were measured in this study; however, reduction of ferric GLB-6 with dithionite also occurred instantaneously, indicating rapid reduction kinetics as well). The low affinity for exogenous ligands would be critical in maintaining a constant redox potential physiologically, which could be severely altered upon ligand binding to the heme site.
The fact that the redox potential of GLB-6 is lower than that of O2/O2•− redox couple suggests that GLB-6 may directly interact with O2 to generate superoxide, a reactive oxygen species which has been shown to play an important role in physiological responses such as transcriptional regulation, protein activation, bioenergy output, cell proliferation, and apoptosis (55, 56). GLB-6 is implicated in O2 dependent behavior in C. elegans, in which the overexpression of GLB-6 suppresses worm aggregation (personal communication, P. McGrath and C. Bargmann). GLB-6 is expressed in the RMG hub interneuron (57) where the aggregation and related behaviors are controlled. Thus, it is possible that GLB-6 could serve as a redox sensor that responds to oxidative stress, either directly or indirectly by the change in O2 concentration. With rapid oxidation kinetics, GLB-6 would be ideal as a redox sensing protein.
Notably, other C. elegans globins have been suggested to serve as sensory proteins. As mentioned in the introduction, GLB-5 has been shown to be a bis-histidyl ligated globin protein involved in O2 sensing (25). Most recently, another bis-histidyl ligated globin, GLB-26, has been characterized (along with GLB-1, a penta-coordinated globin that reversibly binds O2). GLB-26 was shown to have very low affinity for exogenous ligands, suggestive of a role in O2 reduction and redox reactions, as seen with GLB-6 (23). However, GLB-26 is reported to be expressed exclusively in the head mesodermal and stomato-intestinal muscle cells (23), in contrast to GLB-6, which is expressed in neurons. Thus, it appears that C. elegans has evolved to utilize globins for various sensory functions in different cells and tissues, which also explains the unusually high number of globin genes present in these worms.
Finally, although it was not the focus of present study, the non-heme domain of GLB-6 (residues 1–186) may also play a role in facilitating signal transduction. This domain currently, however, has no known function. Studies are underway to gain a better understanding of the non-heme domain of GLB-6, the possible cooperativity between the non-heme and heme domains, and the physiological role of the protein in O2 sensing in C. elegans.
Supplementary Material
ACKNOWLEDGMENT
We thank Professor Cornelia Bargmann and Dr. Patrick McGrath of the Rockefeller University for providing us with the cDNA of GLB-6 and sharing valuable information concerning the physiological function of GLB-6. We thank P. Adams, J. Kuriyan, and beamline scientists at ALS 8.3.1 and 5.0.3 for helpful discussions during data collection and structure determination. We also thank the Marletta lab for critical reading of the manuscript.
ABBREVIATIONS
- Mb
myoglobin
- Hb
hemoglobin
- Ngb
neuroglobin
- Cgb
cytoglobin
- GsGCS
globin-coupled sensor
Footnotes
This study was funded by National Institutes of Health Grant GM077365 (M.A.M) and supported by the America Heart Association Western States Affiliate Postdoctoral Fellowship Program (J.Y.).
All attempts to express and purify distal and proximal histidine mutants were not successful. The histidine mutants either did not express well (H254G and H286G) or did not contain heme (H254L/V/Q/M and H286M).
The non-zero y-intercept (0.075 s−1) in the O2 titration reflects the small experimental error in the GLB-6 autoxidation rate. This error represents only 6% of the rate measured at ambient O2 levels (1.23 s−1).
SUPPORTING INFORMATION
Supporting Information Available: MALDI-TOF spectra of GLB-6. This material is available free of charge via the Internet at http://pubs.acs.org.
PDB Accession Code: 3MVC
REFERENCES
- 1.de Sanctis D, Dewilde S, Pesce A, Moens L, Ascenzi P, Hankeln T, Burmester T, Bolognesi M. Crystal Structure of cytoglobin: the fourth globin type discovered in man displays heme hexa-coordination. J Mol Biol. 2004;336:917–927. doi: 10.1016/j.jmb.2003.12.063. [DOI] [PubMed] [Google Scholar]
- 2.Pesce A, Dewilde S, Nardini M, Moens L, Ascenzi P, Hankeln T, Burmester T, Bolognesi M. Human brain neuroglobin structure reveals a distinct mode of controlling oxygen affinity. Structure. 2003;11:1087–1096. doi: 10.1016/s0969-2126(03)00166-7. [DOI] [PubMed] [Google Scholar]
- 3.Vallone B, Nienhaus K, Matthes A, Brunori M, Nienhaus GU. The structure of carbonmonoxy neuroglobin reveals a heme-sliding mechanism for control of ligand affinity. Proc Natl Acad Sci USA. 2004;101:17351–17356. doi: 10.1073/pnas.0407633101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vallone B, Nienhaus K, Brunori M, Nienhaus GU. The structure of murine neuroglobin: Novel pathways for ligand migration and binding. Proteins. 2004;56:85–92. doi: 10.1002/prot.20113. [DOI] [PubMed] [Google Scholar]
- 5.Dewilde S, Kiger L, Burmester T, Hankeln T, Baudein-Crueza V, Aerts T, Marden MC, Caubergs R, Moens L. Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family. J Biol Chem. 2001;276:38949–38955. doi: 10.1074/jbc.M106438200. [DOI] [PubMed] [Google Scholar]
- 6.Sawai H, Kawada N, Yoshizato K, Nakajima H, Aono S, Shiro Y. Characterization of the heme environmental structure of cytoglobin, a fourth globin in humans. Biochemistry. 2003;42:5133–5142. doi: 10.1021/bi027067e. [DOI] [PubMed] [Google Scholar]
- 7.Burmester T, Weich B, Reinhardt S, Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- 8.Burmester T, Ebner B, Weich B, Hankeln T. Cytoglobin: a novel globin type ubiquitously expressed in vertebrate tissues. Mol Biol Evol. 2002;19:416–421. doi: 10.1093/oxfordjournals.molbev.a004096. [DOI] [PubMed] [Google Scholar]
- 9.Khan AA, Wang Y, Sun Y, Mao XO, Miles E, Graboski J, Chen S, Ellerby LM, Jin K, Greenberg DA. Neuroglobin-overexpressing transgenic mice are resistant to cerebral and myocardial ischemia. Proc Natl Acad Sci USA. 2006;103:17944–17948. doi: 10.1073/pnas.0607497103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, Jin K, Mao XO, Zhu Y, Greenberg DA. Neuroglobin is up-regulated by and protects neurons from hypoxic-ischemic injury. Proc Natl Acad Sci USA. 2001;98:15306–15311. doi: 10.1073/pnas.251466698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y, Jin K, Peel A, Mao XO, Xie L, Greenberg DA. Neuroglobin protects the brain from experimental stroke in vivo. Proc Natl Acad Sci USA. 2003;100:3497–3500. doi: 10.1073/pnas.0637726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber RE, Vinogradov SN. Nonvertebrate hemoglobins: functions and molecular adaptations. Physiol Rev. 2001;81:569–628. doi: 10.1152/physrev.2001.81.2.569. [DOI] [PubMed] [Google Scholar]
- 13.Vinogradov S, Moens L. Diversity of globin function: enzymatic, transport, storage, and sensing. J Biol Chem. 2008;283:8773–8777. doi: 10.1074/jbc.R700029200. [DOI] [PubMed] [Google Scholar]
- 14.Mowat CG, Gazur B, Campbell LP, Chapman SK. Flavin-containing heme enzymes. Arch Biochem Biophys. 2010;493:37–52. doi: 10.1016/j.abb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Nunoshiba T, Derojas-Walker T, Tannenbaum SR, Demple B. Roles of nitric oxide in inducible resistance of Escherichia coli to activated murine macrophages. Infect Immun. 1995;63:794–798. doi: 10.1128/iai.63.3.794-798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Membrillo-Hernandez J, Coopamah MD, Anjum MF, Stevanin TM, Kelly A, Hughes MN, Poole RK. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a "Nitric oxide Releaser," and paraquat and is essential for transcriptional responses to oxidative stress. J Biol Chem. 1999;274:748–754. doi: 10.1074/jbc.274.2.748. [DOI] [PubMed] [Google Scholar]
- 17.Gardner PR, Gardner AM, Martin LA, Salzman AL. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci U S A. 1998;95:10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Phillips GN., Jr Structure of the oxygen sensor in Bacillus subtilis: signal transduction of chemotaxis by control of symmetry. Structure. 2003;11:1097–1110. doi: 10.1016/s0969-2126(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 19.Hou S, Larsen RW, Boudko D, Riley CW, Karatan E, Zimmer M, Ordal GW, Alam M. Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature. 2000;403:540–544. doi: 10.1038/35000570. [DOI] [PubMed] [Google Scholar]
- 20.Hou S, Freitas T, Larsen RW, Piatibratov M, Sivozhelezov V, Yamamoto A, Meleshkevitch EA, Zimmer M, Ordal GW, Alam M. Globin-coupled sensors: a class of heme-containing sensors in Archaea and Bacteria. Proc Natl Acad Sci USA. 2001;98:9353–9358. doi: 10.1073/pnas.161185598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoogewijs D, Geuens E, Dewilde S, Moens L, Vierstraete A, Vinogradov S, Vanfleteren JR. Genome-wide analysis of the globin gene family of C. elegans. IUBMB Life. 2004;56:697–702. doi: 10.1080/15216540500037562. [DOI] [PubMed] [Google Scholar]
- 22.Hoogewijs D, Geuens E, Dewilde S, Vierstraete A, Moens L, Vinogradov S, Vanfleteren JR. Wide diversity in structure and expression profiles among members of the Caenorhabditis elegans globin protein family. BMC Genomics. 2007;8:356. doi: 10.1186/1471-2164-8-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geuens E, Hoogewijs D, Nardini M, Vinck E, Pesce A, Kiger L, Fago A, Tilleman L, De Henau S, Marden MC, Weber RE, Van Doorslaer S, Vanfleteren J, Moens L, Bolognesi M, Dewilde S. Globin-like proteins in Caenorhabditis elegans in vivo localization, ligand binding and structural properties. BMC Biochem. 2010;11:17. doi: 10.1186/1471-2091-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, Bargmann CI. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persson A, Gross E, Laurent P, Busch KE, Bretes H, De Bono M. Natural variation in a neural globin tunes oxygen sensing in wild Caenorhabditis elegans. Nature. 2009;458:1030–1033. doi: 10.1038/nature07820. [DOI] [PubMed] [Google Scholar]
- 26.Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 27.Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol. 2006;4:e274. doi: 10.1371/journal.pbio.0040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski A, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination, Acta Crystallogr. Sect. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 30.Lamzin VS, Perrakis A, Wilson KS. The ARP/WARP suite for automated construction and refinement of protein models. In: Rossmann MG, Arnold E, editors. International Tables for Crystallography. Dordrecht: Kluwer Academic Publishers; 2001. pp. 720–722. [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics, Acta Crystallogr. Sect. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 34.Boon EM, Huang SH, Marletta MA. A molecular basis for NO selectivity in soluble guanylate cyclase. Nature Chem Biol. 2005;1:53–59. doi: 10.1038/nchembio704. [DOI] [PubMed] [Google Scholar]
- 35.Dutton PL. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 36.Ozols J, Strittmatter P. The interaction of porphyrins and metalloporphyrins with apocytochrome b5. J Biol Chem. 1964;239:1018–1023. [PubMed] [Google Scholar]
- 37.Duff SMG, Wittenberg BA, Hill RD. Expression, purification, and properties of recombinant barley (Hordeum sp.) hemoglobin. Optical spectra and reactions with gaseous ligands. J Biol Chem. 1997;272:16746–16752. doi: 10.1074/jbc.272.27.16746. [DOI] [PubMed] [Google Scholar]
- 38.Arredondo-Peter R, Hargrove MS, Sarath G, Moran JF, Lohrman J, Olson JS, Klucas RV. Rice hemoglobins. Gene cloning, analysis, and O2-binding kinetics of a recombinant protein synthesized in Escherichia coli. Plant Physiol. 1997;115:1259–1266. doi: 10.1104/pp.115.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babcock GT, Widger WR, Cramer WA, Oertling WA, Metz JG. Axial ligands of chloroplast cytochrome b-559: Identification and requirement for a heme-cross-linked polypeptide structure. Biochemistry. 1985;24:3638–3645. doi: 10.1021/bi00335a036. [DOI] [PubMed] [Google Scholar]
- 40.Choi CYH, Cerda JF, Chu H-A, Babcock GT, Marletta MA. Spectroscopioc characterization of the heme-binding sites in Plasmodium falciparum histidine-rich protein 2. Biochemistry. 1999;38:16916–16924. doi: 10.1021/bi991665k. [DOI] [PubMed] [Google Scholar]
- 41.Hu S, Smith KM, Spiro TG. Assignment of protoheme resonance Raman spectrum by heme labeling in myoglobin. J Am Chem Soc. 1996;118:12638–12646. [Google Scholar]
- 42.Kitagawa T. The heme protein structure and the iron-histidine stretching mode. In: Spiro TG, editor. Biological Applications of Raman Spectroscopy: Resonance Raman Spectra of Heme and Metalloproteins. New York: John Wiley & Sons; 1988. pp. 97–131. [Google Scholar]
- 43.Tran R, Boon EM, Marletta MA, Mathies RA. Resonance Raman Spectra of an O2-Binding H-NOX Domain Reveal Heme Relaxation upon Mutation. Biochemistry. 2009;48:8568–8577. doi: 10.1021/bi900563g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antonini E, Brunori M. Hemoglobin and myoglobin in their reactions with ligands. Amsterdam: North-Holland Pub. Co; 1971. [Google Scholar]
- 45.Olson JS, Phillips J, GN Myoglobin discriminates between O2, NO, and CO by electrostatic interactions with the bound ligand. J Biol Inorg Chem. 1997;2:544–552. [Google Scholar]
- 46.Uzan J, Dewilde S, Burmester T, Hankeln T, Moens L, Hamdane D, Marden MC, Kiger L. Neuroglobin and other hexacoordinated hemoglobins show a weak temperature dependence of oxygen binding. Biophys J. 2004;87:1196–1204. doi: 10.1529/biophysj.104.042168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiger L, Uzan J, Dewilde S, Burmester T, Hankeln T, Moens L, Hamdane D, Baudin-Crueza V, Marden MC. Neuroglobin ligand binding kinetics. IUBMB Life. 2004;56:709–719. doi: 10.1080/15216540500037711. [DOI] [PubMed] [Google Scholar]
- 48.Sawyer DT, Valentine JS. How super is superoxide? Acc Chem Res. 1981;14:393–400. [Google Scholar]
- 49.Rotsaert FAJ, Hallberg BM, de Vries S, Moenne-Loccoz P, Divne C, Renganathan V, Gold MH. Biophysical and structural analysis of a novel heme B iron ligation in the flavocytochrome cellobiose dehydrogenase. J Biol Chem. 2003;278:33224–33231. doi: 10.1074/jbc.M302653200. [DOI] [PubMed] [Google Scholar]
- 50.Miller GT, Zhang B, Hardman JK, Timkovich R. Converting a c-type to a b-type cytochrome: Met61 to His61 mutant of Pseudomonas cytochrome c-551. Biochemistry. 2000;39:9010–9017. doi: 10.1021/bi000232a. [DOI] [PubMed] [Google Scholar]
- 51.Raphael AL, Gray HB. Axial ligand replaement in horse heart cytochrome c by semisynthesis. Proteins. 1989;6:338–340. doi: 10.1002/prot.340060316. [DOI] [PubMed] [Google Scholar]
- 52.Dou Y, Admiraal J, Ikeda-Saito M, Krzywda S, Wilkinson AJ, Li T, Olson JS, Prince RC, Pickering IJ, George GN. Alteration of axial coordination by protein engineering in myoglobin. Bisimidazole ligation in the His64-->Val/Val68-->His double mutant. J Biol Chem. 1995;270:15993–16001. doi: 10.1074/jbc.270.27.15993. [DOI] [PubMed] [Google Scholar]
- 53.Pesce A, Thijs L, Nardini M, Desmet F, Sisinni L, Gourlay L, Bolli A, Coletta M, Van Doorslaer S, Wan X, Alam M, Ascenzi P, Moens L, Bolognesi M, Dewilde S. HisE11 and HisF8 provide bis-histidyl heme hexa-coordination in the globin domain of Geobacter sulfurreducens globin-coupled sensor. J Mol Biol. 2009;386:246–260. doi: 10.1016/j.jmb.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 54.Mauk AG, Moore GR. Control of metalloprotein redox potentials: what does site-directed mutagenesis of hemoproteins tell us? J Biol Inorg Chem. 1997;2:119–125. [Google Scholar]
- 55.Brown DI, Griendlin KK. Nox proteins in signal transduction. Free Radical Biol Med. 2009;47:1239–1253. doi: 10.1016/j.freeradbiomed.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RAJC, Bargmann CI. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berman MC, Adnams CM, Ivanetich KM, Kench JE. Autoxidation of soluble trypsin-cleaved microsomal ferrocytochrome b5 and formation of superoxide radicals. Biochem J. 1976;157:237–246. doi: 10.1042/bj1570237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hargrove MS, Barry JK, Brucker EA, Berry MB, Phillips GN, Jr, Olson JS, Arredondo-Peter R, Dean JM, Klucas RV, Sarath G. Characterization of recombinant soybean leghemoglobin a and apolar distal histidine mutants. J Mol Biol. 1997;266:1032–1042. doi: 10.1006/jmbi.1996.0833. [DOI] [PubMed] [Google Scholar]
- 60.Smagghe BJ, Kundu S, Hoy JA, Halder P, Weiland TR, Savage A, Venugopal A, Goodman M, Premer S, Hargrove MS. Role of phenylalanine B10 in plant nonsymbiotic hemoglobins. Biochemistry. 2006;45:9735–9745. doi: 10.1021/bi060716s. [DOI] [PubMed] [Google Scholar]
- 61.Brantley RE, Jr, Smerdon SJ, Wilkinson AJ, Singleton EW, Olson JS. The mechanism of autooxidation of myoglobin. J Biol Chem. 1993;268:6995–7010. [PubMed] [Google Scholar]
- 62.Mansouri A, Winterhalter KH. Nonequivalence of chains in hemoglobin oxidation. Biochemistry. 1973;12:4946–4949. doi: 10.1021/bi00748a020. [DOI] [PubMed] [Google Scholar]
- 63.Fago A, Hundahl C, Dewilde S, Gilany K, Moens L, Weber RE. Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin. Molecular mechanisms and physiological significance. J Biol Chem. 2004;279:44417–44426. doi: 10.1074/jbc.M407126200. [DOI] [PubMed] [Google Scholar]
- 64.Halder P, Trent JT, III, Hargrove MS. Influence of the protein matrix on intramolecular histidine ligation in ferric and ferrous hexacoordinate hemogloins. Proteins. 2007;66:172–182. doi: 10.1002/prot.21210. [DOI] [PubMed] [Google Scholar]
- 65.Walker FA, Emrick D, Rivera JE, Hanquet BJ, Buttlaire DH. Effect of heme orientation on the reduction potential of cytochrome b5. J Am Chem Soc. 1988;110:6234–6240. doi: 10.1021/ja00226a045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.