Extraocular muscles accumulate cytochrome c oxidase (COX)-deficient fibers from an earlier age and at a much faster rate than do skeletal muscles, suggesting an accelerated aging process. Most of these COX-deficient fibers harbor high levels of clonally expanded, somatic mitochondrial DNA deletions.
Abstract
Purpose.
Mitochondrial function and the presence of somatic mitochondrial DNA (mtDNA) defects were investigated in extraocular muscles (EOMs) collected from individuals covering a wide age range, to document the changes seen with normal aging.
Methods.
Cytochrome c oxidase (COX) and succinate dehydrogenase (SDH) histochemistry was performed on 46 EOM samples to determine the level of COX deficiency in serial cryostat muscle sections (mean age, 42.6 years; range, 3.0–96.0 years). Competitive three-primer and real-time PCR were performed on single-fiber lysates to detect and quantify mtDNA deletions. Whole-genome mitochondrial sequencing was also performed to evaluate the contribution of mtDNA point mutations to the overall mutational load.
Results.
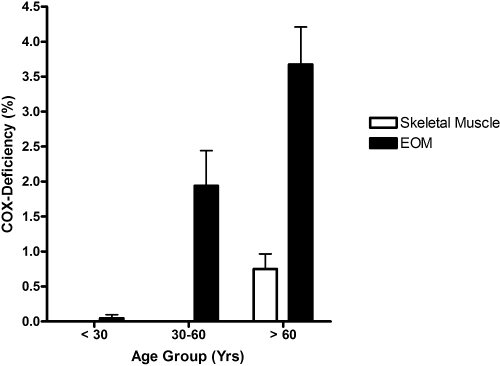
COX-negative fibers were seen in EOMs beginning in the third decade of life, and there was a significant age-related increase: <30 years, 0.05% (n = 17); 30 to 60 years, 1.94% (n = 13); and >60 years, 3.34% (n = 16, P = 0.0001). Higher levels of COX deficiency were also present in EOM than in skeletal muscle in all three age groups (P < 0.0001). Most of the COX-negative fibers harbored high levels (>70%) of mtDNA deletions (206/284, 72.54%) and the mean deletion level was 66.64% (SD 36.45%). The mutational yield from whole mitochondrial genome sequencing was relatively low (1/19, 5.3%), with only a single mtDNA point mutation identified among COX-negative fibers with low deletion levels ≤70%.
Conclusions.
The results show an exponential increase in COX deficiency in EOMs beginning in early adulthood, which suggests an accelerated aging process compared with other postmitotic tissues.
Mitochondria are unique in having their own genetic material in the form of a double-stranded circular molecule ∼16,569 bp long, and a cell can contain many thousands of mitochondrial DNA (mtDNA) molecules, depending on their energy demand.1 mtDNA codes for 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and 13 essential structural subunits of the mitochondrial respiratory chain, which are responsible for generating adenosine triphosphate (ATP) through oxidative phosphorylation. Mitochondrial genetic disorders are increasingly recognized as an important cause of chronic human disease, with a minimum prevalence of ∼1 in 5000,2 but the frequency of pathogenic mtDNA mutations in the general population is even higher, at ∼1 in 200.3,4 As mitochondria possess a high-copy-number genome, mutant and wild-type mtDNA molecules often co-exist within the same cell at various degrees of heteroplasmy. However, the deleterious effect of most mtDNA mutations on oxidative phosphorylation and cellular function becomes apparent only when the proportion of the mutant species exceeds a critical threshold of 60% to 80%.5–7
Both mtDNA point mutations and deletions accumulate with age in a range of tissues including brain, liver, colon, and skeletal and cardiac muscle.8,9 Within individual cells, these mtDNA defects expand clonally to reach sufficiently high levels to trigger a biochemical defect,10,11 which can be observed histochemically as cytochrome c oxidase (COX) deficiency.12–16 Although the association with normal aging and late-onset, sporadic neurodegenerative diseases is still under intense investigation, recent mouse models exhibiting premature aging phenotypes were shown to harbor high levels of somatic mtDNA defects, supporting a causal link.17 However, the rates of accumulation between different tissues and the regional distribution, even within the same tissue, are not always uniform.12,14,18–21 These tissue-specific aging patterns suggest an explanation for the selective involvement of certain organ systems in mitochondrial genetic disorders, with the expression and clonal expansion of the pathogenic mtDNA defect being influenced by the same local factors.
In a study of postmortem extraocular muscle (EOM) specimens, COX-deficient fibers were present from the third decade of life, and there was a marked age-related increase.22 Remarkably, the level of COX deficiency was five to six times higher than in skeletal, diaphragmatic, and cardiac muscle specimens,23,24 but due to the technical limitations at the time of the study, the molecular basis for this impairment in mitochondrial oxidative metabolism was not investigated. To further explore the hypothesis of an accelerated mitochondrial aging process in EOMs, we performed a histochemical and molecular study of aging human EOMs, using sensitive assays for the detection and quantification of mtDNA deletions and point mutations.
Materials and Methods
Tissue Collection
EOMs were collected from donor eyes within 24 hours of death (n = 21) and from resections for nonparalytic strabismus surgery (n = 25): medial rectus (n = 11), lateral rectus (n = 8), superior oblique (n = 1), inferior oblique (n = 1), and unknown muscle (n = 25). All included individuals had no personal or family history of mitochondrial and neuromuscular diseases, evidence of neoplasia or active viral infections (e.g., HIV), or previous exposure to cytotoxic or antiretroviral therapy. EOM samples were frozen in melting liquid isopentane (−150°C) and then stored at −80°C. The study had the relevant institutional ethics committee approval and complied with the Declaration of Helsinki.
Histochemistry
EOM blocks were serially sectioned and mounted on both glass and membrane slides with a cryostat (Microm HM560; Thermo Fisher, Berlin, Germany) at a thickness of 20 μm. The tissue sections were then stained by using standard protocols for COX, succinate dehydrogenase (SDH), and dual COX-SDH.25 The latter is a well-validated method for accurately quantifying the proportion of fibers in a tissue section that exhibit a mitochondrial respiratory chain defect. Normal COX-positive fibers stain brown, whereas COX-negative fibers are stained blue by the SDH counterstain as a result of impaired complex IV activity.26 We also compared the histochemical findings in this EOM cohort with a previously studied group of quadriceps skeletal muscle biopsies from 69 healthy control subjects with a mean age of 56.3 years (range, 21.0–95.0 years; SD 24.7 years).27
DNA Isolation from Single EOM Fibers
Single EOM fibers were initially dissected with fine glass capillaries held onto a manipulator (Micromanipulator; Leica Microsystems, Wetzlar, Germany) and subsequently with a laser dissecting microscope (LMD 6000; Leica Microsystems), when the latter became available. The individual fibers were collected into a 0.5 mL microcentrifuge cap (Eppendorf, UK) and incubated in a 10 μL lysis solution containing proteinase K at 55°C for 2 hours and 95°C for 10 minutes.26 The molecular investigations were then performed on the single-fiber lysate within 24 hours.
mtDNA Deletion Assays
The presence of the 4977-bp common deletion was specifically assessed by using a well-established competitive three-primer assay.28 The mtDNA copy number was quantified using a real-time PCR protocol based on the fact that the MTND1 gene is only rarely involved in large-scale rearrangements, whereas the MTND4 gene is removed in >97% of all reported mtDNA deletions.6,7,29 Fluorogenic probes (TaqMan; Applied Biosystems, Warrington, UK) were designed to amplify mtDNA fragments within the MTND1 and MTND4 genes on a real-time sequence-detection system (GeneAmp 5700; Applied Biosystems).30 Both reactions had previously been optimized, and they were confirmed by the standard curve method to be linear over an appropriate concentrate range for each run. All single-fiber lysates were analyzed in triplicate, and the mtDNA deletion level was derived from the difference in threshold cycle value (Ct) between these two probes, using the 2−ΔCt method. There was no significant difference in the distribution of mtDNA deletion levels for both COX-positive (P = 0.2839) and COX-negative fibers (P = 0.5344), when the manual and laser microdissection techniques (data not shown) were compared. These two datasets were therefore combined in the final analysis.
The total number of mtDNA molecules (total copy number) present in laser-microdissected fiber sections was derived from the MTND1 Ct values and the number of wild-type mtDNA molecules (wild-type copy number) from the MTND4 Ct values.7,30 Both total and wild-type mtDNA contents were calculated by dividing the absolute copy number by fiber area (in square micron), which was obtained from the dissecting microscope software (LMD 6000; Leica Microsystems) before laser microdissection. For the purpose of mtDNA copy number measurements, COX-positive and COX-negative fibers were laser-microdissected from membrane slides that had been stained only for SDH.
Long-Range PCR
A 10,843-bp fragment (129-5875), encompassing the mutational hotspots along the major arc of the mitochondrial genome and MTND4, was amplified with a two-step, long-range PCR method.14,31 The PCR products were electrophoresed in a 0.7% agarose gel at 40 V for 3 hours before being visualized under ultraviolet light. Deleted mtDNA products selected for additional breakpoint analysis were gel extracted (QIAquick kit; Qiagen, Crawley, UK), and sequenced (ABI 3100 Genetic Analyzer; Applied Biosystems), by using a primer walking method.14,31
Whole Mitochondrial Genome Sequencing
The whole mitochondrial genome was PCR-amplified in nine overlapping fragments of ∼2 kb which were then used as the templates for a second round of PCR, each first-round product generating four overlapping fragments of ∼450 to 750 bp.32 DNA polymerase (AmpliTaq Gold; Applied Biosystems) was used for our two-round PCR strategy, and all 36 fragments generated were purified and then sequenced (BigDye Termination Chemistry on ABI 3100 genetic analyzer; Applied Biosystems). Sequencing results were then compared to the revised Cambridge reference sequence33 (SeqScape software ver. 2.1; Applied Biosystems). Levels of mtDNA heteroplasmy, if present, were quantified from the relative peaks of each genotype on the sequencing chromatogram. The detection threshold for heteroplasmic variants with this technique has been shown to be ∼30%.26,32 Both first and second rounds of PCR were repeated from the original single-fiber lysate for all identified heteroplasmic mtDNA variants and possible pathogenic mtDNA point mutations. For homoplasmic mtDNA variants, which are unlikely to account for the observed biochemical COX defect, only the second round of PCR was repeated, followed by resequencing of the amplified product.
Statistical Analysis
The χ2 analysis, independent sample t-test, one- and two-way ANOVA were used for group comparisons, as required (GraphPad ver. 4; San Diego, CA).
Results
Level of COX Deficiency in EOMs
EOM samples were analyzed for 46 individuals with a mean age of 42.6 years (range, 3.0–96.0; SD 28.1). The youngest individual with COX-negative fibers was a 26-year-old man (0.81%), and the highest level of COX deficiency (7.60%) was found in a 96-year-old man, the oldest individual in our cohort. Two different EOMs were harvested from the same globe of two individuals, a 74-year-old woman (M46) and a 91-year-old woman (M49). There was no statistically significant difference in COX deficiency levels between the two EOMs collected from the same individual: M46: 3.92% (34/868) vs. 5.54% (27/487), average = 4.73% (P = 0.1861), and M49: 6.10% (26/426) vs. 5.24% (39/744), average = 5.67% (P = 0.5587). No COX-positive, ragged-red fibers were identified in the EOM samples studied.
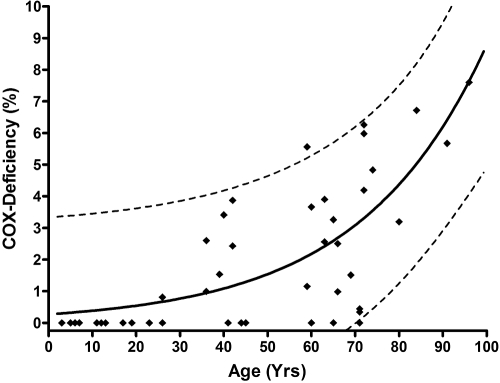
There was a significant age-related increase in COX deficiency: <30 years, 0.05% (n = 17); 30 to 60 years, 1.94% (n = 13); and >60 years, 3.34% (n = 16) (P = 0.0001; Fig. 1). Furthermore, the proportion of COX-negative fibers in the EOMs was significantly higher that that in the skeletal muscle biopsies in all three age groups (main effect of muscle type, P < 0.0001; main effect of age group, P < 0.0001; muscle type × age group interaction, P = 0.0013; Fig. 1). A simulated curve was generated to model an exponential increase in COX-negative EOM fibers with normal aging (r2 = 0.5753) and to predict the upper and lower 95% confidence range (Fig. 2).
Figure 1.
Mean percentage of COX-negative fibers found in EOM and skeletal muscle samples. Error bars, SEM.
Figure 2.
Distribution of COX-deficient EOM fibers with age. The simulated curve models an exponential increase in COX deficiency with normal aging (r2 = 0.5753); dotted lines: predicted upper and lower 95% confidence range (n = 46).
mtDNA Deletion Load
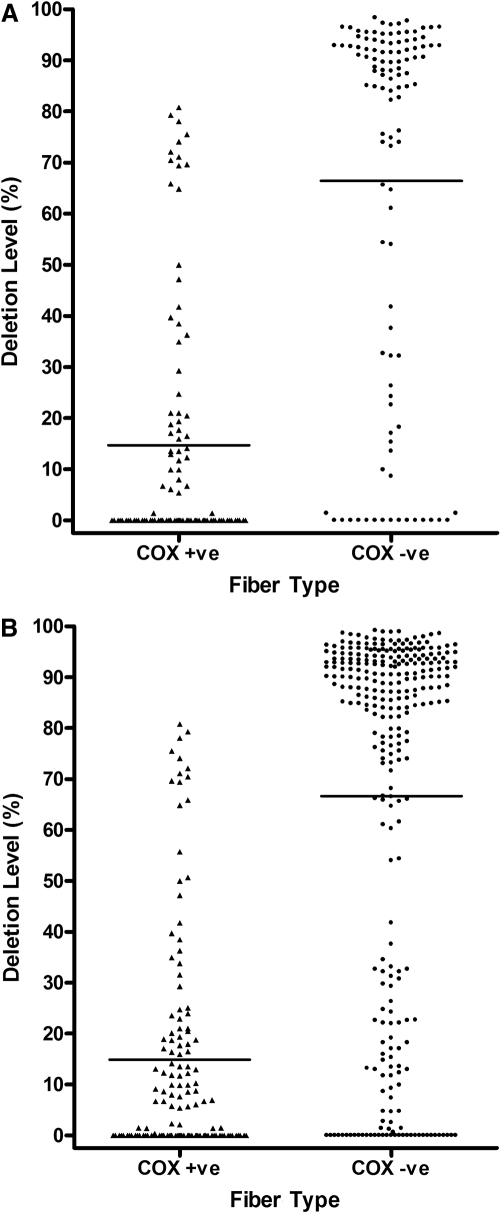
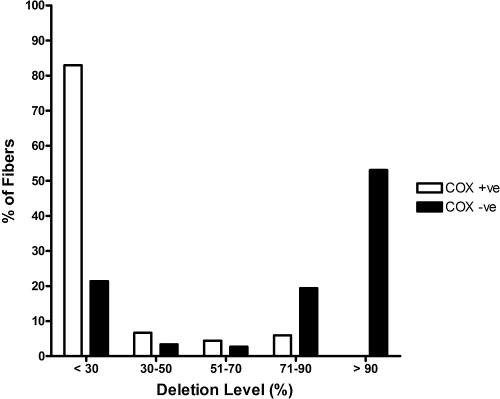
The 4977-bp common deletion was not detected in 67 COX-positive fibers and was identified in only 2 (3.39%) of 59 COX-negative fibers. These two fibers belonged to a 67-year-old man with an overall COX deficiency level of 2.50%, and both fibers harbored high mutation loads (∼95%). Quantitative real-time PCR data were available for 135 COX-positive fibers and 284 COX-negative fibers from 12 individuals (Table 1), including an initial batch of 103 COX-positive and 109 COX-negative fibers (Fig. 3A). The mean deletion level in the COX-positive fibers (14.86%, SD 22.00%, n = 135) was significantly lower than that in the COX-negative fibers (66.64%, SD 36.45%, n = 284, P < 0.0001; Fig. 3B). The majority of the COX-positive fibers (112/135, 82.96%) were below the 30% detection threshold for the real-time assay, whereas nearly three-fourths of the COX-negative fibers (206/284, 72.54%) had mtDNA deletion levels >70% (Fig. 4).
Table 1.
EOM Samples Used for Quantitative Real-Time PCR
| EOM Code | Age | COX Deficiency (%) |
|---|---|---|
| M4 | 36 | 2.60 |
| M5 | 66 | 0.98 |
| M8 | 65 | 3.26 |
| M9 | 60 | 3.66 |
| M11 | 42 | 2.43 |
| M12 | 69 | 1.51 |
| M14 | 59 | 5.56 |
| M15 | 39 | 1.53 |
| M16 | 96 | 7.60 |
| M26 | 36 | 0.99 |
| M46* | 74 | 4.73 |
| M49* | 91 | 5.67 |
Average value of two different EOMs harvested from the same globe.
Figure 3.
Level of mtDNA deletion in single COX-positive and COX-negative EOM fibers. (A) Initial group; (B) total group. Horizontal line: mean deletion value. (A) COX-positive, 14.71% (SD 24.13%, n = 103); COX-negative, 66.46% (SD 36.66%, n = 109); (B) COX-positive, 14.86% (SD 22.0%, n = 135); COX-negative, 66.64% (SD 36.45%, n = 284). There was no significant difference in the distribution of mtDNA deletion levels for both COX-positive (P = 0.9581) and COX-negative fibers (P = 0.9634), when comparing the initial and total groups.
Figure 4.
Proportion of COX-positive and COX-negative EOM fibers with different mtDNA deletion load.
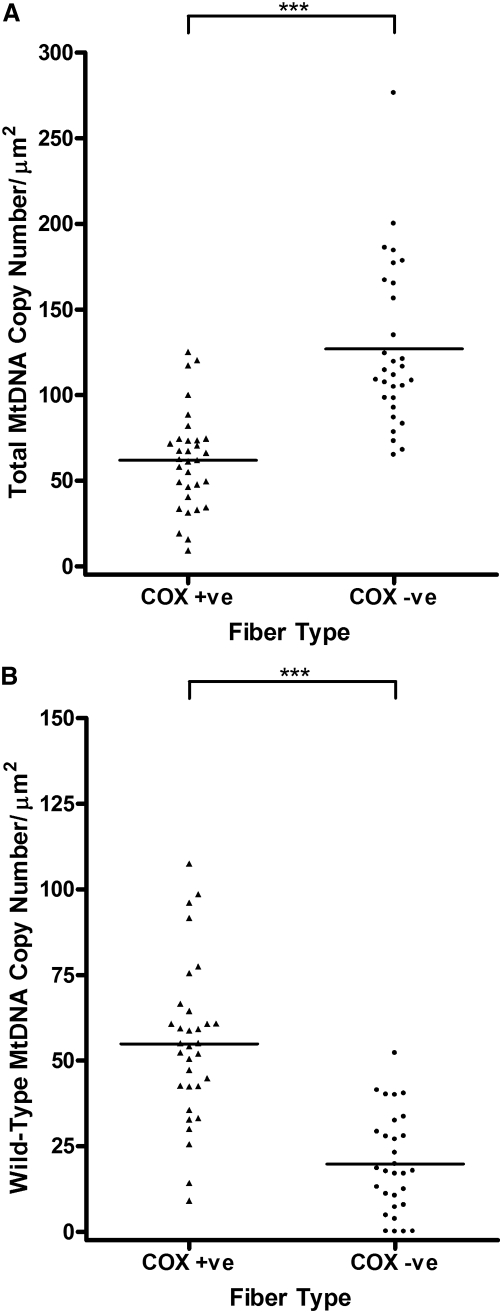
There was evidence of significant mtDNA proliferation in the COX-negative fibers (mean total mtDNA content, 127.00 copies/μm2, SD 47.40 copies/μm2; n = 30), with a ratio of 2.05 compared with that in the COX-positive fibers (mean total mtDNA content, 61.93 copies/μm2, SD 28.55 copies/μm2; n = 32, P < 0.0001; Fig. 5A). A marked reduction in the number of wild-type mtDNA molecules was observed in COX-negative fibers (mean wild-type mtDNA content, 19.77 copies/μm2, SD 14.40 copies/μm2; n = 30), compared with the number in COX-positive fibers (mean wild-type mtDNA content, 54.90/μm2, SD 22.90 copies/μm2; n = 32) (P < 0.0001; Fig. 5B).
Figure 5.
(A) Total and (B) wild-type mtDNA copy number/μm2 in single 20 μm–thick COX-positive (n = 32) and COX-negative (n = 30) EOM fiber sections (***P < 0.0001).
Characterization of mtDNA Deletions
Long-range PCR analysis of six COX-negative fibers with deletion levels >70% produced no full-length product, but a single, smaller amplicon was detected in each case. The deletion sizes ranged from 3 to 7 kb, and breakpoint analysis was performed on the gel-extracted product from three fibers. The analysis revealed that two fibers had a previously reported 6,990-bp deletion (7,808-14,799) with a 6-bp (TCATCG) direct repeat (class I),31 whereas one fiber had a novel 3,546-bp deletion (9,588-13,135) with no flanking repeats (class III).
Mitochondrial Genome Variants
The whole mitochondrial genome was sequenced from the serial sections of 19 COX-deficient fibers with low deletion levels (≤70%) on real-time PCR, and 6 COX-positive fibers: M8 (COX-positive, n = 2; COX-negative, n = 9), M9 (COX-negative, n = 3), and M12 (COX-positive, n = 4; COX-negative, n = 7). Several homoplasmic and heteroplasmic mtDNA variants were identified that diverged from the revised Cambridge reference sequence,33 and they were also not found in other fibers from the same EOM sample: rRNA (n = 4), tRNA (n = 1), complex subunit genes (n = 8), and the D-loop noncoding region (n = 5; Table 2).
Table 2.
Mitochondrial Genome Variants Identified in Single EOM Fibers
| Patient/Fiber | Variant | Gene | Level | AA Change | Conservation | Databases |
|---|---|---|---|---|---|---|
| M8 | ||||||
| F1 +ve | m.195_196insA | Noncoding | Homoplasmic | — | — | Not reported |
| F3 +ve | m.195_196insA | Noncoding | Homoplasmic | — | — | Not reported |
| m.3472T>C | MTND1 | Heteroplasmic (60%) | p.Phe56Leu | Poor | mtDB: 1/2704 | |
| F3 −ve | m.564G>A* | Noncoding | Homoplasmic | — | — | Not reported |
| F8 −ve | m.901G>A | MTRNR1 | Heteroplasmic (80%) | — | — | Not reported |
| F16 −ve | m.564G>A* | Noncoding | Homoplasmic | — | — | Not reported |
| M9 | ||||||
| F15 −ve | m.73A>G | Noncoding | Homoplasmic | — | — | Polymorphism |
| m.3089A>C | MTRNR2 | Homoplasmic | — | — | Not reported | |
| m.5547A>G | MTTW | Homoplasmic | — | High | Not reported | |
| F16 −ve | m.482_565del* | Noncoding | Homoplasmic | — | — | Not reported |
| M12 | ||||||
| F1 +ve | m.72T>C | Noncoding | Homoplasmic | — | — | Polymorphism |
| F3 +ve | m.2405C>T | MTRNR2 | Homoplasmic | — | — | Not reported |
| F1 −ve | m.5459C>T | MTND2 | Homoplasmic | p.Ile330lle | Poor | mtDB: 1/2703 |
| m.8089A>G | MTCO2 | Homoplasmic | p.Leu168Leu | High | Not reported | |
| m.8251G>A | MTCO2 | Homoplasmic | p.Gly222Gly | Poor | Polymorphism | |
| m.8855C>T | MTATP6 | Homoplasmic | p.Ala110Val | Poor | mtDB: 1/2703 | |
| m.8994G>A | MTATP6 | Homoplasmic | p.Leu156Leu | High | Polymorphism | |
| F4 −ve | m.2386C>T | MTRNR2 | Homoplasmic | — | — | mtDB: 1/2704 |
| m.10972G>A | MTND4 | Homoplasmic | p.Trp71Trp | High | mtDB: 2/2704 | |
| F7 −ve | m.72T>C | Noncoding | Homoplasmic | — | — | Polymorphism |
| F8 −ve | m.4035A>G | MTND1 | Homoplasmic | p.Leu243Leu | Poor | Not reported |
The variants listed were not found in other single EOM fibers sequenced from the same patient, and the following databases were checked to determine whether these had been reported as mtDNA single nucleotide polymorphisms: MITOMAP29 (http://www.mitomap.org/Center for Molecular and Mitochondrial Medicine and Genetics, University of California, Irvine, CA); mtDB34 (http://www.genpat.uu.se/mtDB/ Department of Genetics and Pathology, University of Uppsala, Sweden); and mtDNA databases35 (MitoKor, San Diego, CA). The evolutionary conservation of altered amino acid (AA) residues was assessed with the PIR-International Protein Sequence Database (http://pir.georgetown.edu/ Bioinformatics Graduate Program, Georgetown University, Washington, DC)36 and the m.5547A>G tRNA variant was analyzed with the Mamit-tRNA database (http://mamit-trna.u-strasbg.fr/tables.asp?aminoacid = 25/ Institute of Molecular and Cellular Biology, University of Strasbourg, France). Ala, alanine; F, fiber; Gly, glycine; Ile, isoleucine; Leu, leucine; Phe, phenylalanine; Trp, tryptophan; Val, valine.
Within the major H-strand promoter region (545–567) of the noncoding control region.
Discussion
COX deficiency is an important biomarker of mitochondrial dysfunction. This deficiency was evident in EOMs from the third decade of life, with a significant age-related increase. Our results confirm an earlier study by Muller-Hocker et al.,22 in which a similar age-related increase in COX-negative EOM fibers was found, with an average defect density ranging from ∼1 (<50 years) to ∼4 (>70 years) mm2. This finding is in sharp contrast to skeletal muscle from healthy control subjects in whom COX deficiency was observed only in those >60 years of age and at relatively low levels (<1%).27 Increased COX deficiency has also been reported in various neuronal populations as part of normal aging,14,37,38 but again the deficiency levels only reached ∼1% in the more elderly group (i.e., significantly lower than in EOMs). Although there is a scatter of data points, our modeling experiments supported an exponential increase in the proportion of COX-negative EOM fibers with age. Except for one specimen, the level of COX deficiency measured in the remaining 45 EOMs fell within the predicted upper and lower 95% confidence range (Fig. 2). This normative dataset is a valuable resource that could be applied for diagnostic purposes when analyzing the significance of COX-deficient fibers identified in EOMs resected at the time of corrective strabismus surgery in patients with suspected mitochondrial disorders. In a parallel investigation of 13 patients with chronic progressive external ophthalmoplegia (CPEO) due to mtDNA disease, we have shown that EOM samples from these patients had levels of COX deficiency an order of magnitude higher than that in the normal aging range predicted in the current study.39
There are major biological differences between EOMs and skeletal muscle that very likely account for the earlier age at which COX deficiency becomes manifest in EOMs and the greater deficiency seen in healthy elderly subjects relative to that in skeletal muscle. EOMs have a distinct pattern of embryonic development, arising from prechordal plate and rostral mesodermal condensations, compared with the somitic and lateral plate mesodermal origins of skeletal muscle.40 As a consequence, EOM and skeletal muscle fibers exhibit fundamental differences in their fiber type composition, their myosin heavy chain isoform expression, and the basic organization of their motor units.41,42 Of importance, because of the need for sustained muscular contractions and fatigue resistance,43 EOMs have a higher mitochondrial content than do skeletal muscles,44 and they possess a richer vascular supply to support these greater metabolic requirements.45 It is also important to note that a population of activated satellite cells has been identified in normal EOMs, resulting in a slow but continuous myonuclear addition to existing myofibers, unlike skeletal muscle where satellite cells remain quiescent in the noninjured state.46–48
Although the 4977-bp common deletion is found in up to a third of all patients with mitochondrial disorders who have single mtDNA deletions,49,50 it was found in only 2 (3.39%) of 59 COX-negative fibers. This specific finding in EOMs is consistent with previous studies that have reported the 4977-bp common deletion in only a very small proportion of COX-negative skeletal muscle fibers.13,51,52 However, a more sensitive real-time PCR assay made it apparent that the majority of the COX-negative fibers (72.54%) harbored high deletion levels (>70%), with evidence of significant mitochondrial proliferation. This compensatory mechanism was insufficient to maintain the critical number of wild-type mtDNA molecules necessary to maintain normal oxidative phosphorylation, triggering a cellular energetic deficit. Using a two-step, long-range PCR method, we also showed clonal expansion of a single deleted mtDNA species in COX-negative fibers, in agreement with previous observations in aging skeletal muscle13,53 and other postmitotic tissues.14,16
The entire mitochondrial genome of 19 COX-negative fibers with low deletion levels (≤70%) was sequenced to evaluate the role of mtDNA point mutations in aging EOMs. The majority of the substitutions within mitochondrial structural genes were synonymous (6/8): The m.3472T>C variant (complex I) was heteroplasmic (60%) and was found in a COX-positive fiber, whereas the m.8855C>T variant (complex V) is likely to be a neutral polymorphism, converting a poorly conserved alanine to valine and having been previously reported on the mtDB database.34 Only one nucleotide substitution within a tRNA gene was identified, a novel homoplasmic m.5547A>G variant (MTTW). It is likely to be pathogenic, as it alters a highly conserved base within the tRNA tryptophan anticodon loop, and it is adjacent to other positions implicated in mitochondrial disease: m.5545C>T54 and m.5549G>A.55 The overall mutational yield was relatively low (1/19, 5.3%), but comparable to that found by Fayet et al.,52 who identified pathogenic mtDNA point mutations in only 8 (7.3%) of 109 COX-negative skeletal muscle fibers. Our results are also in agreement with the current literature which suggests that somatic mtDNA deletions tend to accumulate in postmitotic tissues, whereas point mutations show a predilection for rapidly dividing mitotic tissues.12,14,15,19,31,56 Because of the limitations of our real-time assay, a proportion of mtDNA deletions will have been missed, and although additional studies are required, it is possible that nuclear defects involving complex subunits and their assembly factors account for the COX-deficient status of some EOM fibers.57
What is the relevance of our findings in EOMs to disease and the aging process? EOMs are a useful tissue for investigating the contribution of mitochondrial dysfunction in aging, as they are tonically active in comparison to skeletal muscle, where reduced oxidative phosphorylation could be due to physical inactivity and not an age-related phenomenon per se.27 Our study has confirmed an accelerated rate of accumulation of COX deficiency in EOMs compared with skeletal muscle, with the majority of the COX-deficient fibers harboring high levels of clonally expanded, somatic mitochondrial DNA deletions. This phenomenon provides an attractive explanation for the high prevalence of CPEO among mitochondrial patients, and further studies into the mechanisms involved are crucial for determining the possibility of genetic manipulation for the treatment of this progressive and debilitating ocular disorder.
Footnotes
Supported in part by a Wellcome Trust program grant, the Medical Research Council Translational Muscle Centre, the Newcastle upon Tyne Hospitals NHS Trust, and the UK National Commissioning Group for Rare Mitochondrial Disorders of Adults and Children (RWT, DMT). PYWM is an MRC Research Fellow in Neuro-ophthalmology and LCG is supported by Newcastle University Centre for Brain Ageing and Vitality.
Disclosure: P. Yu-Wai-Man, None; J. Lai-Cheong, None; G.M. Borthwick, None; L. He, None; G.A. Taylor, None; L.C. Greaves, None; R.W. Taylor, None; P.G. Griffiths, None; D.M. Turnbull, None
References
- 1.Schapira AHV. Mitochondrial disease. Lancet 2006;368:70–82 [DOI] [PubMed] [Google Scholar]
- 2.Schaefer AM, McFarland R, Blakely EL, et al. Prevalence of mitochondrial DNA disease in adults. Ann Neurol 2008;63:35–39 [DOI] [PubMed] [Google Scholar]
- 3.Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet 2008;83:254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manwaring N, Jones MM, Wang JJ, et al. Population prevalence of the MELAS A3243G mutation. Mitochondrion 2007;7:230–233 [DOI] [PubMed] [Google Scholar]
- 5.Shoubridge EA, Karpati G, Hastings KEM. Deletion mutants are functionally dominant over wild-type mitochondrial genomes in skeletal-muscle fiber segments in mitochondrial disease. Cell 1990;62:43–49 [DOI] [PubMed] [Google Scholar]
- 6.Bua EA, McKiernan SH, Wanagat J, McKenzie D, Aiken JM. Mitochondrial abnormalities are more frequent in muscles undergoing sarcopenia. J Appl Physiol 2002;92:2617–2624 [DOI] [PubMed] [Google Scholar]
- 7.Durham SE, Samuels DC, Cree LM, Chinnery PF. Normal levels of wild-type mitochondrial DNA maintain cytochrome c oxidase activity for two pathogenic mitochondrial DNA mutations but not for m. 3243A→G. Am J Hum Genet 2007;81:189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinnery PF, Samuels DC, Elson J, Turnbull DM. Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: is there a common mechanism? Lancet 2002;360:1323–1325 [DOI] [PubMed] [Google Scholar]
- 9.Krishnan KJ, Greaves LC, Reeve AK, Turnbull D. The ageing mitochondrial genome. Nucleic Acids Res 2007;35:7399–7405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinnery PF, Samuels DC. Relaxed replication of mtDNA: a model with implications for the expression of disease. Am J Hum Genet 1999;64:1158–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elson JL, Samuels DC, Turnbull DM, Chinnery PF. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am J Hum Genet 2001;68:802–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nekhaeva E, Bodyak ND, Kraytsberg Y, et al. Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues. Proc Natl Acad Sci U S A 2002;99:5521–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bua E, Johnson J, Herbst A, et al. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am J Hum Genet 2006;79:469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet 2006;38:515–517 [DOI] [PubMed] [Google Scholar]
- 15.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet 2006;38:518–520 [DOI] [PubMed] [Google Scholar]
- 16.Reeve AK, Krishnan KJ, Turnbull D. Age related mitochondrial degenerative disorders in humans. BioTechnol J 2008;3:750–756 [DOI] [PubMed] [Google Scholar]
- 17.Trifunovic A, Wredenberg A, Falkenberg M, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 2004;429:417–423 [DOI] [PubMed] [Google Scholar]
- 18.Corral-debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial-DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet 1992;2:324–329 [DOI] [PubMed] [Google Scholar]
- 19.Liu VWS, Zhang CF, Nagley P. Mutations in mitochondrial DNA accumulate differentially in three different human tissues during ageing. Nucleic Acids Res 1998;26:1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murdock DG, Christacos NC, Wallace DC. The age-related accumulation of a mitochondrial DNA control region mutation in muscle, but not brain, detected by a sensitive PNA-directed PCR clamping based method. Nucleic Acids Res 2000;28:4350–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Michikawa Y, Mallidis C, et al. Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication. Proc Natl Acad Sci U S A 2001;98:4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller-Hocker J, Schneiderbanger K, Stefani FH, Kadenbach B. Progressive Loss of cytochrome-c-oxidase in the human extraocular-muscles in aging: a cytochemical-immunohistochemical study. Mutat Res 1992;275:115–124 [DOI] [PubMed] [Google Scholar]
- 23.Muller-Hocker J. Cytochrome-c-oxidase deficient cardiomyocytes in the human heart–an age-related phenomenon: a histochemical ultracytochemical study. Am J Pathol 1989;134:1167–1173 [PMC free article] [PubMed] [Google Scholar]
- 24.Muller-Hocker J. Cytochrome c oxidase deficient fibres in the limb muscle and diaphragm of man without muscular disease: an age-related alteration. J Neurol Sci 1990;100:14–21 [DOI] [PubMed] [Google Scholar]
- 25.Johnson MA, Barron MJ. Muscle biopsy analysis. In: Lane RJM. ed. Handbook of Muscle Disease New York: Marcel Dekker; 1996:61–79 [Google Scholar]
- 26.Taylor RW, Barron MJ, Borthwick GM, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest 2003;112:1351–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brierley EJ, Johnson MA, James OFW, Turnbull DM. Effects of physical activity and age on mitochondrial function. Q J Med 1996;89:251–258 [DOI] [PubMed] [Google Scholar]
- 28.Sciacco M, Bonilla E, Schon EA, DiMauro S, Moraes CT. Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum Mol Genet 1994;3:13–19 [DOI] [PubMed] [Google Scholar]
- 29.Brandon MC, Lott MT, Nguyen KC, et al. MITOMAP: a human mitochondrial genome database. Nucleic Acids Res 2005;33:D611–D613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He L, Chinnery PF, Durham SE, et al. Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res 2002; 2002;30(14):e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reeve AK, Krishnan KJ, Elson JL, et al. Nature of mitochondrial DNA deletions in substantial nigra neurons. Am J Hum Genet 2008;82:228–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor RW, Taylor GA, Durham SE, Turnbull DM. The determination of complete human mitochondrial DNA sequences in single cells: implications for the study of somatic mitochondrial DNA point mutations. Nucleic Acids Res 2001;29:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999;23:147. [DOI] [PubMed] [Google Scholar]
- 34.Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res 2006;34:D749–D751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrnstadt C, Elson JL, Fahy E, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 2002;70:1152–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu CH, Yeh LSL, Huang HZ, et al. The protein information resource. Nucleic Acids Res 2003;31:345–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cottrell DA, Blakely EL, Johnson MA, Ince PG, Borthwick GM, Turnbull DM. Cytochrome c oxidase deficient cells accumulate in the hippocampus and choroid plexus with age. Neurobiol Aging 2001;22:265–272 [DOI] [PubMed] [Google Scholar]
- 38.Barron MJ, Johnson MA, Andrews RM, et al. Mitochondrial abnormalities in ageing macular photoreceptors. Invest Ophthalmol Vis Sci 2001;42:3016–3022 [PubMed] [Google Scholar]
- 39.Greaves LC, Yu-Wai-Man P, Blakely EL, et al. Mitochondrial DNA defects and selective extraocular muscle involvement in CPEO. Invest Ophthalmol Vis Sci 2010:51:3340–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porter JD, Baker RS, Ragusa RJ, Brueckner JK. Extraocular muscles: basic and clinical aspects of structure and function. Surv Ophthalmol 1995;39:451–484 [DOI] [PubMed] [Google Scholar]
- 41.Yu Wai Man CY, Chinnery PF, Griffiths PG. Extraocular muscles have fundamentally distinct properties that make them selectively vulnerable to certain disorders. Neuromuscul Disord 2005;15:17–23 [DOI] [PubMed] [Google Scholar]
- 42.Spencer RF, Porter JD. Biological organization of the extraocular muscles. Prog Brain Res 2006;151:43–80 [DOI] [PubMed] [Google Scholar]
- 43.Fuchs AF, Binder MD. Fatigue resistance of human extraocular muscles. J Neurophysiol 1983;49:28–34 [DOI] [PubMed] [Google Scholar]
- 44.Carry MR, Ringel SP, Starcevich JM. Mitochondrial morphometrics of histochemically identified human extraocular-muscle fibers. Anat Rec 1986;214:8–16 [DOI] [PubMed] [Google Scholar]
- 45.Wooten GF, Reis DJ. Blood flow in extraocular muscle of cat. Arch Neurol 1972;26:350–352 [DOI] [PubMed] [Google Scholar]
- 46.McLoon LK, Wirtschafter JD. Continuous myonuclear addition to single extraocular myofibers in uninjured adult rabbits. Muscle Nerve 2002;25:348–358 [DOI] [PubMed] [Google Scholar]
- 47.McLoon LK, Wirtschafter J. Activated satellite cells in extraocular muscles of normal adult monkeys and humans. Invest Ophthalmol Vis Sci 2003;44:1927–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLoon LK, Rowe J, Wirtschafter J, McCormick KM. Continuous myofiber remodeling in uninjured extraocular myofibers: myonuclear turnover and evidence for apoptosis. Muscle Nerve 2004;29:707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schon EA, Rizzuto R, Moraes CT, Nakase H, Zeviani M, Dimauro S. A direct repeat is a hotspot for large-scale deletion of human mitochondrial-DNA. Science 1989;244:346–349 [DOI] [PubMed] [Google Scholar]
- 50.Yamashita S, Nishino I, Nonaka I, Goto YI. Genotype and phenotype analyses in 136 patients with single large-scale mitochondrial DNA deletions. J Hum Genet 2008;53:598–606 [DOI] [PubMed] [Google Scholar]
- 51.Brierley EJ, Johnson MA, Lightowlers RN, James OFW, Turnbull DM. Role of mitochondrial DNA mutations in human aging: implications for the central nervous system and muscle. Ann Neurol 1998;43:217–223 [DOI] [PubMed] [Google Scholar]
- 52.Fayet G, Jansson M, Sternberg D, et al. Ageing muscle: clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function. Neuromuscul Disord 2002;12:484–493 [DOI] [PubMed] [Google Scholar]
- 53.Cao ZJ, Wanagat J, McKiernan SH, Aiken JM. Mitochondrial DNA deletion mutations are concomitant with ragged red regions of individual, aged muscle fibers: analysis by laser-capture microdissection. Nucleic Acids Res 2001;29:4502–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sacconi S, Salviati L, Nishigaki Y, et al. A functionally dominant mitochondrial DNA mutation. Hum Mol Genet 2008;17:1814–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson I, Hanna MG, Alsanjari N, Scaravilli F, Morganhughes JA, Harding AE. A new mitochondrial-DNA mutation associated with progressive dementia and chorea: a clinical, pathological, and molecular-genetic study. Ann Neurol 1995;37:400–403 [DOI] [PubMed] [Google Scholar]
- 56.Reeve AK, Krishnan KJ, Taylor G, et al. The low abundance of clonally expanded mitochondrial DNA point mutations in aged substantia nigra neurons. Aging Cell 2009;8:496–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muller-Hocker J, Seibel P, Schneiderbanger K, Kadenbach B. Different in situ hybridization patterns of mitochondrial-DNA in cytochrome-c oxidase-deficient extraocular-muscle fibers in the elderly. Virchows Arch 1993;422:7–15 [DOI] [PubMed] [Google Scholar]