The authors identified a new genetic factor, SLC16A12, which encodes a monocarboxylate transporter, as involved in age-related cataract. Sequence alterations in its 5′untranslated region affect translational efficiency, a potential mechanism to challenge homeostasis within the lens.
Abstract
Purpose.
Knowledge of genetic factors predisposing to age-related cataract is very limited. The aim of this study was to identify DNA sequences that either lead to or predispose for this disease.
Methods.
The candidate gene SLC16A12, which encodes a solute carrier of the monocarboxylate transporter family, was sequenced in 484 patients with cataract (134 with juvenile cataract, 350 with age-related cataract) and 190 control subjects. Expression studies included luciferase reporter assay and RT-PCR experiments.
Results.
One patient with age-related cataract showed a novel heterozygous mutation (c.-17A>G) in the 5′untranslated region (5′UTR). This mutation is in cis with the minor G-allele of the single nucleotide polymorphism (SNP) rs3740030 (c.-42T/G), also within the 5′UTR. Using a luciferase reporter assay system, a construct with the patient's haplotype caused a significant upregulation of luciferase activity. In comparison, the SNP G-allele alone promoted less activity, but that amount was still significantly higher than the amount of the common T-allele. Analysis of SLC16A12 transcripts in surrogate tissue demonstrated striking allele-specific differences causing 5′UTR heterogeneity with respect to sequence and quantity. These differences in gene expression were mirrored in an allele-specific predisposition to age-related cataract, as determined in a Swiss population (odds ratio approximately 2.2; confidence intervals, 1.23–4.3).
Conclusions.
The monocarboxylate transporter SLC16A12 may contribute to age-related cataract. Sequences within the 5′UTR modulate translational efficiency with pathogenic consequences.
Cataract is the clouding of the eye's lens that impairs normal vision. It is estimated that cataract accounts for 17 million cases of blindness worldwide, with approximately half of all cases occurring in Asia and Africa.1,2 Different criteria—age of onset, morphologic features, and mode of inheritance—can be used to classify the various forms of cataracts. Based on the age of onset, one distinguishes between childhood (congenital and juvenile) cataract and age-related cataract (ARC), but this criterion does not necessarily indicate etiology.3 Genetic predisposition plays a crucial role in childhood cataract.1,2 Congenital and juvenile forms of cataract show wide heterogeneity with respect to genetic and phenotypic aspects.4 A number of mutations in approximately 20 genes have been described as causing childhood cataract.3,5–7
Approximately 80% of all cataracts are age-related and idiopathic. Depending on the location of the opacity within the lens, ARC is termed cortical, nuclear, or subcapsular. There are also forms of mixed cataract that feature more than one morphological sign. In general, maintenance of an intact, transparent lens requires balanced homeostasis of metabolic components.8 ARC is considered a multifactorial disease in which environmental components and genetic predisposition contribute to the development of the pathologic condition. Interactions between these factors are likely, and knowledge of the cause of ARC may provide crucial information for the prevention of and potential therapy for the disease. Among environmental risk factors are smoking, exposure to UV-B radiation, and alcohol.9 In addition, physiological conditions such as age, sex, diabetes, high body mass index, persistent intraocular inflammation, prolonged corticosteroid administration, and oxidative damage seem to promote the development of ARC.9 In light of this complexity, knowledge of genetic risk factors is still scarce. Variants of the detoxifying enzymes arylamine N-acetyltransferase-2 and glutathione-S-transferase (GST) were found to be associated with ARC.10–12 Furthermore, two sequence variants in the vicinity of the 3′ end of the gene for Eph-receptor tyrosine kinase type A2 (EPHA2) were shown to associate with both childhood and age-related forms of cataract.13 Recently, a mutation in the gene encoding αA-crystallin was reported to be associated with ARC because of the loss of chaperone-like activity.14 SNP-based allele frequencies and, consequently, association with a disease phenotype vary often among different ethnic groups, exemplified by a sequence variant in the gene encoding galactokinase, an enzyme involved in galactose metabolism15,16 and GST.10,11,17 Similarly, heat-shock transcription factor 4 may be involved in ARC in an Asian population.18
To the list of genes involved in childhood cataract19 we recently added SLC16A12, which encodes a monocarboxylate transporter.6 Although the substrate of this transporter is not yet known, its importance for the establishment or maintenance of homeostasis was suggested because a premature termination codon in SLC16A12 leads to juvenile cataract and renal glucosuria.6 Based on this proposed function of the transporter, we speculated that insufficient activity could interfere with the maintenance of homeostatic conditions within the lens and could lead to ARC. Now we report the effects on ARC of two sequence alterations in the 5′untranslated region (5′UTR) of SLC16A12. The importance of 5′UTR sequences for translational regulation has been demonstrated in several other genes.20–23 Approximately 10% of all coding genes are regulated at the mRNA level and, hence, influence translational efficiency. Disturbance of the regulation of the translational machinery leads to perturbed cellular metabolism and may tilt the physiological balance from healthy to diseased states, as occurs in breast cancer, Alzheimer's disease, bipolar affective disorder, fragile X-syndrome, and others.20,22 Our work focuses on sequence alterations within the 5′UTR of SLC16A12 and offers an explanation for the development of ARC.
Materials and Methods
Patients
Patients with childhood cataract and ARC, including cortical, nuclear, posterior subcapsular, and mixed types of cataract, were seeking ophthalmologic examination in Switzerland. Subjects from among the general population in Switzerland served as controls. Information on patient's condition of hypertension, diabetes mellitus, smoking behavior, alcohol consumption, and exposure to UV-B radiation was not available. Patients gave written informed consent for participation in scientific research. All experiments involving human subjects were conducted according to the principles expressed in the Declaration of Helsinki.
DNA Analysis
DNA was prepared either by the precipitation method (Gentra Kit; (Qiagen, Hilden, Germany) or by magnetic bead technology (Chemagen, Aachen, Germany). For PCR, approximately 50 ng template DNA and primers (Table 1) were cycled 35 times with annealing and extension temperatures of 60°C and 72°C, respectively, lasting 1 minute each. DNA sequencing was performed with commercially available technology (Applied Biosystems, Rotkreuz, Switzerland).
Table 1.
Primer Characteristics
| Primer Name | Sequence (5′-3′) | Purpose |
|---|---|---|
| Intronic primers 3F | gtctgccccagtctagtattca | Genomic DNA sequencing |
| Intronic primers 3R | cggaaatacacacacaccaca | Genomic DNA sequencing |
| CL forward | ATGCaagcttGCCAGGTAGCGTTCTATGCC | Cloning |
| CL reverse | CGGAAATACACACACACCACA | Cloning |
| SLC16A12RT_ex1_2f | cggtggctcagATACAGGAT | RT-PCR |
| SLC16A12RT_ex4r | aaacagccagccacaatcat | RT-PCR |
| RTPRC_2F | GTGTGACCATGCTCTGTGct | RT-PCR |
| RTPRC_4R | AAGACAAAGCCCCCAAGAAT | RT-PCR |
| RTPCR_3F | caggaagtcactggacagca | RT-PCR |
| RTPCR_5R | gcatgattcccacttgacag | RT-PCR |
| cSLC16A12_1F | CCCTCTTCCCTCTCCCTGA | RT-PCR |
| cSLC16A12_3R | TGTGCAGATGGTAACAAGGAAAC | RT-PCR |
| SLC16A12RTqex2f | TTAGCCTCCCAAAGTGATGG | RT-PCR |
| SLC16A12RTqex4r | CGTTTGTGCGTAATCCTGAG | RT-PCR |
RNA Analysis
RNA from vascular smooth muscle cell (VSMC) cultures was isolated (All Prep DNA RNA Mini Kit; Qiagen, Hilden, Germany). RNA was evaluated with a bioanalyzer (2100 Bioanalyzer; Agilent Technologies, Palo Alto, CA). Two-step RT-PCR required cDNA synthesis (Superscript III; Invitrogen, Basel, Switzerland). Standard RT-PCR conditions were applied (Hotfire Taq Polymerase; Solis Biodyne, Tartu, Estonia) for 1 minute at an annealing temperature of 58°C, 1 minute elongation time at 72°C, and 39 cycles. PCR products were analyzed by 1% agarose gel electrophoresis. Quantitative sequencing of the SLC16A12 c.-42T>G variant was performed as described.24 Briefly, to determine the correction factors, genomic DNA (gDNA) was amplified in duplicate, and each amplicon was sequenced in eight different reactions. For RNA, one-step RT-PCR (Qiagen) was performed in triplicate, and each amplicon was sequenced eight times. Potential splice sites were sought with the online tool ESEfinder 3.0 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home).25,26
In Silico Analysis of 5′UTR Variants on RNA Folding
Putative RNA folding structures were predicted using Mfold with standard settings (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi).27 RNA structures were predicted for SLC16A12 (CL reverse NM_213606) and for its 5′UTR alone.
Plasmids, Vectors, and Cloning
Exon 3 of SLC16A12 contains part of the 5′UTR and the initiation ATG followed by 36 encoded amino acids. A fragment of the 118-bp SLC16A12 sequence (5′-GCCAGGTAGCGTTCTATGCCAACCTTGAATGCCATCAGGAAGTCACTGGACAGCAAACTCTTCCAAGATCATAACTT[G/T]GGCTGTTGGAGCAACCTGGAAAAG[A/G]AGAAAAAAGAAAAACC-3′ was cloned in front of the luciferase gene in pGL3 control vector (Promega, Madison, WI) with HindIII and NcoI restriction enzymes. For this purpose, exon 3 was PCR amplified using an upstream primer (Table 1) that contained a HindIII restriction site at its 5′ end and a downstream primer (Table 1). Genomic DNA heterozygous for the SNP rs3740030 and genomic DNA from the cataract patient with c.-42G/T and c. -17A/G served as templates. PCR amplicons and vector pGL3 were digested with restriction enzymes HindIII and NcoI and were ligated with DNA ligase (Promega). Constructs were verified by DNA sequence analysis.
Mammalian Cell Culture Experiments
Cells (104 HEK293T) were seeded on 96-well plates in 0.5 mL DMEM, 10% FBS, and 1% penicillin/streptomycin and were transfected 24 hours later. Plasmid DNA (615 ng [600 ng pFirefly construct, 15 ng pRenilla construct]) dissolved in 2 μL calcium chloride (2.5 M) and 10 μL HeBS (2×) was used for each transfection (20 μL transfection mix/well). Cells were harvested 48 hours after transfection, and Luciferase activities were measured with a Luciferase reporter assay system (Dual Glo; Promega Dual Glo) and a luminometer (Luminoskan Ascent; Thermo-Labsystems, Egelsbach, Germany).
Isolation and Cultivation of Vascular Smooth Muscle Cells
Vascular smooth muscle cells were isolated from radial arteries of patients undergoing coronary artery bypass grafting, as previously described.28 Identification was made by immunofluorescent staining for smooth muscle α-actin (no. 1148818; Roche Diagnostics, Mannheim, Germany). Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and were used up to passage 10.
Statistical Analysis
DNA Analysis.
For statistical calculation of odds ratio and significance, an open access Internet portal was used (http://faculty.vassar.edu/lowry/odds2x2.html; June 2009). Calculations were based on a P = 0.05 level of significance.
RNA Analysis.
For the arithmetic mean of replicates (gDNA, n = 4; cDNA, n = 6), upper and lower confidence limits were determined using critical values of paired t-test distribution.
Mammalian Cell Culture Experiments.
Averages and confidence intervals (paired t-test) were calculated and normalized on T-allele activity. Tissue culture experiments were performed twice using three technical replicas per measurement.
Results
Identification of Mutation in the 5′UTR of SLC16A12 and Patient Characteristics
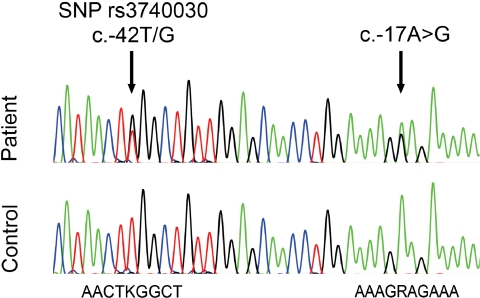
For mutation screening, we investigated genomic DNA of 350 patients with ARC and 134 patients with childhood cataract. DNA sequences from all exons and adjacent intron regions of SLC16A12 were analyzed. A 79-year-old woman with ARC was found to carry a heterozygous sequence alteration in the 5′UTR (c.-17A>G; Fig. 1), but coding and approximately 50 nucleotides flanking intron sequences were unchanged. The sequence change was not found in any of the other 483 patients or in the 380 alleles of control subjects. This uniqueness makes the change unlikely to be a normal sequence variant; rather, it indicates pathogenicity. Hence, we will refer to it from now on as mutation. The patient was also heterozygous at an annotated SNP rs3740030 (c.-42T/G) that lies just upstream of the mutation. Cloning and sequencing of the patient's exon 3 showed that the mutation and the G-allele are on the same chromosome.
Figure 1.
Electropherogram showing genomic DNA of 5′UTR section from control subject and ARC patient. Nucleotides of the immediate surroundings of SNP rs3740030 (c.-42) and the mutation (c.-17) are given. K = T and G; R = A and G.
The patient was seen by an ophthalmologist for cataract. The lens of her left eye showed nuclear and subcapsular opacities plus pseudoexfoliation of the lens capsule that was removed when she was 75. At age 79, her right eye was also subjected to surgery. The right lens showed nuclear brunescent sclerosis with pulverulent anterior and posterior cortical opacities plus pseudoexfoliation of the lens capsule. No information about other family members was available.
Quantitative Assessment of SLC16A12 Transcripts
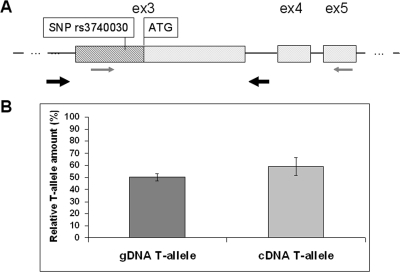
Because the minor G-allele and the mutation reside within the 5′UTR of SLC16A12, it is likely that these changes affect translational rather than transcriptional efficiency. To assess the latter possibility, we determined the amount of SLC16A12 mRNA by quantitative sequencing experiments, a method that requires RNA from heterozygous tissue. It is based on comparing allele frequencies of complementary DNA (cDNA) with those of gDNA from the same sample.24 Given that no tissue was available from the cataract patient, we focused our attention on measuring SNP allele-specific transcripts in surrogate cells. Among available tissues known to express SLC16A12, vascular smooth muscle cells from heterozygous donors served for this purpose. RNA was subjected to one-step RT-PCR using primers in exons 3 and 5, to encompass the SNP (Fig. 2A). As expected, the contribution of the T-allele in genomic DNA was 50% (± 3%). No significant difference was measured for the T-allele in the cDNA (57% ± 9%; Fig. 2B), indicating that the two SNP alleles were present at comparable amounts. We concluded that regulatory mechanisms of translation rather than differential transcription were affected by the patient's sequence alterations.
Figure 2.
Quantitative sequencing analysis. (A) Drawing of exons (ex) 3, 4, and 5 of SLC16A12. Exon 3 contains the coding region beginning with ATG (light gray box) and 5′UTR (hatched box) containing SNP rs3740030. gDNA and transcript representing cDNA were obtained from vascular smooth muscle cells of a subject heterozygous for SNP rs3740030 (c.-42T/G). RT-PCR primers are indicated by thin arrows in exons 3 and exon 5. Locations of primers for genomic DNA amplification are indicated with thick arrows in intronic regions flanking exon 3. (B) Quantitative analysis expressed as relative amount of T-allele containing transcripts, given in percentages, for gDNA and mRNA (cDNA). Confidence intervals are shown for eight technical replicas.
Influence of SLC16A12 5′UTR Sequences at c.-42 and c.-17 on Translational Efficiency
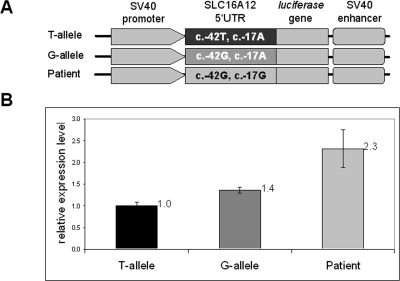
To investigate whether the SNP and the patient's c.-17A>G mutation had effects on translation, constructs containing 5′UTR sequences in front of the luciferase gene were tested in HEK cells, by measuring luciferase reporter activity (Fig. 3). This assay is well suited to simulate translational activity changes because all constructs contain the same promoter, making it likely that transcriptional efficiency is the same among all constructs (Fig. 3A). Statistically significant differences in luciferase activity were observed among the three SLC16A12 constructs (Fig. 3B). The construct that mimicked the patient's haplotype yielded the highest luciferase activity compared with both SNP alleles (2.315 ± 0.433-fold higher). Furthermore, the SNP alleles yielded significantly different luciferase activities such that the minor G-allele led to higher levels than did the more common T-allele (1.362 ± 0.07-fold higher). These results demonstrate a significant effect of 5′UTR sequences and suggest an enhanced potential of SLC16A12 translation.
Figure 3.
Luciferase reporter activity. (A) Schematic representation of cloned constructs. T-allele (c.-42T) and G-allele (c.-42G) contain 122 bp of SLC16A12 5′UTR. Patient construct contains c.-42G and c.-17G. (B) Relative luciferase activity. Values obtained for the T-allele were set to 1, to which other values were normalized. Displayed are the means (numerically) and confidence intervals (graphically) of three technical replicas. Results are from two independent experiments. The presence of the G-allele alone shows 1.4-fold and the presence of the G-allele in combination with the mutation (patient) shows 2.3-fold elevated luciferase expression levels.
Effects of 5′UTR on RNA Folding by In Silico Analysis
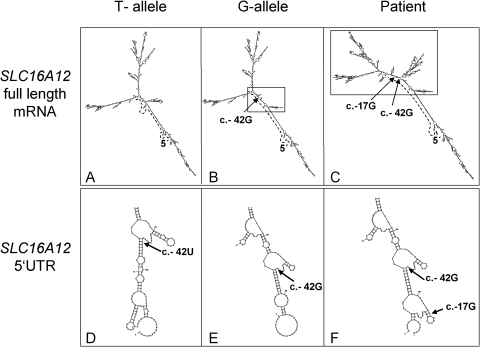
One possible mechanism to achieve such regulation is through RNA folding. We simulated the influence of the 5′UTR sequence variants on secondary structures using the method Mfold,27 which predicts a structure based on free energy minimization. RNA structures of the entire SLC16A12 mRNA, (5′UTR, coding sequences, and 3′UTR) were predicted (Figs. 4A–C). Differences between the T- and G-alleles of SNP rs3740030 affect some branches near the centers of the molecules (Figs. 4A, 4B). The patient's mutation in combination with the SNP G-allele caused a strikingly different RNA structure (Fig. 4C). In addition, 5′UTR sequences alone folded in an allele-specific manner (Figs. 4D–F). Taken together, our experimental data, in combination with the bioinformatic predictions, indicate an effect on nucleotide-specific RNA folding properties.
Figure 4.
Predicted SLC16A12 RNA foldings. T-allele and G-allele refer to the SNP rs3740030 (c.-42T/G). Patient has sequences c.-42G and c.-17G. (A–C) RNA foldings for the entire SLC16A12 transcript. Differences in mRNA structure are highlighted (box). The 5′-end is marked (5′). Dotted line: 5′UTR. Arrows: positions c.-42G, c.42U, and c.-17G. (D–F) Predicted RNA foldings for 5′UTR sequences only.
SNP Allele-Specific Sequence Heterogeneity of 5′UTR SLC16A12 Transcripts
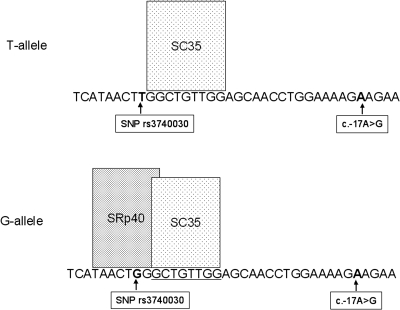
The 5′UTR contains sequences from exons 1 and 2 and part of exon 3. To assess potential splicing effects on the generation of the 5′UTR, we initially applied the online tool ESEfinder 3.0. Splice factor SC35 was predicted to bind to the region immediately adjacent to the SNP, independently of the allele-specific nucleotide (Fig. 5). Interestingly, splice factor SRp40 was predicted to bind exclusively to G-allele transcripts and not to the T-allele transcripts. No binding sites for any of the ESEfinder 3.0–specific splice factors were found at the c.-17.
Figure 5.
Prediction of splice factor binding site for SLC16A12 5′UTR. ESEfinder predictions were applied to cDNA from T- and G-alleles of SNP rs3740030. Factor SC35 binds independently of the allele sequence. In contrast, the binding of splice factor SRp40 is predicted only for the G-allele sequence. At the c.-17 position, no splice factor binding sites are predicted for G or A.
These observations supported the existence of alternative and allele-specific splicing of SLC16A12 transcripts generating 5′UTR heterogeneity.
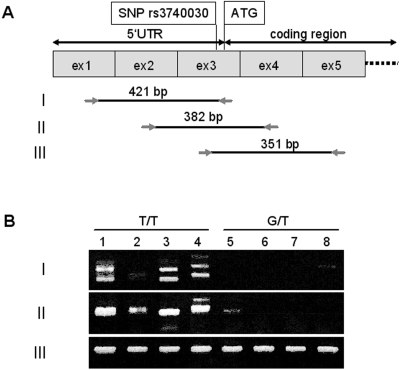
To test these predictions, we performed RT-PCR experiments with various primer combinations on cDNA from vascular smooth muscle cells of donors who were either homozygous T/T or heterozygous G/T with respect to the SNP rs3740030 (Fig. 6). Primer combination III (exons 3–5) yielded amplicons of highly similar intensity (Fig. 6), confirming our quantitative transcript data that showed no allele-specific effect (Fig. 2). SNP nucleotide identity, T or G/T, was confirmed by sequencing. Amplification with primer combinations I yielded fragments displaying heterogeneities in size, sequence, and intensity. Surprisingly, in heterozygous samples, these were greatly reduced, and only two samples showed weakly staining single fragments (Fig. 6B). DNA sequence analysis of sample 5 (primer combination II) revealed the expected presence of exons 2, 3, and 4 but unexpectedly only the T nucleotide of the SNP. Sample 8 (primer combination I) contained a short fragment intron 1 sequence in addition to exons 1, 2, and 3. At the SNP position, only the G-nucleotide was found. The weak staining of this fragment suggested reduced levels of this transcript, which may be the reason for chance amplification. Sequence heterogeneity in the homozygous samples resulted from some fragments containing exons 1, 2, and 3, some had an additional short segment of intron 1, and yet others lacked exon 2. Primer combination II amplified fragments containing sequences from exons 2, 3, and 4 (DNA sequence data not shown). These striking differences between the homozygous and heterozygous samples within the 5′UTR region indicated the possibility of an allele-specific splicing mechanism that led to severe reductions in transcripts containing exons 1 and 2 in heterozygous tissues.
Figure 6.
SLC16A12 transcript analysis by RT-PCR. (A) Schematic representation of exons 1 through 5 (size not to scale). Position of SNP rs3740030 and the ATG initiation codon as well as the 5′UTR and coding regions are indicated above the exons. Primer combinations (I, II, III) and their respective sizes (bp) are shown. (B) Electrophoretically separated RT-PCR products from homozygous T/T (1–4) and heterozygous G/T (5–8) donors of vascular smooth muscle cells.
Affect of Allele Specificity on the Risk for Cataract
Assessment of the genotype for SNP rs3740030 in 484 cataract patients (350 with ARC, 134 with childhood cataract), most of whom were of Caucasian background, and in 190 ethnically matched control subjects confirmed an overrepresentation of the T-allele (Table 2), as had been reported for Caucasian populations of European descent (HapMap: T-allele frequency of 91.6%). Furthermore, we noticed overrepresentation of the G-allele in patient groups compared with control subjects (juvenile cataract, 6.72%; ARC, 8.71%, control subjects, 4.21%) that was statistically significant only for the ARC patient group (P = 0.00601768; χ2 = 7.545). To assess the extent to which the G-allele could increase the risk for cataract, odds ratios were calculated. Persons with either the G-allele only or the G/T or G/G genotype are at an approximately 2.2-fold increased risk for ARC (Table 3). Conversely, the T-allele or the T/T genotype confers protection against ARC. Given that the control population originated from the general Swiss population, we cannot exclude the possibility that an individual of this control population may develop ARC. In such case, we would predict that the odds ratio shifts even further toward a risk association with the G-allele. Odds ratios for juvenile cataract were statistically not significant (Table 3).
Table 2.
Genotype and Allele Frequencies of SNP rs3740030
| Genotypes |
Alleles |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| TT | TG | GG | Total | T | G | Total | T (%) | G (%) | |
| Control | 175 | 14 | 1 | 190 | 364 | 16 | 380 | 95.79 | 4.21 |
| Juvenile | 116 | 18 | 0 | 134 | 250 | 18 | 268 | 93.28 | 6.72 |
| Age-related | 291 | 57 | 2 | 350 | 639 | 61 | 700 | 91.29 | 8.71 |
Number of subjects identified with respective genotypes and alleles. Allele frequencies are given in percentages.
Table 3.
Association of SNP rs3740030 with Cataract
| Allele |
Genotype |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Odds Ratio | CI | P | χ2 | Odds Ratio | CI | P | χ2 | |||
| Juvenile | 1.638 | 0.8196 | 3.2736 | 0.158341 | 1.99 | 1.8103 | 0.8774 | 3.7352 | 0.104862 | 2.63 |
| Age-related | 2.1718 | 1.2339 | 3.8225 | 0.006001 | 7.55 | 2.3654 | 1.3021 | 4.297 | 0.003836 | 8.36 |
Odds ratio, confidence interval (CI), P, and χ2 are shown. Statistical significance was reached if odds ratio and CI were >1, P < 0.05, and χ2 > 3.84. Analyzed were 190 control subjects, 134 patients with juvenile cataract, and 350 patients with age-related cataract.
Discussion
The complex disease ARC is influenced by a multitude of environmental and genetic factors. Knowledge of the identity of the genetic contributions is limited and focuses on the identification of rare variants or the association of sequence variants in affected families and patient populations11–14,16,18 and on whole genome scans.29 Previously, we showed that a mutation in the monocarboxylate transporter SLC16A12 causes juvenile cataract, likely through a disturbance of solute homeostasis.6 Given that maintenance of homeostasis within the aging lens is likely to be essential,8 this transporter is a prime candidate for ARC. The data presented here support this hypothesis. First, a point mutation and an SNP within the 5′UTR was found in a 79-year-old woman with ARC. Second, in vitro and ex vivo experiments provide evidence that these sequences affect expression of SLC16A12 by modulating translational efficiency. Third, the SNP within the 5′UTR is likely associated with ARC within a Swiss population.
The c.-17A>G alteration in the patient with ARC was unique among our patient cohort (n = 484) and was also absent in our control population (n = 180), suggesting that it is a mutation rather than an SNP. Supportively, no sequence variation at position c.-17 of SLC16A12 has been reported in available DNA sequence databases to date. The second alteration found in cis in the age-related patient is the only annotated exonic SNP rs3740030 (c.-42T/G). Its allele frequency is population dependent. In contrast to a Caucasian population of European descent in which the minor allele is relatively seldom (8.4%), its frequency is higher in a Han Chinese population (27.4%) (HapMap, January 2010). It would be intriguing to investigate whether this is related to the incidence rate of cataract in the Chinese population.30 Based on our data in a Swiss population, the minor G-allele was more frequent in ARC patients than in the control group. This increased frequency translates into an approximately 2.2-fold increased risk and renders SNP rs3740030 a potential modifier in the development of ARC. Numerous examples from genome-wide association studies demonstrate a population-specific effect. ARC is no exception. For example, variants of GST have been investigated in various ethnically different cataract patient populations with varying results. Although no association was found in an Italian population,17 the opposite was true for an Estonian11 and a Turkish10 population. The SNP in SLC16A12 studied here may underlie similar conditions because the frequency in the Swiss control population was lower than that reported by HapMap. In addition, in preliminary studies, we were unable to verify the association in a British control population.
Based on the location of the opacity within the lens, one distinguishes between nuclear (the most frequent form), cortical, posterior subcapsular, and mixed-type cataract.31 Some risk factors have been reported as associated specifically with one of the subtypes of ARC,10,11,13 whereas association of the N-acetyltransferase 2 isoform “slow” acetylator12 and of kinesin light-chain KCL1 sequence variants32 have not been. We did not differentiate between subtypes; therefore, the association of SNP rs3740030 in SLC16A12 could account for all ARC in our study, but we cannot exclude that the increased risk would also be subtype specific.
The underlying molecular basis for ARC can be explained by altered expression levels of SLC16A12. Luciferase activity increases from the common T-allele to the minor G-allele to the patient haplotype (G-allele and mutation), thus offering an explanation for predisposition to and manifestation of the disease. Most likely the pathogenic mechanism acts posttranscriptionally because equal amounts of the SNP allele-specific SLC16A12 transcripts were found and 5′UTR regions are known to harbor control elements that regulate translation. This regulation may occur by way of RNA folding, upstream open-reading frames (uORFs), differential splicing, or specific sequence elements. We found no evidence for the latter. In contrast, the RNA-folding models demonstrated visible allele-specific differences, with a most drastic change for the patient allele. These structural changes correlate with changes in luciferase reporter activity. In all likelihood, no single mechanism is responsible for the observed effects. In addition, or complementary, to RNA folding, uORFs may be involved. Alterations in uORFs of other genes (TPO, HTR3A, BRAC1, TGFß3) were reported to cause various diseases by enhancing gene expression through elevation of the level of translation.22,33 The SLC16A12 5′UTR contains four uORFs. One of them, which is not in frame with SLC16A12, harbors both sequence alterations discussed here. Given that this uORF expands into the coding frame, it may have an inhibitory effect on guiding ribosomes beyond the initiation of the ATG codon.34–43 The additional feature that the SNP G-allele changes the coding frame of that uORF slightly (p.L21W) could also influence translational efficiency.
Differential splicing of the 5′UTR may also explain the allele-specific effects. Examples have been reported for the TFII-I transcription factor44 and for the estrogen receptor β gene.45 For the SNP rs3740030, the action of splice factor SRp40, which is predicted to bind exclusively to transcripts generated from the minor G-allele, may cause the observed allele-specific 5′UTR sequence heterogeneity. Furthermore, the skipping of exon 2 causes shortening of the 5′UTR and elimination of another uORF, thereby increasing the likelihood of influencing the translational regulation. Taken together, at least three different mechanisms may apply, singly or in concert, to explain the altered translational efficiency associated with the SNP and the mutation within the 5′UTR.
Knowledge of the exact function of the monocarboxylate transporter SLC16A12 requires identification of its substrates, which is currently under investigation. Nevertheless, the SLC16A12 nonsense mutation leads to juvenile cataract6 and nucleotide changes in the 5′UTR associate with ARC, demonstrating the importance of this transporter. The adult lens structure demands a well-functioning transport system for metabolites to supply nutrients and to remove waste products,8 and the solute carrier SLC16A12 may have an important function in that activity. Complete removal of the transporter leads to congenital cataract and renal glucosuria.6 In contrast, alterations in the amount of the transporter, as described here, may lead to imbalanced solute homeostasis, which over time leads to ARC. Given SLC16A12 expression in tissues other than the lens, it seems feasible that other organs could experience such consequences. Renal glucosuria is a prime candidate of such a condition but other, as yet not described, physiological conditions may become manifest.
We have provided initial examples that SLC16A12 5′UTR alterations are involved in the manifestation of and the predisposition to ARC by modifying translational efficiency. Physiological consequences are likely to challenge tissue homeostasis and to transform a healthy lens into a diseased lens.
Acknowledgments
The authors thank Helen Greutert for help with cell cultures, Walther Hänseler for help with cloning, and Gabor Matyas for sharing his knowledge of DNA sequencing and for maintaining the sequencing facility.
Footnotes
Supported by the National Institute for Health Research Biomedical Research Centre for Ophthalmology and The Special Trustees of Moorfields Eye Hospital London (SB); Wellcome Trust and National Institute for Health Research (UK); Moorfields Eye Hospital BMRC (ATM); and Swiss National Science Foundation Grant 320030_127558 (FLM, DFS).
Disclosure: J. Zuercher, None; J. Neidhardt, None; I. Magyar, None; S. Labs, None; A.T. Moore, None; F.C. Tanner, None; N. Waseem, None; D.F. Schorderet, None; F.L. Munier, None; S. Bhattacharya, None; W. Berger, None; B. Kloeckener-Gruissem, None
References
- 1.Brian G, Taylor H. Cataract blindness—challenges for the 21st century. Bull World Health Organ 2001;79:249–256 [PMC free article] [PubMed] [Google Scholar]
- 2.Congdon NG, Friedman DS, Lietman T. Important causes of visual impairment in the world today. JAMA 2003;290:2057–2060 [DOI] [PubMed] [Google Scholar]
- 3.Shiels A, Hejtmancik JF. Genetic origins of cataract. Arch Ophthalmol 2007;125:165–173 [DOI] [PubMed] [Google Scholar]
- 4.Francis PJ, Berry V, Bhattacharya SS, Moore AT. The genetics of childhood cataract. J Med Genet 2000;37:481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiels A, Bennett TM, Knopf HL, et al. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am J Hum Genet 2007;81:596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kloeckener-Gruissem B, Vandekerckhove K, Nurnberg G, et al. Mutation of solute carrier SLC16A12 associates with a syndrome combining juvenile cataract with microcornea and renal glucosuria. Am J Hum Genet 2008;82:772–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamieson RV, Farrar N, Stewart K, et al. Characterization of a familial t(16;22) balanced translocation associated with congenital cataract leads to identification of a novel gene, TMEM114, expressed in the lens and disrupted by the translocation. Hum Mutat 2007;28:968–977 [DOI] [PubMed] [Google Scholar]
- 8.Truscott RJ. Age-related nuclear cataract: a lens transport problem. Ophthalmic Res 2000;32:185–194 [DOI] [PubMed] [Google Scholar]
- 9.Hodge WG, Whitcher JP, Satariano W. Risk factors for age-related cataracts. Epidemiol Rev 1995;17:336–346 [DOI] [PubMed] [Google Scholar]
- 10.Guven M, Unal M, Sarici A, Ozaydin A, Batar B, Devranoglu K. Glutathione-S-transferase M1 and T1 genetic polymorphisms and the risk of cataract development: a study in the Turkish population. Curr Eye Res 2007;32:447–454 [DOI] [PubMed] [Google Scholar]
- 11.Juronen E, Tasa G, Veromann S, et al. Polymorphic glutathione S-transferases as genetic risk factors for senile cortical cataract in Estonians. Invest Ophthalmol Vis Sci 2000;41:2262–2267 [PubMed] [Google Scholar]
- 12.Tamer L, Yilmaz A, Yildirim H, et al. N-acetyltransferase 2 phenotype may be associated with susceptibility to age-related cataract. Curr Eye Res 2005;30:835–839 [DOI] [PubMed] [Google Scholar]
- 13.Shiels A, Bennett TM, Knopf HL, et al. The EPHA2 gene is associated with cataracts linked to chromosome 1p. Mol Vis 2008;14:2042–2055 [PMC free article] [PubMed] [Google Scholar]
- 14.Bhagyalaxmi SG, Srinivas P, Barton KA, et al. A novel mutation (F71L) in αA-crystallin with defective chaperone-like function associated with age-related cataract. Biochim Biophys Acta 2009;1792:974–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maraini G, Hejtmancik JF, Shiels A, et al. Galactokinase gene mutations and age-related cataract: lack of association in an Italian population. Mol Vis 2003;9:397–400 [PubMed] [Google Scholar]
- 16.Okano Y, Asada M, Fujimoto A, et al. A genetic factor for age-related cataract: identification and characterization of a novel galactokinase variant, ‘Osaka,’ in Asians. Am J Hum Genet 2001;68:1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alberti G, Oguni M, Podgor M, et al. Glutathione S-transferase M1 genotype and age-related cataracts: lack of association in an Italian population. Invest Ophthalmol Vis Sci 1996;37:1167–1173 [PubMed] [Google Scholar]
- 18.Shi Y, Shi X, Jin Y, et al. Mutation screening of HSF4 in 150 age-related cataract patients. Mol Vis 2008;14:1850–1855 [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen L, Mikkelsen A, Nurnberg P, et al. Comprehensive mutational screening in a cohort of Danish families with hereditary congenital cataract. Invest Ophthalmol Vis Sci 2009;50:3291–3303 [DOI] [PubMed] [Google Scholar]
- 20.Reynolds PR. In sickness and in health: the importance of translational regulation. Arch Dis Child 2002;86:322–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet 2007;8:711–723 [DOI] [PubMed] [Google Scholar]
- 22.Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell 2009;101:251–262 [DOI] [PubMed] [Google Scholar]
- 23.Cazzola M, Skoda RC. Translational pathophysiology: a novel molecular mechanism of human disease. Blood 2000;95:3280–3288 [PubMed] [Google Scholar]
- 24.Magyar I, Colman D, Arnold E, et al. Quantitative sequence analysis of FBN1 premature termination codons provides evidence for incomplete NMD in leukocytes. Hum Mutat 2009;30:1355–1364 [DOI] [PubMed] [Google Scholar]
- 25.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a Web resource to identify exonic splicing enhancers. Nucleic Acids Res 2003;31:3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 2006;15:2490–2508 [DOI] [PubMed] [Google Scholar]
- 27.Zuker M. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003;31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss S, Frischknecht K, Greutert H, et al. Different migration of vascular smooth muscle cells from human coronary artery bypass vessels: role of Rho/ROCK pathway. J Vasc Res 2007;44:149–156 [DOI] [PubMed] [Google Scholar]
- 29.Iyengar SK, Klein BE, Klein R, et al. Identification of a major locus for age-related cortical cataract on chromosome 6p12–q12 in the Beaver Dam Eye Study. Proc Natl Acad Sci U S A 2004;101:14485–14490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu WM, Cheng CY, Liu JH, Tsai SY, Chou P. Prevalence and causes of visual impairment in an elderly Chinese population in Taiwan: the Shihpai Eye Study. Ophthalmology 2004;111:62–69 [DOI] [PubMed] [Google Scholar]
- 31.Klein BE, Klein R, Linton KL. Prevalence of age-related lens opacities in a population: the Beaver Dam Eye Study. Ophthalmology 1992;99:546–552 [DOI] [PubMed] [Google Scholar]
- 32.Andersson ME, Zetterberg M, Tasa G, et al. Variability in the kinesin light chain 1 gene may influence risk of age-related cataract. Mol Vis 2007;13:993–996 [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet 2006;22:119–122 [DOI] [PubMed] [Google Scholar]
- 34.Alderete JP, Jarrahian S, Geballe AP. Translational effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J Virol 1999;73:8330–8337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beffagna G, Occhi G, Nava A, et al. Regulatory mutations in transforming growth factor-β3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc Res 2005;65:366–373 [DOI] [PubMed] [Google Scholar]
- 36.Degnin CR, Schleiss MR, Cao J, Geballe AP. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J Virol 1993;67:5514–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill JR, Morris DR. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA: dependence on translation and coding capacity of the cis-acting upstream open reading frame. J Biol Chem 1993;268:726–731 [PubMed] [Google Scholar]
- 38.Mize GJ, Ruan H, Low JJ, Morris DR. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J Biol Chem 1998;273:32500–32505 [DOI] [PubMed] [Google Scholar]
- 39.Niesler B, Flohr T, Nothen MM, et al. Association between the 5′ UTR variant C178T of the serotonin receptor gene HTR3A and bipolar affective disorder. Pharmacogenetics 2001;11:471–475 [DOI] [PubMed] [Google Scholar]
- 40.Parola AL, Kobilka BK. The peptide product of a 5′ leader cistron in the β2 adrenergic receptor mRNA inhibits receptor synthesis. J Biol Chem 1994;269:4497–4505 [PubMed] [Google Scholar]
- 41.Wang Z, Fang P, Sachs MS. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol Cell Biol 1998;18:7528–7536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werner M, Feller A, Messenguy F, Pierard A. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell 1987;49:805–813 [DOI] [PubMed] [Google Scholar]
- 43.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 2000;20:8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makeyev AV, Bayarsaihan D. Alternative splicing and promoter use in TFII-I genes. Gene 2009;433:16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith L. Post-transcriptional regulation of gene expression by alternative 5′-untranslated regions in carcinogenesis. Biochem Soc Trans 2008;36:708–711 [DOI] [PubMed] [Google Scholar]