Characterization of HTRA1 physiological substrates in the context of RPE extracellular milieu reveals pathways linking HTRA1 to complement pathway and age-related macular degeneration.
Abstract
Purpose.
To define the role of the serine protease HTRA1 in age-related macular degeneration (AMD) by examining its expression level and identifying its potential substrates in the context of primary RPE cell extracellular milieu.
Methods.
Primary RPE cell cultures were established from human donor eyes and screened for CFH, ARMS2, and HTRA1 risk genotypes by using an allele-discrimination assay. HTRA1 expression in genotyped RPE cells was determined by using real-time PCR and quantitative proteomics. Potential HTRA1 substrates were identified by incubating RPE-conditioned medium with or without human recombinant HTRA1. Selectively cleaved proteins were quantified by using the differential stable isotope labeling by amino acids in cell culture (SILAC) strategy.
Results.
HTRA1 mRNA levels were threefold higher in primary RPE cells homozygous for the HTRA1 promoter risk allele than in RPE cells with the wild-type allele, which translated into a twofold increase in HTRA1 secretion by RPE cells with the risk genotype. A total of 196 extracellular proteins were identified in the RPE secretome, and only 8 were found to be selectively cleaved by the human recombinant HTRA1. These include fibromodulin with 90% cleavage, clusterin (50%), ADAM9 (54%), vitronectin (54%), and α2-macroglobulin (55%), as well as some cell surface proteins including talin-1 (21%), fascin (40%), and chloride intracellular channel protein 1 (51%).
Conclusions.
Recombinant HTRA1 cleaves RPE-secreted proteins involved in regulation of the complement pathway (clusterin, vitronectin, and fibromodulin) and of amyloid deposition (clusterin, α2-macroglobulin, and ADAM9). These findings suggest a link between HTRA1, complement regulation, and amyloid deposition in AMD pathogenesis.
Major advances in our understanding of age-related macular degeneration (AMD) genetics have occurred over the past few years.1–9 Three strong genetic loci predisposing to AMD have been identified to date, the complement factor H variant (rs1061170, CFH Y402H), the serine protease high temperature requirement A1 (HTRA1) promoter variant (rs11200638, −512 G>A), and the age-related maculopathy susceptibility 2 (ARMS2) variant (rs10490924, A69S).1–9 However, it is not clear how these functionally unrelated genes lead to the predominant pathologic features in AMD10 (e.g., drusen formation and/or choroidal neovascularization [CNV]). In addition, the HTRA1 and ARMS2 polymorphisms are in high linkage disequilibrium (LD), making it difficult to pinpoint the causal gene.5–8 Although the original studies showed that an HTRA1 promoter variant results in increased expression of HTRA1 protein,7,8 later studies have challenged these findings and suggested that an ARMS2 variant is the causal factor.9,11 In this study, we sought to examine the status of HTRA1 expression and define its potential substrates in the context of the extracellular milieu of the RPE cells, which approach may provide insights into the role of HTRA1 in AMD pathogenesis.
HTRA1 is a member of the high temperature requirement A family of serine proteases comprising three other members: HTRA2, HTRA3, and HTRA4. The exact role of these different types of HTRA is not well understood. However, all seem to exhibit a nonspecific protease activity. HTRA1 is a 50-kDa secreted protein composed of a signaling peptide, an insulin growth factor binding domain, a Kazal-like protease inhibitor domain, a conserved serine protease domain, and a PDZ domain (Fig. 1). It is expressed in a variety of tissues and organs, with the highest levels in placenta and mature epidermis.12 Altered expression of this protein has been associated with several pathologic conditions, including AMD.8,13 Although low levels of HTRA1 are reported in many cancer cell lines and tumors,14–17 high levels are reported in rheumatoid arthritis,18 osteoarthritis,18–20 Alzheimer's disease,21 and Duchenne muscular dystrophy.22 Moreover, a single nucleotide polymorphism (SNP) in the promoter region of HTRA1 was recently identified as a major genetic risk factor for AMD,7,8 with odds ratios similar to or even greater than those reported for CFH.8 In vitro studies have shown that HTRA1 is involved in the degradation of several proteins, including tubulins,23 insulin growth factor binding protein-5,24 amyloid precursor,21 fibronectin,18 and aggrecan.25 However, little is known about the expression of this protein and its substrate specificity in the context of RPE extracellular environment.
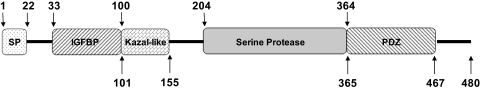
Figure 1.
Structure of the full-length human HTRA1 protein. Boxes represent protein domains and numbers indicate amino acid positions. SP, signal peptide domain (1-22) cleaved in mature secreted protein; IGFBP, insulin growth factor–binding protein domain (33-100); Kazal-like: Kazal-like domain, with a structure similar to that of Kazal-type serine protease inhibitors. This domain may be responsible for the inhibition of autocleavage of HTRA1 (101-155). Serine protease domain: contains HDS, a catalytic triad (204-364); PDZ, postsynaptic density protein (psd95), drosophila disc large tumor suppressor (dlga), and zonula occludens-1 protein (zo-1) domain (365-467) recognize a broad range of hydrophobic polypeptides in the HTRA1 substrate.
Materials and Methods
Establishment of Primary RPE Cell Cultures from Donor Eyes
Globes or poles of Caucasian donor eyes were received through the National Disease Research Interchange (NDRI, Philadelphia, PA). Donors included both males and females, with and without a clinical history of AMD. Eyes of donors with diabetes or other known ocular disease were excluded from the study. Eye globes were obtained on average 9 hours after death and delivered to our laboratory in less than 24 hours. Serology tests of each donor were performed by NDRI to exclude samples with potential infectious diseases. Primary RPE cell cultures were prepared according to Engelmann and Valtink26 with slight modifications. Briefly, the anterior segment from each donor eye was removed by cutting around the iris. The eye cup was opened flat by five incisions along the sclera. A 1-cm2 portion of the central retinal region was excised for the experiment. The neurosensory retina was removed from the excised piece with forceps, and the RPE/choroid layer was separated from the sclera and washed gently with PBS containing penicillin (100 U/mL) and streptomycin (100 μg/mL). The RPE/choroid was then incubated in 0.5 mg/mL of collagenase mixture IA+IV (1:1) for 5 hours. The suspension containing RPE cells was carefully transferred to a sterile centrifuge tube and pelleted by centrifugation at 100g for 5 minutes. The cell pellet was resuspended in DMEM/F12 supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin and was seeded on a 0.1% gelatin-coated plate. Growing cultures were tested for their epithelial cell content by using both immunochemical staining and flow cytometry (FACS; BD Biosciences, Franklin Lakes, NJ) analysis for cytokeratin 18.27–29 All these cultures were frozen at the second passage, and cultures with more than 80% epithelial content were used in the study (Supplementary Fig. S1; all supplementary material is available at http://www.iovs.org/cgi/content/full/51/7/3379/DC1).
Screening of RPE Cell Cultures for the AMD Risk Genotypes HTRA1, ARMS2, and CFH
DNA was extracted from each of the RPE cultures and screened for HTRA1, ARMS2, and CFH gene variants by using an allele-discrimination assay (TaqMan; Applied Biosystems, Inc., Foster City, CA).28 An allele-specific PCR reaction for each SNP was performed with 30 ng of genomic DNA, 900 nM of forward and reverse PCR primers, and 200 nM of fluorescent and allele-discrimination probes (VIC and FAM; BD Biosciences) in a final volume of 8 μL. The PCR reaction was performed at 95°C for 11 minutes and 44 cycles of 15 seconds at 92°C and 1 minute at an annealing temperature of 60°C. Reactions were performed with a robot (MWG Biotech, Ebersberg, Germany), and fluorescence ratios and allele calls were measured with a real-time PCR system (model 7900; Applied Biosystems, Inc. [ABI] Foster City, CA). For the HTRA1 variant (rs11200638; −512 G>A), the following forward and reverse primers (F: 5′-CTGCCAGCTCCGCGGACGCT-3′, R: 5′ CCCCGCGGTCAGGGTCCCG-3′) and the following fluorescent allele discrimination probes (WT-VIC-5′-TCGTCCGGCCGCA-3′; MT-FAM-5′-TCGTCCAGCCGCA-3′) were used. The following forward and reverse PCR primers (F: 5′-CTTATTTATTTATCATTGTTATGGTCCTTAGGA-AAATGTTATTT-3′ and R: 5′-GGCAGGCAACGTCTATAGATTTACC-3′) and the following fluorescent allele-discrimination probes (WT: VIC-5′-TTTCTTCCATAATTTTG-3′; MT-FAM-5′- TTTCTTCCATGATTTTG-3′) were used to screen for the CFH variant (rs1061170; 1227 T>C). For the ARSM2 variant (rs10490924; 270 G>T), predesigned primers and probes (Assay ID C_29934973_20; ABI) were used.
Measurements of HTRA1 Expression in RPE Cells
Quantitative PCR.
HTRA1 expression at the mRNA level was quantified by extracting total RNA from RPE cultures with wild-type genotype GG (n = 11), heterozygous risk allele GA (n = 7), and homozygous risk allele AA (n = 3) for the HTRA1 promoter region (TRIzol; Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. Each extracted RNA sample was treated with DNase-I and cDNA was synthesized (SuperScript III first-Strand Synthesis kit; Invitrogen) with oligo dT primer. Real-time PCR was performed (7900; ABI) with 20× HTRA1 primers and probe (Assay ID Hs01016151_m1; ABI) and 20× glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers and probe for the control. Relative quantification was performed using the comparative threshold (Ct) method after determining the Ct values for reference (GAPDH) and target (HTRA1) genes in each sample sets according to the 2−ΔΔCt method,30 as described by the manufacturer.
Western Blot Analysis.
HTRA1 expression at the protein level was quantified by culturing RPE cells with heterozygous or homozygous risk alleles, as well as RPE cells with the wild-type genotype, for the HTRA1 promoter region, in 10-cm dishes. After confluence, the cultures were washed six times with sterile PBS and incubated with serum-free medium for another 24 hours. The conditioned medium was collected, filtered through 0.2 μm polyethersulfone syringe filter (Corning, Inc, Corning, NY), and assayed for protein concentration by using the Bradford assay (Pierce, Rockford, IL). Equal amounts of proteins (10 μg) from each sample were concentrated by vacuum centrifugation and resuspended in 14 μL of 50% Laemmli buffer (Bio-Rad, Hercules, CA). Proteins were separated by SDS-PAGE under reducing conditions and transferred to nitrocellulose membrane (GE Healthcare, Piscataway, NJ) for Western blot analysis. Nonspecific binding sites were blocked with 5% milk (Bio-Rad) and membrane was incubated overnight with rabbit polyclonal antibody against human HTRA1 (1:1K, Abcam, Cambridge, MA) in 2.5% milk. The next day, the membrane was rinsed with Tris buffer containing salt and Tween and incubated with goat anti-rabbit IgG (1:3K, Bio-Rad) conjugated with horseradish peroxidase. Immunoblots were visualized by chemiluminescence (GE Healthcare), and Ponceau red was used to control for protein loading.
Targeted Mass Spectrometry Quantification of RPE-Secreted HTRA1.
First, the concentration of stable-isotope–labeled HTRA1 protein was determined by mass spectrometry after spiking conditioned medium of SILAC (stable isotope labeling with amino acids in cell culture)-labeled RPE cells grown in the presence of 13C6-Arg, 13C6,15N2-Lys with known amounts of human recombinant HTRA1. The stock of stable-isotope–labeled HTAR1 was then used as a standard to spike the conditioned medium collected from RPE cultures with wild-type, heterozygous, or homozygous genotype for the HTRA1 promoter risk allele. Spiked samples were resolved on SDS-PAGE, and the bands in the 50-kDa region were excised and in-gel digested with trypsin. The resulting peptides were extracted and analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS).
Proteomics Analysis of HTRA1 Substrates
To define HTRA1 substrates, we collected conditioned medium from RPE culture with the homozygous wild-type genotype for both the CFH and the HTRA1 promoters and incubated it in the presence or absence of human recombinant HTRA1 (Invitek, Berlin, Germany). Differential SILAC strategy was used to unambiguously detect potential HTRA1 substrates. One set of cells was grown in 13C6-Arg, 13C6,15N2-Lys–containing medium, while the other set was grown in regular medium containing 12C6-Arg, 12C6, 14N2-Lys.28 Aliquots of unlabeled conditioned medium containing 50 μg total proteins were concentrated four times to reduce the volume by vacuum centrifugation and then incubated with recombinant HTRA1 at 37°C for 4 hours (enzyme to total proteins ratio, 1:100, wt/wt) and for 20 hours (enzyme to total proteins ratio, 1:50, wt/wt). The amount of HTRA1 was doubled for the 20-hour incubation to account for enzyme self degradation. Similar aliquots of SILAC-labeled medium were incubated under the same conditions but in the absence of HTRA1. To validate the data, a reverse experiment was performed where SILAC-labeled samples were treated with HTRA1, whereas unlabeled samples were kept untreated.31 Treated and untreated sample pairs were then mixed at 1:1, further concentrated by using a 3-kDa cutoff filter (Millipore Corp., Billerica, MA), and processed for SDS-PAGE and mass spectrometry analysis. After the gel was stained (Bio-Safe Coomassie; Bio-Rad), each lane was sliced into 65 bands, and each band was in-gel digested with trypsin and the resulting peptides analyzed by LC-MS/MS. To validate the proteomic result, human recombinant clusterin (R&D Systems, Minneapolis, MN) and human purified CFH, C3b, and C3 (Complement Technology, Tyler, TX) were incubated with HTRA1 for 6 hours (enzyme to recombinant protein ratio, 1:25).
Liquid Chromatography-Tandem Mass Spectrometry
LC-MS/MS was performed on a nano-HPLC system (NanoLC 2D; Eksignet, Dublin, CA) connected to a hybrid mass spectrometer (LTQ/Orbitrap; Thermo Fisher Scientific, Inc., San Jose, CA). Eluting peptides were introduced into the mass spectrometer via a 10-μm silica tip (New Objective Inc., Ringoes, NJ) adapted to a nano-electrospray source (Thermo Fisher Scientific, Inc.). The spray voltage was set at 1.3 kV and the heated capillary at 180°C. The mass spectrometer was operated in data-dependent mode in which one cycle of experiments consisted of one full-MS survey followed by three sequential pairs of intercalated MS/MS experiments. The targeted ion counts in the ion trap during full-MS and MS/MS were 30,000 and 10,000, respectively. MS measurements were performed with the orbitrap spectrometer at 60,000 resolution with accuracy better than 5 ppm. Peptides were fragmented in the linear ion trap by using collision-induced dissociation with the collision gas (helium) pressure set at 1.3 mTorr and the normalized collision energy set at 35%.
Database Search and Quantification
Protein identification was performed against the human SwissProt database (UniProtKB/Swiss-Prot release 15.4 of June 2009; http://www.ebi.ac.uk/swissprot/ provided in the public domain by the European Bioinformatics Institute, Heidelberg, Germany) using a software package (Bioworks 3.3 equipped with the Sequest search engine; Thermo Fisher Scientific, Inc., San Jose, CA). The database was indexed with assumptions for fully enzymatic tryptic cleavage with two missed cleavages and the following possible protein modifications: 16-Da shift for oxidized Met and 6- and 8-Da shifts for stable isotope labeled Arg and Lys, respectively. The search result was filtered with DTASelect (http://fields.scripps.edu/DTASelect/ provided in the public domain by The Scripps Research Institute, La Jolla, CA) according to the following acceptance criteria: DeltaCn (ΔCn) > 0.08, a variable threshold of Xcorr versus charge state (Xcorr ≥ 1.8 for z = 1, Xcorr ≥ 2.5 for z = 2, Xcorr ≥ 3.5 for z = 3, and Xcorr ≥ 4.5 for z = 4). MS spectra for each identified peptide were extracted from raw data by using Raw Xtractor. Census, a proteomics quantification software, was used to generate an extracted ion chromatogram (XIC) for each peptide and to measure protein ratios in SILAC-labeled versus unlabeled samples. Both of these tools are publicly available at the Scripps Research Institute (http://fields.scripps.edu/?q=content/download). Data were checked for validity by spectral visualization.32
Statistical Analysis
Two comparisons (GG versus AG group, and GG versus AA group) were made for both mRNA and protein expression with independent t-tests. The resulting P-values were then adjusted for multiple comparisons using the Sidak33 method.
Results
Human Primary RPE Cell Cultures and Genotyping
We collected eyes of 34 donors and established 30 primary RPE cultures. The donors' mean age was 65 years, and average time from death to enucleation was 9 hours. Cells were subcultured to validate their epithelial origin and to screen for AMD-associated SNPs (CFH, HTRA1, and ARMS2), as described in the Methods section. Detailed information about the sex, age, clinical history, and genotype of each donor are reported in Supplementary Table S1. The overall genotype distribution of the 30 established RPE cultures is shown in Table 1. Only eight of the donors were found to harbor the wild-type genotype for CFH, HTRA1, and ARMS2. The remaining 22 donors were heterozygous and/or homozygous for at least one of the three candidate genes. Again HTRA1 and ARMS2 variants were found to be in complete LD and in agreement with previous studies.34 Seven of the donors had a clinical history of AMD, as indicated in parentheses under each group in Table 1. Six of the seven clinically documented AMD donors harbored at least one of the at-risk gene variants. The single donor carrying a diagnosis of AMD but without any currently identified risk allele was reported to have smoked two packs of cigarettes per day for 40 years. The degree of the disease in each AMD donor was not evaluated in this study, because most of the macula was used to establish a sufficient amount of primary RPE cultures.
Table 1.
Distribution of AMD Risk Genotypes among the Established Human Primary RPE Cell Cultures
n = 30. Numbers in parentheses indicate the number of donors with clinically documented AMD within each group. For example, among the eight donors with the homozygous wild-type genotype for both CFH and HTAR1/ARMS2 one had clinically documented AMD.
Wild-type genotype.
Heterozygote risk genotype.
Homozygote risk genotype.
Quantitative Analyses of HTRA1 Expression in Genotyped RPE Cells
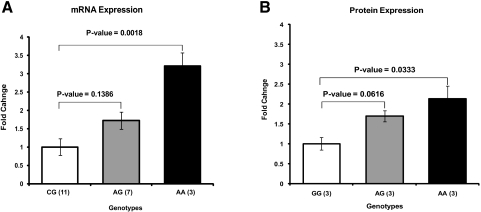
To examine the effect of the HTRA1 promoter polymorphism on the expression levels of HTRA1, we selected 21 actively growing RPE cultures including 11 with the wild-type genotype GG, 7 with the heterozygous genotype GA, and 3 with the homozygous genotype AA for the risk allele in the promoter region of the HTRA1 gene. HTRA1 expression was examined in these cells both at the mRNA and protein levels. The HTRA1 mRNA levels were 3- and 1.5-fold higher in primary RPE cultures with homozygous and heterozygous risk alleles for the HTRA1 promoter, respectively, than in primary RPE cultures with the wild-type homozygous genotype (Fig. 2A). This finding was confirmed by Western blot analysis of HTRA1 protein in the conditioned media collected from RPE cultures with the different HTRA1 promoter genotypes (Fig. 2B).
Figure 2.
Expression levels of HTRA1 mRNA and protein in human RPE cell cultures derived from donors with wild-type and at-risk genotypes for the HTRA1 promoter region. (A) qRT PCR of HTRA1 mRNA levels in primary RPE cultures. Samples were prepared from RPE cultures with wild-type (n = 11), heterozygous (n = 7), and homozygous (n = 3) genotypes for the HTRA1 promoter risk allele (G is the wild-type allele and A is the risk allele). Total RNA was isolated from cultured RPE cells. cDNA was prepared by reverse transcription. qRT-PCR was performed with forward and reverse primers that recognize exons 6 and 7 of the HTRA1 gene. HTRA1 mRNA transcript levels were measured. All values were normalized to wild-type GG genotype. (B) Western blot analysis of HTRA1 protein levels in the conditioned media of RPE cultures. RPE cells with wild-type genotype GG (n = 3), at-risk heterozygous genotype AG (n = 3), and at-risk homozygous genotype AA (n = 3) were grown in serum-containing medium. After the cells reached confluence, the medium was replaced with serum-free medium for 24 hours. Media were collected, and an equal amount of proteins (10 μg) from each sample was concentrated and resolved by SDS-PAGE for Western blot analysis. HTRA1 band intensities were normalized to total loaded proteins. Error bars, SE.
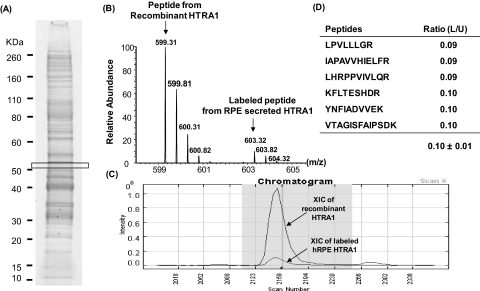
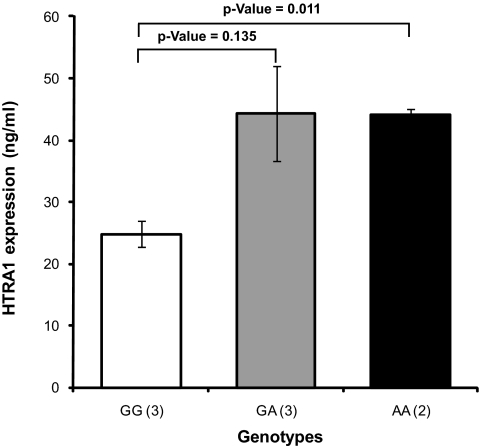
To further validate our Western blot analysis results, we performed absolute quantitative analysis of the HTRA1 protein with stable-isotope–labeled HTRA1 used as a standard for mass spectrometry. We first determined the concentration of HTRA1 secreted by SILAC-labeled RPE cells grown in 13C6-Arg, 13C6, 15N2-Lys by spiking their conditioned medium with 200 ng/mL of recombinant unlabeled HTRA1 (Fig. 3). The average intensity ratio of unlabeled and labeled peptide pairs from the recombinant HTRA1 and the RPE-secreted HTRA1, respectively, was found to be equal to 0.1 ± 0.01 and was used to calculate the amount of RPE-secreted HTRA1. This RPE cell culture secreted approximately 20 ± 1.49 ng/mL of HTRA1 (Fig. 3D). Having validated the approach, a known amount of the established stable-isotope–labeled HTRA1 standard was spiked into the conditioned media collected from unlabeled primary RPE cultures with the different genotypes of the HTRA1 promoter risk allele. HTRA1 levels were elevated approximately twofold in the RPE cells with the homozygous genotype for the HTRA1 promoter risk allele (44 ± 1.13 ng/mL) and in the RPE cells with the heterozygous genotype for the HTRA1 prompter risk allele (44 ± 13.34 ng/mL) compared with that in the RPE cells with the wild-type genotype for the HTAR1 promoter (24 ± 3.55 ng/mL; Fig. 4).
Figure 3.
Stable-isotope–labeled standard for absolute quantification of HTRA1 protein. Conditioned medium collected from a fully labeled RPE culture grown in medium containing 13C6-Arg, 13C6,15N2-Lys was spiked with human recombinant unlabeled HTRA1 at a final concentration of 200 ng/mL. Samples were run on SDS-PAGE, bands were sliced and digested by trypsin, and the resulting peptides were analyzed by LC-MS/SM. (A) SDS-PAGE of total secreted proteins collected from a 24-hour RPE culture grown in SILAC-labeled, serum-free medium. As expected, HTRA1 protein was identified with six different peptides ∼50 kDa located in the boxed area. (B) Example of a mass spectrum for the YNFIADVVEK peptide of HTRA1. This peptide was detected as a pair of unlabeled (599.31 m/z) and labeled peptide (603.31 m/z), both as double-charged ions. Both these peptides were sequenced, and the difference in mass of 8 Da (4 Da for the double-charged ions) agrees with a Lys residue at the C-terminal. (C) The intensity ratio of heavy to light peak was determined from an XIC for unlabeled and labeled peptide pairs using Census software and reflects the ratio of endogenous RPE HTRA1 to the recombinant HTRA1. (D) A total of six peptide pairs were detected for HTRA1, and the ratios between unlabeled and labeled peptide pairs was measured. Since the amount of recombinant HTRA1 added to the conditioned medium is known, it allowed us to calculate that 10-cm culture dish containing 5 × 106 RPE cells secrete approximately 20 ± 1.49 ng/mL of HTRA1.
Figure 4.
Effect of the HTRA1 promoter genotype on HTRA1 expression in RPE cultures. Stable-isotope–labeled HTRA1 (20 ng/mL) was used as a standard to spike conditioned media collected from fully confluent RPE cultures with the homozygous wild-type genotype GG (n = 3), the heterozygous at-risk genotype GA (n = 3), and homozygous at-risk genotype AA (n = 2) for the HTRA1 promoter. Quantities of secreted HTRA1 were calculated from intensity ratios of light and heavy peptide pairs detected from the unlabeled RPE-secreted HTRA1 and the stable-isotope–labeled HTRA1 standard. The number of unique peptides used to determine HTRA1 in each genotype group was as follows: eight peptides for the GG variant group, four for the GA variant, and four for the AA variant. Error bars, SE between replicate samples analyzed for each genotype.
Screening for HTRA1 Substrate in RPE Cell-Conditioned Medium
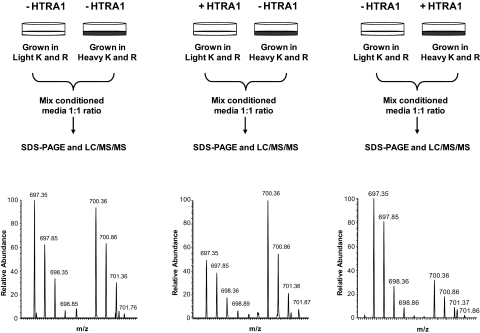
Differential SILAC strategy35 in combination with SDS-PAGE and LC-MS/MS allowed both detection and quantification of proteins that were selectively cleaved by the recombinant HTRA1. A total of 196 extracellular proteins were identified and quantified in the primary RPE-conditioned medium (Supplementary Table S2). All 196 proteins had a ratio close to 1 in the control sample pairs with an average mean of 0.98 and SD of ±0.15. Ratios outside 3 SD of this average value (0.54–1.40) in the HTRA1-treated and untreated sample pairs were considered significant. According to this criterion, only 8 of the 196 proteins were found to be selectively cleaved by HTRA1, and they are reported in Table 2 with accession numbers and ratios in HTRA1-treated versus untreated samples. An example of such HTRA1 substrate is illustrated for clusterin protein in Figure 5. The clusterin ratio between SILAC-labeled and unlabeled sample remained at 1:1 when both samples were untreated, but decreased to 0.5 in HTRA1-treated versus untreated samples, demonstrating that HTRA1 degraded clusterin under the experimental conditions used. Specific degradation of clusterin was further confirmed by the detection of clusterin peptides in lower bands (∼ 35 Da) in HTRA1-treated samples versus untreated samples (Supplementary Fig. S2).
Table 2.
List of Potential HTRA1 Substrates Identified in the RPE Conditioned Medium
| Peptide (n) | Control* (−)HTRA1/(−)HTRA1 | 4 h (+)HTRA1/(−)HTRA1 | 20 h (+)HTRA1/(−)HTRA1 | ||
|---|---|---|---|---|---|
| Fibromodulin | Q06828 | 5 | 1.13 ± 0.15 | 0.32 ± 0.01 | 0.13 ± 0.00 |
| Talin-1 | Q9Y490 | 15 | 0.95 ± 0.13 | 0.74 ± 0.14 | 0.21 ± 0.04 |
| Fascin | Q16658 | 8 | 0.98 ± 0.12 | 0.97 ± 0.15 | 0.40 ± 0.04 |
| α2-Macroglobulin | P01023 | 5 | 0.90 ± 0.02 | 0.16 ± 0.00 | 0.45 ± 0.00 |
| Vitronectin | P04004 | 4 | 1.06 ± 0.42 | 1.07 ± 0.10 | 0.46 ± 0.05 |
| ADAM9 | Q13443 | 2 | 0.97 ± 0.07 | 1.01 ± 0.10 | 0.46 ± 0.11 |
| Clusterin | P10909 | 15 | 1.00 ± 0.08 | 0.81 ± 0.00 | 0.48 ± 0.09 |
| CLIC1 | O00299 | 3 | 1.03 ± 0.06 | 0.73 ± 0.00 | 0.51 ± 0.07 |
The coefficient of variation was determined from the control data and was found to be <14% for the 196 protein measured. Thus, experimental values that are 3 SD (±42%) of the control mean value were considered significant. Errors are associated with forward and reverse SILAC experiments and the number of peptides detected for each protein. Peptide (n), number of peptides used to measure ratios in control and experimental samples; ADAM 9, disintegrin and metalloproteinase domain-containing protein 9; CLIC1, chloride intracellular channel protein 1.
Figure 5.
Differential SILAC strategy for screening for HTRA1 substrates in RPE-conditioned medium. Top: SILAC-labeled or unlabeled conditioned media were incubated with (+HTRA1) or without (−HTRA1) for 20 hours at 37°C. Treated and untreated conditioned media were then mixed at 1:1 ratio and processed for SDS-PAGE and LC-MS/MS. Clusterin was identified by 15 labeled and unlabeled peptide pairs in the RPE secretome. Bottom: mass spectra for one of the clusterin peptide detected as a pair of SILAC-labeled and unlabeled peptides. The intensity ratio of these peaks was equal to 1 when both samples were untreated with HTRA1 (left), but decreased to 0.5 in treated versus untreated sample pairs (center and right), clearly indicating that clusterin was degraded by HTRA1.
Among the eight proteins cleaved by HTRA1, fibromodulin was the most affected protein since its levels deceased by 70% and by >90% after treatment with HTRA1 for 4 and 20 hours, respectively. Similarly clusterin, vitronectin, α2-macroglobulin, talin-1, fascin, and chloride intracellular channel protein 1 decreased by more than 50% after 20 hours treatment with HTRA1.
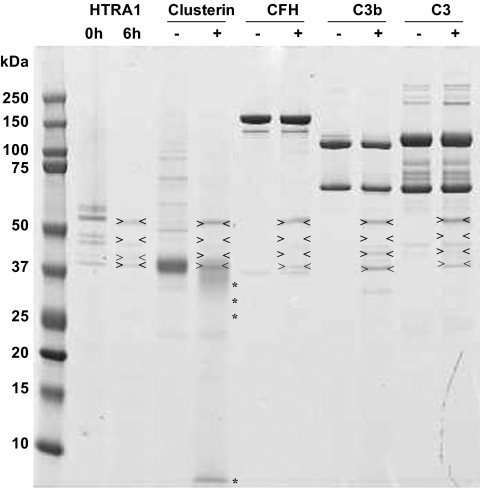
To evaluate whether cleavage of clusterin by HTRA1 is specific, we incubated four different human purified proteins including clusterin, CFH, C3 and C3b in the presence or absence of HTRA1 (Fig. 6). Two micrograms of each protein were incubated with and without recombinant HTRA1 (80 ng) for 6 hours at 37°C followed by SDS-PAGE analysis. Whereas clusterin was significantly degraded by HTRA1 (>50% degradation), CFH, C3, and C3b remained intact, clearly demonstrating that clusterin is a specific substrate for HTRA1. To ensure that degradation of these proteins is not a result of contamination of recombinant HTRA1 with other proteases, we excised the multiple bands seen in the SDS-PAGE for the HTRA1 recombinant protein alone and processed them for mass spectrometry. The data showed that all these bands corresponded to human HTRA1 migrating at different position (Supplementary Fig. S3). These multiple gel bands are most likely the result of HTRA1 autodegradation and/or incomplete reduction of cysteine residues. Indeed, HTRA1 has a total of 16 cysteine residues, and it is well known that cysteine-rich proteins are often oxidized, resulting in an altered migration pattern during gel electrophoresis.
Figure 6.
Clusterin was a specific substrate for HTRA1. The results of a shotgun proteome profiling analysis suggested that clusterin is selectively cleaved by HTRA1. To validate this finding, three recombinant proteins regulating the complement pathway, including human recombinant clusterin and purified human CFH, C3, and C3b were incubated with human recombinant HTRA1. Each protein (2 μg) was incubated with (+) and without (−) recombinant HTRA1 (0.08 μg) for 6 hours at 37°C (enzyme to protein ratio, 1:25, wt/wt) in 4 μL solution of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 5 mM CaCl2. Whereas clusterin was significantly degraded by HTRA1 (> 50% degradation), the other proteins—CFH, C3, and C3b—remained intact. The first two lines represent HTRA1 alone after 0 and 6 hours' incubation at 37°C in the same buffer. Arrows: HTRA1 itself along with the autolysis product. *HTRA1-cleaved products.
Discussion
HTRA1 expression was significantly increased in RPE cells harboring the homozygous risk genotype for the HTRA1 promoter compared with that in RPE cells with the wild-type genotype. This finding agrees with that in the study by Yang et al.,8 who showed that HTRA1 mRNA levels were three times higher in the lymphocytes collected from AMD donors than in those collected from age-matched control donors. Others have demonstrated a similar increase in the HTRA1 mRNA levels in the macular region of eyes collected from AMD donors with the at-risk HTRA1 promoter genotype.13 These data are in contrast to those in the report by Kanda et al.,9 who showed that polymorphism in the promoter region of HTRA1 does not affect its expression level and concluded that the ARMS2 polymorphism, which is in high LD with the HTRA1 polymorphism is the causal SNP. ARMS2 is a poorly characterized protein, and there are ongoing debates about its expression and its cellular localization.11 In an earlier study, ARMS2 was localized to mitochondria,9 whereas more recent studies suggested a cytosolic34 or even extracellular localization.36 In this study, we did not succeed in detecting ARMS2 protein intracellularly or in the conditioned media, perhaps because of its low expression levels. However, HTRA1 was readily expressed and secreted by cultured RPE cells.
Local secretion of HTRA1 by RPE cells prompted us to screen for potential substrates in the context of the RPE extracellular milieu. Our SILAC strategy readily facilitated high-throughput screening for such substrates among several hundred other proteins. The cleaved proteins included fibromodulin, clusterin, α2-macroglobulin, ADAM9, and vitronectin (Supplementary Table S2). Fibronectin, which has been reported to be a substrate for HTRA1 in arthritis tissues,18 was not affected by HTRA1 treatment in the RPE extracellular milieu (Supplementary Fig. S4). Other proteins, such as chloride channel protein 1, fascin, and talin-1, were also degraded by HTRA1. However, the significance of their degradation by HTRA1 remains to be further investigated, since HTRA1 is readily secreted protein, and these proteins are normally localized at the cell periphery connected to the cytoskeleton, as in the case of talin-1 and fascin. Their presence in the conditioned medium could be due to cell shedding or leakage. At this time and because HTRA1 is physiologically secreted outside the cell, we focused on its interaction with other secreted proteins, but it also may be of interest in the future to look at the effect of HTRA1 on peripheral proteins.
HTRA1 Cleavage of Proteins Involved in the Complement Cascade
Vitronectin and clusterin have been suggested to act as fluid-phase regulators of the complement pathway by inhibiting the formation of the terminal complex.37,38 In addition, fibromodulin, which was selectively degraded by HTRA1, is also reported to be involved in the regulation complement pathway by inhibiting the alternative pathway by directly binding to CFH.39 The binding affinity of fibromodulin to CFH was higher in the Y402 variant than in the H402 variant.40 Degradation of these complement pathway regulators by HTRA1 clearly supports a link between HTRA1 and the complement pathway in AMD pathogenesis. However, further protein–protein interaction studies are needed to confirm this network.
HTRA1 Cleavage of Proteins Involved in the Regulation of Amyloid Deposition
Other interesting proteins that were found to be selectively cleaved by HTRA1 in the RPE extracellular milieu are those involved in the regulation of amyloid deposition. Indeed, both α2-macroglobulin and ADAM9 are involved in processing of the amyloid protein precursor. Although ADAM9 cleaves the amyloid precursor at the amyloid β (A-β) domain and releases a neuroprotective ectodomain,41,42 α2-macroglobulin has recently been reported to inhibit amyloid fibril formation,43 probably via sequestering the A-β polypeptide.44 Moreover, a recent study showed that ADAM9 knockout leads to retinal degeneration in mice45 supporting our hypothesis that levels of ADAM9 may be decreased in the presence of high levels of HTRA1, thus leading to amyloid accumulation in the sub-RPE extracellular space. Clusterin has also been reported to be a protective chaperone protein against amyloid deposition,46,47 and its degradation by HTRA1 may also contribute to uncontrolled amyloid deposition. The fact that HTRA1 cleaves these proteins strongly supports the indirect implication of HTRA1 in drusen formation. Indeed, amyloid, which is readily secreted by RPE cells (Supplementary Table S2), is a major component in drusen48,49 and may be more likely to accumulate in cases in which HTRA1 expression is increased.
Although preliminary, these data strongly suggest a role for HTAR1 in AMD pathogenesis and its link to both the complement pathway and amyloid deposition. We suggest that increased expression of HTRA1 in RPE cells with a risk genotype directly affects factors involved in the regulation of the complement pathway (e.g., clusterin, vitronectin, and fibromodulin) as well as factors involved in the regulation of amyloid deposition (clusterin, α2-macroglobulin, and ADAM9). However, these pathways remain to be directly verified in human donor eyes and/or in AMD animal models when they become available.
Supplementary Material
Acknowledgments
The authors thank all the donors and their families for participating, Kristy Brown for help with the mass spectrometry analysis, Stephanie Duguez for help with the flow cytometry, Eric P. Hoffman and Joseph M. Devaney for help with the genetic analyses, and Mary Rose and Anamaris M. Colberg-Poley for help in editing the manuscript.
Footnotes
Supported by National Institutes of Health/National Eye Institute (NIH/NEI) Grant R21EY016723, NEI ARRA administrative supplement R21EY016723-02S109, and partially by National Institute of Child Health and Development (NICHD)/NIH Core Grants 5R24HD050846 and 5P30HD040677. Eunkyung An is a predoctoral student in the Biochemistry and Molecular Genetics Program of the Institute for Biomedical Sciences at the George Washington University. This work is from a dissertation to be presented to the above program in partial fulfillment of the requirements of the PhD degree.
Disclosure: E. An, None; S. Sen, None; S.K. Park, None; H. Gordish-Dressman, None; Y. Hathout, None
References
- 1.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005;308:385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science 2005;308:419–421 [DOI] [PubMed] [Google Scholar]
- 3.Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A 2005;102:7227–7232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science 2005;308:421–424 [DOI] [PubMed] [Google Scholar]
- 5.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet 2005;14:3227–3236 [DOI] [PubMed] [Google Scholar]
- 6.Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet 2005;77:389–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 2006;314:989–992 [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Camp NJ, Sun H, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science 2006;314:992–993 [DOI] [PubMed] [Google Scholar]
- 9.Kanda A, Chen W, Othman M, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci U S A 2007;104:16227–16232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henkind P. Senile macular degeneration. Annu Rev Med 1971;22:95–102 [DOI] [PubMed] [Google Scholar]
- 11.Fritsche LG, Loenhardt T, Janssen A, et al. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet 2008;40:892–896 [DOI] [PubMed] [Google Scholar]
- 12.De Luca A, De Falco M, Severino A, et al. Distribution of the serine protease HtrA1 in normal human tissues. J Histochem Cytochem 2003;51:1279–1284 [DOI] [PubMed] [Google Scholar]
- 13.Chan CC, Shen D, Zhou M, et al. Human HtrA1 in the archived eyes with age-related macular degeneration. Trans Am Ophthalmol Soc 2007;105:92–97, discussion 97–98 [PMC free article] [PubMed] [Google Scholar]
- 14.Zumbrunn J, Trueb B. Primary structure of a putative serine protease specific for IGF-binding proteins. FEBS Lett 1996;398:187–192 [DOI] [PubMed] [Google Scholar]
- 15.Chien J, Staub J, Hu SI, et al. A candidate tumor suppressor HtrA1 is downregulated in ovarian cancer. Oncogene 2004;23:1636–1644 [DOI] [PubMed] [Google Scholar]
- 16.Baldi A, De Luca A, Morini M, et al. The HtrA1 serine protease is down-regulated during human melanoma progression and represses growth of metastatic melanoma cells. Oncogene 2002;21:6684–6688 [DOI] [PubMed] [Google Scholar]
- 17.Esposito V, Campioni M, De Luca A, et al. Analysis of HtrA1 serine protease expression in human lung cancer. Anticancer Res 2006;26:3455–3459 [PubMed] [Google Scholar]
- 18.Grau S, Richards PJ, Kerr B, et al. The role of human HtrA1 in arthritic disease. J Biol Chem 2006;281:6124–6129 [DOI] [PubMed] [Google Scholar]
- 19.Hu SI, Carozza M, Klein M, Nantermet P, Luk D, Crowl RM. Human HtrA, an evolutionarily conserved serine protease identified as a differentially expressed gene product in osteoarthritic cartilage. J Biol Chem 1998;273:34406–34412 [DOI] [PubMed] [Google Scholar]
- 20.Wu J, Liu W, Bemis A, et al. Comparative proteomic characterization of articular cartilage tissue from normal donors and patients with osteoarthritis. Arthritis Rheum 2007;56:3675–3684 [DOI] [PubMed] [Google Scholar]
- 21.Grau S, Baldi A, Bussani R, et al. Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proc Natl Acad Sci U S A 2005;102:6021–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakay M, Zhao P, Chen J, Hoffman EP. A web-accessible complete transcriptome of normal human and DMD muscle. Neuromuscul Disord 2002;12(suppl 1):S125–S141 [DOI] [PubMed] [Google Scholar]
- 23.Chien J, He X, Shridhar V. Identification of tubulins as substrates of serine protease HtrA1 by mixture-based oriented peptide library screening. J Cell Biochem 2009;107:253–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou J, Clemmons DR, Smeekens S. Expression and characterization of a serine protease that preferentially cleaves insulin-like growth factor binding protein-5. J Cell Biochem 2005;94:470–484 [DOI] [PubMed] [Google Scholar]
- 25.Chamberland A, Wang E, Jones AR, et al. Identification of a novel HTRA1-susceptible cleavage site in human aggrecan: evidence for the involvement of htra1 in aggrecan proteolysis in vivo. J Biol Chem 2009;284(40):27352–27359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelmann K, Valtink M. RPE cell cultivation. Graefes Arch Clin Exp Ophthalmol 2004;242:65–67 [DOI] [PubMed] [Google Scholar]
- 27.McKechnie NM, Boulton M, Robey HL, Savage FJ, Grierson I. The cytoskeletal elements of human retinal pigment epithelium: in vitro and in vivo. J Cell Sci 1988;91:303–312 [DOI] [PubMed] [Google Scholar]
- 28.An E, Lu X, Flippin J, et al. Secreted proteome profiling in human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. J Proteome Res 2006;5:2599–2610 [DOI] [PubMed] [Google Scholar]
- 29.Hathout Y, Flippin J, Fan C, Liu P, Csaky K. Metabolic labeling of human primary retinal pigment epithelial cells for accurate comparative proteomics. J Proteome Res 2005;4:620–627 [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl M. Relative quantification. In: Dorak T. ed. Real Time PCR Oxford, UK: Talyor & Francis; 2006:63–82 [Google Scholar]
- 31.An E, Gordish-Dressman H, Hathout Y. Effect of TNF-alpha on human ARPE-19-secreted proteins. Mol Vis 2008;14:2292–2303 [PMC free article] [PubMed] [Google Scholar]
- 32.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods 2008;5:319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sidak A. Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc JASA 1967;62:626–633 [Google Scholar]
- 34.Wang G, Spencer KL, Court BL, et al. Localization of age-related macular degeneration-associated ARMS2 in cytosol, not mitochondria. Invest Ophthalmol Vis Sci 2009;50:3084–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002;1:376–386 [DOI] [PubMed] [Google Scholar]
- 36.Kortvely E, Hauck SM, Duetsch G, et al. ARMS2 is a constituent of the extracellular matrix providing a link between familial and sporadic age-related macular degenerations. Invest Ophthalmol Vis Sci 2010;51(1):79–88 [DOI] [PubMed] [Google Scholar]
- 37.Jenne DE, Lowin B, Peitsch MC, Bottcher A, Schmitz G, Tschopp J. Clusterin (complement lysis inhibitor) forms a high density lipoprotein complex with apolipoprotein A-I in human plasma. J Biol Chem 1991;266:11030–11036 [PubMed] [Google Scholar]
- 38.Podack ER, Muller-Eberhard HJ. Isolation of human S-protein, an inhibitor of the membrane attack complex of complement. J Biol Chem 1979;254:9808–9814 [PubMed] [Google Scholar]
- 39.Sjoberg A, Onnerfjord P, Morgelin M, Heinegard D, Blom AM. The extracellular matrix and inflammation: fibromodulin activates the classical pathway of complement by directly binding C1q. J Biol Chem 2005;280:32301–32308 [DOI] [PubMed] [Google Scholar]
- 40.Sjoberg AP, Trouw LA, Clark SJ, et al. The factor H variant associated with age-related macular degeneration (His-384) and the non-disease-associated form bind differentially to C-reactive protein, fibromodulin, DNA, and necrotic cells. J Biol Chem 2007;282:10894–10900 [DOI] [PubMed] [Google Scholar]
- 41.Hotoda N, Koike H, Sasagawa N, Ishiura S. A secreted form of human ADAM9 has an alpha-secretase activity for APP. Biochem Biophys Res Commun 2002;293:800–805 [DOI] [PubMed] [Google Scholar]
- 42.Allinson TM, Parkin ET, Turner AJ, Hooper NM. ADAMs family members as amyloid precursor protein alpha-secretases. J Neurosci Res 2003;74:342–352 [DOI] [PubMed] [Google Scholar]
- 43.Yerbury JJ, Kumita JR, Meehan S, Dobson CM, Wilson MR. alpha2-Macroglobulin and haptoglobin suppress amyloid formation by interacting with prefibrillar protein species. J Biol Chem 2009;284:4246–4254 [DOI] [PubMed] [Google Scholar]
- 44.Fabrizi C, Businaro R, Lauro GM, Fumagalli L. Role of alpha2-macroglobulin in regulating amyloid beta-protein neurotoxicity: protective or detrimental factor? J Neurochem 2001;78:406–412 [DOI] [PubMed] [Google Scholar]
- 45.Parry DA, Toomes C, Bida L, et al. Loss of the metalloprotease ADAM9 leads to cone-rod dystrophy in humans and retinal degeneration in mice. Am J Hum Genet 2009;84:683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boggs LN, Fuson KS, Baez M, et al. Clusterin (Apo J) protects against in vitro amyloid-beta (1-40) neurotoxicity. J Neurochem 1996;67:1324–1327 [DOI] [PubMed] [Google Scholar]
- 47.McHattie S, Edington N. Clusterin prevents aggregation of neuropeptide 106-126 in vitro. Biochem Biophys Res Commun 1999;259:336–340 [DOI] [PubMed] [Google Scholar]
- 48.Dentchev T, Milam AH, Lee VM, Trojanowski JQ, Dunaief JL. Amyloid-beta is found in drusen from some age-related macular degeneration retinas, but not in drusen from normal retinas. Mol Vis 2003;9:184–190 [PubMed] [Google Scholar]
- 49.Luibl V, Isas JM, Kayed R, Glabe CG, Langen R, Chen J. Drusen deposits associated with aging and age-related macular degeneration contain nonfibrillar amyloid oligomers. J Clin Invest 2006;116:378–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.