The authors purified amacrine cells, and analyzed their developmental gene expression profile and survival signaling. They also compared the gene expression profile of two neighboring retinal cells during development: amacrine and retinal ganglion cells.
Abstract
Purpose.
To describe how developing amacrine cells and retinal ganglion cells (RGCs) differ in survival signaling and global gene expression.
Methods.
Amacrine cells were immunopurified and processed for gene microarray analysis. For survival studies, purified amacrine cells were cultured at low density in serum-free medium, with and without peptide trophic factors and survival pathway inhibitors. The differences in gene expression between amacrine cells and RGCs were analyzed by comparing the transcriptomes of these two cell types at the same developmental ages.
Results.
The amacrine cell transcriptome was very dynamic during development. Amacrine cell gene expression was remarkably similar to that of RGCs, but differed in several gene ontologies, including polarity- and neurotransmission-associated genes. Unlike RGCs, amacrine cell survival in vitro was independent of cell density and the presence of exogenous trophic factors, but necessitated Erk activation via MEK1/2 and AKT signaling. Finally, comparison of the gene expression profile of amacrine cells and RGCs provided a list of polarity-associated candidate genes that may explain the inability of amacrine cells to differentiate axons and dendrites as RGCs do.
Conclusions.
Comparison of the gene expression profile between amacrine cells and RGCs may improve our understanding of why amacrine cells fail to differentiate axons and dendrites during retinal development and of what makes amacrine cells differ in their resistance to neurodegeneration. Switching RGCs to an amacrine cell-like state could help preserve their survival in neurodegenerative diseases like glaucoma, and amacrine cells could provide a ready source of replacement RGCs in such optic neuropathies.
Amacrine cells are retinal interneurons essential for visual function, as they modulate retinal signaling on retinal ganglion cells (RGCs).1 More than 30 types of amacrine cells in the mammalian retina can be classified by morphology, physiology, stratification patterns, or expression of specific markers.2–5 In the developing retina, amacrine cells are born at the same time as RGCs, and many of them even migrate to the same layer of the retina.6,7 Interestingly, amacrine cells appear to resist neurodegeneration after either photoreceptor or RGC death.8
The signaling of RGC survival has been well-studied9–12; however, little is known about amacrine cell biology. For example, what is the molecular basis for their resistance to degeneration upon loss of their targets (RGCs)? Why do they not differentiate their neurites into axons and dendrites as RGCs do? In this study, we characterized amacrine cell biology in vivo and in vitro, using highly purified cultures of amacrine cells. These data present a comprehensive comparative analysis of two neighboring central nervous system (CNS) neurons, and demonstrate fundamental differences between RGCs and amacrine cells in gene expression and survival signaling.
Methods
Animals
Sprague-Dawley rats were used for these experiments in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and in accordance with institutional animal care and use committee review and approval.
Amacrine Cell and RGC Purification
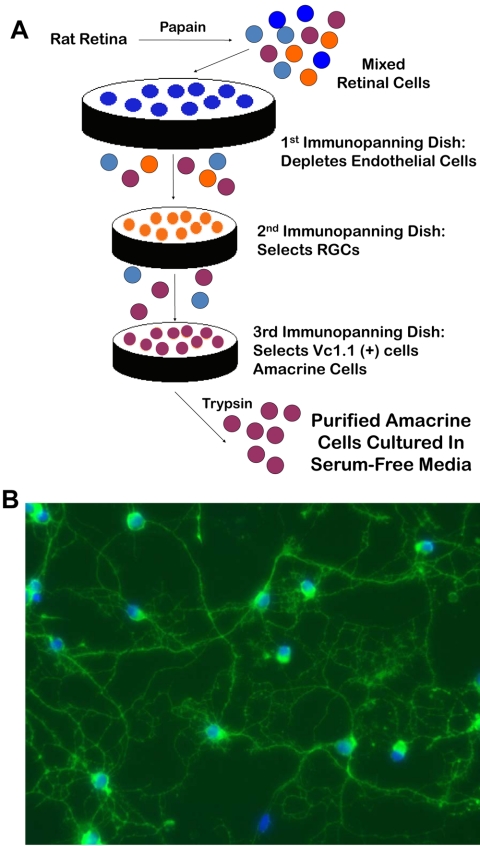
Amacrine cells and RGCs were purified by immunopanning as previously described.9,11 Briefly, embryonic and postnatal rat retinas were dissociated with papain (Worthington, Lakewood, NJ) and mechanically triturated to obtain a single-cell suspension. Enrichment of amacrine cells to 88% purity was achieved after depleting rat macrophages (1:75, AI A51240; Accurate Chemical, Westbury, NY) and T11d7- and Ox7-positive cells (including RGCs) and immunopanning for Vc1.1-positive cells13 (see Fig. 1 and Supplementary Table S1; all Supplementary Tables are available at http://www.iovs.org/cgi/content/full/51/7/3800/DC1).
Figure 1.
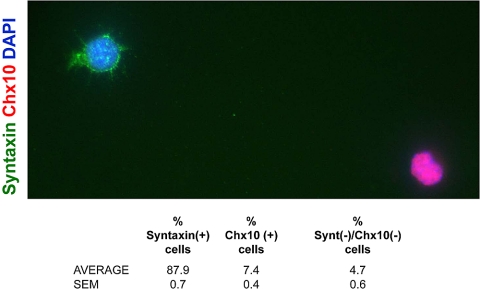
Purification of amacrine cells by immunopanning. (A) Acutely dissected retinas from embryonic and early postnatal rats were dissociated in papain and triturated to obtain a single-cell suspension. After depletion of macrophages and RGCs from the retinal suspension, the amacrine cells were selected with the monoclonal antibody Vc1.1. Subsequent trypsinization yielded amacrine cells at least ∼88% pure, which were subsequently cultured in serum-free medium. (B) Immunostaining of purified amacrine cells after 2 days in vitro with anti-syntaxin antibody revealed a typical pattern of neurite outgrowth.
RNA Preparation, Microarray Hybridization, and Data Analysis
Amacrine cells from embryonic (E20) and early postnatal (P5, P11) rats were acutely purified.11 Total RNA was extracted (RNeasy; Qiagen, Valencia, CA) and shipped to the NIH Neuroscience Microarray Consortium (at the University of California Los Angeles), where it was amplified and processed for hybridization onto rat genome arrays (RAE 230 2.0 GeneChip; Affymetrix, Santa Clara, CA). Three microarrays were used for each postnatal amacrine cell age (P5, P11), and four were used for E20 amacrine cells. RNA collected from independent samples obtained on different days served as the starting material for each microarray.
Raw data files were analyzed (Microarray Suite 5.0; Affymetrix), and statistical analysis was performed (Excel; Microsoft, Redmond, WA; and NetAffx Analysis Center; Affymetrix) as described in the Results section. Amacrine cell microarray data have been deposited in the NIH Neuroscience Microarray Consortium database (http://np2.ctrl.ucla.edu/np2/home.do) and are also available in Supplementary Table S1.
Comparison of Amacrine Cell and RGC Gene Expression Profiles
We compared the gene expression profile of amacrine cells to previously published data on RGCs.14 Since the microarray platforms used for amacrine cells and RGCs were different (RAE 230 2.0 and RG34U A-C, respectively; Affymetrix), we developed a method for probe set matching. Using the “Array Comparison Spreadsheets” from Affymetrix (http://www.affymetrix.com/support/technical/manual/comparison_spreadsheets_manual.pdf), we found that of 31,099 probe sets in the RAE 230 2.0 array and 26,379 probe sets in the RG34U A-C array set, 16,749 rat genes were probed by both platforms, allowing us to analyze cross-platform data representing 54% of the amacrine cell probes and 63% of the RGC probes.
One of the caveats in the use of the Good Match Spreadsheets is that the relation of probe sets between RAE 230 2.0 and RG 34UA-C arrays is not one to one, but rather many to one.15 We eliminated duplicates in our datasets by removing the probes with the lowest percentage “present” call (Affymetrix algorithm) across samples and then the lowest expression levels across all ages. According to these criteria, we also removed probes with the same UniGene identification numbers,16 which yielded a final pool of 14,457 shared unique probes between the amacrine and RGC datasets (http://www.ncbi.nlm.nih.gov/UniGene; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD). The final probe comparison spreadsheet is available in Supplementary Table S2.
To compare gene expression levels for the full probe set between amacrine cells and RGCs (as in Fig. 5), analysis of the gene expression profiles was performed independent of expression levels, to avoid the hazards of cross-platform normalization. We generated a ranking system, where the average of three to four biological replicates for each probe within one dataset was ranked according to its expression level compared with the rest of the probes within that dataset. The highest expressed probe in amacrine cells received a rank of 1 and so forth; the same methods was used for the RGCs. Rankings within an ontology were averaged for each cell type at each age studied, creating the index used in Figure 5.
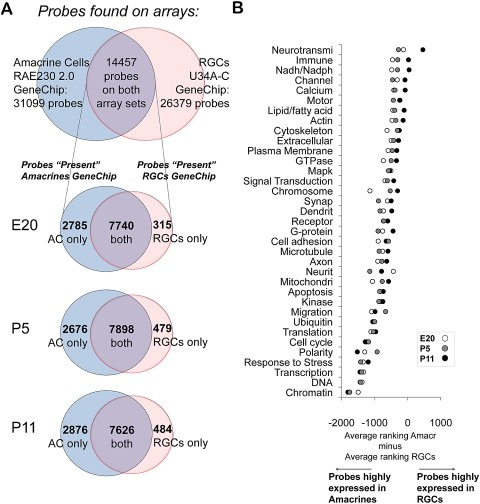
Figure 5.
Differentially expressed gene ontologies between RGCs and amacrine cells. (A) Venn diagrams describing gene representation on the two different platforms (top) and gene expression at E20, P5, and P11 between amacrine cells and RGCs. After eliminating duplicate probes for the same gene, 14,457 probes were represented in both datasets. Of these, most were “present” in both RGCs and amacrine cells at all three ages, but over 2600 were expressed uniquely by amacrine cells, and over 300 were expressed uniquely by RGCs, comparing at each age. The area of the circles is not exactly proportional to the number of probes they contain. (B) Average difference in ranked expression between amacrine cells and RGCs in 35 overlapping gene ontologies. Only probes that were “present” in both datasets were included in the analysis. (See the Methods and Results section for average ranking details; the datasets and the ranking for each probe can be found in Supplementary Table S2.)
When directly comparing expression levels of a subset of genes (as in Fig. 6), we normalized the datasets by comparing the average of 14,457 probes for amacrine cells and RGCs at each developmental age. The adjusted RGC absolute expression levels were calculated by multiplying the RGC expression levels by a ratio factor generated by dividing the normalized, average expression level of all amacrine cell probes by the normalized, average expression level of all RGC probes.
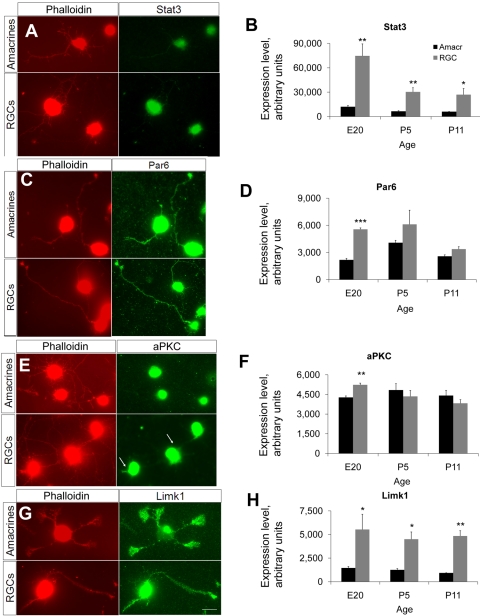
Figure 6.
Immunostaining of polarity-associated genes and corresponding microarray expression levels. (A, C, E, G) Acutely purified amacrine cells and RGCs were cultured on glass coverslips and immunostained at 3 DIV with antibodies as shown, and rhodamine-conjugated phalloidin to counterstain actin filaments and highlight growth cones. Scale bar, 20 μm. (B, D, F, H) Levels of expression of polarity-associated probes from the microarray data for amacrine cells and RGCs during development. Expression levels were adjusted between the two datasets by multiplying the RGC probes by the ratio of the means for amacrine cells and RGCs at each developmental age.
Quantitative Real-Time Reverse Transcription–PCR
To validate the microarray data, we performed quantitative real-time PCR (qRT-PCR) and compared the expression levels of a subset of genes of interest between E20 and P11 amacrine cells. Total RNA was extracted from acutely purified cells as described above, of which 1 μg was reversed transcribed into cDNA (iScript; Bio-Rad, Hercules, CA). Equal amounts of cDNA were further amplified by real time-PCR (iQ SYBR Green Supermix; Bio-Rad). We used 18S ribosomal RNA as an internal reference gene. Primers used were as follows: Calb1 (forward): TGCAGGCACGAAAGAAGGCTGG; Calb1 (reverse): CGGTGGGTAAGACATGGGCCAAC; Dapk1 (forward): CTGATGGGCGCCAACGTGGA; Dapk1 (reverse): CCGCAGTCTTGCCAGGAGCC; Rasgrf1 (forward): GCGCTGCGTGACAGAGTGGA; Rasgrf1 (reverse): TGGCCCCCAGGGCTTCTCAG; C1q-L1 (forward): CGGCCAGCGGCAAGTTTACA; C1q-L1 (reverse): GCAATGGCACTGGCCCGCAC; and 18s (forward): GAACTGAGGCCATGATTAAGAG; 18s (reverse): CATTCTTGGCAAATGCTTTC. The change ratio was calculated by using the ΔΔCt method.17 Reactions were run with five replicates per primer pair and repeated at least three times on different days.
Immunofluorescence
For quantification of the purity of amacrine cell cultures, amacrine cells were acutely purified, plated on PDL-coated glass coverslips, and immunostained at 1 day in vitro (DIV). Briefly, the cells were fixed with 4% PFA for 5 minutes, rinsed three times in PBS, and permeabilized for 5 minutes with 1% Triton X-100. After another round of rinses in PBS, the cells were blocked and permeabilized with 20% donkey serum and 0.1% Triton X-100 in antibody buffer (150 mM NaCl, 50 mM Tris base, 1% BSA, 100 mM l-lysine, 0.04% Na azide [pH 7.4]). Sheep anti-Chx10 (1:200, AB9016; Millipore, Billerica, MA) and mouse anti-syntaxin (1:200, ab3265; Abcam, Cambridge, MA) antibodies were incubated overnight at 4°C. Secondary antibodies (donkey anti-mouse Alexa 488 and donkey anti-sheep Alexa 594; Invitrogen, Carlsbad, CA) were used at a dilution 1:500 and incubated for 4 hours in the dark at room temperature. Coverslips were later mounted on glass slides (Vectashield with DAPI; Vector Laboratories, Burlingame, CA) and examined with an inverted fluorescence microscope (Carl Zeiss Meditec, Dublin, CA).
For immunostaining of polarity genes, RGCs and amacrine cells acutely purified from the same animals were cultured on PDL-coated glass coverslips with or without laminin (Trevigen, Gaithersburg, MD), respectively, in serum-free medium. At 3 DIV, cells were incubated at 37°C with PFA (2% final concentration) for 30 minutes, postfixed with 4% PFA for 10 minutes at room temperature and the coverslips were rinsed three times with PBS and blocked for 30 minutes with 20% normal goat serum and 0.2% Triton X-100 in antibody buffer. Primary antibodies were incubated overnight at 4°C as follows: rabbit anti-Par6 (1:100, sc-25,525, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-atypical PKC (1:100, sc-216, Santa Cruz Biotechnology), mouse anti-Limk1 (1:500, L13020; BD Biosciences, Mississauga, ON Canada), and mouse anti-Stat3 (1:100, 9139, Cell Signaling Technology). Secondary detection was performed with fluorescent antibodies at a 1:500 (Alexa-488; Invitrogen). TRITC-conjugated phalloidin (3.3 μg/mL; P1951; Sigma-Aldrich, St. Louis, MO) was added together with the secondary antibodies and incubated overnight at 4°C. The coverslips were mounted on glass slides and examined with an inverted fluorescence microscope (Carl Zeiss Meditec, Inc.). Cells immunostained with the same antibody were exposed to the same fluorescence intensity. Exposure was carefully controlled to maximize comparison between amacrine and RGC immunofluorescence and to better capture the expression of polarity genes in the neurites.
Amacrine Cell Culture and Survival Assays
Acutely purified amacrine cells were plated at different densities (8, 47, or 156 cells/mm2) in tissue-culture wells coated with PDL (70 kDa, 10 μg/mL; Sigma-Aldrich) in serum-free medium (Neurobasal; Invitrogen-Gibco) as described9,12 containing insulin (50 ng/mL), forskolin (5 mM; Sigma-Aldrich), CNTF (10 ng/mL), BDNF (50 ng/mL; Peprotech, Rocky Hill, NJ), and a modified version of B2718 with antibiotics (1X Pen-Strep; Invitrogen-Gibco). The cultures were maintained at 37°C in a humidified incubator with 10% CO2. To evaluate cell survival as a function of cell density (see Fig. 7A), we used calcein-AM (1 μM; Invitrogen, Carlsbad, CA) and the nuclear dye DAPI. Initially, we intended to use DAPI as a nuclear maker for all cells and calcein as a marker of living cells. This method would allow unequivocal counting of cell viability. To our surprise, when imaging the cells under a fluorescence microscope, we saw exclusion of DAPI by calcein-positive cells. We then added a third dye (propidium iodide) and confirmed this result. Invitrogen states that DAPI is cell impermeant when added at low doses. We used DAPI at a final concentration of 30 ng/mL. Cells that were calcein+/DAPI− were considered to be alive; dead cells were DAPI+. For the rest of the survival experiments (Fig. 7B, 7C), the cells were plated at the lowest density (8 cells/mm2). We used calcein-AM (1 μM) and the nuclear dyes Hoechst (1:5000; Invitrogen) and propidium iodide (PI; 1:5000; Sigma-Aldrich) were added to the media for 30 to 45 minutes. Live cells were defined as calcein+/PI–; all PI+ cells were classified as dead. The cells were manually counted, and at least three wells per condition were averaged within each experiment. The experiments were repeated at least five times to confirm the results.
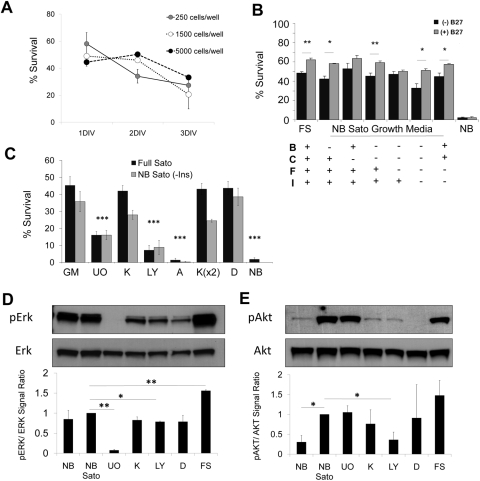
Figure 7.
Amacrine cell survival in vitro. (A) Amacrine cells were purified and plated at different densities in serum-free medium. Cells were counted at 1, 2, or 3 DIV after adding calcein-AM and the nuclear dye DAPI. Survival was calculated as the percentage of cells that were calcein+ of the total number of calcein+ and DAPI+ cells. (N = 2; n > 150 for each condition. Error bars: SEM). (B, C) Acutely purified P8 amacrine cells were plated at low density in growth medium containing peptide trophic factors and forskolin, with and without pharmacologic inhibitors (see below). Survival was quantified at 3 DIV with calcein/PI (N > 3; n > 150 for each condition; ***P < 0.001, one-way ANOVA with the Tukey post hoc test; **P < 0.01, *P < 0.05, paired t-test. Error bars: SEM). (D, E) Amacrine cells were purified and incubated for 2 hours in growth medium with or without inhibitors as labeled, after which they were centrifuged and processed for protein extraction. Example Westerns are shown; graphs are the average of at least two experiments, normalized to NB Sato (*P < 0.05, **P < 0.005; unpaired t-test. Error bars: SEM). B, BDNF; C, CNTF; F, forskolin; I, insulin; NB, Neurobasal+penicillin/streptomycin; NB Sato, Neurobasal+Sato stock+pyruvate+penicillin/streptomycin; GM, growth medium; U0, U0126; K, K252a; LY, LY294002; A, AG490; D, DMSO; FS, full Sato.
Immunoblot and Densitometric Analysis
Acutely purified amacrine cells were resuspended in serum-free medium, with or without various peptide trophic factors and pharmacologic inhibitors, and rotated for 2 hours at 37°C, after which the cells were processed for protein extraction according to standard protocols. The pharmacologic inhibitors used were U0126 (10 μM), K252a (400 nM), AG490 (100 μM), PD98059 (20–30 μM; all from Calbiochem, San Diego, CA), and LY294002 (50 μM; Cell Signaling Technology, Danvers, MA). The antibodies used for Western blot were: rabbit anti-phospho-Mapk antiserum (1:1000; E7028; Sigma-Aldrich), goat anti-Erk1 antiserum (1:1000; sc-93; Santa Cruz Biotechnology), rabbit anti-phospho-Akt antiserum (1:1500; 9271), and rabbit anti-Akt antiserum (1:1000; 9272 Cell Signaling Technology, Danvers, MA). Secondary antibodies conjugated to horseradish peroxidase (1:2000; Santa Cruz Biotechnology) were incubated for 2 hours at room temperature and revealed with enhanced chemiluminescence (Pierce Biotechnology, Rockford, IL). Densitometric analysis of the Western blots was then conducted (Photoshop software; Adobe, San Jose, CA). Experiments were repeated at least three times to confirm the results.
Results
Amacrine Cell Purification
Amacrine cells from E20 and P8 rats were acutely purified by immunopanning with the Vc1.1 antibody19 and cultured in serum-free medium (Fig. 1A). Since amacrine cells only make up to 9% to 12% of the cells in the retina12 and ∼40% of the cells in the inner nuclear layer of the mouse retina,3,20 we performed sequential immunopanning to obtain a high amacrine cell yield (Fig. 1A).12 To confirm the purity of the cultured cells, we performed double immunostaining using both the monoclonal antibody HPC-1 (Fig. 1B), which recognizes syntaxin, an amacrine cell marker21,22 and an antibody against the pan-bipolar cell marker Chx10.23 We found that 88% of the purified cells were immunopositive for HPC-1 and 7% were immunopositive for Chx10, whereas no cells were immunoreactive for both antibodies (Fig. 2). Thus, the immunopanning technique yielded a culture highly enriched in amacrine cells, although data from postnatal amacrine cell cultures did reflect slight contamination from bipolar cells.
Figure 2.
Immunostaining and quantification of amacrine cell culture purity. P7 amacrine cells were acutely purified and plated on glass coverslips in serum-free medium. At 1 DIV, the cells were immunostained for the amacrine cell marker syntaxin (HPC-1) and the bipolar cell marker Chx10. The table shows the percentage of cells that were immunopositive for each marker and represents the average of three coverslips (n > 90 cells per coverslip).
Amacrine Cell Gene Expression Profile
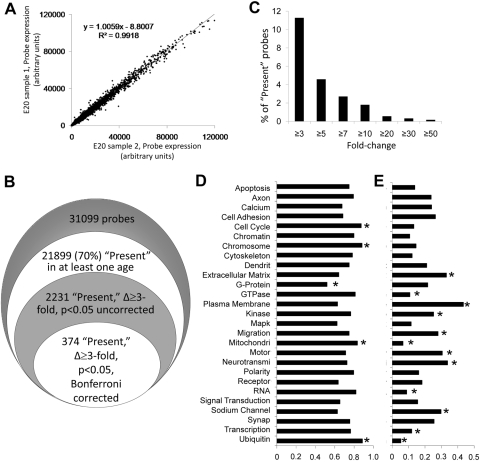
What genes do amacrine cells express during development? Recent analyses of amacrine cell gene expression at the single cell level have yielded beautiful pictures of the molecular diversity of these cells,24,25 but we had undertaken an overview of amacrine cell gene profiling at a population level through perinatal development. Amacrine cell mRNAs isolated from acutely purified E20, P5, and P11 rats were hybridized to expression arrays (Rat Genome 230 2.0; Affymetrix) containing 31,099 probes representing more than 28,000 rat genes. Three to four biological replicates were collected for each age, and we found little intersample variability within the same age group (Pearson r2 ≥ 0.90 for all pairwise comparisons; Fig. 3A).
Figure 3.
Amacrine cells' gene expression profile. Amacrine cells from E20, P5, and P11 rats were purified by immunopanning. Three to four biological replicates were independently processed for microarray analysis (RAE 230 2.0; Affymetrix), and data were analyzed (Microarray Suite 5.0; Affymetrix; National Center for Biotechnology and Information [NCBI]; Gene Ontology databases; and Excel, Microsoft). (A) Shown is a sample of gene expression data from two biological replicates of E20 amacrines, demonstrating high replicability. (B) Amacrine cells expressed ∼70% of the 31,099 probes in the microarray. Of those, 2231 probes changed at least threefold during development. The number of probes that changed significantly decreased after Bonferroni correction. (C) Frequency of changes in the dataset. Of the 21,899 present probes in at least one age, ∼10% were developmentally regulated and ∼2% changed at least 10-fold. (D, E) We classified 31,099 probe sets by gene ontology and fit 16,753 (54%) probes into 27 partially overlapping categories. (D) Amacrine cells expressed 52% to 89% of the probes in each category. (E) Fraction of probes in these gene ontologies that changed threefold or more during amacrine cell development. *Significantly higher or lower expressed (D) or developmentally changing (E) ontology (by pair-wise χ2 test at P < 0.05 after Bonferroni correction).
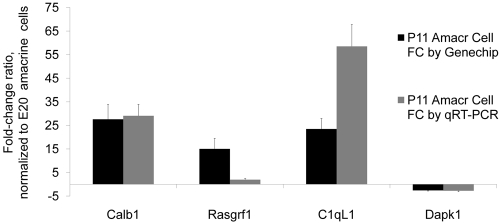
The amacrine cells expressed 21,899 (70%) of the 31,099 probes in at least two or more of the samples at one or more ages (Fig. 3B), and 16,247 probes (52%) were expressed at all ages, according to Affymetrix's “present” call algorithm. Of those 21,899 probes present in at least two or more of the samples at one or more ages, 2468 probes (11%) changed at least threefold during development, 344 (2%) changed at least 10-fold, and 120 (0.5%) changed at least 20-fold (Fig. 3C, Table 1). When ANOVA was used to calculate probes that changed more than threefold with a P < 0.05, 2231 probes (∼10%) changed between E20 and P11 (without correction), and 374 probes (1.7%) changed if a Bonferroni correction was used (Fig. 3B). These represent two extremes of statistical validation; the true number of changing probes probably lies between these two numbers. To validate these results, we performed quantitative reverse transcription real-time PCR (qRT-PCR) on several genes that changed more than 10-fold by microarray (Calb1, Rasgrf1, and C1ql1) and a survival-associated gene, Dapk1. We confirmed the consistency between microarray and qRT-PCR data (Fig. 4), Consistent with our previous work,14 there were differences in the change ratio between microarray and RT-PCR data; however, the direction of the change was the same. These data, together with the immunostaining for polarity genes presented below (see Fig. 6), help validate the microarray data.
Table 1.
Twenty Largest Gene Changes during Amacrine Cell Development
| Probe | Gene Symbol | Gene Name | Change Ratio (Max/Min) |
|---|---|---|---|
| 1368145_at | Pcp4 | Purkinje cell protein 4 | 35.6 |
| 1370201_at | Calb1 | Calbindin 1 | 27.6 |
| 1378045_at | C1ql1 | Complement component 1, q subcomponent-like 1 | 23.5 |
| 1368247_at | Hspa1a III Hspa1b III Hspa1l | Heat shock 70kD protein 1A III heat shock 70kD protein 1B (mapped) III heat shock 70kD protein 1-like (mapped) | 21.3 |
| 1383075_at | Ccnd1 | Cyclin D1 | 20.1 |
| 1384533_at | — | — | 18.1 |
| 1371450_at | — | — | 16.4 |
| 1388944_at | — | — | 15.3 |
| 1370912_at | Hspa1b | Heat shock 70kD protein 1B (mapped) | 15.3 |
| 1370996_at | Rasgrf1 | RAS protein-specific guanine nucleotide-releasing factor 1 | 15.0 |
| 1395473_at | Gnb3 | Guanine nucleotide binding protein (G protein), beta 3 | 14.9 |
| 1377867_at | RGD1562284 | Similar to glutaminyl-peptide cyclotransferase precursor (QC) | 14.6 |
| 1371643_at | Ccnd1 | Cyclin D1 | 14.5 |
| 1373326_at | — | — | 14.4 |
| 1391464_at | — | — | 14.3 |
| 1368864_at | Synpr | Synaptoporin | 14.1 |
| 1393263_at | — | — | 13.8 |
| 1380552_at | — | — | 12.9 |
| 1383210_at | — | — | 12.8 |
| 1383887_at | RGD1306991 | Similar to protein C20orf103 precursor | 12.4 |
Max, maximum expression level at all ages; Min, lowest expression level at all ages.
Figure 4.
Validation of the amacrine cell microarray data. Relative change ratio of P11 amacrine cells by microarray and by qRT-PCR, normalized to E20 amacrine cells.
To further classify the gene expression profile of amacrine cells, we distributed the 31,099 probes of the RAE 230 2.0 arrays into overlapping gene ontology categories by using data available from Affymetrix's Netaffx Analysis Center (http://www.affymetrix.com/analysis/index.affx) and the Gene Ontology Consortium database (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi).26 We fit 16,753 (54%) probes into 27 partially overlapping categories, and we found that amacrine cells express 52% to 89% of the probes in each category. Categories such as mitochondria, cell cycle, chromosome, and ubiquitin were overrepresented and the category G-protein was underrepresented (χ2; Fig. 3D). We next analyzed the fraction of probes within each category that changed at least threefold during development. We found that 11 of the 27 categories were significantly regulated during development (χ2; Fig. 3E), including neurotransmission/tter, plasma membrane, and migration. These results may reflect the vast differentiation that amacrine cells undergo between E20 and P11,27–31 when they start secreting neurotransmitters and there are changes in the levels of their receptors on the plasma membrane. At this same developmental stage, amacrine cells migrate to populate the INL and GCL.32 Conversely, probes in categories such as cell cycle, polarity, and apoptosis were less likely to change during development, as the peak of amacrine cell generation occurs embryonically.6,20,28,33,34 It is possible that some of the probes in these categories also change at the protein level during development by posttranslational regulation. Compartmentalization, binding with inactivating partners or changes in the phosphorylation levels are not reflected in these microarray analyses (Fig. 3E, Table 2). These data suggest that the amacrine cell transcriptome is very dynamic during development, similar to our prior findings with purified RGCs.14
Table 2.
Neurotransmi-Genes in Amacrine Cells and RGCs
| Gene Symbol | Gene Name | Mean E20 |
Mean P5 |
Mean P11 |
|||
|---|---|---|---|---|---|---|---|
| Amacr | RGCs | Amacr | RGCs | Amacr | RGCs | ||
| Unique expression in RGCs by present call | |||||||
| Chrna7 | Cholinergic receptor, nicotinic, alpha polypeptide 7 | 673 | 918 | 509 | 671 | 869 | 669 |
| Aldh5a1 | Aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 546 | 574 | 709 | 730 | 899 | 473 |
| Unique expression in amacrine cells by Affymetrix “Present”call: | |||||||
| — | — | 9,632 | 1,692 | 20,623 | 957 | 25,627 | 403 |
| — | — | 9,332 | 46 | 16,645 | 113 | 13,509 | 110 |
| Slc1a3 | Solute carrier family 1 (glial high affinity glutamate transporter), member 3 | 6,296 | 199 | 10,550 | 207 | 12,565 | 211 |
| Exoc5 | Exocyst complex component 5 | 5,579 | 419 | 5,683 | 362 | 4,984 | 308 |
| Lin7c | Lin-7 homolog C (C. elegans) | 5,066 | 22 | 3,494 | 24 | 3,118 | 20 |
| Slc32a1 | Solute carrier family 32 (GABA vesicular transporter), member 1 | 4,660 | 1,586 | 14,460 | 1,076 | 14,566 | 2,437 |
| Incenp | Inner centromere protein | 1,832 | 1,488 | 1,518 | 1,012 | 679 | 969 |
| Chrna4 | Cholinergic receptor, nicotinic, alpha polypeptide 4 | 1,330 | 2,952 | 1,323 | 3,994 | 1,221 | 3,943 |
| Kcnh2 | Potassium voltage-gated channel, subfamily H (eag-related), member 2 | 1,266 | 1,572 | 1,445 | 1,186 | 2,960 | 1,010 |
| Th | Tyrosine hydroxylase | 1,130 | 1,117 | 2,815 | 1,193 | 2,737 | 857 |
| Rims1 | Regulating synaptic membrane exocytosis 1 | 1,056 | 105 | 1,936 | 63 | 3,922 | 93 |
| Grm2 | Glutarnate receptor, metabotropic 2 | 993 | 570 | 2,480 | 559 | 3,854 | 293 |
| Gabra3 | Gamma-aminobutyric acid (GABA-A) receptor, subunit alpha 3 | 889 | 336 | 1,927 | 455 | 2,468 | 548 |
| Stx3 | Syntaxin 3 | 714 | 107 | 375 | 215 | 1,336 | 167 |
| Rab15 | RAB15, member RAS oncogene family | 707 | 121 | 382 | 221 | 707 | 213 |
| Nedd1 | Neural precursor cell expressed, developmentally down-regulated gene 1 | 671 | 175 | 937 | 84 | 318 | 98 |
| Cacna1a | Calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | 581 | 3,794 | 653 | 2,995 | 1,153 | 3,504 |
| Cln3 | Ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) | 506 | 638 | 484 | 1,512 | 668 | 944 |
| Lin7b | Lin-7 homolog b (C. elegans) | 488 | 172 | 620 | 659 | 1,553 | 763 |
| Slc6a9 | Solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | 453 | 984 | 2,036 | 818 | 7,792 | 1,447 |
| Drd4 | Dopamine receptor D4 | 439 | 582 | 273 | 230 | 490 | 425 |
| Htr3a | 5-hydroxytryptamine (serotonin) receptor 3a | 388 | 83 | 304 | 184 | 1,096 | 181 |
| Nos1ap | Nitric oxide synthase 1 (neuronal) adaptor protein | 330 | 1,668 | 296 | 1,216 | 330 | 1,710 |
| Gabrg3 | Gamma-aminobutyric acid (GABA) A receptor, subunit gamma 3 | 230 | 79 | 227 | 123 | 320 | 62 |
| Grin2a | Glutamate receptor, ionotropic, N-methyl D-aspartate 2A | 210 | 107 | 142 | 41 | 192 | 97 |
Changes in amacrine cell gene expression could be due in part to a change in the subpopulations of amacrine cells purified by immunopanning at embryonic and postnatal ages, although we have yields of well over 50%, suggesting that we are purifying a broad swath of amacrine cell subtypes at all these ages. To further validate our dataset, we compared our E20 amacrine cell data against recently published P0 amacrine cell and RGC single-cell expression profiling.24,25 Consistent with their results, we found that amacrine cells (and not RGCs) expressed high levels of the transcription factor AP-2β (TCFAP-2β) and that all the samples of amacrine cells expressed high levels of neuronal leucine-rich repeat protein-3 (Lrrn3), two recently validated amacrine cell markers.24 We also observed amacrine cell expression of probe sets for gamma synuclein (Sncg), early B-cell factor 3 (Ebf3), and neurofilament 68 (Nefl), genes previously described to be highly enriched in RGCs.35–37 These discrepancies may be explained by several experimental differences, including posttranscriptional control of expression (not explored here), neuronal age and species (E20 rat versus P0 mouse), starting RNA material (millions of purified amacrine cells including different subtypes pooled together versus single cells analyzed separately), potential amplification artifacts (less amplification is necessary for pooled cells), and potential for contamination or cellular misidentification in either dataset.
Comparison of Amacrine Cell and RGC Gene Expression through Development
We next compared our gene expression datasets of amacrine cells and RGCs.14 We identified 14,457 probes shared between the different microarray platforms of the two datasets—more than half of the total data. The manipulation necessary for this comparison across different platforms (see the Methods section) may have decreased the robustness of these analyses, but the large number of shared, cross-platform probes would be expected to compensate for this limitation. Of the 14,457 probes shared between the two array types (see the Methods section; Fig. 5A), 8575 (59%) were “present” by Affymetrix algorithm in at least two samples of one age in both gene chip datasets; 2640 (18%) were present only in amacrine cells, and 432 (3%) were expressed only in RGCs. The remaining 2910 (20%) were absent from both cell types. A breakdown of these numbers at E20, P5, and P11 is shown in Figure 5A. Of the 3072 probes expressed at any age either by amacrine cells or by RGCs but not by both cell types, amacrine cells expressed more unique genes than RGCs at all ages (Fig. 5A) and across all 35 gene ontologies analyzed. We found 25 neurotransmitter-associated genes that were exclusively expressed by amacrine cells at all ages but not by RGCs (Table 2) and only two specific to RGCs. Occasionally, probes had higher absolute expression levels in RGCs than in amacrine cells, but the Affymetrix algorithm called them “absent,” as commonly occurs when in analysis of probes with low absolute levels of expression. The higher number of neurotransmitter-associated probes expressed exclusively by amacrine cells and not RGCs is consistent with the transmitter variety described in amacrine cells as a whole.2,38–40
We next compared the gene expression levels for the probes expressed by both RGCs and amacrine cells at different ages (Fig. 5A). Since the absolute gene expression levels for amacrine cells and for RGCs are on different, arbitrary scales, we converted absolute gene expression levels to ranked gene expression levels for each cell type, assigning the gene with the highest amacrine cell expression at a given age a rank of 1, and continuing down to a rank of 10,525 for E20, 10,574 for P5, and 10,502 for P11 (the number of genes present at each of those ages) and repeated the process for the RGC genes. Since RGCs expressed fewer genes at every age than did the amacrine cells, the RGC rank list was scaled to 10,525 for E20, and so on, to match the length of the amacrine cell gene list. The genes “present” only in amacrine cells were given terminal rankings (e.g., 10,526) in RGCs, and vice versa, at each age. We then compared the ranked expression for each gene between the amacrine cells and the RGCs, to create a simple index. The average of the differences between rankings of amacrine cells and RGCs compared at E20, P5, and P11 of all the probes within a given gene ontology is shown in Figure 5B, where the scale shows genes ontologies with genes that are more highly expressed in RGCs at one end and more highly expressed in amacrine cells at the other end. For example, RGCs at all ages expressed higher levels of genes within the neurotransmi- and immune ontologies (Fig. 5B, top; Table 2). This difference could be because amacrine cells secrete a variety of neurotransmitters and RGCs only a few but at higher levels, thus demonstrating greater differences in gene expression rankings.2,38–40 On the bottom of Figure 5B, amacrine cells expressed higher levels of genes in the categories chromatin, DNA, and transcription. The index identified differences between amacrine cells and RGCs that were largely consistent across E20, P5, and P11 ages in most ontologies, except for a few ontologies such as polarity, neurit-, chromosome, and neurotransmi- which demonstrated greater developmental variability across this age range between these two neuron types.
Comparing the expression profile of more than 50 polarity-associated genes, we found that some are differentially regulated during amacrine cell development or differ between amacrine cells and RGCs (Table 3, Figs. 6B, 6D, 6F, 6H). For example, microtubule affinity-regulating kinase 2 (Mark2), which negatively regulates dendrite development in cultured hippocampal neurons,41 was expressed at moderate levels by amacrine cells but was not detected in RGCs. Conversely, signal transducer and activator of transcription 3 (Stat3), which when activated can promote neurite outgrowth in primary sensory neurons,42 was expressed fivefold more in RGCs than in amacrine cells (Fig. 6B). Consistent with previous findings,43 RGCs were immunopositive for Stat3, but we found that amacrine cell immunostaining for Stat3 was barely detectable above background (Fig. 6A), consistent with the microarray data (Fig. 6B).
Table 3.
Polarity-Associated Genes in Amacrine Cells and RGCs
| Amacrine Cells Probes | RGCs Probes | Gene Symbol | Gene Name | Change Ratio Amacr/RGCs | Present (P) Algorithm |
|---|---|---|---|---|---|
| 1368710_at | Z83869CDS_at | Mark2 | MAP/microtubule affinity-regulating kinase 2 | 42.04 | P in amacr only |
| 1388657_at | rc_AI060199_at | Apc | Adenomatosis polyposis coli | 12.62 | P in both cell types |
| 1392916_at | rc_AI144722_at | Kif2c | Kinesin family member 2C | 12.21 | P in amacr only |
| 1374912_at | rc_AI060200_at | Mtap7 | Microtubule-associated protein 7 | 9.83 | P in both cell types |
| 1385636_at | rc_AI029226_at | Lcp1 | Lymphocyte cytosolic protein 1 | 5.83 | P in both cell types |
| 1392926_at | rc_AA875154_at | Hes1 | Hairy and enhancer of split 1 (Drosophila) | 4.98 | P in both cell types |
| 1387837_at | D38629_at | Ezr | Ezrin | 4.63 | P in both cell types |
| 1387036_at | D13417_G_at | — | — | 4.02 | P in both cell types |
| 1392667_at | rc_AI070793_at | Fat1 | FAT tumor suppressor homolog 1 (Drosophila) | 3.43 | P in both cell types |
| 1389210_at | rc_AI012958_at | Pten | Phosphatase and tensin homolog | 3.25 | P in both cell types |
| 1368053_at | AB005549_at | Lims1 | LIM and senescent cell antigen-like domains 1 | 2.70 | P in amacr only |
| 1377753_at | rc_AA894088_at | Clasp1 | Cytoplasmic linker associated protein 1 | 2.59 | P in both cell types |
| 1374156_at | rc_AI031043_at | Lama1 | Laminin, alpha 1 | 2.58 | P in amacr only |
| 1371326_at | rc_AI169125_at | Mpp5 | Membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) | 2.45 | P in both cell types |
| 1370875_at | X67788_at | Csnk1a1 | Casein kinase-1, alpha-1 | 2.37 | P in both cell types |
| 1370112_at | rc_AA963447_at | Vangl2 | Vang-like 2 (van gogh, Drosophila) | 2.28 | P in both cell types |
| 1374486_at | rc_AA901314_at | Cfl1 | cofilin 1, non-muscle | 2.22 | P in both cell types |
| 1370234_at | X05834_at | Csnk2a2 | Casein kinase 2, alpha prime polypeptide | 2.17 | P in both cell types |
| 1370832_at | U06434_at | Cab39 | Calcium binding protein 39 | 1.97 | P in both cell types |
| 1368123_at | L29232_at | Gsk3b | Glycogen synthase kinase 3 beta | 1.88 | P in both cell types |
| 1373537_at | rc_AI072819_at | Igf1r | Insulin-like growth factor 1 receptor | 1.86 | P in both cell types |
| 1370267_at | X73653_at | Fzd3 | Frizzied homolog 3 (Drosophila) | 1.85 | P in amacr only |
| 1388816_at | rc_AI144591_at | RGD1309453 | Similar to hypothetical protein FLJ32884 | 1.83 | P in both cell types |
| 1375935_at | rc_AI230547_at | Scd2 | Stearoyl-Coenzyme A desaturase 2 | 1.80 | P in both cell types |
| 1367654_at | rc_AI103370_at | Ilk | Integrin linked kinase | 1.76 | P in both cell types |
| 1372244_at | rc_AA849965_at | Cdc42 | Cell division cycle 42 homolog (S. cerevisiae) | 1.73 | P in both cell types |
| 1399105_at | rc_AA928279_at | Pard3 | Par-3 (partitioning defective 3) homolog (C. elegans) | 1.61 | P in both cell types |
| 1375994_at | rc_AA946326_at | Fn1 | Fibronectin 1 | 1.39 | P in both cell types |
| 1371429_at | rc_AI170693_at | Prkci | Protein kinase C, iota | 1.27 | P in both cell types |
| 1370197_a_at | M18332_S_at | Clasp2 | CLIP associating protein 2 | 1.23 | P in both cell types |
| 1369149_at | D31873_G_at | Ctnna1 | Catenin (cadherin associated protein), alpha 1 | 1.16 | P in both cell types |
| 1390156_a_at | rc_AA800882_G_at | Llgl1 | Lethal giant larvae homolog 1 (Drosophila) | 1.10 | P in both cell types |
| 1381526_at | rc_AI044894_at | Prkcz | Protein kinase C, zeta | 1.07 | P in both cell types |
| 1369997_at | rc_AI101690_at | Dag1 | Dystroglycan 1 | 1.03 | P in both cell types |
| 1375685_at | rc_AA850743_at | Dchs1 | Dachsous 1 (Drosophila) | 1.01 | P in both cell types |
| 1387777_at | rc_AI176814_at | Cfl1 | Cofilin 1, non-muscle | −1.10 | P in both cell types |
| 1370287_a_at | M60666_S_at | Dlgn1 | Discs, large homolog 1 (Drosophila) | −1.13 | P in both cell types |
| 1371725_at | rc_AA944803_at | Prickle2 | Prickle-like 2 (Drosophila) | −1.13 | P in both cell types |
| 1379369_at | rc_AI171526_at | Nckap1 | NCK-associated protein 1 | −1.18 | P in both cell types |
| 1387871_at | rc_AI235500_at | Cap1 | CAP, adenytate cyclase-associated protein 1 (yeast) | −1.22 | P in both cell types |
| 1371921_at | rc_AI177170_at | Tpm1 | Tropomyosin 1, alpha | −1.28 | P in both cell types |
| 1374646_at | rc_AA955819_G_at | Pard6a | Par-6 (partitioning detective 6,) homolog alpha (C. elegans) | −1.34 | P in both cell types |
| 1373047_at | rc_AI179206_at | Bin3 | Bridging integrator 3 | −1.54 | P in both cell types |
| 1368099_at | rc_AI175481_at | Pak1 | P21 (CDKN1A)-activated kinase 1 | −1.85 | P in both cell types |
| 1368809_at | rc_AA850040_at | Dvl1 | Dishevelled, dsh homolog 1 (Drosophila) | −1.86 | P in both cell types |
| 1389093_at | rc_AI175989_at | — | — | −2.30 | P in both cell types |
| 1367668_a_at | rc_AA875269_at | Myh9 | Myosin, heavy polypeptide 9, non-muscle | −2.37 | P in both cell types |
| 1370825_a_at | rc_AA925473_G_art | — | — | −2.80 | P in both cell types |
| 1370184_at | rc_AI177598_F_at | Prickle1 | Prickle-like 1 (Drosophila) | −2.82 | P in both cell types |
| 1370224_at | rc_AI008865_S_at | Limk1 | LIM domain kinase 1 | −3.26 | P in both cell types |
| 1371407_at | rc_AI105087_at | Ccl4 | Chemokine (C-C motif) ligand 4 | −3.38 | P in both cell types |
| 1385430_at | rc_AA799637_at | Stat3 | Signal transducer and activator of transcription 3 | −5.33 | P in both cell types |
We next investigated the expression and localization of a few other polarity proteins between the two cell types during neurite growth in vitro. All immunostaining performed trended in the same direction as the microarray data, adding confidence to the data's reliability. We did not detect any differences in the expression or localization of some proteins, including Par6 (Fig. 6C). Others, however, were differentially expressed or localized. For example, atypical protein kinase C (aPKC), which is a part of the Par3/Par6/aPKC complex necessary to establish cell polarity in mammalian epithelial cells,44 and whose activity is necessary for neurite polarization and axon formation in hippocampal neurons,45 was localized in the cytoplasm and along initial neurite segments in RGCs, but it was only present in the cytoplasm of amacrine cells (Fig. 6E). Limk1, a kinase that regulates actin cytoskeleton dynamics46,47 and controls growth cone motility,48 was highly localized to the growth cones in RGCs and also present in the lamellipodia of amacrine cells (Fig. 6G). Of interest, although in RGCs Limk1 has a ubiquitous distribution in the cell body; it is clearly excluded from the nucleus in amacrine cells, which may suggest a difference in its biological functioning between these two cell types.49 Although the functional role of these expressed polarity genes in amacrine cells remains to be tested, taken together, these data suggest that amacrine cells express many of the genes important in neurite differentiation (that is, axon versus dendrite differentiation), and that differential protein localization may explain the differences in axon/dendrite polarization in these cell types.
Amacrine Cell Survival Signaling Pathways
It is a general tenet in neuroscience that neurons are dependent on their targets for survival, both during development and in the adult.10 In vivo in optic neuropathies, however, amacrine cells appear to resist degeneration after RGC death.8 Although the molecular signals sufficient to promote RGC survival have been extensively studied and characterized,9,11,50–55 little is known about the cues that regulate amacrine cell survival. We analyzed survival-related genes with gene ontologies containing apoptosis, MAPK, PI3K, and a few hand-picked additional genes. We identified the subset of these genes that differed between amacrine cells and RGCs. Of 112 probes found in both datasets, 84 (75%) were “present” in both the amacrine cells and RGCs, 19 (17%) probes were “present” only in amacrine cells and 7 (7%) probes were unique to RGCs (Supplementary Table S3.) Several interesting candidate genes differed, including ciliary neurotrophic factor receptor (Cntfr) and huntingtin (Htt), which were detected in amacrine cells but not in RGCs. Htt may play a role in survival signaling by enhancing BDNF axonal transport,56 and is one of the binding partners of optineurin,57 a protein with mutations that have been associated with primary open-angle glaucoma.58 Although optineurin expression in RGCs increases during development,14,59,60 it would be interesting to explore whether the deficiency of its binding partner Htt may underlie the susceptibility of RGCs to neurodegeneration. Among the many genes expressed by both cell types were most of the bcl-2 family. However, RGCs expressed higher levels of the proapoptotic molecules Bax and Bad, raising the hypothesis that RGCs are more susceptible to death than are amacrine cells, because they have a balance in bcl-2 family members that is shifted toward apoptosis.14
Do the same signaling pathways mediate the survival of RGCs and amacrine cells? To address this question, we purified primary amacrine cells from P8 to P9 rats, cultured them in a variety of defined growth media without serum for up to 3 days, and quantified their survival by using a live/dead assay (see Methods). First, we found that amacrine cell survival was not enhanced by increasing cell density (Fig. 7A), in contrast to RGC survival,9 suggesting that amacrine cells may not secrete factors into the media that enhance their own survival in a paracrine fashion. Exogenous addition of brain-derived neurotrophic factor (BDNF) and ciliary neurotrophic factor (CNTF), two of the trophic peptides that most strongly support RGC survival,9,61 had no effect on amacrine cell survival (Fig. 7B). Indeed, at cell culture densities that are most likely too low for amacrine cells to condition the medium with paracrine signaling molecules, amacrine cell survival was remarkably high, even in the absence of peptide trophic factors. There was a modest effect of adding a modified B27 supplement containing hormones and antioxidants,18 particularly in the absence of other peptide trophic factors (Fig. 7B). This high, constitutive survival was observed solely from the constituents of the Sato supplement,62 which includes putrescine, transferrin, progesterone, selenite, and albumin, plus pyruvate, glutamine, thyroid hormone, and n-acetyl cysteine (Fig. 7B). These data suggest that amacrine cells depend more on hormone and antioxidant signals than on peptide trophic factors for their survival.
To explore which intracellular signaling pathways mediate the observed survival in vitro, we cultured amacrine cells in full Sato (FS) medium containing pharmacologic inhibitors for a subset of known survival signaling pathways. When amacrine cells were treated with the MEK1/2 specific inhibitor U0126, there was a threefold decrease in survival (Fig. 7C). PI3K and JAK/STAT inhibitors (LY294002 and AG490, respectively) also decreased amacrine cell survival in vitro (Fig. 7C). Although amacrine cells expressed TrkA, -B, and -C by microarray analysis, addition of the Trk inhibitor K252a did not affect survival, consistent with the modest effect of exogenous BDNF on survival in these experiments (Figs. 7B, 7C), suggesting that autocrine Trk signaling is not responsible for amacrine cell survival. We confirmed the specificity of the U0126, K252a, and LY294002 inhibitors by Western blot with amacrine cells acutely purified and allowed to recover for 2 hours at 37°C without peptide trophic factors in the presence of the inhibitors (Figs. 7D, 7E). Again, addition of K252a did not affect ERK1/2 signaling compared with the control, consistent with its minimal effect on survival. The addition of either U0126 or LY294002 inhibited the activation of ERK1/2 and AKT, respectively, and impaired amacrine cell survival (Figs. 7D, 7E). Taken together, these results show that amacrine cells require MAPK, PI3K, and JAK/STAT activation for survival, but we do not know what upstream signals are responsible for activating these pathways.
Discussion
Amacrine Cells' Transcriptome Changes during Early Retinal Development
We and others have previously reported on the transcriptomes of identified cell populations in the CNS, including RGCs, striatal and corticospinal motor neurons,63,64 and genetic labeling techniques have considerably expanded the scope of CNS neurons amenable to such isolation and analysis, either by cell-sorting fluorescent cells65 or by directly purifying translated RNAs from neuronal subpopulations that express RNA binding proteins.66 We highly purified amacrine cells by immunopanning and provide a database for the transcriptome of amacrine cells at three developmental ages (E20, P5 and P11). Previous datasets have been generated to study developmental changes in the gene expression of neighboring neurons,63,64 and this database, together with the RGC transcriptome,14 yields a comprehensive description at a molecular level of two synaptically closely related CNS neurons. In addition, these data add significantly to the snapshot of single-cell gene expression profiling performed on 32 amacrine cells mostly from P0 and P5.25 With at least 25 to 30 different morphologic subtypes of amacrine cells previously described,67,68 a population-level profile of amacrine cell gene expression certainly adds value to the single cell data currently available. These data may provide opportunities in the future for better understanding retinal development, wiring, and degeneration, and ultimately CNS neurologic diseases.
Patterns of Gene Expression in Amacrine Cells and RGCs
Two cells that come from a common progenitor, migrate (in part) to the same retinal layer, and synaptically integrate in the same retinal layer, may be expected to share common gene expression. Indeed, we found that most genes (>74%) were expressed at similar levels in RGCs and amacrine cells at embryonic and postnatal ages. With an interest in pursuing the observed differences in the cell biology of amacrine cells and RGCs, however, we focused mainly on their differences in gene expression.
We found that RGCs more highly expressed genes associated with the terms immune and neurotransmitter/mission and metabolic pathways, such as Nadh/Nadph. The role of molecules of the immune system in the CNS was first thought to be limited to inflammation, injury, and autoimmune disorders. Recent findings, in particular in the visual system, have shown that major histocompatibility complex (MHC) class I molecules and proteins of the complement cascade, such as C1q and C3, have an unexpected role in CNS development and plasticity.69–71 Our data provide a significant list of immune system genes expressed by both amacrine cells and RGCs, to help build hypotheses for the molecular pathway of synapse formation and elimination during development and after injury.
Similarly, the high expression levels of genes associated with ontologies such as DNA, chromatin, and transcription by amacrine cells compared with RGCs could underlie amacrine cell heterogeneity or plasticity during retinal development. With a more dynamic chromatin remodeling capacity, amacrine cells may express more varied sets of transcription factors or other downstream targets at different times, which could contribute to their cellular diversity.
Amacrine Cells' Resistance to Neurodegeneration
Both CNS and PNS neurons are strongly dependent on target-derived peptide trophic signals for their survival, and competition for target-derived signals is intimately related to neuronal survival and synapse formation during development.72,73 Most of the data for these observations have been derived from studies of long-projection neurons, and it is not known whether the same principles apply to locally integrated interneurons. In optic neuropathies such as glaucoma, when most of the RGCs die, amacrine cells are largely not affected, despite the loss of their major synaptic target.8,74 Furthermore, in rat optic nerve transection experiments in which >90% of the RGCs die within 2 to 4 weeks, amacrine cells continue to survive and express calbindin, calretinin, and GABAergic cell markers after 3 months.8 Similarly, GABAergic amacrine cell subpopulations are resistant to retinal ischemia in a rat ischemia–reperfusion model.75
Results of a study have suggested that amacrine cells require high concentrations of insulin for survival,76 or that antagonizing TrkB receptor signaling in retinal explants decreases the number of nuclei in the inner nuclear layer77 and, in vivo, reduces the number of parvalbumin-positive amacrine cells in the retina.78 Interpretation of such experiments may be confounded by a decrease in the expression of phenotypic cell markers without cell death.78,79 We used our ability to highly purify and culture amacrine cells in defined, serum-free medium to ask whether the same trophic factors that strongly promote RGC survival9 similarly promote amacrine cell survival in vitro. We found that peptide trophic factors did not significantly contribute to amacrine cell survival in vitro, even when amacrine cells were cultured at clonal density, despite expression of trkB, CNTF-Ra, LIF-R, and IGF-R1 detected on the microarrays. Blocking of MEK1/2 or PI3K signaling pathways significantly impaired survival, suggesting that these intracellular signaling pathways are necessary for amacrine cell survival and raising the question of how these pathways are being activated. The Western blot experiments in Figure 5 suggest that without the addition of exogenous peptide trophic factors, there is a basal activity of MEK1/2 and PI3K that is further increased when cells are grown in Sato supplement–containing medium.
Taken together, these data demonstrate that although RGC and amacrine cell survival are mediated through the same intracellular signaling pathways,11 their requirement for exogenous peptide trophic factors differs. Survival of amacrine cells in our low-density cultures in the absence of exogenous peptide trophic factors suggests potential regulation by autocrine signaling80 or by hormone or antioxidant support alone. Thus, these retinal interneurons may not depend on target RGCs for peptide trophic support.
Involvement of Amacrine Cell Properties in the CNS for Repair and Regeneration
To overcome the failure of adult mammalian CNS regeneration, neurons must survive and reextend their axons to their targets. It is intriguing to consider whether amacrine cells, which exhibit some survival independence from peptide trophic factors and which survive degenerative diseases affecting RGCs, may represent a residual retinal population of neurons that could be induced to replace the visual functions of lost RGCs. Understanding the molecular components that could allow amacrine cells to extend and differentiate axons may allow these cells to serve as a local source of RGC replacement in designing therapies targeted to cure blindness.
Supplementary Material
Acknowledgments
The authors thank Colin Barnstable for generously providing the Vc1.1 antibody; Sawsan Khuri, Camilo Valdes, and Willie Buchser for assistance with data analysis and presentation; and Yan Shi, Travis Rice-Stitt, and Raul Corredor for technical assistance.
Footnotes
Supported by National Institute for Neurological Disorders and Stroke (NINDS) Grant NS061348 (JLG); NINDS T32 Training Grant NS007044 (MVA); National Eye Institute Grant P30 EY014801; The Glaucoma Foundation (JLG); and an unrestricted grant from Research to Prevent Blindness to the University of Miami.
Disclosure: N.J. Kunzevitzky, None; M.V. Almeida, None; J.L. Goldberg, None
References
- 1.Nelson R, Kolb H, Robinson MM, Mariani AP. Neural circuitry of the cat retina: cone pathways to ganglion cells. Vision Res 1981;21:1527–1536 [DOI] [PubMed] [Google Scholar]
- 2.Masland RH. Neuronal diversity in the retina. Curr Opin Neurobiol 2001;11:431–436 [DOI] [PubMed] [Google Scholar]
- 3.Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci 1998;18:8936–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolb H, Nelson R. Amacrine cells of the cat retina. Vision Res 1981;21:1625–1633 [DOI] [PubMed] [Google Scholar]
- 5.Kolb H, Nelson R, Mariani A. Amacrine cells, bipolar cells and ganglion cells of the cat retina: a Golgi study. Vision Res 1981;21:1081–1114 [DOI] [PubMed] [Google Scholar]
- 6.Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol 2004;474:304–324 [DOI] [PubMed] [Google Scholar]
- 7.Lee MY, Shin SL, Han SH, Chun MH. The birthdates of GABA-immunoreactive amacrine cells in the rat retina. Exp Brain Res 1999;128:309–314 [DOI] [PubMed] [Google Scholar]
- 8.Kielczewski JL, Pease ME, Quigley HA. The effect of experimental glaucoma and optic nerve transection on amacrine cells in the rat retina. Invest Ophthalmol Vis Sci 2005;46:3188–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer-Franke A, Kaplan MR, Pfrieger FW, Barres BA. Characterization of the signaling interactions that promote the survival and growth of developing retinal ganglion cells in culture. Neuron 1995;15:805–819 [DOI] [PubMed] [Google Scholar]
- 10.Goldberg JL, Barres BA. The relationship between neuronal survival and regeneration. Annu Rev Neurosci 2000;23:579–612 [DOI] [PubMed] [Google Scholar]
- 11.Goldberg JL, Espinosa JS, Xu Y, Davidson N, Kovacs GT, Barres BA. Retinal ganglion cells do not extend axons by default: promotion by neurotrophic signaling and electrical activity. Neuron 2002;33:689–702 [DOI] [PubMed] [Google Scholar]
- 12.Goldberg JL, Klassen MP, Hua Y, Barres BA. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science 2002;296:1860–1864 [DOI] [PubMed] [Google Scholar]
- 13.Arimatsu Y, Naegele JR, Barnstable CJ. Molecular markers of neuronal subpopulations in layers 4, 5, and 6 of cat primary visual cortex. J Neurosci 1987;7:1250–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang JT, Kunzevitzky NJ, Dugas JC, Cameron M, Barres BA, Goldberg JL. Disease gene candidates revealed by expression profiling of retinal ganglion cell development. J Neurosci 2007;27:8593–8603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang KB, Kong SW, Greenberg SA, Park PJ. Combining gene expression data from different generations of oligonucleotide arrays. BMC Bioinformatics 2004;5:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo WP, Jenssen TK, Butte AJ, Ohno-Machado L, Kohane IS. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics 2002;18:405–412 [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Stevens B, Chang J, Milbrandt J, Barres BA, Hell JW. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods 2008;171:239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnstable CJ, Hofstein R, Akagawa K. A marker of early amacrine cell development in rat retina. Brain Res 1985;352:286–290 [DOI] [PubMed] [Google Scholar]
- 20.Young RW. Cell differentiation in the retina of the mouse. Anat Rec 1985;212:199–205 [DOI] [PubMed] [Google Scholar]
- 21.Bennett MK, Calakos N, Scheller RH. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 1992;257:255–259 [DOI] [PubMed] [Google Scholar]
- 22.Tagaya M, Toyonaga S, Takahashi M, et al. Syntaxin 1 (HPC-1) is associated with chromaffin granules. J Biol Chem 1995;270:15930–15933 [DOI] [PubMed] [Google Scholar]
- 23.Liu IS, Chen JD, Ploder L, et al. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron 1994;13:377–393 [DOI] [PubMed] [Google Scholar]
- 24.Trimarchi JM, Stadler MB, Roska B, et al. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J Comp Neurol 2007;502:1047–1065 [DOI] [PubMed] [Google Scholar]
- 25.Cherry TJ, Trimarchi JM, Stadler MB, Cepko CL. Development and diversification of retinal amacrine interneurons at single cell resolution. Proc Natl Acad Sci U S A 2009;106:9495–9500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson J, Tian N, Caywood MS, Reimer RJ, Edwards RH, Copenhagen DR. Vesicular neurotransmitter transporter expression in developing postnatal rodent retina: GABA and glycine precede glutamate. J Neurosci 2003;23:518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voinescu PE, Kay JN, Sanes JR. Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J Comp Neurol 2009;517:737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redburn DA, Rowe-Rendleman C. Developmental neuro-transmitters: signals for shaping neuronal circuitry. Invest Ophthalmol Vis Sci 1996;37:1479–1482 [PubMed] [Google Scholar]
- 30.Casini G, Brecha NC. Postnatal development of tyrosine hydroxylase immunoreactive amacrine cells in the rabbit retina: I. Morphological characterization. J Comp Neurol 1992;326:283–301 [DOI] [PubMed] [Google Scholar]
- 31.Kay JN, Roeser T, Mumm JS, et al. Transient requirement for ganglion cells during assembly of retinal synaptic layers. Development 2004;131:1331–1342 [DOI] [PubMed] [Google Scholar]
- 32.Prada C, Puelles L, Genis-Galvez JM, Ramirez G. Two modes of free migration of amacrine cell neuroblasts in the chick retina. Anat Embryol (Berl) 1987;175:281–287 [DOI] [PubMed] [Google Scholar]
- 33.Reese BE, Colello RJ. Neurogenesis in the retinal ganglion cell layer of the rat. Neuroscience 1992;46:419–429 [DOI] [PubMed] [Google Scholar]
- 34.Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A 1996;93:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mu X, Beremand PD, Zhao S, et al. Discrete gene sets depend on POU domain transcription factor Brn3b/Brn-3.2/POU4f2 for their expression in the mouse embryonic retina. Development 2004;131:1197–1210 [DOI] [PubMed] [Google Scholar]
- 36.Surgucheva I, Weisman AD, Goldberg JL, Shnyra A, Surguchov A. Gamma-synuclein as a marker of retinal ganglion cells. Mol Vis 2008;14:1540–1548 [PMC free article] [PubMed] [Google Scholar]
- 37.Mu X, Fu X, Sun H, Beremand PD, Thomas TL, Klein WH. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev Biol 2005;280:467–481 [DOI] [PubMed] [Google Scholar]
- 38.Massey SC, Redburn DA. Transmitter circuits in the vertebrate retina. Prog Neurobiol 1987;28:55–96 [DOI] [PubMed] [Google Scholar]
- 39.Kolb H. Amacrine cells of the mammalian retina: neurocircuitry and functional roles. Eye 1997;11:904–923 [DOI] [PubMed] [Google Scholar]
- 40.Wassle H. Parallel processing in the mammalian retina. Nat Rev Neurosci 2004;5:747–757 [DOI] [PubMed] [Google Scholar]
- 41.Terabayashi T, Itoh TJ, Yamaguchi H, et al. Polarity-regulating kinase partitioning-defective 1/microtubule affinity-regulating kinase 2 negatively regulates development of dendrites on hippocampal neurons. J Neurosci 2007;27:13098–13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miao T, Wu D, Zhang Y, et al. Suppressor of cytokine signaling-3 suppresses the ability of activated signal transducer and activator of transcription-3 to stimulate neurite growth in rat primary sensory neurons. J Neurosci 2006;26:9512–9519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller A, Hauk TG, Fischer D. Astrocyte-derived CNTF switches mature RGCs to a regenerative state following inflammatory stimulation. Brain 2007;130:3308–3320 [DOI] [PubMed] [Google Scholar]
- 44.Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K. Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol 2004;6:328–334 [DOI] [PubMed] [Google Scholar]
- 45.Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 2003;112:63–75 [DOI] [PubMed] [Google Scholar]
- 46.Matsui S, Matsumoto S, Adachi R, et al. LIM kinase 1 modulates opsonized zymosan-triggered activation of macrophage-like U937 cells: possible involvement of phosphorylation of cofilin and reorganization of actin cytoskeleton. J Biol Chem 2002;277:544–549 [DOI] [PubMed] [Google Scholar]
- 47.Rosso S, Bollati F, Bisbal M, et al. LIMK1 regulates Golgi dynamics, traffic of Golgi-derived vesicles, and process extension in primary cultured neurons. Mol Biol Cell 2004;15:3433–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Endo M, Ohashi K, Sasaki Y, et al. Control of growth cone motility and morphology by LIM kinase and Slingshot via phosphorylation and dephosphorylation of cofilin. J Neurosci 2003;23:2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang N, Mizuno K. Nuclear export of LIM-kinase 1, mediated by two leucine-rich nuclear-export signals within the PDZ domain. Biochem J 1999;338:793–798 [PMC free article] [PubMed] [Google Scholar]
- 50.Mansour-Robaey S, Clarke DB, Wang YC, Bray GM, Aguayo AJ. Effects of ocular injury and administration of brain-derived neurotrophic factor on survival and regrowth of axotomized retinal ganglion cells. Proc Natl Acad Sci U S A 1994;91:1632–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mey J, Thanos S. Intravitreal injections of neurotrophic factors support the survival of axotomized retinal ganglion cells in adult rats in vivo. Brain Res 1993;602:304–317 [DOI] [PubMed] [Google Scholar]
- 52.Klocker N, Braunling F, Isenmann S, Bahr M. In vivo neurotrophic effects of GDNF on axotomized retinal ganglion cells. Neuroreport 1997;8:3439–3442 [DOI] [PubMed] [Google Scholar]
- 53.Schmeer C, Straten G, Kugler S, Gravel C, Bahr M, Isenmann S. Dose-dependent rescue of axotomized rat retinal ganglion cells by adenovirus-mediated expression of glial cell-line derived neurotrophic factor in vivo. Eur J Neurosci 2002;15:637–643 [DOI] [PubMed] [Google Scholar]
- 54.Peinado-Ramon P, Salvador M, Villegas-Perez MP, Vidal-Sanz M. Effects of axotomy and intraocular administration of NT-4, NT-3, and brain-derived neurotrophic factor on the survival of adult rat retinal ganglion cells. A quantitative in vivo study. Invest Ophthalmol Vis Sci 1996;37:489–500 [PubMed] [Google Scholar]
- 55.Cui Q, Lu Q, So KF, Yip HK. CNTF, not other trophic factors, promotes axonal regeneration of axotomized retinal ganglion cells in adult hamsters. Invest Ophthalmol Vis Sci 1999;40:760–766 [PubMed] [Google Scholar]
- 56.Gauthier LR, Charrin BC, Borrell-Pages M, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 2004;118:127–138 [DOI] [PubMed] [Google Scholar]
- 57.Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet 1998;7:1463–1474 [DOI] [PubMed] [Google Scholar]
- 58.Rezaie T, Child A, Hitchings R, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science 2002;295:1077–1079 [DOI] [PubMed] [Google Scholar]
- 59.Rezaie T, Sarfarazi M. Molecular cloning, genomic structure, and protein characterization of mouse optineurin. Genomics 2005;85:131–138 [DOI] [PubMed] [Google Scholar]
- 60.De Marco N, Buono M, Troise F, Diez-Roux G. Optineurin increases cell survival and translocates to the nucleus in a Rab8-dependent manner upon an apoptotic stimulus. J Biol Chem 2006;281:16147–16156 [DOI] [PubMed] [Google Scholar]
- 61.Jo SA, Wang E, Benowitz LI. Ciliary neurotrophic factor is and axogenesis factor for retinal ganglion cells. Neuroscience 1999;89:579–591 [DOI] [PubMed] [Google Scholar]
- 62.Bottenstein JE, Sato GH. Growth of a rat neuroblastoma cell line in serum-free supplemented medium. Proc Natl Acad Sci U S A 1979;76:514–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci 2006;9:443–452 [DOI] [PubMed] [Google Scholar]
- 64.Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 2005;45:207–221 [DOI] [PubMed] [Google Scholar]
- 65.Sugino K, Hempel CM, Miller MN, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci 2006;9:99–107 [DOI] [PubMed] [Google Scholar]
- 66.Heiman M, Schaefer A, Gong S, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 2008;135:738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.MacNeil MA, Heussy JK, Dacheux RF, Raviola E, Masland RH. The shapes and numbers of amacrine cells: matching of photofilled with Golgi-stained cells in the rabbit retina and comparison with other mammalian species. J Comp Neurol 1999;413:305–326 [PubMed] [Google Scholar]
- 68.MacNeil MA, Masland RH. Extreme diversity among amacrine cells: implications for function. Neuron 1998;20:971–982 [DOI] [PubMed] [Google Scholar]
- 69.Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Functional requirement for class I MHC in CNS development and plasticity. Science 2000;290:2155–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stevens B, Allen NJ, Vazquez LE, et al. The classical complement cascade mediates CNS synapse elimination. Cell 2007;131:1164–1178 [DOI] [PubMed] [Google Scholar]
- 71.Goddard CA, Butts DA, Shatz CJ. Regulation of CNS synapses by neuronal MHC class I. Proc Natl Acad Sci U S A 2007;104:6828–6833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oppenheim RW. Cell death during development of the nervous system. Annu Rev Neurosci 1991;14:453–501 [DOI] [PubMed] [Google Scholar]
- 73.Purves D. Body and Brain: A Trophic Theory of Neural Connections Cambridge, MA: Harvard University Press; 1988:231. [DOI] [PubMed] [Google Scholar]
- 74.Jakobs TC, Libby RT, Ben Y, John SW, Masland RH. Retinal ganglion cell degeneration is topological but not cell type specific in DBA/2J mice. J Cell Biol 2005;171:313–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dijk F, van Leeuwen S, Kamphuis W. Differential effects of ischemia/reperfusion on amacrine cell subtype-specific transcript levels in the rat retina. Brain Res 2004;1026:194–204 [DOI] [PubMed] [Google Scholar]
- 76.Politi LE, Rotstein NP, Salvador G, Giusto NM, Insua MF. Insulin-like growth factor I is a potential trophic factor for amacrine cells. J Neurochem 2001;76:1199–1211 [DOI] [PubMed] [Google Scholar]
- 77.Cusato K, Bosco A, Linden R, Reese BE. Cell death in the inner nuclear layer of the retina is modulated by BDNF. Brain Res Dev Brain Res 2002;139:325–330 [DOI] [PubMed] [Google Scholar]
- 78.Rickman DW, Bowes Rickman C. Suppression of trkB expression by antisense oligonucleotides alters a neuronal phenotype in the rod pathway of the developing rat retina. Proc Natl Acad Sci U S A 1996;93:12564–12569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cellerino A, Arango-Gonzalez BA, Kohler K. Effects of brain-derived neurotrophic factor on the development of NADPH-diaphorase/nitric oxide synthase-positive amacrine cells in the rodent retina. Eur J Neurosci 1999;11:2824–2834 [DOI] [PubMed] [Google Scholar]
- 80.Acheson A, Conover JC, Fandl JP, et al. A BDNF autocrine loop in adult sensory neurons prevents cell death. Nature 1995;374:450–453 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.