Abstract
This article presents several covalent inhibitors, including examples of successful drugs, as well as highly selective, irreversible inhibitors of emerging therapeutic targets, such as fatty acid amide hydolase. Covalent inhibitors have many desirable features, including increased biochemical efficiency of target disruption, less sensitivity toward pharmacokinetic parameters and increased duration of action that outlasts the pharmacokinetics of the compound. Safety concerns that must be mitigated include lack of specificity and the potential immunogenicity of protein–inhibitor adduct(s). Particular attention will be given to recent technologies, such as activity-based protein profiling, which allow one to define the proteome-wide selectivity patterns for covalent inhibitors in vitro and in vivo. For instance, any covalent inhibitor can, in principle, be modified with a ‘clickable’ tag to generate an activity probe that is almost indistinguishable from the original agent. These probes can be applied to any living system across a broad dose range to fully inventory their on and off targets. The substantial number of drugs on the market today that act by a covalent mechanism belies historical prejudices against the development of irreversibly acting therapeutic small molecules. Emerging proteomic technologies offer a means to systematically discriminate safe (selective) versus deleterious (nonselective) covalent inhibitors and thus should inspire their future design and development.
Brief history & examples of covalent inhibitors
The design of selective covalent inhibitors is conceptually very attractive but in practice hard to achieve. That is because it is difficult to strike the right balance between reactivity and selectivity. In many cases, a highly electrophilic species (e.g., α-halo ketone, α,β-unsaturated ketone, fluorophosphonate (FP) or cyanamide) needs to be incorporated into the inhibitor to achieve covalent modification of a protein target [1]. Alkylation of other macromolecules can take place in vivo, leading to deleterious effects, or the reactive species may be scavenged by ubiquitous low-molecular-weight nucleophiles such as glutathione. Indeed many researchers avoid covalent inhibitors owing to the potential toxicity associated with the protein adduct(s), especially if the covalent modification is not selective. However, in cases where selectivity can be achieved and mechanism-based toxicity is not a concern, the increased biochemical efficiency associated with an irreversible mechanism can actually lead to heightened therapeutic margins, as lower drug concentrations are required for efficacy [2,3]. As a testament to the validity of this strategy, there are several examples of successful drugs incorporating tempered or masked electrophiles leading to covalent modification of their protein target (Figures 1 & 2). In fact, of the 74 enzymes that are inhibited by marketed drugs, 19 are irreversibly inhibited via covalent modification [4,5]. While this article will focus on covalent irreversible inhibitors, it should be noted that another important nonequilibrium binding mechanism involves slow dissociation binding kinetics, which leads to pseudo-irreversible or insurmountable inhibition. This mechanism is important to the drug action of the angiotensin II receptor antagonist candesartan, the muscarinic M3 receptor antagonist tiotropium, the histamine H1 receptor antagonist desloratadine, the CCR5 antagonist maraviroc and the HIV-1 protease inhibitor darunavir [2,3,6–8].
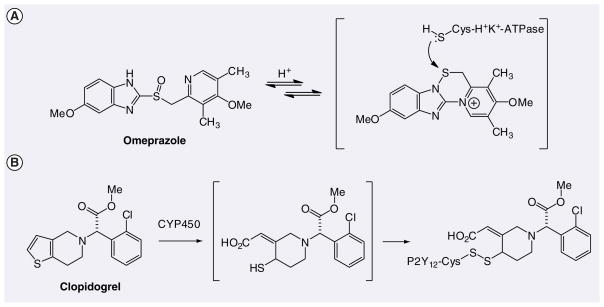
Figure 1.
Mechanism-based covalent inhibition via disulfide adduct formation.
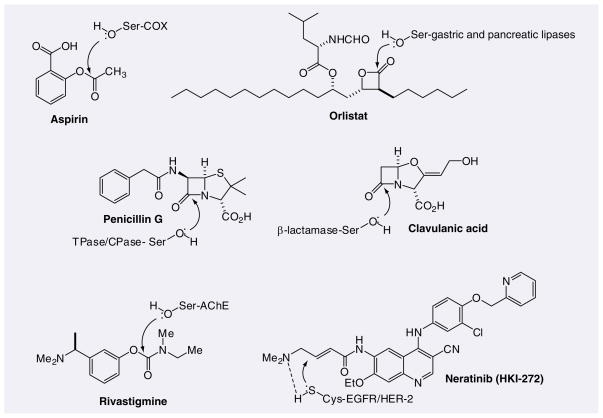
Figure 2.
Examples of covalent inhibitors including their protein target(s) with active-site nucleophile. The arrow indicates the position of attack by the nucleophile on the drug resulting in covalent modification of the target.
In the 1970s considerable effort was put into the design of mechanism-based enzyme inactivators or suicide substrates as an approach to develop highly selective enzyme inhibitors as drugs [9–11]. This approach avoids the direct use of a highly reactive species that can indiscriminately react with various macromolecules and instead aims to start with a relatively innocuous substrate analog, which is activated by the target enzyme to generate an electrophilic species that is attacked by a nucleophile in the active site, leading to irreversible inhibition of the enzyme. This approach is very challenging and some of the most notable successes were not originally designed as irreversible inhibitors; rather, their mechanism of action was discovered serendipitously. For example, omeprazole is a prodrug that covalently modifies gastric H+/K+-ATPase, the enzyme responsible for proton transport as the final step in gastric acid secretion [12]. It is converted under acidic conditions in the stomach to a tetracyclic sulfenamide intermediate that binds covalently to cysteine residues of the H+/K+-ATPase to form disulfide adduct(s) (Figure 1A) [13–15]. Clopidogrel is a prodrug that covalently binds to the adenosine 5′-diphosphate receptor P2Y12 resulting in irreversible inhibition of platelet aggregation [16]. It undergoes hepatic metabolism to an active metabolite (Act-Met) containing a free thiol, which forms a covalent disulfide adduct with a cysteine of P2Y12 (Figure 1B) [17–19].
There are several examples of covalent inhibitors that are successful drugs, and representative examples are shown in Figure 2 [2,3,6–8]. These examples should encourage medicinal chemists to consider this strategy when the biochemical mechanism supports such an approach. Aspirin is a NSAID that irreversibly acetylates an active site serine residue of the cyclooxygenases COX-1 (Ser-529) and COX-2 (Ser-516) (Figure 2) [20,21]. The covalent adduct results in a distortion of the arachidonic acid docking site, thereby blocking the approach of the substrate to the active site and leading to inhibition of COX-1 and COX-2 [22]. Tetrahydrolipstatin is a semisynthetic derivative of lipstatin that inhibits fat absorption [23]. It is a covalent inhibitor of gastric and pancreatic lipases, resulting from β-lactone reaction with the serine nucleophiles of the lipases to form stable ester bonds [24]. β-lactam antibiotics acylate the active site serine of penicillin-binding proteins (PBPs) and kill bacteria by inhibiting the final step of cell wall biosynthesis [25,26]. Class A and B PBPs are transpeptidases that catalyze the formation of peptide crosslinks between adjacent peptidoglycan strands and class C PBPs are D-Ala carboxypeptidases that may modulate the degree of crosslinking by removing the terminal D-Ala of the peptide. Bacteria acquire resistance to β-lactam antibiotics by producing highly mutated PBPs or by producing β-lactamases that catalyze the hydrolysis of the β-lactam ring, preventing their interaction with PBPs [27–29]. Clavulanate is a naturally occurring β-lactam [30] that forms a kinetically stable acyl-enzyme intermediate and inactivates β-lactamase [31,32]. Nucleophilic attack by the active-site Ser-70 opens the β-lactam ring of clavulanate and the resulting oxazolidine ring opens to generate an imine adduct that is rapidly decarboxylated, revealing a covalently bound trans-α,β-eneamine adduct [32]. Therefore, clavulanate is used in combination with approved β-lactam antibiotics to overcome resistance in bacteria that secrete β-lactamase.
Rivastigmine is a carbamate inhibitor of the serine hydrolase acetylcholinesterase, the principal enzyme that degrades acetylcholine at cholinergic synapses and is used for the symptomatic treatment of Alzheimer’s disease. The crystal structure of rivastigmine with Torpedo acetylcholinesterase revealed that the catalytic serine nucleophile (Ser-200) was carbamylated, with the phenol leaving group being retained in the active site [33]. The decarbamylation of the adduct was found to be unusually slow, which may be explained by a movement of His-440 of the catalytic triad, such that nucleophilic attack of a water molecule is not permitted.
Neratinib is an irreversible inhibitor of the human EGF receptor tyrosine kinases EGFR and HER-2 and is in Phase III clinical trials for breast cancer [34–36]. It contains a 4-(dimethylamino)-crotonamide Michael acceptor that forms a covalent bond with a conserved cysteine residue, Cys-773 in EGFR and Cys-805 in HER-2. It is proposed that the Michael addition of the cysteine is accelerated owing to intramolecular general base catalysis by the dimethylamino group. Importantly, neratinib retains activity against tumors that have developed resistance to the noncovalent EGFR inhibitors gefitinib or erlotinib [35,37]. For instance, the T790M mutation in EGFR causes resistance to gefitinib. It has been shown that the T790M mutants retain low nanomolar affinity for gefitinib, but have an affinity for ATP that is increased by more than an order of magnitude [38]. It is postulated that this increase in ATP-binding affinity is the reason the mutants become resistant to reversible binding drugs such as gefitinib, but not to neratinib, since irreversible inhibitors are noncompetitive with ATP.
Despite these successful examples, there still appears to be reluctance toward developing irreversible inhibitors in the pharmaceutical industry. This largely stems from the perception that covalent drugs lack selectivity and the resulting protein adducts lead to toxicity. Until recently, no method existed to closely monitor the selectivity of covalent drugs or differentiate which protein adducts cause toxic effects. However, modern chemical proteomics has begun to provide technologies to experimentally address these concerns in relevant model systems [39]. For example, as will be discussed later, covalent inhibitors can be readily modified with clickable tags resulting in activity-based probes that are almost indistinguishable from the original agent. These probes can be applied to any living system across a broad dose range to fully inventory their on and off targets.
Potential advantages of covalent inhibitors
In recent years, the pursuit of covalent inhibitors as a medicinal chemistry strategy [40,41] has been reinvigorated by an increased emphasis on the biochemical mechanism and efficiency of drug action required for success [2], importance of residence time [6,8,42] and the advent of techniques to probe the selectivity of covalent binders [43,44].
Selective covalent binding of a drug candidate to the desired target can be beneficial owing to the increased biochemical efficiency associated with the nonequilibrium-binding mechanism (Box 1). The nonequilibrium binding of irreversible inhibitors limits the competition with high endogenous ligand concentrations, allowing the desired pharmacological effect to be achieved at lower drug concentrations/doses. In fact, 80% of marketed drugs have to compete with an endogenous ligand [7], therefore perhaps it is not surprising that approximately 30% of marketed drugs that act on an enzyme target are irreversible [4].
Box 1. Pros and cons of covalent inhibitors.
Pros
-
Increased biochemical efficiency [2,3]:
Nonequilibrium binding limits the competition with high endogenous substrate/ligand concentrations.
Less sensitive to pharmacokinetic parameters (i.e., clearance and protein binding).
-
Potential longer duration of action dependent on the synthesis of new enzyme:
Pharmacodynamic effect outlasts pharmacokinetics of inhibitor;
Long residence time [6];
Less frequent dosing.
Most efficient strategy when complete inactivation of target is required.
-
Potential for improved therapeutic index assuming no mechanism-based toxicity:
Lower drug concentrations required for efficacy (reduced dosages);
If the drug inactivates the target and is eliminated quickly, off-target toxicities and drug–drug interactions will decrease.
Cons
Potential immunogenicity of protein adduct leading to an allergic response or drug hypersensitivity reaction (idiosyncratic) [45–48].
Higher risk if covalent inhibitor lacks specificity (nonspecific covalent binding should be avoided).
Not optimal for targets when the mechanism of action requires short residence time, transient inhibition or partial inhibition [49,50].
Furthermore, less than desirable PK properties can often be tolerated as the pharmaco-dynamic action of covalent inhibitors usually outlasts measurable plasma drug levels. Once covalently inactivated, the target is neutralized and the activity can only be recovered by synthesis of new protein. Therefore, as long as resynthesis of the protein is not too fast, only enough drug exposure to inactivate the target is necessary and sustained systemic exposure of the drug (long half-life) may not be required. If the drug inactivates the target and is eliminated from the circulation quickly, the potential for off-target toxicities and drug–drug interactions will decrease, which can lead to increased therapeutic margins.
There are several instances where the use of irreversible inhibitors is particularly advantageous. For example, when the biochemical mechanism of target inhibition involves buildup of substrate, covalent inhibitors are attractive because they prevent the achievement of mass-action equilibrium between the inhibitor and substrate.
Covalent inhibitors may also be preferred when complete and sustained target inactivation is required. The target activity can only be recovered by synthesis of new protein.
Potential disadvantages of covalent inhibitors
Much of the negative connotations surrounding covalent protein adducts can be traced to literature on drug candidates that undergo bioactivation to form a chemically reactive metabolite, which can covalently bind to target proteins [45–59]. Protein covalent binding emerged as a mechanism of drug toxicity in the early 1970s and many marketed drugs that have been associated with idiosyncratic adverse drug reactions (ADRs) are known to form reactive metabolites that are capable of covalently modifying proteins [52–54]. Immune-mediated ADRs are thought to be caused by an abnormal immune reaction triggered by an immunogenic drug–protein adduct [46–48]. According to the hapten hypothesis, drugs are too small to stimulate an immune response; however, the drug or metabolite can act as a hapten upon covalent binding to a protein. These drug–protein adducts provide antigenic determinants for the immune response, but it remains unclear what factors determine individual susceptibility for immunological tolerance versus immune reaction [45,55]. In one of the early examples, it was discovered that acetaminophen can be metabolized to a reactive metabolite, which covalently binds to microsomal proteins that can cause hepatotoxicity [56]. Although a clear causal relationship between covalent binding and toxicity is lacking, most pharmaceutical companies have instituted screens during lead optimization to weed out compounds that form reactive metabolites in efforts to decrease attrition [52,53]. This is logical, since idiosyncratic ADRs cannot be predicted from preclinical toxicology assessments and we have a limited understanding of which protein adducts are immunogenic and which are not. For covalent inhibitors that target an intracellular or membrane-bound protein, one could speculate that the resulting protein adducts would have a lower risk of immunogenicity compared with an extracellular protein adduct (extracellular antigen), which would probably have more effective antigen presentation to major histocompatibility complex and/or T-cell receptors, as well as be more likely to produce an antibody response. More work will be necessary to determine whether the localization of the protein could be a factor in differentiating a toxic drug–protein adduct from a nontoxic one [57]. Chemical biology tools and analytical technologies now exist such that the relationship between specific protein adducts and toxicity can begin to be systematically characterized [44].
It is interesting to note that idiosyncratic ADRS (IADRs) are more frequently associated with compounds used at high daily doses. Therefore, the risk of IADRs from reactive metabolites can be mitigated if a low dose of the drug can be used. In fact, there are no examples of drugs that are dosed below 10 mg/day that cause IADRs [58,59]. In an analogous fashion, it could be inferred that the risk associated with developing a covalent inhibitor could be minimized if the dose was less than 10 mg/day. Fortunately, as pointed out in the previous section, covalent inhibitors often have increased biochemical efficiency and are less sensitive to pharmacokinetic parameters, which together, favor efficacy at lower doses.
In many cases, the potential benefits of a covalent inhibitor could outweigh the potential risk inherent in forming a protein adduct and this risk could in fact be minimal if a low dose (<10 mg/day) is achievable. However, covalent binding may not be optimal for targets when the mechanism of action requires short residence time, transient inhibition or partial inhibition [49,50]. For example, memantine is a NMDA receptor antagonist that has weak binding and short residence time [49]. These properties are desirable to achieve a clinically tolerated antagonist for this mechanism, because it is necessary to block excessive activation of the NMDA receptor, while leaving normal function relatively intact to avoid side effects. In this case, an irreversible inhibitor would not be appropriate because excessive blockade of the NMDA receptor leads to clinically unacceptable side effects. Another example where an irreversible inhibitor with long residence time might lead to mechanism-based toxicity would be for treatments aimed at use-dependent ion channels [3,42]. In this case it is desirable to design compounds that block the channel in the open state and rapidly dissociate from the channel in the resting state. Lastly, some G-protein-coupled receptors that are subject to internalization may not be ideal candidates for covalent drugs [41,60].
Activity-based protein profiling to characterize the selectivity of covalent inhibitors
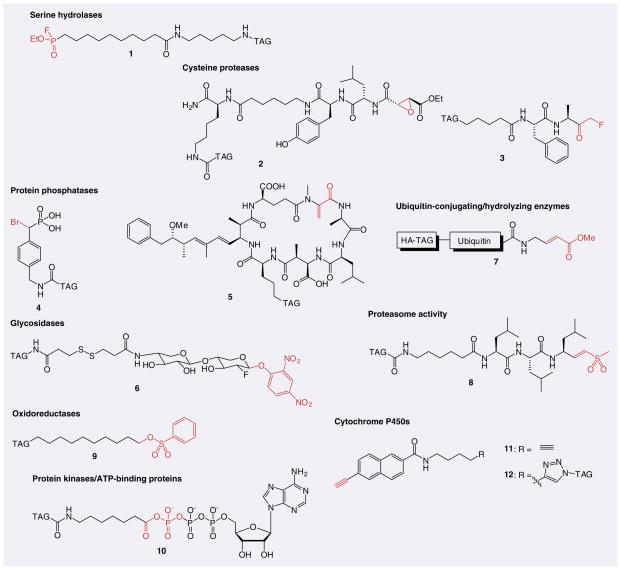
Activity-based protein profiling (ABPP) has emerged as a powerful chemoproteomic tool to characterize the selectivity of enzyme inhibitors on a global scale [43,61,62]. ABPP is a chemical strategy that utilizes active site-directed covalent probes to profile the functional state of enzymes in complex proteomes. Activity-based probes (ABPs) contain a reactive group to covalently modify the active site of enzymes in proteomes and a reporter group (typically a rhodamine or biotin) for detection and identification of protein targets (Figure 3A) [63]. Gel-based ABPP technologies enable visualization of labeling events using SDS-PAGE separation followed by either in-gel fluorescence (fluorescent reporter tag) or avidin blotting (biotin reporter tag). MS platforms, such as ABPP-MudPIT, enable the enrichment and identification of probe-labeled proteins from a complex proteome [64–66]. ABPs (1–12) have been developed for a number of enzyme classes [62], including serine hydrolases [67–69], cysteine proteases [70–72], serine/threonine [73] and tyrosine phosphatases [74], glycosidases [75,76], ubiquitin-conjugating/-hydrolyzing enzymes [77–79], proteasomes [80], oxidoreductases [81,82], ATP-binding enzymes (e.g., kinases) [83–85] and cytochrome P450s (Figure 4) [86,87].
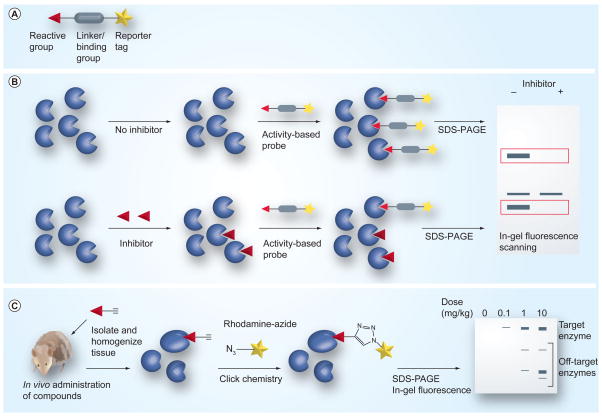
Figure 3. Assessment of global selectivity of covalent inhibitors by activity-based protein profiling (ABPP).
(A) Representative structure of an activity-based probe (ABP), which contains a reactive group, a linker or binding group and a reporter tag. (B) Competitive ABPP to determine the selectivity of an inhibitor against an enzyme family that is targeted by a particular ABP (with fluorescent reporter tag in this example). Probe-labeled proteins are analyzed by SDS-PAGE (in-gel fluorescence) and those that show significant reductions in fluorescent intensity in the presence of inhibitor are scored as targets of the inhibitor. (C) Click chemistry ABPP profiling to characterize the selectivity of covalent inhibitors in vivo. Covalent inhibitors are converted to activity-based probes via incorporation of an alkyne handle and these probes are administered to living systems (cells or animals). Probe-labeled proteins are conjugated to rhodamine-azide using click chemistry and analyzed by SDS-PAGE (in-gel fluorescence).
Figure 4. Representative activity-based probes for individual enzyme families or subfamilies.
Reactive groups are highlighted. Tag: Biotin, rhodamine, TAMRA, BODIPY or HA.
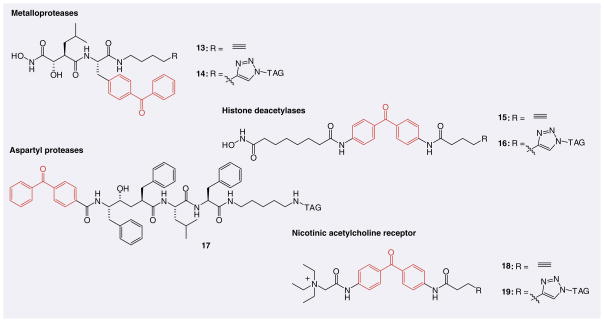
Many potential targets do not possess a nucleophilic active-site residue (Ser, Cys or Lys) for covalent labeling by electrophilic ABPs. A possible solution to this limitation is to incorporate a photoaffinity group into an inhibitor scaffold to create a covalent adduct with the target upon exposure to UV light. This strategy has been successfully employed to create photoreactive ABPP probes (13–19) for metalloproteases [88–91], histone deacetylases [92,93], aspartyl proteases [94–96], Abl kinase [97] and the nicotinic acetylcholine receptor (Figure 5) [98].
Figure 5. Representative photoreactive activity-based probes that achieve target selectivity through binding affinity and covalent labeling is accomplished by exposure to UV light.
Tag: Biotin, rhodamine or TAMRA.
Competitive ABPP
A competitive ABPP platform can be employed to identify protein targets and assess the selectivity of an enzyme inhibitor in native biological systems by measuring the ability of an inhibitor to slow the rate of reaction of the enzyme with a particular ABP [99–101]. Briefly, inhibitor-treated total tissue or cell extracts are subject to profiling with a relevant ABP, and IC50 values for inhibitor targets can be measured as a decrease in enzyme labeling by the ABP (Figure 3B). Competitive ABPP assays can be performed in complex proteome mixtures, enabling the simultaneous evaluation of inhibitor potency and selectivity within a relevant native proteome.
Click chemistry-ABPP
Original protocols for ABPP required the homogenization of cells and tissues prior to treatment with the ABPs, with the drawback of removing proteins from their native environment and disrupting specific activities. This limitation was circumvented by integration of click chemistry (CC) [102,103] and ABPP, resulting in the creation of smaller, more versatile probes using an alkyne or azide group as a latent reporter tag [104–107]. Replacing bulky reporter tags with alkyne groups enables the probe-labeling step to occur in vivo within live cells and organisms. CC is applied to append an azide-functionalized reporter tag to the labeled proteins after cell lysis and homogenization (Figure 3c).
An important application of CC-ABPP is the evaluation of target selectivity of covalent inhibitors in whole cells and animals [108,109]. In many cases, an alkyne can be incorporated into a covalent inhibitor with minimal disruption to the cell permeability and binding interactions of the parent small molecule. The alkyne analog of the covalent inhibitor can be administered to mice, the tissue of interest can be harvested and subjected to CC conjugation with the desired reporter tag for identification of the target proteins. Several examples that utilize CC-ABPP to identify protein targets and compound selectivity will be discussed in the following sections.
Highly selective covalent inhibitors for emerging therapeutic targets: fatty acid amide hydrolase as a case study
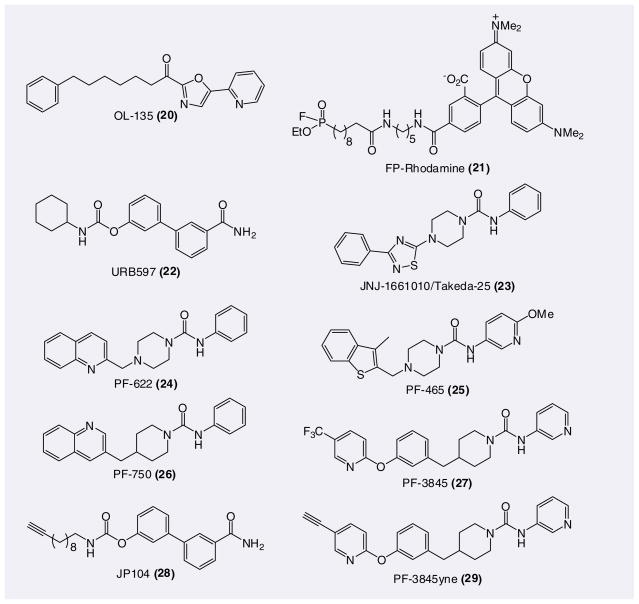
Fatty acid amide hydrolase (FAAH) is an integral membrane enzyme that degrades the fatty acid amide family of signaling lipids, including the endocannabinoid anandamide (AEA) [110,111]. Genetic or pharmacological inactivation of FAAH leads to analgesic and anti-inflammatory phenotypes in rodents without showing the undesirable side effects observed with direct cannabinoid receptor agonists. Selective pharmacological blockade of FAAH elevates the levels and prolongs the effects of anandamide (and other FAAs) only when and where it is synthesized and released on demand [112,113]. Therefore, there is much interest in developing selective FAAH inhibitors as a strategy to discern the endogenous functions of AEA-mediated endocannabinoid pathways and FAAH may represent an attractive therapeutic target for the treatment of inflammatory pain [114]. Several classes of FAAH inhibitors have been reported, including reversibly (e.g., tri-fluoromethyl ketones and α-ketoheterocycles; 20) and irreversibly (e.g., FPs; 21, carbamates; 22 and ureas; 23–27) acting agents (Figure 6) [114,115]. Reversible inhibitors, such as the α-ketoheterocycle OL-135 (20), have been found to display good in vitro potency and selectivity for FAAH relative to other serine hydrolases in mammalian proteomes [99,116,117], but produce only transient elevations in AEA in vivo [118]. The submaximal efficacy of reversible FAAH inhibitors may be due to their rapid metabolism, as well as the fact that near complete (>85%) blockade of FAAH activity is required to maintain elevated AEA levels in vivo [119]. For targets such as FAAH where inhibition leads to elevated levels of substrates, a further potential drawback of reversible inhibitors is that their efficiency and potency can be diminished by mass-action competition with endogenous substrates [2]. Irreversible inhibitors overcome this problem, but selectivity remains an important issue. Considering that FAAH is a serine hydrolase and that there are at least 200 members of this enzyme class in the human proteome, assessing and optimizing inhibitor selectivity represent major challenges. To help address this issue, ABPs against the serine hydrolase class of enzymes have been developed by linking a reactive FP group to a fluorophore or biotin reporter tag [67–69] and these probes have been utilized to profile the proteomic selectivity of FAAH inhibitors [99–101,109,120]. In the absence of an inhibitor, the FP probe labels all the serine hydrolases in the proteome. Serine hydrolases that show significant reductions in probe labeling intensity in the presence of inhibitor are scored as targets of the compound.
Figure 6.
Covalent fatty acid amide hydrolase inhibitors; OL-135 is reversible.
Ureas as selective covalent FAAH inhibitors
Recently, we [100,109,120] and others [121,122] have reported piperazine/piperidine aryl ureas as an emerging class of FAAH inhibitors. In 2007, we reported that the quinoline piperidine urea PF-750 (26) inhibited FAAH in a time-dependent manner (IC50= 52 nM with 30 min preincubation) by covalently modifying the enzyme active site serine nucleophile [100]. PF-750 was confirmed to be covalently attached to the Ser-241 of FAAH through a carbamate linkage by the PF-750-h/rFAAH crystal structure [123]. The irreversible covalent inhibition by PF-750 was rather surprising considering the stability of the urea functional group. Despite the covalent mechanism, PF-750 selectively inhibited FAAH relative to other mammalian serine hydrolases in vitro as determined by competitive ABPP (Figure 3B) [100]. Similarly, no off targets were observed for the benzothiophene piperazine urea PF-465 (25) [120]. By contrast, multiple serine hydrolase off-targets were observed for URB-597 (22) and OL-13 (20), particularly amongst FP-labeled proteins migrating between 55 and 65 kDa [99–101,120]. To confirm that the different selectivity profiles of FAAH inhibitors determined in vitro were also observed in vivo, mice were treated with PF-750 or URB-597 for 1 h, then sacrificed and tissue was removed for competitive ABPP analysis with FP-rhodamine (21) [100]. Serine hydrolase targets of PF-750 and URB-597 were detected by SDS-PAGE and in-gel fluorescence scanning. At each dose tested, both URB-597 and PF-750 selectively targeted FAAH in the brain. PF-750 showed no detectable off-target activity in peripheral tissues (e.g., liver) either, however, URB-597 was found to block FP labeling of several liver serine hydrolases between the molecular masses of 55 and 65 kDa. A proposed mechanism that could explain this exquisite selectivity is a specific binding-induced activation of the urea in the FAAH active site, which renders the reactivity of urea similar to an amide.
Elucidation of the irreversible mode of action of the piperidine/piperazine ureas prompted us to modify the FAAH assay so that inhibitor potencies could be measured as kinact/Ki values. Unlike IC50 values, kinact/Ki values do not change with various preincubation times and have been described as the best measure of potencies for irreversible inhibitors [51]. Using this measure, PF-750 (26) was determined to have a moderate potency (kinact/Ki = 791 M−1s−1) for FAAH. More recently, a series of biaryl ether urea analogs with improved potency has been reported [109]. PF-3845 (27) was the most potent inhibitor (kinact/Ki = 14,310 M−1s−1) reported. Structural studies support that PF-3845 gains its potency from a more extended set of van der Waals interactions between the biaryl ether piperidine moiety and the hydrophobic acyl chain-binding pocket of FAAH based on a crystal structure of a PF-3845-h/rFAAH complex.
In vivo selectivity of the urea PF-3845yne & carbamate JP104 using CC-ABPP
Following confirmation that the carbamate URB597 (22) and the urea PF-3845 (27) covalently modified the serine nucleophile of FAAH, the alkynyl analogs JP104 (28) and PF-3845yne (29) were synthesized and their protein targets were directly analyzed in vivo by CC-ABPP [108,109]. Administration of these probes to FAAH+/+ and FAAH−/− mice, followed by tissue homogenization and conjugation of a rhodamine reporter tag to probe-labeled proteins by CC, revealed their proteome-wide in vivo target selectivity (Figure 3c). The carbamate JP104 was selective for FAAH in the nervous system, but labeled several additional enzymes in peripheral tissues, including multiple carboxyesterases, whereas the urea PF-3845yne was completely selective for FAAH in both the nervous system and peripheral tissue. PF3845yne and JP104 selectively reacted with a single protein in mouse brain that was confirmed as FAAH based on its absence in FAAH−/− mice. In liver, however, PF3845yne and JP104 showed strikingly different profiles, with the former agent once again showing selective reactivity with FAAH and the latter inhibitor labeling a number of proteins that were found in both FAAH+/+ and FAAH−/− mice.
Further examples of selective, covalent inhibitors emerging from chemoproteomic endeavors
Carbamate inhibitors for the serine hydrolase family
Selective and potent covalent inhibitors for several members of the serine hydrolase family have recently emerged. These inhibitors are based on a carbamate scaffold that results in irreversible carbamylation of the active site serine nucleophile. The carbamate chemotype has emerged as a privileged scaffold for potent serine hydrolase inhibitors owing to its tempered electrophilicity and hydrolytic stability following carbamylation. The FP probe can be applied in a competitive ABPP platform to screen carbamate libraries against large numbers of serine hydrolases [124], circumventing the need for protein purification and substrate assays. These chemoproteomic endeavors have resulted in the development of inhibitors for both annotated and unannotated members of this enzyme family.
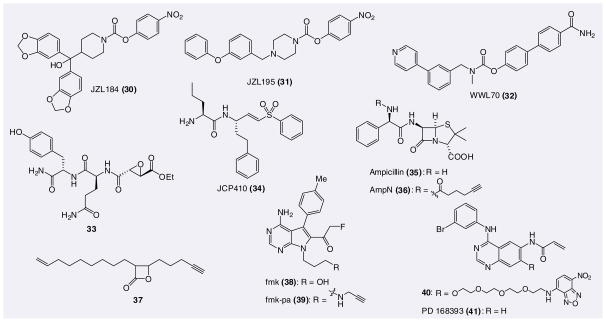
Recently, competitive ABPP screening of a carbamate library led to the development of a selective pharmacological agent against monoacylglycerol lipase (MAGL) [125,126]. MAGL is thought to be the primary enzyme responsible for hydrolyzing the endocannabinoid 2-arachidonoylglycerol. Using competitive ABPP, a potent and selective covalent inhibitor for MAGL, JZL184 (30), was obtained (Figure 7). JZL184 is based on a piperidine–carbamate scaffold and demonstrates high in vivo potency resulting in near-complete blockage of MAGL activity at 4 mg/kg with minimal effects on other brain serine hydrolases, including FAAH. The structural similarity between the piperidine-carbamate MAGL inhibitors (i.e., 30) and piperazine/piperidine–urea FAAH inhibitors (24–27) also inspired the use of competitive ABPP to develop dual FAAH–MAGL inhibitors such as JZL195 (31) [127]. The development of highly selective as well as polypharmacological probes, such as JZL184/195, provides researchers with valuable tools to dissect the roles of the endocannabinoids in a variety of biological systems.
Figure 7.
Covalent inhibitors and clickable covalent probes emerging from chemoproteomic endeavors.
Similar competitive ABPP methods were used to identify a carbamate inhibitor of the uncharacterized serine hydrolase, α/β-hydrolase 6 [124]. This inhibitor, WWL70 (32), exhibited an IC50 value of 70 nM in brain membranes and was demonstrated to be highly selective for ABHD6 relative to 27 other serine hydrolase activities present in these proteomes. The development of potent covalent inhibitors of uncharacterized enzymes, facilitated by ABPP, generates valuable tools for annotating novel enzyme function.
Epoxide & vinyl-sulfone inhibitors for the cysteine protease family
One of the earliest applications of ABPP to inhibitor discovery was the use of cysteine protease probes to identify a selective inhibitor of cathepsin B. The cysteine protease-selective probe, DCG-04 (2), was used in a competitive ABPP strategy to monitor the potency and selectivity of a library of epoxy-succinyl small molecules in rat liver extracts [128]. This study identified a selective covalent inhibitor (33) of cathepsin B, a protease that is implicated in tumor invasion. The synthesis of more elaborate epoxy-succinyl libraries introduced binding groups on either side of the epoxide and resulted in the discovery of covalent inhibitors for other papain fold cysteine proteases [129,130]. Detailed in vivo studies in mice, facilitated by ABPP tools, demonstrated that these compounds show overall rapid clearance in serum, which circumvented problems of nonspecificity induced by compound accumulation in tissues of interest.
Activity-based protein profiling and covalent inhibitors have also played a vital role in the identification of protein activities critical for the invasion and rupture of eukaryotic cells by the malaria parasite Plasmodium falciparum. A library of chloroisocoumarins and peptide vinyl sulfones were screened to identify compounds that block the release of the parasite from host red blood cells [131]. These studies identified a compound JCP410 (34), which contained a vinyl sulfone known to covalently modify cysteine nucleophiles on cysteine proteases. The protein target of JCP410 was identified as dipeptidyl peptidase 3 and a competitive ABPP platform using the broad spectrum cysteine protease probe, DCG-04 (2), confirmed the target of the compound and additionally identified cross-reactivity with several members of the related falcipain family of proteases.
β-lactones & β-lactams as antibiotics
In addition to competitive ABPP platforms, CC-ABPP has found a similar niche in the area of small-molecule target discovery. CC-ABPP relies on alkyne functionalization of covalent inhibitors with minimal disruption to structure, binding affinity and cell permeability. Alkyne-functionalized inhibitors are administered to live cells or organisms and CC is used to tag inhibitor-modified proteins with a reporter group after cell lysis and homogenization (Figure 3c). A study by Sieber et al. highlights the use of click chemistry for protein target identification of covalent inhibitors [132,133]. Well-established antibiotics such as cephalosporin, ampicillin (35) and aztreonam are β-lactams that covalently modify their protein targets. Alkyne-functionalized versions of these antibiotics, for example AmpN (36), were synthesized and their protein targets were investigated using CC-ABPP methods. These compounds were shown to target a diverse number of PBPs both in vitro and in vivo. These tools enabled the investigation of the protein targets of these common antibiotics in a variety of bacterial strains at different concentrations. Similar studies were carried out using a library of alkyne-functionalized β-lactones [134]. These studies identified selective inhibitors (e.g., 37) for the bacterial caseinolytic protein protease, a serine protease that is crucial for virulence of many bacterial pathogens [135]. CC-ABPP facilitates the discovery of protein targets of bioactive small molecules and provides a tool to study potency and selectivity of these molecules in a variety of biological systems.
Inhibitors for protein kinase subfamilies
p90 ribosomal protein S6 kinases (RSKs) are members of the serine/threonine protein kinase family. Although ATP binding sites on protein kinases are highly conserved, the RSK family contains a threonine and a cysteine residue that act as selectivity filters to distinguish the RSK ATP binding sites from other kinases. With this information in hand, a fluoromethylketone inhibitor, fmk (38), was developed that potently inactivates RSK1 and RSK2 in mammalian cells [83]. In order to assess the selectivity of 38, the alkyne-functionalized variant fmk-pa (39) was synthesized [84]. Unlike the fluorophore or biotin tagged analogs of 38, the alkyne-variant 39 demonstrated high cellular potency and, using CC, it was demonstrated that 39 achieves selective and saturable modification of endogenous RSK1 and RSK2 in mammalian cells. Furthermore, the fluorescent covalent probe 40 has been developed based on the irreversible EGFR inhibitor PD 168393 (41). This probe was used to show that there is a linear correlation between inhibition of EGFR kinase activity and inhibition of downstream cellular signaling events [136].
These examples highlight the utility of competitive ABPP platforms and CC-ABPP to identify novel covalent inhibitors, as well as to assess the selectivity of covalent inhibitors in complex proteomes in vitro and in vivo. As the repertoire of available ABPs expands to novel enzyme families, these chemical proteomic technologies will facilitate the development of highly selective covalent inhibitors for as yet untargeted proteins.
Future perspective
Almost 30% of the marketed drugs whose molecular targets are enzymes act by irreversible inhibition [4]. This high percentage is rather surprising considering the strong historical bias against developing irreversible inhibitors as clinical candidates in the pharmaceutical industry. One of the main rationales for this bias is derived from the high inherent reactivity of functional groups generally associated with covalent modifications of proteins. Excessively reactive covalent modifiers can form covalent bonds with a large number of enzymes/proteins, often within (or even extending beyond) the same mechanistic class. Compounding these concerns, there has historically been no direct way to evaluate the selectivity of covalent inhibitors against a large number of proteins in native biological systems. Traditional approaches for testing selectivity have involved setting up individual substrate-based assays with a limited number of candidate ‘off-target’ enzymes. However, this approach excludes the analysis of uncharacterized enzymes, owing to the lack of substrate-based assays. Moreover, it does not take into account the often unpredictable relationship between compound efficacy and selectivity, which is affected by many variables, including target (and off-target) location, concentration and PK properties of the inhibitor and, therefore, must be empirically established in vivo. Functional proteomic methods, such as ABPP, have recently emerged that enable the selectivity of inhibitors to be evaluated against numerous enzymes in parallel directly in native cells and tissues. ABPP can also be combined with CC to create probes capable of fully surveying the direct targets of covalent inhibitors in living systems [104,105,108,109]. As discussed previously, these studies have already revealed covalent inhibitors that display a surprisingly high level of selectivity in the proteome [84,100,109,120,125].
This perspective is not advocating the broad application of covalent inhibitors for all types of targets. Rather, we argue that one should not rule out this approach, especially for enzyme targets where complete inactivation of the target is both desired and tolerated (i.e., no mechanism-based toxicity). Furthermore, the risk of developing a covalent inhibitor should be minimized if the compound is selective for the desired target. Selectivity can be achieved in several ways:
Activation of a compound toward nucleophilic attack only within the target active site as proposed for the urea FAAH inhibitors [100,109,120] and β-lactam antibiotics [137];
Selective distribution of a ‘reactive’ compound to the target tissue of interest, as is the case with orlistat, where the action of the drug is localized to the lumen with negligible concentrations in the plasma;
Mechanism-based unveiling of a reactive group leading to covalent modification at the target site of action, as is the case with omeprazole;
Combination of a tempered electrophile with a selective noncovalent binder.
The last approach sounds very attractive but in practice is quite challenging. Chemical biology methodology such as CC-ABPP should help advance this approach and successful examples have already emerged in the literature [84,109]. We must learn from our past successes and failures and, at the same time, be opportunistic in our application of new technologies to help guide the design of safe and efficacious drugs, whether they be covalent or noncovalent in mechanism.
Executive summary.
Selective covalent binding of a drug candidate to the desired target can be beneficial owing to the increased biochemical efficiency associated with the nonequilibrium-binding mechanism.
Beyond their potential use as drugs, selective covalent inhibitors represent highly versatile pharmacological tools for assessing protein function in vivo.
Chemical proteomic technologies, such as activity-based protein profiling, allow one to define the proteome-wide selectivity patterns for covalent inhibitors in vitro and in vivo.
Covalent inhibitors can be readily modified with clickable tags resulting in activity-based probes that can be applied to any living system across a broad dose range to inventory their on and off targets.
Ongoing clinical studies with an emerging cadre of highly selective covalent inhibitors should further clarify their therapeutic utility and possible risks.
Key term
- Chemical proteomics
Involves the use of chemical tools for identifying small molecule protein interactions in complex biological systems and is generally based on activity-based protein profiling using covalent activity-based probes or compound-immobilized affinity chromatography
- Immunogen
Molecule that can initiate an immune response. All immunogens are antigens, but not all antigens are immunogens
- Antigen
Substance that can be bound by an antibody or surface receptor on T cells
- Activity-based protein profiling
Applies chemical probes to profile the functional state of enzymes in complex proteomes. An activity-based probe can distinguish active enzyme from inactive zymogen or inhibitor-bound forms
- Activity-based probe
Typical activity-based probes comprise a reactive group to covalently modify the active site of a particular enzyme class and a reporter group for detection and isolation of probe-labeled proteins
- Click chemistry
Bioorthogonal reaction that applies the copper catalyzed, stepwise version of Huisgen’s 1,3-dipolar cycloaddition reaction to form a stable triazole linkage between an azide and alkyne
- kinact/Ki
When characterizing covalent inhibitors, it is important to take both the equilibrium binding (Ki) and the rate of covalent bond formation (kinact) into account. The best measure of inhibitory potency for an irreversible inhibitor is the second order rate constant obtained from the ratio kinact/Ki. Unlike IC50values, the ratio of kinact/Ki is independent of preincubation time and enzyme and substrate concentrations
Footnotes
For reprint orders, please contact reprints@future-science.com
Financial & competing interests disclosure
Douglas Johnson is an employee and shareholder of Pfizer Global Research and Development. Benjamin Cravatt is a professor at The Scripps Research Institute in La Jolla, CA, USA, and has served as a consultant with Pfizer Global Research and Development since 2003. This work was supported in part by the NIH, the Helen L Dorris Institute for the Study of Neurological and Psychiatric Disorders in Children and Adolescents, and The Skaggs Institute for Chemical Biology. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
- 1.Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 2.Swinney DC. Biochemical mechanisms of drug action: what does it take for success? Nat Rev Drug Discov. 2004;3:801–808. doi: 10.1038/nrd1500. [DOI] [PubMed] [Google Scholar]
- 3.Swinney DC. The role of binding kinetics in therapeutically useful drug action. Curr Opin Drug Discov Devel. 2009;12:31–39. [PubMed] [Google Scholar]
- 4.Robertson JG. Mechanistic basis of enzyme-targeted drugs. Biochemistry. 2005;44:5561–5571. doi: 10.1021/bi050247e. [DOI] [PubMed] [Google Scholar]
- 5.Robertson JG. Enzymes as a special class of therapeutic target: clinical drugs and modes of action. Curr Opin Struct Biol. 2007;17:674–679. doi: 10.1016/j.sbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Copeland RA, Pompliano DL, Meek TD. Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 7.Swinney DC. Biochemical mechanisms of new molecular entities (NMEs) approved by United States FDA during 2001–2004. mechanisms leading to optimal efficacy and safety. Curr Top Med Chem. 2006;6:461–478. doi: 10.2174/156802606776743093. [DOI] [PubMed] [Google Scholar]
- 8.Tummino PJ, Copeland RA. Residence time of receptor-ligand complexes and its effect on biological function. Biochemistry. 2008;47:5481–5492. doi: 10.1021/bi8002023. [DOI] [PubMed] [Google Scholar]
- 9.Penning TM. Design of suicide substrates: an approach to the development of highly selective enzyme inhibitors as drugs. Trends Pharmacol Sci. 1983;4:212–217. [Google Scholar]
- 10.Rando RR. New modes of enzyme inactivator design. Trends Pharmacol Sci. 1980;1:168–171. [Google Scholar]
- 11.Walsh C. Suicide substrates: mechanism-based enzyme inactivators. Tetrahedron. 1982;38:871–909. [Google Scholar]
- 12.Olbe L, Carlsson E, Lindberg P. A proton-pump inhibitor expedition: the case histories of omeprazole and esomeprazole. Nat Rev Drug Discov. 2003;2:132–139. doi: 10.1038/nrd1010. [DOI] [PubMed] [Google Scholar]
- 13.Im WB, Sih JC, Blakeman DP, McGrath JP. Omeprazole, a specific inhibitor of gastric (H+-K+)-ATPase, is a H+-activated oxidizing agent of sulfhydryl groups. J Biol Chem. 1985;260:4591–4597. [PubMed] [Google Scholar]
- 14.Lindberg P, Nordberg P, Alminger T, Braendstroem A, Wallmark B. The mechanism of action of the antisecretory agent omeprazole. J Med Chem. 1986;29:1327–1329. doi: 10.1021/jm00158a001. [DOI] [PubMed] [Google Scholar]
- 15.Shin JM, Cho YM, Sachs G. Chemistry of covalent inhibition of the gastric (H+, K+)-ATPase by proton pump inhibitors. J Am Chem Soc. 2004;126:7800–7811. doi: 10.1021/ja049607w. [DOI] [PubMed] [Google Scholar]
- 16.Bhatt DL, Topol EJ. Scientific and therapeutic advances in antiplatelet therapy. Nat Rev Drug Discov. 2003;2:15–28. doi: 10.1038/nrd985. [DOI] [PubMed] [Google Scholar]
- 17.Pereillo JM, Maftouh M, Andrieu A, et al. Structure and stereochemistry of the active metabolite of clopidogrel. Drug Metab Dispos. 2002;30:1288–1295. doi: 10.1124/dmd.30.11.1288. [DOI] [PubMed] [Google Scholar]
- 18.Savi P, Pereillo JM, Uzabiaga MF, et al. Identification and biological activity of the active metabolite of clopidogrel. Thromb Haemost. 2000;84:891–896. [PubMed] [Google Scholar]
- 19.Savi P, Zachayus JL, Delesque-Touchard N, et al. The active metabolite of Clopidogrel disrupts P2Y12 receptor oligomers and partitions them out of lipid rafts. Proc Natl Acad Sci USA. 2006;103:11069–11074. doi: 10.1073/pnas.0510446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth GJ, Stanford N, Majerus PW. Acetylation of prostaglandin synthase by aspirin. Proc Natl Acad Sci USA. 1975;72:3073–3076. doi: 10.1073/pnas.72.8.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Der Ouderaa FJ, Buytenhek M, Nugteren DH, Van Dorp DA. Acetylation of prostaglandin endoperoxide synthetase with acetylsalicylic acid. Eur J Biochem. 1980;109:1–8. doi: 10.1111/j.1432-1033.1980.tb04760.x. [DOI] [PubMed] [Google Scholar]
- 22.DeWitt DL, el-Harith EA, Kraemer SA, et al. The aspirin and heme-binding sites of ovine and murine prostaglandin endoperoxide synthases. J Biol Chem. 1990;265:5192–5198. [PubMed] [Google Scholar]
- 23.Guerciolini R. Mode of action of orlistat. Int J Obes Relat Metab Disord. 1997;21(Suppl 3):S12–S23. [PubMed] [Google Scholar]
- 24.Hadvary P, Sidler W, Meister W, Vetter W, Wolfer H. The lipase inhibitor tetrahydrolipstatin binds covalently to the putative active site serine of pancreatic lipase. J Biol Chem. 1991;266:2021–2027. [PubMed] [Google Scholar]
- 25.Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yocum RR, Waxman DJ, Rasmussen JR, Strominger JL. Mechanism of penicillin action: penicillin and substrate bind covalently to the same active site serine in two bacterial D-alanine carboxypeptidases. Proc Natl Acad Sci USA. 1979;76:2730–2734. doi: 10.1073/pnas.76.6.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim D, Strynadka NCJ. Structural basis for the β-lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat Struct Biol. 2002;9:870–876. doi: 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]
- 28.Strynadka NCJ, Adachi H, Jensen SE, et al. Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 A resolution. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 29.Wilke MS, Lovering AL, Strynadka NCJ. β-Lactam antibiotic resistance: a current structural perspective. Curr Opin Microbiol. 2005;8:525–533. doi: 10.1016/j.mib.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Howarth TT, Brown AG, King TJ. Clavulanic acid, a novel β-lactam isolated from Streptomyces clavuligerus: x-ray crystal structure analysis. J Chem Soc Chem Commun. 1976:266–267. [Google Scholar]
- 31.Hugonnet JE, Blanchard JS. Irreversible inhibition of the Mycobacterium tuberculosis β-lactamase by clavulanate. Biochemistry. 2007;46:11998–12004. doi: 10.1021/bi701506h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tremblay LW, Hugonnet JE, Blanchard JS. Structure of the covalent adduct formed between Mycobacterium tuberculosis β-lactamase and clavulanate. Biochemistry. 2008;47:5312–5316. doi: 10.1021/bi8001055. [DOI] [PubMed] [Google Scholar]
- 33.Bar-On P, Millard CB, Harel M, et al. Kinetic and structural studies on the interaction of cholinesterases with the anti-Alzheimer drug rivastigmine. Biochemistry. 2002;41:3555–3564. doi: 10.1021/bi020016x. [DOI] [PubMed] [Google Scholar]
- 34.Tsou HR, Overbeek-Klumpers EG, Hallett WA, et al. Optimization of 6,7-disubstituted-4-(arylamino)quinoline-3-carbonitriles as orally active, irreversible inhibitors of human epidermal growth factor receptor-2 kinase activity. J Med Chem. 2005;48:1107–1131. doi: 10.1021/jm040159c. [DOI] [PubMed] [Google Scholar]
- 35.Wissner A, Mansour TS. The development of HKI-272 and related compounds for the treatment of cancer. Arch Pharm. 2008;341:465–477. doi: 10.1002/ardp.200800009. [DOI] [PubMed] [Google Scholar]
- 36.Mukherji D, Spicer J. Second-generation epidermal growth factor tyrosine kinase inhibitors in non-small cell lung cancer. Expert Opin Investig Drugs. 2009;18:293–301. doi: 10.1517/13543780902762843. [DOI] [PubMed] [Google Scholar]
- 37.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon GM, Cravatt BF. Challenges for the ‘chemical-systems’ biologist. Nat Chem Biol. 2008;4:639–642. doi: 10.1038/nchembio1108-639. [DOI] [PubMed] [Google Scholar]
- 40.Potashman MH, Duggan ME. Covalent modifiers: an orthogonal approach to drug design. J Med Chem. 2009;52:1231–1246. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- 41.Smith AJ, Zhang X, Leach AG, Houk KN. Beyond picomolar affinities: quantitative aspects of noncovalent and covalent binding of drugs to proteins. J Med Chem. 2009;52:225–233. doi: 10.1021/jm800498e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R, Monsma F. The importance of drug-target residence time. Curr Opin Drug Discov Devel. 2009;12:488–496. [PubMed] [Google Scholar]
- 43.Barglow KT, Cravatt BF. Activity-based protein profiling for the functional annotation of enzymes. Nat Methods. 2007;4:822–827. doi: 10.1038/nmeth1092. [DOI] [PubMed] [Google Scholar]
- 44.Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chem Res Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lavergne SN, Park BK, Naisbitt DJ. The roles of drug metabolism in the pathogenesis of T-cell-mediated drug hypersensitivity. Curr Opin Allergy Clin Immunol. 2008;8:299–307. doi: 10.1097/ACI.0b013e3283079c64. [DOI] [PubMed] [Google Scholar]
- 46.Park BK, Sanderson JP, Naisbitt DJ. Drugs as haptens, antigens, and immunogens. In: Pichler WJ, editor. Drug Hypersensitivity. Karger, Basel; Switzerland: 2007. pp. 55–65. [Google Scholar]
- 47.Uetrecht J. Idiosyncratic drug reactions: past, present, and future. Chem Res Toxicol. 2008;21:84–92. doi: 10.1021/tx700186p. [DOI] [PubMed] [Google Scholar]
- 48.Uetrecht J. Immune-mediated adverse drug reactions. Chem Res Toxicol. 2009;22:24–34. doi: 10.1021/tx800389u. [DOI] [PubMed] [Google Scholar]
- 49.Lipton SA. Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat Rev Drug Discov. 2006;5:160–170. doi: 10.1038/nrd1958. [DOI] [PubMed] [Google Scholar]
- 50.Ohlson S. Designing transient binding drugs: a new concept for drug discovery. Drug Discov Today. 2008;13:433–439. doi: 10.1016/j.drudis.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Copeland RA. Evaluation of Enzyme Inhibitors in Drug Discovery: a Guide to Medicinal Chemists and Pharmacologists. Wiley; Oxford, UK: 2005. p. 296. [PubMed] [Google Scholar]
- 52.Evans DC, Watt AP, Nicoll-Griffith DA, Baillie TA. Drug–protein adducts: an industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chem Res Toxicol. 2004;17:3–16. doi: 10.1021/tx034170b. [DOI] [PubMed] [Google Scholar]
- 53.Kumar S, Kassahun K, Tschirret-Guth RA, Mitra K, Baillie TA. Minimizing metabolic activation during pharmaceutical lead optimization: progress, knowledge gaps and future directions. Curr Opin Drug Discov Devel. 2008;11:43–52. [PubMed] [Google Scholar]
- 54.Zhou S, Chan E, Duan W, Huang M, Chen YZ. Drug bioactivation, covalent binding to target proteins and toxicity relevance. Drug Metab Rev. 2005;37:41–213. doi: 10.1081/dmr-200028812. [DOI] [PubMed] [Google Scholar]
- 55.Uetrecht J. Evaluation of which reactive metabolite, if any, is responsible for a specific idiosyncratic reaction. Drug Metab Rev. 2006;38:745–753. doi: 10.1080/03602530600959615. [DOI] [PubMed] [Google Scholar]
- 56.Bond GR. Acetaminophen protein adducts: a review. Clin Toxicol. 2009;47:2–7. doi: 10.1080/15563650801941831. [DOI] [PubMed] [Google Scholar]
- 57.Hopkins JE, Naisbitt DJ, Kitteringham NR, Dearman RJ, Kimber I, Park BK. Selective haptenation of cellular or extracellular protein by chemical allergens: association with cytokine polarization. Chem Res Toxicol. 2005;18:375–381. doi: 10.1021/tx049688+. [DOI] [PubMed] [Google Scholar]
- 58.Kalgutkar AS, Gardner I, Obach RS, et al. A comprehensive listing of bioactivation pathways of organic functional groups. Curr Drug Metab. 2005;6:161–225. doi: 10.2174/1389200054021799. [DOI] [PubMed] [Google Scholar]
- 59.Nakayama S, Atsumi R, Takakusa H, et al. A zone classification system for risk assessment of idiosyncratic drug toxicity using daily dose and covalent binding. Drug Metab Dispos. 2009;37:1970–1977. doi: 10.1124/dmd.109.027797. [DOI] [PubMed] [Google Scholar]
- 60.Conn PM, Ulloa-Aguirre A, Ito J, Janovick JA. G protein-coupled receptor trafficking in health and disease: lessons learned to prepare for therapeutic mutant rescue in vivo. Pharmacol Rev. 2007;59:225–250. doi: 10.1124/pr.59.3.2. [DOI] [PubMed] [Google Scholar]
- 61.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 62.Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 63.Sadaghiani AM, Verhelst SHL, Bogyo M. Tagging and detection strategies for activity-based proteomics. Curr Opin Chem Biol. 2007;11:20–28. doi: 10.1016/j.cbpa.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 64.Jessani N, Niessen S, Wei BQ, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Meth. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 65.Sieber SA, Cravatt BF. Analytical platforms for activity-based protein profiling – exploiting the versatility of chemistry for functional proteomics. Chem Commun (Camb) 2006:2311–2319. doi: 10.1039/b600653c. [DOI] [PubMed] [Google Scholar]
- 66.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP) – a general method for mapping sites of probe modification in proteomes. Nat Protoc. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- 67.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40:4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 69.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics. 2001;1:1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 70.Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem Biol. 2000;7:569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 71.Greenbaum DC, Arnold WD, Lu F, et al. Small molecule affinity fingerprinting. A tool for enzyme family subclassification, target identification, and inhibitor design. Chem Biol. 2002;9:1085–1094. doi: 10.1016/s1074-5521(02)00238-7. [DOI] [PubMed] [Google Scholar]
- 72.Kato D, Boatright KM, Berger AB, et al. Activity-based probes that target diverse cysteine protease families. Nat Chem Biol. 2005;1:33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 73.Shreder KR, Liu Y, Nomanhboy T, et al. Design and synthesis of AX7574: a microcystin-derived, fluorescent probe for serine/threonine phosphatases. Bioconjug Chem. 2004;15:790–798. doi: 10.1021/bc0499580. [DOI] [PubMed] [Google Scholar]
- 74.Krishnamurthy D, Barrios AM. Profiling protein tyrosine phosphatase activity with mechanistic probes. Curr Opin Chem Biol. 2009;13:375–381. doi: 10.1016/j.cbpa.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 75.Vocadlo DJ, Bertozzi CR. A strategy for functional proteomic analysis of glycosidase activity from cell lysates. Angew Chem Int Ed Engl. 2004;43:5338–5342. doi: 10.1002/anie.200454235. [DOI] [PubMed] [Google Scholar]
- 76.Hekmat O, Kim YW, Williams SJ, He S, Withers SG. Active-site peptide ‘fingerprinting’ of glycosidases in complex mixtures by mass spectrometry. Discovery of a novel retaining β-1,4-glycanase in Cellulomonas fimi. J Biol Chem. 2005;280:35126–35135. doi: 10.1074/jbc.M508434200. [DOI] [PubMed] [Google Scholar]
- 77.Borodovsky A, Ovaa H, Kolli N, et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 78.Love KR, Catic A, Schlieker C, Ploegh HL. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat Chem Biol. 2007;3:697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- 79.Love KR, Pandya RK, Spooner E, Ploegh HL. Ubiquitin C-terminal electrophiles are activity-based probes for identification and mechanistic study of ubiquitin conjugating machinery. ACS Chem Biol. 2009;4:275–287. doi: 10.1021/cb9000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berkers CR, Verdoes M, Lichtman E, et al. Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nat Methods. 2005;2:357–362. doi: 10.1038/nmeth759. [DOI] [PubMed] [Google Scholar]
- 81.Adam GC, Cravatt BF, Sorensen EJ. Profiling the specific reactivity of the proteome with non-directed activity-based probes. Chem Biol. 2001;8:81–95. doi: 10.1016/s1074-5521(00)90060-7. [DOI] [PubMed] [Google Scholar]
- 82.Adam GC, Sorensen EJ, Cravatt BF. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nat Biotechnol. 2002;20:805–809. doi: 10.1038/nbt714. [DOI] [PubMed] [Google Scholar]
- 83.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cohen MS, Hadjivassiliou H, Taunton J. A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nat Chem Biol. 2007;3:156–160. doi: 10.1038/nchembio859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Patricelli MP, Szardenings AK, Liyanage M, et al. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 86.Wright AT, Cravatt BF. Chemical proteomic probes for profiling cytochrome p450 activities and drug interactions in vivo. Chem Biol. 2007;14:1043–1051. doi: 10.1016/j.chembiol.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wright AT, Song JD, Cravatt BF. A suite of activity-based probes for human cytochrome P450 enzymes. J Am Chem Soc. 2009;131:10692–10700. doi: 10.1021/ja9037609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci USA. 2004;101:10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sieber SA, Niessen S, Hoover HS, Cravatt BF. Proteomic profiling of metalloprotease activities with cocktails of active-site probes. Nat Chem Biol. 2006;2:274–281. doi: 10.1038/nchembio781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chan EW, Chattopadhaya S, Panicker RC, Huang X, Yao SQ. Developing photoactive affinity probes for proteomic profiling: hydroxamate-based probes for metalloproteases. J Am Chem Soc. 2004;126:14435–14446. doi: 10.1021/ja047044i. [DOI] [PubMed] [Google Scholar]
- 91.David A, Steer D, Bregant S, et al. Cross-linking yield variation of a potent matrix metalloproteinase photoaffinity probe and consequences for functional proteomics. Angew Chem Int Ed Engl. 2007;46:3275–3277. doi: 10.1002/anie.200604408. [DOI] [PubMed] [Google Scholar]
- 92.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc Natl Acad Sci USA. 2007;104:1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Salisbury CM, Cravatt BF. Optimization of activity-based probes for proteomic profiling of histone deacetylase complexes. J Am Chem Soc. 2008;130:2184–2194. doi: 10.1021/ja074138u. [DOI] [PubMed] [Google Scholar]
- 94.Li YM, Xu M, Lai MT, et al. Photoactivated γ-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405:689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- 95.Fuwa H, Takahashi Y, Konno Y, et al. Divergent synthesis of multifunctional molecular probes to elucidate the enzyme specificity of dipeptidic γ-secretase inhibitors. ACS Chem Biol. 2007;2:408–418. doi: 10.1021/cb700073y. [DOI] [PubMed] [Google Scholar]
- 96.Shi H, Liu K, Xu A, Yao SQ. Small molecule microarray-facilitated screening of affinity-based probes (AfBPs) for γ-secretase. Chem Commun (Camb) 2009;46:5030–5032. doi: 10.1039/b910611a. [DOI] [PubMed] [Google Scholar]
- 97.Kalesh KA, Sim DS, Wang J, Liu K, Lin Q, Yao SQ. Small molecule probes that target Abl kinase. Chem Commun (Camb) 2009;46:1118–1120. doi: 10.1039/b919888a. [DOI] [PubMed] [Google Scholar]
- 98.Tantama M, Lin WC, Licht S. An activity-based protein profiling probe for the nicotinic acetylcholine receptor. J Am Chem Soc. 2008;130:15766–15767. doi: 10.1021/ja805868x. [DOI] [PubMed] [Google Scholar]
- 99.Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21:687–691. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 100.Ahn K, Johnson DS, Fitzgerald LR, et al. Novel mechanistic class of fatty acid amide hydrolase inhibitors with remarkable selectivity. Biochemistry. 2007;46:13019–13030. doi: 10.1021/bi701378g. [DOI] [PubMed] [Google Scholar]
- 101.Zhang D, Saraf A, Kolasa T, et al. Fatty acid amide hydrolase inhibitors display broad selectivity and inhibit multiple carboxylesterases as off-targets. Neuropharmacology. 2007;52:1095–1105. doi: 10.1016/j.neuropharm.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 102.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 103.Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 104.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 105.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 106.Salisbury CM, Cravatt BF. Click chemistry-led advances in high content functional proteomics. QSAR Comb Sci. 2007;26:1229–1238. [Google Scholar]
- 107.Best MD. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48:6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- 108.Alexander JP, Cravatt BF. Mechanism of carbamate inactivation of FAAH: implications for the design of covalent inhibitors and in vivo functional probes for enzymes. Chem Biol. 2005;12:1179–1187. doi: 10.1016/j.chembiol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ahn K, Johnson DS, Mileni M, et al. Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem Biol. 2009;16:411–420. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–432. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- 111.Ahn K, McKinney MK, Cravatt BF. Enzymatic pathways that regulate endocannabinoid signaling in the nervous system. Chem Rev. 2008;108:1687–1707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Piomelli D. The molecular logic of endocannabinoid signalling. Nat Rev Neurosci. 2003;4:873–884. doi: 10.1038/nrn1247. [DOI] [PubMed] [Google Scholar]
- 113.Di Marzo V. Targeting the endocannabinoid system: to enhance or reduce? Nat Rev Drug Discov. 2008;7:438–455. doi: 10.1038/nrd2553. [DOI] [PubMed] [Google Scholar]
- 114.Ahn K, Johnson DS, Cravatt BF. Fatty acid amide hydrolase as a potential therapeutic target for the treatment of pain and CNS disorders. Expert Opin Drug Discovery. 2009;4:763–784. doi: 10.1517/17460440903018857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Seierstad M, Breitenbucher JG. Discovery and development of fatty acid amide hydrolase (FAAH) inhibitors. J Med Chem. 2008;51:7327–7343. doi: 10.1021/jm800311k. [DOI] [PubMed] [Google Scholar]
- 116.Leung D, Du W, Hardouin C, et al. Discovery of an exceptionally potent and selective class of fatty acid amide hydrolase inhibitors enlisting proteome-wide selectivity screening: concurrent optimization of enzyme inhibitor potency and selectivity. Bioorg Med Chem Lett. 2005;15:1423–1428. doi: 10.1016/j.bmcl.2004.12.085. [DOI] [PubMed] [Google Scholar]
- 117.Mileni M, Garfunkle J, DeMartino JK, Cravatt BF, Boger DL, Stevens RC. Binding and inactivation mechanism of a humanized fatty acid amide hydrolase by α-ketoheterocycle inhibitors revealed from cocrystal structures. J Am Chem Soc. 2009;131:10497–10506. doi: 10.1021/ja902694n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lichtman AH, Leung D, Shelton CC, et al. Reversible inhibitors of fatty acid amide hydrolase that promote analgesia: evidence for an unprecedented combination of potency and selectivity. J Pharmacol Exp Ther. 2004;311:441–448. doi: 10.1124/jpet.104.069401. [DOI] [PubMed] [Google Scholar]
- 119.Fegley D, Gaetani S, Duranti A, et al. Characterization of the fatty acid amide hydrolase inhibitor cyclohexyl carbamic acid 3′-carbamoyl-biphenyl-3-yl ester (URB597): effects on anandamide and oleoylethanolamide deactivation. J Pharmacol Exp Ther. 2005;313:352–358. doi: 10.1124/jpet.104.078980. [DOI] [PubMed] [Google Scholar]
- 120.Johnson DS, Ahn K, Kesten S, et al. Benzothiophene piperazine and piperidine urea inhibitors of fatty acid amide hydrolase (FAAH) Bioorg Med Chem Lett. 2009;19:2865–2869. doi: 10.1016/j.bmcl.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Karbarz MJ, Luo L, Chang L, et al. Biochemical and biological properties of 4-(3-phenyl-[1,2,4] thiadiazol-5-yl)-piperazine-1-carboxylic acid phenylamide, a mechanism-based inhibitor of fatty acid amide hydrolase. Anesth Analg. 2009;108:316–329. doi: 10.1213/ane.0b013e31818c7cbd. [DOI] [PubMed] [Google Scholar]
- 122.Keith JM, Apodaca R, Xiao W, et al. Thiadiazolopiperazinyl ureas as inhibitors of fatty acid amide hydrolase. Bioorg Med Chem Lett. 2008;18:4838–4843. doi: 10.1016/j.bmcl.2008.07.081. [DOI] [PubMed] [Google Scholar]
- 123.Mileni M, Johnson DS, Wang Z, et al. Structure-guided inhibitor design for human FAAH by interspecies active site conversion. Proc Natl Acad Sci USA. 2008;105:12820–12824. doi: 10.1073/pnas.0806121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J Am Chem Soc. 2007;129:9594–9595. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- 125.Long JZ, Li W, Booker L, et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Long JZ, Nomura DK, Cravatt BF. Characterization of monoacylglycerol lipase inhibition reveals differences in central and peripheral endocannabinoid metabolism. Chem Biol. 2009;16:744–753. doi: 10.1016/j.chembiol.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Long JZ, Nomura DK, Vann RE, et al. Dual blockade of FAAH and MAGL identifies behavioral processes regulated by endocannabinoid crosstalk in vivo. Proc Natl Acad Sci USA. 2009;106:20270–20275. doi: 10.1073/pnas.0909411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Greenbaum D, Baruch A, Hayrapetian L, et al. Chemical approaches for functionally probing the proteome. Mol Cell Proteomics. 2002;1:60–68. doi: 10.1074/mcp.t100003-mcp200. [DOI] [PubMed] [Google Scholar]
- 129.Yuan F, Verhelst SH, Blum G, Coussens LM, Bogyo M. A selective activity-based probe for the papain family cysteine protease dipeptidyl peptidase I/cathepsin C. J Am Chem Soc. 2006;128:5616–5617. doi: 10.1021/ja060835v. [DOI] [PubMed] [Google Scholar]
- 130.Sadaghiani AM, Verhelst SH, Gocheva V, et al. Design, synthesis, and evaluation of in vivo potency and selectivity of epoxysuccinyl-based inhibitors of papain-family cysteine proteases. Chem Biol. 2007;14:499–511. doi: 10.1016/j.chembiol.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 131.Arastu-Kapur S, Ponder EL, Fonovic UP, et al. Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat Chem Biol. 2008;4:203–213. doi: 10.1038/nchembio.70. [DOI] [PubMed] [Google Scholar]
- 132.Staub I, Sieber SA. β-Lactams as selective chemical probes for the in vivo labeling of bacterial enzymes involved in cell wall biosynthesis, antibiotic resistance, and virulence. J Am Chem Soc. 2008;130:13400–13409. doi: 10.1021/ja803349j. [DOI] [PubMed] [Google Scholar]
- 133.Staub I, Sieber SA. β-lactam probes as selective chemical-proteomic tools for the identification and functional characterization of resistance associated enzymes in MRSA. J Am Chem Soc. 2009;131:6271–6276. doi: 10.1021/ja901304n. [DOI] [PubMed] [Google Scholar]
- 134.Bottcher T, Sieber SA. β-lactones as privileged structures for the active-site labeling of versatile bacterial enzyme classes. Angew Chem Int Ed Engl. 2008;47:4600–4603. doi: 10.1002/anie.200705768. [DOI] [PubMed] [Google Scholar]
- 135.Bottcher T, Sieber SA. β-lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J Am Chem Soc. 2008;130:14400–14401. doi: 10.1021/ja8051365. [DOI] [PubMed] [Google Scholar]
- 136.Blair JA, Rauh D, Kung C, et al. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat Chem Biol. 2007;3:229–238. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]
- 137.Rando RR. On the mechanism of action of antibiotics which act as irreversible enzyme inhibitors. Biochem Pharmacol. 1975;24:1153–1160. doi: 10.1016/0006-2952(75)90055-6. [DOI] [PubMed] [Google Scholar]