Abstract
Objective
To define the altered gene expression profile of endometriotic lesions in a mouse model of surgically-induced endometriosis
Design
Autologous experimental mouse model.
Setting
Medical school department.
Animals
Adult C57Bl6 mice.
Intervention(s)
Endometriosis was surgically-induced by auto-transplantation of uterine tissue to the intestinal mesentery. Endometriotic lesions and eutopic uteri were recovered at 3 or 29 days post-induction.
Main Outcome Measure(s)
Altered gene expression was measured in the endometriotic lesion relative to the eutopic uterus by genome wide cDNA microarray analysis and was confirmed by real time RT-PCR for six genes. Relevant categories of altered genes were identified using gene ontology analysis to determine groups of genes enriched for altered expression.
Result(s)
The expression of 479 and 114 genes was altered in the endometriotic lesion compared to the eutopic uterus at 3 or 29 days post-induction, respectively. Gene ontology enrichment analysis revealed that genes associated with the extracellular matrix, cell adhesions, immune function, cell growth, and angiogenesis were altered in the endometriotic lesion compared to the eutopic uterus.
Conclusion(s)
Based on gene expression analysis, the mouse model of surgically-induced endometriosis appears to be a good model for studying the pathophysiology and treatment of endometriosis.
Keywords: endometriosis, mouse model, gene expression profile
Introduction
Endometriosis is a chronic estrogen dependent disease characterized by the growth of ectopic endometrial tissue (1). The incidence is estimated to be up to 10% of women of reproductive age and 30-50% of women seeking treatment for infertility (1, 2). Despite the high incidence of endometriosis, the etiology is unknown. The theory proposed by Sampson in 1927 suggests that endometrial tissue is released into the peritoneal cavity via retrograde menstruation (3). The shed tissue then implants and grows ectopically. This theory is supported by the fact that up to 90% of women experience retrograde menstruation (4-6); and yet, endometriosis only affects a much smaller proportion of women.
In accordance with Sampson's hypothesis, endometriosis spontaneously develops only in menstruating animals such as humans and some primate species. However, ethical considerations limit the comprehensive study of endometriosis in humans. For example, repetitive laparoscopies cannot be performed to study disease pathogenesis and pathophysiology in humans. Animal models, however, provide a good alternative to test hypotheses about the time course and progression of the disease, developmental and environmental factors related to the pathophysiology of the disease, and interventions developed as treatments for the disease (7).
Many animal models have been developed to further our understanding of endometriosis. Primate models are clearly desirable, but cost and lack of housing facilities severely limit their use. As an alternative model system, rodents are readily available and comparatively inexpensive. Rodent models are unique in that they can be homologous, with tissues from the same or syngeneic individuals, or they can be heterologous, with explanted human tissue transplanted to immunocompromised animals (7).
Initial homologous rodent models of surgically-induced endometriosis were developed in the rat (8) and the techniques were then transferred to the mouse (9). Both models share similarities to the disease in women. Like human endometriotic lesions, those obtained from mice and rats are cyst like structures containing endometrial glands and stroma, contain numerous adhesions in the peritoneal cavity, often have hemosiderin laden macrophages present, and the growth is estrogen dependent (8-11). Both rat and mouse models are currently employed in endometriosis research (11-18).
Endometriotic lesions resemble eutopic endometrial tissue histologically, but endometriotic lesions have different gene expression profiles than eutopic endometrial tissue (1, 19-22). Although many investigators currently use the mouse model of endometriosis, an in depth analysis of altered gene expression in the mouse endometriotic lesion is lacking (11-15). Therefore, in this study we sought to define for the first time the altered gene expression profile in the mouse model of surgically-induced endometriosis at two time points: an early stage during establishment of endometriotic lesions (3 days) and a later stage after establishment of endometriotic lesions (29 days).
Methods
Animals
Animals were group housed in polysulfone cages in temperature (68-74°), humidity (40-44%), and light controlled (12L:12D) conditions at the University of Missouri – Columbia. All experimental procedures were approved by the University of Missouri Animal Care and Use Committee. Animals were fed Purina Rodent Chow 5008 (St. Louis, MO) and received acidified water ad libitum from polysulfone bottles. Intact mice were brought into estrus 72 hours prior to induction and collection of endometriosis by transferring male bedding to the female cages. Estrus cycle stage was assessed at the time of collection by examining vaginal cytology (23).
At six to seven months of age C57BL6/J female mice underwent surgery to induce endometriosis. A modified method, as previously described in the rat by Vernon and Wilson (8) and in the mouse by Cummings and Metcalf (9), was used. Briefly, animals were anesthetized and the left uterine horn was ligated with 5-0 black silk suture, removed, and opened longitudinally. Three 2 mm diameter sections were obtained from the excised uterine horn using a biopsy punch (Miltex 33-31, York, PA), and each was gently sutured to a blood vessel in the arterial cascade of the intestinal mesentery with 6-0 black nylon suture. The abdomen was closed with 5-0 absorbable suture and the skin was closed with surgical wound clips. Following surgery, buprenorphine was administered and animal health was closely monitored. Sham mice were treated identically to mice receiving surgery except that suture alone (6-0) was placed around an artery in the intestinal mesentery. At each time point, there were N=8 animals in both the surgery and sham groups.
Animals were collected either 3 or 29 days after surgical induction of endometriosis. At collection all animals were euthanized by carbon dioxide asphyxiation and cardiac puncture and tissues were excised. At 29 days post-induction one third of the intact uterine horn and one endometriotic lesion were 1) homogenized immediately in lysis binding solution (Ambion RNAquous, Austin, TX) for RNA isolation, 2) formalin fixed for about two hours for histological evaluation or 3) snap frozen in liquid nitrogen for later analysis. At collection 3 days post-induction, tissues were handled in the same manner except that two endometriotic lesions were homogenized in lysis binding solution, one was formalin fixed, and no endometriotic lesions were snap frozen.
Histology and Immunohistochemistry
Following fixation, tissues were washed for 30 minutes in PBS three times, dehydrated in 70% ethanol, and paraffin embedded (Research Animal Diagnostic Laboratory, University of Missouri-Columbia). Five-micron tissue sections were cut and hematoxylin and eosin stained. Photographs were taken on an Olympus Microscope at 400× magnification.
Immunostaining was carried out as described previously (24). Briefly, five-micron tissue sections were incubated with rabbit anti-rat Ki67 (Neomarkers – RB-1510-P0, Fremont, CA) followed by secondary antiserum, goat anti-rabbit IgG labeled with Alexa 488 (Invitrogen – A11034, Carlsbad, CA), with 7 μM DAPI (Invitrogen). Slides were mounted and 24 hours later pictures of three fields of two consecutive sections were taken with a Zeiss Axiophot microscope with epi-fluorescence at 400× magnification. The negative control was incubated with blocking buffer alone instead of primary antibody. Positively stained cells in the stromal and epithelial compartments were manually counted with the aid of the computer program Metamorph (Molecular Devices, Union City, CA) in three fields each of eutopic uterus, endometriotic lesion, and sham uterus.
Total Ki67 positive cells were averaged over 3 fields. Paired t-tests were used to analyze differences in Ki67 positive cells in the endometriotic lesion and the eutopic uterus because these tissues are excised from the same animal. Independent t-tests were used to analyze differences in Ki67 positive cells in the sham uterus and eutopic uterus. Statistics were carried out using SPSS for Windows (Rel. 16.0.1. 2007. Chicago: SPSS Inc.).
RNA Isolation
At collection tissues were excised (15-20 mg uterus and about 2 mg or 10 mg endometriotic lesion at 3 or 29 days, respectively), immediately homogenized in 500 μl lysis binding solution (Ambion), and stored at -80°C. Total RNA was isolated from homogenates using RNAqueous spin columns (endometriotic lesions from animals collected at 3 days were isolated with RNAqueous-micro), treated with Turbo DNAse (Ambion), ethanol precipitated, and concentrated. RNA quantity and purity was assessed by UV spectrophotometry using a Nanodrop ND1000 spectrophotometer (Wilmington, DE). RNA quality was visualized on an ethidium bromide stained 1% agarose gel. ImageJ 1.38× (National Institute of Mental Health, Bethesda, MD) was used to determine the intensity of the 18S and 28S bands. The ratio of 28S to 18S was used as a measure of RNA quality. Only samples with a ratio ≥ 1.0 were used for further analysis. RNA from one endometriotic lesion at each time point did not meet these requirements and therefore was not included in further analyses.
Illumina Microarray
For samples analyzed by microarray, quality was further assessed using an Agilent 2100 bioanalyzer (University of Missouri DNA Core). All samples used for the microarray had a 28S to 18S bioanalyzer signal ratio of 0.91 or greater and did not appear to be degraded. Three or four samples per group (3 or 29 day eutopic uterus, endometriotic lesion, or sham uterus) were selected based on estrus cycle stage (estrus, diestrus 1, or early diestrus 2) by vaginal cytology at collection and similar uterine weights.
Illumina mouse ref-8 (vs1) microarrays were used for the global analysis of gene expression. Each array probes 24,886 RefSeq transcripts targets largely from the Rockefeller University Mouse SdB3 and NCBI Entrez Gene databases. Microarray sample preparation and image acquisition was carried out at the University of Missouri DNA Core Facility. Five hundred nanograms of total RNA per sample was used to make biotin labeled antisense RNA (aRNA) using the Illumina TotalPrep RNA Amplification Kit (Ambion) as per the manufacturer's recommendations. Briefly, total RNA was reverse transcribed to first strand cDNA with oligo(dT) primers bearing a 5′-T7 promoter using ArrayScript reverse transcriptase (Ambion). The first strand cDNA then underwent second-strand synthesis and clean up to become the template for in vitro transcription. Biotin-labeled aRNA was synthesized using T7 RNA polymerase with biotin-NTP mix and purified. aRNA (0.75 μg) was hybridized to the Illumina MouseRef-8 Expression BeadChip array at 58° C for 20 hours. After hybridization, the chips were washed and stained with streptavidin-C3. The image data was acquired using the BeadArray reader (Illumina, San Diego, CA).
Analysis of microarray gene expression data was primarily performed using the Linear Models for Microarray Data (limma) package (25) and the lumi package (26), available through the Bioconductor project (27). After this pre-processing was completed (see Supplementary Materials), the analysis of differential gene expression was performed using moderated t-statistics applied to the log-transformed (base 2) normalized intensity for each gene using an Empirical Bayes approach (28, 29), in which the standard errors are shrunk towards a common value. Two contrasts were tested and their nature required different models to be fit. Because the measurements from the endometriotic lesion and eutopic uterus are taken from the same animal, a modified mixed linear model that treated each animal as a block accounted for the dependency between measurements. The within-block correlations were constrained to be equal between genes (29), and then information was borrowed across genes to moderate the standard deviations between genes via an empirical Bayes method (28). The comparison between sham uterus and eutopic uterus was analyzed using a more standard linear model since the measurements were independent.
For each contrast of interest, the log fold change was computed along with the aforementioned-moderated t-statistics nominal and adjusted p-values and the estimated log-odds ratios of differential expression. Adjustment for multiple testing was made using the false discovery rate (FDR) method of Benjamini and Hochberg (30). Probes with adjusted p-values less than 5% (which corresponded to controlling the FDR at 5% or less) and fold changes of at least 2 were selected as differentially expressed. To facilitate interpretation in this report, log fold changes were transformed back to fold change on the data scale and log-odds ratios of differential expression were converted into probabilities of differential expression.
Gene Ontology Enrichment
Gene ontology (GO) analyses were conducted on the resulting list of significantly different genes. The purpose of the analyses was to test the association between Gene Ontology Consortium terms (31) and differentially expressed genes. GO analyses were carried out for over-representation of biological process, molecular function, and cellular component ontologies, and for each GO term we computed the nominal hypergeometric probability of observing the number of differentially expressed genes falling within a GO term due to chance (see Supplementary Material for more details). The results were used to assess whether the number of selected genes associated with a given term was larger than expected under the null hypothesis. GO terms containing less than 10 genes from our gene universe were not considered to be reliable indicators. Terms with a p-value <1% were considered significant.
cDNA Synthesis for Real-Time RT-PCR
Two hundred nanograms total RNA (n=6-8 animals per group) were reverse transcribed with SuperScript III First-Strand Synthesis Kit (Invitrogen) as per the manufacturer's recommendations and as previously described in more detail (24). For each sample, a negative control (noRT) was prepared identically but without reverse transcriptase or ribonuclease inhibitor. After cDNA was generated it was diluted 2-fold with ultra pure, nuclease-free water.
Real time RT-PCR
Real time RT-PCR was performed in an ABI 7500 instrument (Applied Biosystems, Foster City, CA) using either the SYBRgreen technology with primers designed in Vector NTI (Invitrogen) and synthesized by IDT DNA (Coralville, IA) (Estrogen receptor alpha (Esr1) (NM_007956) forward primer: 5′-ACCATTGACAAGAACCGGAG-3′, reverse primer: 5′-CCTGAAGCACCCATTTCATT-3′, Tm=84.9, 18s rRNA (K01364) forward primer: 5′-TTCCTTACCTGGTTGATCCTGCCA-3′, reverse primer: 5′-AGCCATTCGCAGTTTCACTGTACC-3′, Tm=81, Mmp10 (NM_019471) forward primer: 5′-TGCAGTTGGAGAACACGGAGA-3′, reverse primer: 5′-CTGAGGGTGCAAGTGTCCATTT-3′, Tm=82.1) or with TaqMan primer and probe Assay on Demand sets from ABI (Hp, Mmp3, Mmp12, Mmp13, and Timp1). SYBRgreen primers were designed to be intron spanning and prior to use were validated by comparing the Ct values of cDNA generated from DNAsed and non-DNAsed samples and by the presence of a single peak in the dissociation curve. Only primer sets with greater than 10 cycles difference between RT and noRT reactions were used. SYBRgreen primer set efficiencies were determined in four 10-fold dilutions of cDNA using the equation E=10ˆ[-1/slope] (32) and were all above 84%. Efficiencies of TaqMan primers were assumed to be 100% as per the manufacturer (33).
SYBRgreen real time RT-PCR reactions were prepared with Platinum SYBR green qPCR SuperMix-UDG with ROX (Invitrogen) with primer concentrations at 70 nM for Esr1 and Mmp10 and 140 nM for 18s rRNA. TaqMan real time RT-PCR reactions were prepared with TaqMan Universal PCR Master Mix (ABI) and primer-probe sets at 1×. The reactions were performed as per manufacturers' instructions except that the total volume was proportionally reduced to 25 μL. Prior to identification of a suitable housekeeping gene for real time RT-PCR gene expression normalization, cDNA was column purified, quantified, and diluted as described previously (24) and 5 ng cDNA or noRT was added to each real time RT-PCR reaction. Once 18s rRNA was identified as an appropriate housekeeping gene, 5 μL of 4-fold-diluted cDNA or noRT was added to each real time RT-PCR reaction and target gene expression was normalized to expression of 18s rRNA. For each sample in each run, two reactions containing cDNA and one reaction containing noRT were performed. Real time RT-PCR was repeated three times for each gene.
Real Time RT-PCR Calculation of Relative Gene Expression
The ΔΔCT method was used to calculate relative changes in gene expression (34, 35). The eutopic uterus was set as the reference for each time point and a fold change of 1 indicates that the gene expression is not different from that in the eutopic uterus. Fold changes were calculated using either the Ct value (if cDNA was quantified) or ΔCt value (if the cDNA was not quantified but was normalized to 18s rRNA). Because the endometriotic lesion and eutopic uterus were collected from the same animal, gene expression, and thus Ct values, for these tissues are dependent. Fold change in the endometriotic lesion was calculated for each animal relative to the individual animal's eutopic uterine gene expression and the geometric mean of the resulting fold changes was taken across triplicate real time RT-PCR runs and across all the endometriotic lesion samples to obtain a fold change value for the endometriotic lesions. Fold change in the sham uterus was calculated in the same manner except that individual animal fold changes were calculated relative to the mean expression in the eutopic uterus due to the independent nature of the tissues.
All statistics were performed on the average Ct values and were carried out using SPSS for Windows (Rel. 16.0.1. 2007. Chicago: SPSS Inc.). Since Ct values are logarithmic in nature, non-parametric tests were used for the statistical analysis. For the paired comparison of endometriotic lesions to eutopic uterus, Ct values were compared using the non-parametric Wilcoxon signed-rank exact significance 1-tailed test. A one-tailed test was used because the directionality of the fold change was hypothesized based on the results from the microarray analysis. For the comparison of gene expression in the sham uterus to that in the eutopic uterus, Ct values were analyzed using the non-parametric Mann-Whitney U exact two-tailed test as is appropriate for independent data points.
Results
Mouse Endometriotic Lesions Resemble Lesions from Women
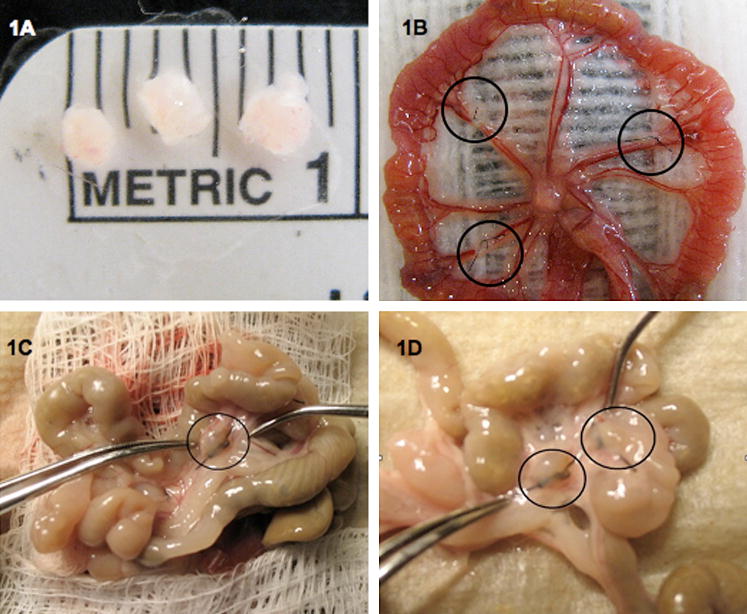
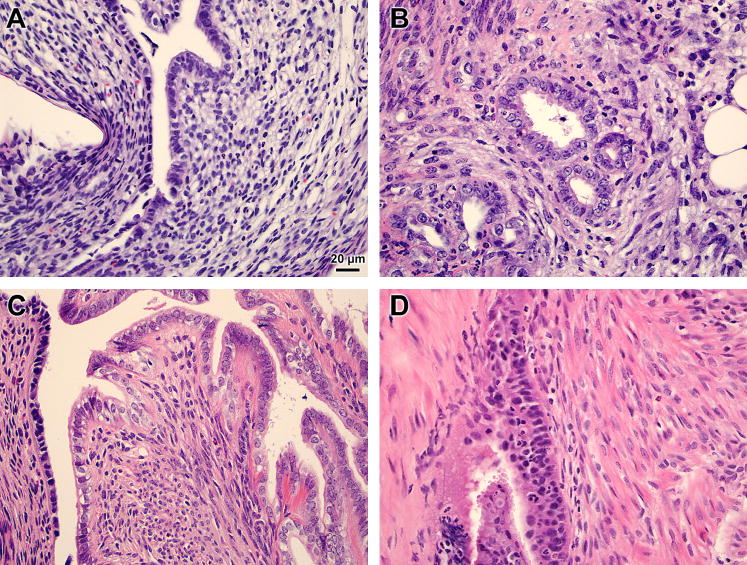
Three days post-induction the endometriotic lesions were hemorrhagic, but by 29 days, the endometriotic lesions had grown and had become cyst-like and encapsulated. At 29 days, the lesions had grown from an average of 1.47 mg at induction to an average of 8.47±3.7 mg and 2.85±1.25 mg with and without fluid, respectively. Most of the endometriotic lesions collected 29 days post-induction were fluid filled and there were numerous adhesions noted in the peritoneum (Fig. 1). Endometriotic lesions in our mouse model contained numerous endometrial glands and stroma similar to those seen in the eutopic uterus (Fig. 2). Sham mice were devoid of ectopic endometrial tissue and peritoneal adhesions.
Figure 1. Surgical induction and collection of endometriosis in the mouse.
Endometriotic lesions were collected 3 or 29 days post-induction from mice surgically-induced to have endometriosis. (A) Three uterine biopsies prior to surgical induction. (B) Three implants sutured to alternating arteries of the intestinal mesentery. (C) An endometriotic lesion 29 days post-induction. (D) Two endometriotic lesions that are surrounded by numerous adhesions 29 days post-induction of endometriosis.
Figure 2. Histology of mouse endometriotic lesion and eutopic uterus.
Mice underwent surgical-induction of endometriosis and eutopic uteri (A, C) and endometriotic lesions (B, D) were excised 3 (A, B) or 29 (C, D) days later. Representative hematoxylin and eosin stained sections at 400× magnification are shown.
Cell Proliferation
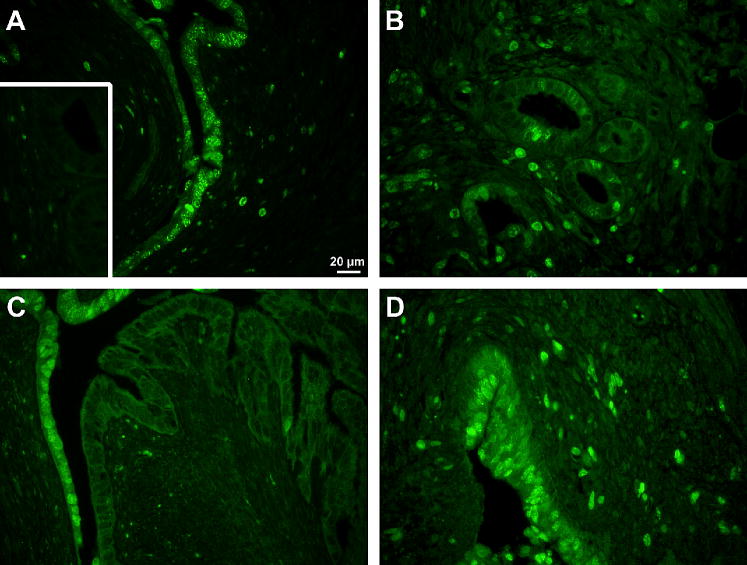
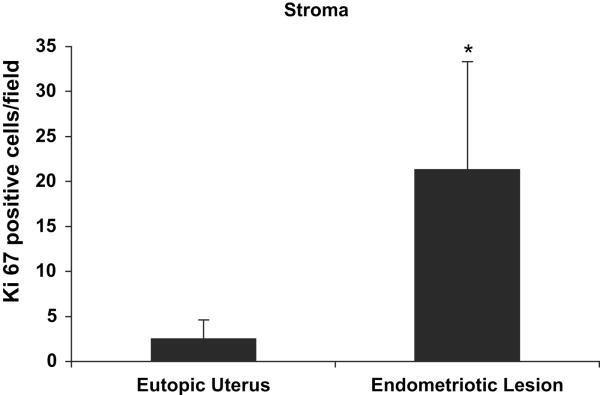
Endometriotic lesions showed strong proliferation 3 days post-induction. In the endometriotic lesion stroma there were 8.3 times as many positively stained cells as in the eutopic uterine stroma, P<0.001 (Figs. 3 and 4). For all other comparisons there were no differences in cell proliferation (data not shown).
Figure 3. Immunohistochemistry of mouse endometriotic lesion and eutopic uterus.
Mice underwent surgical induction of endometriosis and eutopic uteri (A, C) and endometriotic lesions (B, D) were excised 3 (A, B) or 29 (C, D) days later. Representative Ki67 stained sections at 400× magnification are shown. Inset is negative staining control. Pictures are sister sections to those in Fig. 2.
Figure 4. Ki67 quantification.
Tissue sections were immunohistochemically stained with Ki67. Positively stained cells in the stroma were identified and quantified in each section with the aid of the computer program Metamorph. Results shown are average ± standard deviation Ki67 positive cells/field in the eutopic uterus and endometriotic lesion stroma at 3 days post-induction of endometriosis. *Paired t-test, P < 0.05.
Microarray Results
Sentrix Mouse-8 Expression BeadChips (Illumina) whole-genome gene expression arrays were probed with asRNA from eutopic uteri, endometriotic lesions and sham uteri from 3 or 4 animals per group. In the microarray experiment gene expression in the endometriotic lesion was considered altered if the adjusted p-value after multiple testing was <0.05 and the gene expression was 2 fold or greater different relative to gene expression in the eutopic uterus.
The expression of 617 and 159 probes was altered in the endometriotic lesion compared to the eutopic uterus 3 or 29 days post-induction, respectively. This corresponded to 479 and 114 unique and well-annotated genes at each time point. At 3 days post-induction 279 genes were up-regulated and 200 genes were down-regulated (Supplementary Table 1). At 29 days post-induction 95 genes were up-regulated and 19 genes were down-regulated (Supplementary Table 2). There were no genes with significantly altered expression between the sham uterus and eutopic uterus at either time point.
GO Term Enrichment
GO analysis revealed functional terms in which there was overrepresentation of altered gene expression in the endometriotic lesion compared to the eutopic uterus. There were 55 and 28 biological processes, 10 and 5 cellular component, and 19 and 12 molecular function terms overrepresented 3 and 29 days post-induction, respectively. For a complete list of overrepresented terms see Supplementary Tables 3 and 4. There were many enriched terms including those of cell adhesion, collagen catabolism, immune response, cell growth, and angiogenesis. For genes with altered expression in selected overrepresented terms see Tables 1 and 2.
TABLE 1. Selected overrepresented gene ontology terms at 3 days post-induction.
| Term | Odds ratio | Genes |
|---|---|---|
| Positive regulation of adaptive immune response | 11.7 | C3, Cd1d1, Fcer1g, Fcgr3, Tnfsf13b |
| Collagen catabolic process | 11.7 | Mmp10, Mmp13, Mmp14, Mmp3, Mmp9 |
| Leukocyte migration | 8.07 | Csf3r, Scye1, Fcer1g, Fcgr3, Il1b, S100a9, Cxcl15, Selplg |
| Bone mineralization | 7.99 | Cd276, Adrb2, Mmp13, Ptn |
| Regulation of endocytosis | 6.02 | C3, Fcer1g, Fcgr2b, Fcgr3, Fgf10, Mfge8 |
| Locomotory behavior | 5.30 | Csf3r, Fcer1g, Fcgr3, Fgf10, Hoxd10, Cyr61, Il1b, Kit, Xcl1, Pde1b, S100a8, S100a9, Ccl11, Ccl2, Ccl3, Ccl4, Ccl6, Ccl9, Cxcl15, Sema3f, Sod2, Ccl24, Pf4, Rasd2, Atp1a2 |
| Inflammatory response | 4.62 | Chi3l4, C3, Cd14, F2r, Fcer1g, Fcgr3, Fn1, Cxcl1, Il1b, Cfp, Stab1, Saa3, Ccl11, Ccl2, Ccl3, Ccl4, Cxcl15, Tgfb1, Thbs1, Tlr13, Ccl24, Nupr1, Chst1 |
| Angiogenesis | 4.25 | Angpt2, Runx1, Col18a1, Elk3, Eng, Ctgf, Flt1, Cyr61, Anpep, Meis1, Nos3, Notch4, Sema5a, Sox18, Thbs1, Thy1, Tnfaip2, Tnfrsf12a, Egfl7, Robo4 |
| Phosphate transport | 3.75 | Col18a1, Col4a1, Col4a2, Col7a1, C1ql2, 1110001D15Rik, Scara5, C1qtnf3 |
| Regulation of cell motility | 3.75 | Centd3, Col18a1, Pdpn, Lama2, Pdgfb, Pecam1, Egfl7, Robo4 |
| Blood vessel development | 3.72 | Angpt2, Agtr1a, Runx1, Cdh5, Col18a1, Elk3, Eng, Fgf10, Ctgf, Flt1, Gja5, Cyr61, Junb, Anpep, Meis1, Nos3, Notch4, Sema5a, Sox18, Myocd, Thbs1, Thy1, Tnfaip2, Tnfrsf12a, Egfl7, Reck, Robo4 |
| Negative regulation of cell proliferation | 3.51 | Cd276, Axin2, Cdh5, Cdkn1a, Fcgr2b, Fgf10, Gpc3, Klf4, Msx1, Slfn1, Sod2, Tgfb1, Irf6, Nupr1, Cd274 |
| Wound healing | 3.41 | Entpd2, Elk3, F10, F2r, F2rl1, F2rl3, Fn1, Pecam1 |
| Immune response | 3.35 | Spon2, Cd276, Orai1, C3, Cd14, Cd1d1, Ercc1, Fcer1g, Fcgr2b, Fcgr3, Lilrb4, Gpx2, Cxcl1, H2-Aa, H2-Ab1, H2-Eb1, Cd74, Il1b, Xcl1, Il1rl1, Clec4d, Cfp, Ccl11, Ccl2, Ccl3, Ccl4, Ccl6, Ccl9, Cxcl15, Serpina3g, Tgfb1, Thy1, Vav1, Tnfsf13b, Tlr13, Ccl24, Clec4n, Pf4 |
| Cell adhesion | 2.93 | Spon2, Epdr1, Amigo2, Scarb2, Cdh13, Cdh16, Cdh5, Col18a1, Col7a1, Csf3r, Vcan, Efs, Eng, Ctgf, Fn1, Pdpn, Icam2, Cyr61, Itga3, Lama2, Cd93, Mfge8, Pecam1, Stab1, Selplg, Spp1, Thbs1, Thbs2, Tnc, Vav1, Vcam1, Wisp1, Clca5, Tnfrsf12a, Sdk1, Ctnnal1, Dpt, 1110001D15Rik, Esam, Pcdh1, Mfap4, Tnxb, Mcam, Gpnmb |
| Regulation of growth | 2.28 | Adrb2, Socs3, Ctgf, Foxs1, Gas6, Gpc3, Cyr61, Igfbp6, Lgmn, Myocd, Wisp1, Esm1, Htra3, Tcfcp2l1 |
| Organ morphogenesis | 2.28 | Angpt2, Aldh1a1, Runx1, Cebpb, Col18a1, Elk3, Eng, Fgf10, Ctgf, Flt1, Gjb6, Gpc3, Hoxd10, Cyr61, Junb, Anpep, Meis1, Msx1, Ndp, Nos3, Notch4, Tnfrsf11b, Etv4, Sema5a, Sod2, Sox18, Myocd, Tgfb1, Thbs1, Thy1, Tnfaip2, Grem1, Slc26a4, Tnfrsf12a, Egfl7, Chst11, Robo4 |
Pelch. Gene expression in mouse endometriosis. Fertil Steril 2010.
TABLE 2. Selected overrepresented gene ontology terms at 29 days post-induction.
| Term | Odds ratio | Genes |
|---|---|---|
| Acute inflammatory response | 11.4 | Cfd, Serping1, C2, Nupr1 |
| Leukocyte migration | 9.89 | Il1b, Nkx2-3, S100a9 |
| Regulation of blood vessel size | 9.40 | Apoe, Nppa, Gucy1a3 |
| Chemotaxis | 6.93 | Il1b, S100a8, S100a9, Ccl11, Ccl9 |
| Ossification | 6.51 | Chrd, Ctgf, Mgp, Mmp13, Spp1 |
| Response to wounding | 6.20 | Cfd, Apoe, Serping1, C2, F2r, F3, Gpx2, Il1b, Saa3, Ccl11, Serpina3n, Nupr1 |
| Regulation of growth | 4.04 | Apoe, Ctgf, Igfbp2, Wisp1, Wisp2, Tcfcp2l1 |
| Immune response | 3.50 | Il1b, Clec4d, Nkx2-3, Ccl11, Ccl9, Serpina3g, Cxcl14 |
| Tissue development | 3.28 | Apoe, Chrd, Ctgf, Gata6, Klf4, Mgp, Mmp13, Spp1 |
Pelch. Gene expression in mouse endometriosis. Fertil Steril 2010.
Gene Expression Confirmed by Real Time RT-PCR
We determined by microarray that most common housekeeping genes, including Gapdh, Actb, and Rpl13a, had significantly altered gene expression profiles in the endometriotic lesion compared to the eutopic uterus, and thus were not suitable house keeping genes for this set of cDNA (24). The expression of 18s rRNA however, was found by both microarray and real time RT-PCR to be unchanged across all six tissues (eutopic uterus, endometriotic lesion, and sham uterus at both 3 and 29 day time points) and so un-quantified cDNA was normalized to 18s rRNA expression. To validate the microarray data, RNA from 6-8 animals was reverse transcribed to cDNA and real time RT-PCR was performed for six genes known to be altered in the endometriotic tissue of women with endometriosis. These genes were chosen based on our previous experience and our interest in molecules that alter the extracellular matrix.
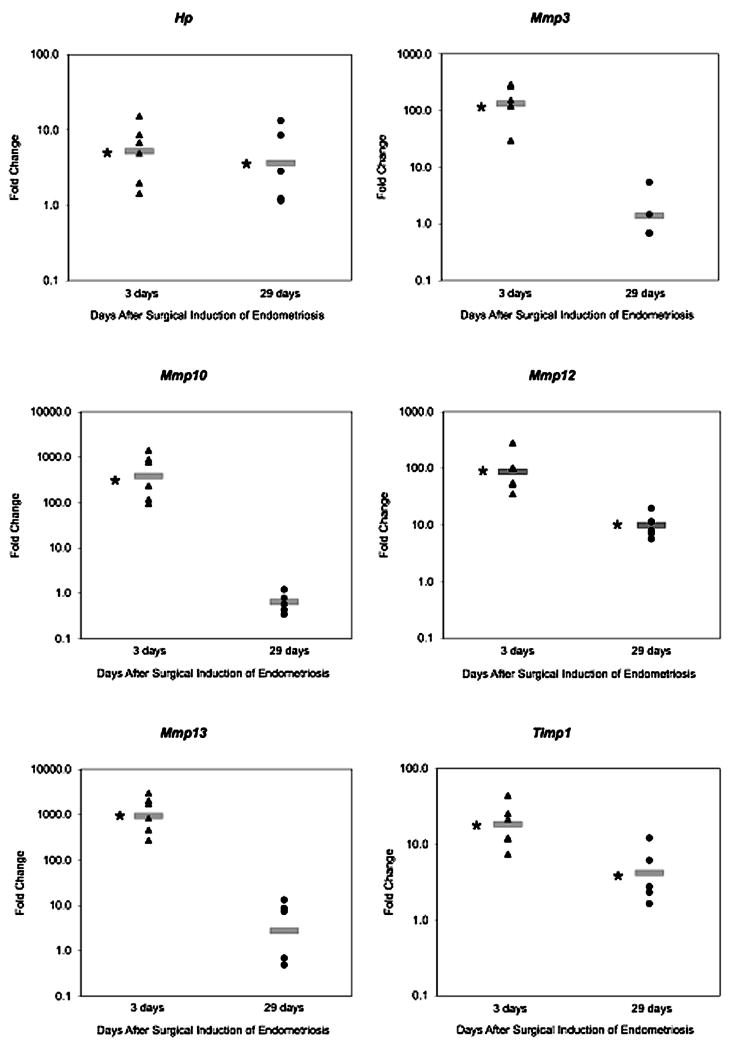
At 3 days post-induction the expression of six genes found to be altered on the microarray was confirmed by real time RT-PCR. Expression of Hp, Mmp3, Mmp10, Mmp12, Mmp13, and Timp1 was altered in the endometriotic lesion by 5.22, 132, 381, 87.0, 938, and 18.5 fold respectively (P < 0.05) (Table 3, Fig. 5).
TABLE 3. Fold change in endometriotic lesion versus eutopic uterus: comparison between Illumina MouseRef-8 microarray and real-time RT-PCR.
| 3 days after induction | 29 days after induction | |||
|---|---|---|---|---|
| Gene | Microarray | Real time RT-PCR | Microarray | Real time RT-PCR |
| Hp | 2.42a | 5.22a | 2.67a | 3.15a |
| Mmp3 | 51.0a | 132a | 1.56 | 1.30 |
| Mmp10 | 31.9a | 381a | 1.06 | 0.58 |
| Mmp12 | 7.54a | 87.0a | 3.19a | 9.88a |
| Mmp13 | 2.52a | 938a | 2.69a | 2.12 |
| Timp1 | 12.6a | 18.5a | 5.01a | 3.93a |
Note: Illumina MouseRef-8 chips were probed with RNA from 3 to 4 animals per group. Real-time RT-PCR was performed on 6–8 animals per group. Data were analyzed as described in Material and Methods. Average fold change in endometriotic lesions relative to eutopic tissue at each time point is shown. Gene expression was considered altered in endometriotic lesions if the fold change was ≤0.5 or ≥2 and P<.05.
P<.05 for endometriotic lesion vs. eutopic uterus within each time point.
Pelch. Gene expression in mouse endometriosis. Fertil Steril 2010.
Figure 5. RT-PCR confirmation of microarray results for selected genes.
Endometriotic lesions and eutopic uteri were excised from mice surgically-induced to have endometriosis after 3 or 29 days. RNA was isolated, cDNA generated, and RT-PCR performed and analyzed using the ΔΔCt method (35). Scatter plots show representative fold changes for each animal collected at 3 or 29 days post-induction. Fold change is relative to the eutopic uterus at each time point, and a fold change of 1 indicates there is no difference between gene expression levels in the endometriotic lesion and eutopic uterus. The gray bar represents the average fold change in the endometriotic lesion at each time point. *P<0.05 for average gene expression in endometriotic lesion versus eutopic uterus.
At 29 days post–induction the results of the microarray analysis were confirmed for five genes by real time RT-PCR. Expression of Hp, Mmp12, and Timp1 was altered in the endometriotic lesion by 3.15, 9.88, and 3.93 fold respectively (P<0.05). Expression of Mmp3 and Mmp10 was unaltered in the endometriotic lesion in both the microarray and real time RT-PCR analyses (Table 3, Fig. 5). Of the six genes examined, only Mmp13 expression was not confirmed by real time RT-PCR. Expression of Mmp13 in the endometriotic lesion was altered on the array by 2.69-fold (P < 0.02) but by real time RT-PCR the fold change of 2.12 was not significant (p = 0.16).
For all genes tested by real time RT-PCR, only Esr1 and Hp at 3 days post-induction were significantly different between the sham uterus and the eutopic uterus (1.59 and 3.12 respectively, P=0.04) (data not shown).
Of the six genes examined at the two time points tested, we confirmed the expression of 11 out of 12 targets, or 92%, which is very similar to previous reports (36). Spearman's rank correlation of the fold changes observed at 29 days post-induction for the microarray and real time RT-PCR was 93% (P<0.01). At 3 days post-induction Spearman's rank correlation of the microarray and real time RT-PCR data was 57%. However, the microarray data for Mmp13 at 3 days post-induction yields two dramatically different fold changes (2.52 and 21.1). In the microarray analysis, if two probes for the same gene were reported as “altered,” the result for the probe located closest to the ABI primer-probe set was reported (applies to results for Hp, Mmp3, and Mmp13 expression at 3 days post-induction). Of all the genes in which there were two or more altered probes, Mmp13 is the only gene in which there was a large discrepancy between the two fold changes. When the alternate (21.1) fold change is considered, Spearman's rank correlation at 3 days post-induction is 82% (P<0.05).
Esr1 expression was not found to be altered during the microarray analysis (0.53 and 0.80-fold at 3 and 29 days post-induction, respectively) but because ESR1 expression has been shown to be significantly down-regulated in women with endometriosis it was examined by real time RT-PCR (37). The expression of Esr1 by real time RT-PCR was 0.39 and 0.52-fold in the endometriotic lesion compared to the eutopic uterus at 3 or 29 days post-induction, respectively (P<0.05).
Discussion
The current work shows for the first time that the altered gene expression profile in this mouse model of endometriosis mirrors that observed in endometriotic lesions from women. This study used a genome-wide microarray analysis to examine the altered gene expression profile at two time points in a mouse model of surgically-induced endometriosis. Importantly, we found many similarities in the altered gene expression profile observed in endometriotic lesions from mice and from women (20). Genes that are known to be mis-expressed in human endometriosis were identified as well as several novel genes. We confirmed the differential expression of six genes by real time RT-PCR. More genes were differentially expressed at 3 days post-induction than at 29 days post-induction. This is in agreement with previous studies suggesting that early endometriotic lesions are more biologically active (38).
Current and previous work has shown that mouse and human endometriotic tissue progresses from a more to a less biologically active form over time. Endometriotic lesions exhibit various phenotypes including red, white, or black/blue (39, 40). Repeated laparoscopies at one, four and 10 months after induction of endometriosis in the baboon, indicated that endometriotic lesions progress through these colors (39, 40). In our model, endometriotic lesions collected 3 days post-induction were small in size and hemorrhagic. Based on the color of the endometriotic lesions collected and their altered gene expression profile, we hypothesize that the collection at 3 days post-induction mimics red lesions in women and should be useful for studying the early endometriotic lesion establishment. In contrast, endometriotic lesions collected from mice 29 days post-induction were clear, fluid filled (cystic), surrounded by extensive adhesions, and mimic established typical endometriotic lesions in women.
Tissue invasion into the mesothelial cell layer at ectopic sites and the development of a new blood supply require significant modification of the extracellular matrix. Endometriotic lesions from women with endometriosis have been shown to secrete many proteins that alter the extracellular space, such as matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) (41, 42). Evidence suggests MMPs and TIMPs, which are highly regulated throughout the menstrual cycle in eutopic endometrium in women without endometriosis, are constitutively expressed in endometriotic lesions (reviewed in (10)) perhaps due to progesterone resistance (43). Additionally, misexpression of MMPs and TIMPs is known to increase tissue invasion in cancer cell lines (44-46)
In the current study, the expression of several extracellular modifying genes was altered in endometriotic lesions. At 3 days post-induction several GO terms associated with protein and collagen catabolism were overrepresented. In the mouse model of endometriosis the expression of Mmps 2, 3, 9, 10, 12, 13, and 14 was increased in the endometriotic lesion relative to the eutopic uterus. Additionally, the expression of Timp 1 and 3 was increased in the endometriotic lesion. At 3-days post induction the microarray results indicated that Mmp3 and Mmp10 were the most over-expressed genes in the endometriotic lesion relative to the eutopic uterus (Supplementary Table 1), yet these two genes were no longer significantly altered at 29 days post-induction (Table 3, Fig. 5). This further supports our hypothesis that the collection of endometriotic lesions at 3 days post-induction represents a time when the tissue is becoming established.
There are many reports demonstrating altered immune system function in women with endometriosis, and the strong inflammatory response seen in the peritoneal cavity of endometriosis patients is believed to contribute to the pathophysiology of the disease (47, 48). For example, HP is upregulated in endometriotic lesions from women and has been found to bind to and interfere with macrophage phagocytic function and to have angiogenic properties (49-51). This suggests a plausible mechanism for failure of peritoneal immune cells to clear ectopic endometrial tissue from the peritoneal cavity in women with endometriosis. In the current study the expression of immunomodulatory genes was altered in the endometriotic lesion compared to the eutopic uterus at both 3 and 29 days post-induction. In the GO analysis, the large GO terms “immune response” and “inflammatory response” as well as several smaller terms including those pertaining to leukocyte migration and regulation of phagocytosis were overrepresented. Specifically, in parallel with observations in women, we found that Hp was up-regulated in the endometriotic lesion at both 3 and 29 days post-induction.
In order to grow and become established, endometriotic cell proliferation must occur. It is unclear, however if there are differences in proliferation between human endometriotic lesions and eutopic tissue (52-54). Iwabe et al. previously reported that endometriotic stromal cells responded to the inflammatory cytokines tumor necrosis factor alpha (TNF) and interleukin 8 (IL8) with increased proliferation in culture (55). TNF and IL8 protein levels are increased in peritoneal fluid of endometriosis patients and it is likely that the cytokine milieu in the peritoneal environment is stimulating stromal cell proliferation (55). Consistent with this, we noted an 8.3-fold increase in stromal cell proliferation in endometriotic lesions relative to eutopic uterus at 3 days post-induction.
The current study is in strong agreement with a recently published human microarray study by Eyster et al. (20). Both studies show many of the same genes altered in endometriotic lesions relative to eutopic uterus. Importantly, genes in gene ontology categories thought to be particularly important to endometriosis were altered in the same direction in both the human study and the current mouse study. Some examples include cadherins, fibronectin, integrins, cell growth genes, THBS1, and TGFB and its receptor ENG. Both studies demonstrate a general lack of altered expression of genes related to apoptosis or estrogen and progesterone synthesis and signaling. For example, in the current study the only apoptosis related gene altered in endometriotic lesions was Bcl2a1a at 3 days post-induction and the only estrogen or progesterone synthesis or signaling gene altered was Esr1. While in strong agreement with the work by Eyster et al. this is in contrast to other reports that show altered expression of other apoptosis (56) and steroid hormone signaling genes (37, 57). The lack of altered expression of steroid hormone metabolizing genes may be related to the relatively short amount of time the lesions were allowed to grow. Fazleabas and colleagues found that in the baboon model of endometriosis, over expression of aromatase did not occur until 9 to 12 months post induction (39).
Gene Expression in the Eutopic Uterus
The previous discussion has focused on the differences in gene expression between the endometriotic lesion and the eutopic uterus. There are also known differences in gene expression in the eutopic endometrium of women with endometriosis compared to control endometrium from women without the disease (58). It is currently not known, however, if the altered gene expression in the eutopic endometrium is due to some inherent difference in gene expression between patients and controls, or alternatively, if the existence of endometriotic lesions generates an altered peritoneal environment that acts on the eutopic endometrium and thus results in aberrant eutopic gene expression.
In the current study, the use of syngeneic animals did not allow us to assess the possibility that there were inherent differences in eutopic uterine gene expression. By including sham control mice, however, we could note any alterations in gene expression in the eutopic uteri that might result from the presence of the endometriotic lesions. In the real time RT-PCR analysis Hp expression in the sham uterus at 3 days post-induction was significantly increased relative to the eutopic uterus; however, the microarray analysis did not reveal any genes with greater than two-fold altered expression between the sham uterus and the eutopic uterus. This finding is in contrast to a recent study by Lee at al. using a similar surgically-induced mouse model of endometriosis that reported altered expression of five of six genes tested in the eutopic uterus of mice compared to control uterus from sham animals (59). This difference may be attributable to the greater amount of tissue implanted in the peritoneum, different location of the implants, or to the longer length of time the endometriotic lesions were allowed to grow post-induction (98 days) in the experiment by Lee et al. (59).
Conclusions
The use of animal models to further our understanding of the pathophysiology of endometriosis has brought new discoveries to the field (7, 10). The present study is in strong agreement with pervious reports of altered gene expression in the rat model of surgically-induced endometriosis and with the altered gene expression profile recently described by Flores et al. (16, 60, 61). These studies support the validity and use of both rodent models. The mouse model of surgically-induced endometriosis presents many advantages for furthering our knowledge of the disease processes in endometriosis. Compared to other model systems, mice are inexpensive, have a short reproductive cycle, and their use in research is considered ethical by most standards. Additionally, a complete time course evaluation of altered endometriotic gene expression spanning many months can be performed in the mouse, and the effect of developmental factors and interventions on disease etiology and progression can be examined. These advantages and the finding that the gene expression profile in the mouse endometriotic lesion mirrors that seen in women makes the mouse model a useful tool for studying endometriosis pathogenesis, pathophysiology, and treatment.
Supplementary Material
Supplementary Table 1: Altered gene expression by microarray in the endometriotic lesion at 3 days post-induction.
Supplementary Table 2: Altered gene expression by microarray in the endometriotic lesion at 29 days post-induction.
Supplementary Table 3: Gene ontology terms overrepresented at 3 days post-induction.
Supplementary Table 4: Gene ontology terms overrepresented at 29 days post-induction.
Acknowledgments
Special thanks to Jacob Redel and Alison Ghormley for their assistance in developing the model system in our laboratory, to Joseph Beeman for his assistance during the inductions and collection, to Stacey Winkler for her assistance in photographing H&E sections and counting glands, to Bridget Niebruegge for her assistance during the inductions and collections as well as for her extensive work quantifying proliferating cells, to Mingyi Zhou and the UMC DNA Core for their work on the microarray study and to Dr. Danny Schust for his editorial suggestions.
Financial support: Support to SCN from University of Missouri Research Council and Research Board Grants, NIH (KO1 DK60567-01) and March of Dimes Basal O'Connor Starter Awards. Support to JWD from March of Dimes Basal O'Connor Starter Award. Support to ALS from the Endocrine Society and KEP from NIH T90 Clinical Biodetectives Training Grant.
Footnotes
Where the work was done: University of Missouri-Columbia, Columbia, Missouri 65203
Disclosure Statement for Authors: No authors have conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–99. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- 2.Cramer DW, Missmer SA. The epidemiology of endometriosis. Ann N Y Acad Sci. 2002;955:11–22. doi: 10.1111/j.1749-6632.2002.tb02761.x. discussion 34-6, 396-406. [DOI] [PubMed] [Google Scholar]
- 3.Sampson JA. Peritoneal endometriosis due to the menstrual dissemination of endometrial tisse into the peritoneal cavity. Am J Obstet Gynecol. 1927;14 [Google Scholar]
- 4.Kruitwagen RF, Poels LG, Willemsen WN, de Ronde IJ, Jap PH, Rolland R. Endometrial epithelial cells in peritoneal fluid during the early follicular phase. Fertil Steril. 1991;55:297–303. doi: 10.1016/s0015-0282(16)54119-3. [DOI] [PubMed] [Google Scholar]
- 5.Bartosik D, Jacobs SL, Kelly LJ. Endometrial tissue in peritoneal fluid. Fertil Steril. 1986;46:796–800. doi: 10.1016/s0015-0282(16)49813-4. [DOI] [PubMed] [Google Scholar]
- 6.Halme J, Hammond MG, Hulka JF, Raj SG, Talbert LM. Retrograde menstruation in healthy women and in patients with endometriosis. Obstet Gynecol. 1984;64:151–4. [PubMed] [Google Scholar]
- 7.Grummer R. Animal models in endometriosis research. Hum Reprod Update. 2006;12:641–9. doi: 10.1093/humupd/dml026. [DOI] [PubMed] [Google Scholar]
- 8.Vernon MW, Wilson EA. Studies on the surgical induction of endometriosis in the rat. Fertil Steril. 1985;44:684–94. [PubMed] [Google Scholar]
- 9.Cummings AM, Metcalf JL. Induction of endometriosis in mice: a new model sensitive to estrogen. Reprod Toxicol. 1995;9:233–8. doi: 10.1016/0890-6238(95)00004-t. [DOI] [PubMed] [Google Scholar]
- 10.Sharpe-Timms KL. Using rats as a research model for the study of endometriosis. Ann N Y Acad Sci. 2002;955:318–27. doi: 10.1111/j.1749-6632.2002.tb02792.x. discussion 40-2, 96-406. [DOI] [PubMed] [Google Scholar]
- 11.Lee B, Du H, Taylor HS. Experimental Murine Endometriosis Induces DNA Methylation and Altered Gene Expression in Eutopic Endometrium. Biol Reprod. 2008 doi: 10.1095/biolreprod.108.070391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Efstathiou JA, Sampson DA, Levine Z, Rohan RM, Zurakowski D, Folkman J, et al. Nonsteroidal antiinflammatory drugs differentially suppress endometriosis in a murine model. Fertil Steril. 2005;83:171–81. doi: 10.1016/j.fertnstert.2004.06.058. [DOI] [PubMed] [Google Scholar]
- 13.Fang Z, Yang S, Gurates B, Tamura M, Simpson E, Evans D, et al. Genetic or enzymatic disruption of aromatase inhibits the growth of ectopic uterine tissue. J Clin Endocrinol Metab. 2002;87:3460–6. doi: 10.1210/jcem.87.7.8683. [DOI] [PubMed] [Google Scholar]
- 14.Lin YJ, Lai MD, Lei HY, Wing LY. Neutrophils and macrophages promote angiogenesis in the early stage of endometriosis in a mouse model. Endocrinology. 2006;147:1278–86. doi: 10.1210/en.2005-0790. [DOI] [PubMed] [Google Scholar]
- 15.Cummings AM, Metcalf JL, Birnbaum L. Promotion of endometriosis by 2,3,7,8-tetrachlorodibenzo-p-dioxin in rats and mice: time-dose dependence and species comparison. Toxicol Appl Pharmacol. 1996;138:131–9. doi: 10.1006/taap.1996.0106. [DOI] [PubMed] [Google Scholar]
- 16.Flores I, Rivera E, Ruiz LA, Santiago OI, Vernon MW, Appleyard CB. Molecular profiling of experimental endometriosis identified gene expression patterns in common with human disease. Fertil Steril. 2007;87:1180–99. doi: 10.1016/j.fertnstert.2006.07.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stilley JA, Woods-Marshall R, Sutovsky M, Sutovsky P, Sharpe-Timms KL. Reduced Fecundity in Female Rats with Surgically Induced Endometriosis and in Their Daughters: A Potential Role for Tissue Inhibitors of Metalloproteinase 1. Biol Reprod. 2009;80 doi: 10.1095/biolreprod.108.073411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobo VL, Soares JM, Jr, de Jesus Simoes M, Simoes Rdos S, de Lima GR, Baracat EC. Does gestrinone antagonize the effects of estrogen on endometrial implants upon the peritoneum of rats? Clinics. 2008;63:525–30. doi: 10.1590/S1807-59322008000400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y, Kajdacsy-Balla A, Strawn E, Basir Z, Halverson G, Jailwala P, et al. Transcriptional characterizations of differences between eutopic and ectopic endometrium. Endocrinology. 2006;147:232–46. doi: 10.1210/en.2005-0426. [DOI] [PubMed] [Google Scholar]
- 20.Eyster KM, Klinkova O, Kennedy V, Hansen KA. Whole genome deoxyribonucleic acid microarray analysis of gene expression in ectopic versus eutopic endometrium. Fertil Steril. 2007;88:1505–33. doi: 10.1016/j.fertnstert.2007.01.056. [DOI] [PubMed] [Google Scholar]
- 21.Borghese B, Mondon F, Noel JC, Fayt I, Mignot TM, Vaiman D, et al. Gene expression profile for ectopic versus eutopic endometrium provides new insights into endometriosis oncogenic potential. Mol Endocrinol. 2008;22:2557–62. doi: 10.1210/me.2008-0322. [DOI] [PubMed] [Google Scholar]
- 22.Matsuzaki S, Canis M, Pouly JL, Botchorishvili R, Dechelotte PJ, Mage G. Differential expression of genes in eutopic and ectopic endometrium from patients with ovarian endometriosis. Fertil Steril. 2006;86:548–53. doi: 10.1016/j.fertnstert.2006.02.093. [DOI] [PubMed] [Google Scholar]
- 23.Goldman JM, Murr AS, Cooper RL. The rodent estrous cycle: characterization of vaginal cytology and its utility in toxicological studies. Birth Defects Res B Dev Reprod Toxicol. 2007;80:84–97. doi: 10.1002/bdrb.20106. [DOI] [PubMed] [Google Scholar]
- 24.Schroder AL, Pelch KE, Nagel SC. Estrogen modulates expression of putative housekeeping genes in the mouse uterus. Endocrine. 2009 doi: 10.1007/s12020-009-9154-6. In press. [DOI] [PubMed] [Google Scholar]
- 25.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. [Google Scholar]
- 26.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 27.R Development Core Team. R: A language and environment for statisical computing. Vienna, Austria: R Foundation for Statistical Computing; 2006. [Google Scholar]
- 28.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–75. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 29.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 30.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–88. [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amplification Efficiency of TaqMan Gene Expression Assays. Applied Biosystems Application Note. 2006;127AP05-03 [Google Scholar]
- 34.Bookout AL, Cummins CL, Kramer MF, Pesola JM, Magngelsdorf DJ. High-Throughput Real-Time Quantiative Reverse Transcription PCR. In: SAusubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al., editors. Current Protocols in Mollecular Biology. Vol. 3. John Wiley & Sons, Inc.; 2006. pp. 15.8.1–.8.28. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Dallas PB, Gottardo NG, Firth MJ, Beesley AH, Hoffmann K, Terry PA, et al. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RT-PCR -- how well do they correlate? BMC Genomics. 2005;6:59. doi: 10.1186/1471-2164-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bukulmez O, Hardy DB, Carr BR, Word RA, Mendelson CR. Inflammatory status influences aromatase and steroid receptor expression in endometriosis. Endocrinology. 2008;149:1190–204. doi: 10.1210/en.2007-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vernon MW, Beard JS, Graves K, Wilson EA. Classification of endometriotic implants by morphologic appearance and capacity to synthesize prostaglandin F. Fertil Steril. 1986;46:801–6. [PubMed] [Google Scholar]
- 39.Fazleabas AT, Brudney A, Gurates B, Chai D, Bulun S. A modified baboon model for endometriosis. Ann N Y Acad Sci. 2002;955:308–17. doi: 10.1111/j.1749-6632.2002.tb02791.x. discussion 40-2, 96-406. [DOI] [PubMed] [Google Scholar]
- 40.D'Hooghe TM, Bambra CS, Raeymaekers BM, Koninckx PR. Serial laparoscopies over 30 months show that endometriosis in captive baboons (Papio anubis, Papio cynocephalus) is a progressive disease. Fertil Steril. 1996;65:645–9. [PubMed] [Google Scholar]
- 41.Osteen KG, Bruner KL, Sharpe-Timms KL. Steroid and growth factor regulation of matrix metalloproteinase expression and endometriosis. Semin Reprod Endocrinol. 1996;14:247–55. doi: 10.1055/s-2007-1016334. [DOI] [PubMed] [Google Scholar]
- 42.Zhou HE, Nothnick WB. The relevancy of the matrix metalloproteinase system to the pathophysiology of endometriosis. Front Biosci. 2005;10:569–75. doi: 10.2741/1552. [DOI] [PubMed] [Google Scholar]
- 43.Bruner-Tran KL, Eisenberg E, Yeaman GR, Anderson TA, McBean J, Osteen KG. Steroid and cytokine regulation of matrix metalloproteinase expression in endometriosis and the establishment of experimental endometriosis in nude mice. J Clin Endocrinol Metab. 2002;87:4782–91. doi: 10.1210/jc.2002-020418. [DOI] [PubMed] [Google Scholar]
- 44.Sato N, Maehara N, Su GH, Goggins M. Effects of 5-aza-2′-deoxycytidine on matrix metalloproteinase expression and pancreatic cancer cell invasiveness. J Natl Cancer Inst. 2003;95:327–30. doi: 10.1093/jnci/95.4.327. [DOI] [PubMed] [Google Scholar]
- 45.Fernandes T, de Angelo-Andrade LA, Morais SS, Pinto GA, Chagas CA, Maria-Engler SS, et al. Stromal cells play a role in cervical cancer progression mediated by MMP-2 protein. Eur J Gynaecol Oncol. 2008;29:341–4. [PubMed] [Google Scholar]
- 46.Simi L, Andreani M, Davini F, Janni A, Pazzagli M, Serio M, et al. Simultaneous measurement of MMP9 and TIMP1 mRNA in human non small cell lung cancers by multiplex real time RT-PCR. Lung Cancer. 2004;45:171–9. doi: 10.1016/j.lungcan.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Gazvani R, Templeton A. Peritoneal environment, cytokines and angiogenesis in the pathophysiology of endometriosis. Reproduction. 2002;123:217–26. doi: 10.1530/rep.0.1230217. [DOI] [PubMed] [Google Scholar]
- 48.Siristatidis C, Nissotakis C, Chrelias C, Iacovidou H, Salamalekis E. Immunological factors and their role in the genesis and development of endometriosis. J Obstet Gynaecol Res. 2006;32:162–70. doi: 10.1111/j.1447-0756.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 49.Sharpe-Timms KL, Zimmer RL, Ricke EA, Piva M, Horowitz GM. Endometriotic haptoglobin binds to peritoneal macrophages and alters their function in women with endometriosis. Fertil Steril. 2002;78:810–9. doi: 10.1016/s0015-0282(02)03317-4. [DOI] [PubMed] [Google Scholar]
- 50.Dobryszycka W. Biological functions of haptoglobin--new pieces to an old puzzle. Eur J Clin Chem Clin Biochem. 1997;35:647–54. [PubMed] [Google Scholar]
- 51.Sharpe-Timms KL, Ricke EA, Piva M, Horowitz GM. Differential expression and localization of de-novo synthesized endometriotic haptoglobin in endometrium and endometriotic lesions. Hum Reprod. 2000;15:2180–5. doi: 10.1093/humrep/15.10.2180. [DOI] [PubMed] [Google Scholar]
- 52.Nisolle M, Casanas-Roux F, Donnez J. Immunohistochemical analysis of proliferative activity and steroid receptor expression in peritoneal and ovarian endometriosis. Fertil Steril. 1997;68:912–9. doi: 10.1016/s0015-0282(97)00341-5. [DOI] [PubMed] [Google Scholar]
- 53.Wingfield M, Macpherson A, Healy DL, Rogers PA. Cell proliferation is increased in the endometrium of women with endometriosis. Fertil Steril. 1995;64:340–6. doi: 10.1016/s0015-0282(16)57733-4. [DOI] [PubMed] [Google Scholar]
- 54.Jurgensen A, Mettler L, Volkov NI, Parwaresch R. Proliferative activity of the endometrium throughout the menstrual cycle in infertile women with and without endometriosis. Fertil Steril. 1996;66:369–75. doi: 10.1016/s0015-0282(16)58502-1. [DOI] [PubMed] [Google Scholar]
- 55.Iwabe T, Harada T, Tsudo T, Nagano Y, Yoshida S, Tanikawa M, et al. Tumor necrosis factor-alpha promotes proliferation of endometriotic stromal cells by inducing interleukin-8 gene and protein expression. J Clin Endocrinol Metab. 2000;85:824–9. doi: 10.1210/jcem.85.2.6335. [DOI] [PubMed] [Google Scholar]
- 56.Harada T, Taniguchi F, Izawa M, Ohama Y, Takenaka Y, Tagashira Y, et al. Apoptosis and endometriosis. Front Biosci. 2007;12:3140–51. doi: 10.2741/2302. [DOI] [PubMed] [Google Scholar]
- 57.Bulun SE, Gurates B, Fang Z, Tamura M, Sebastian S, Zhou J, et al. Mechanisms of excessive estrogen formation in endometriosis. J Reprod Immunol. 2002;55:21–33. doi: 10.1016/s0165-0378(01)00132-2. [DOI] [PubMed] [Google Scholar]
- 58.Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR, et al. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007;148:3814–26. doi: 10.1210/en.2006-1692. [DOI] [PubMed] [Google Scholar]
- 59.Lee B, Du H, Taylor HS. Experimental murine endometriosis induces DNA methylation and altered gene expression in eutopic endometrium. Biol Reprod. 2009;80:79–85. doi: 10.1095/biolreprod.108.070391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharpe-Timms KL, Piva M, Ricke EA, Surewicz K, Zhang YL, Zimmer RL. Endometriotic lesions synthesize and secrete a haptoglobin-like protein. Biol Reprod. 1998;58:988–94. doi: 10.1095/biolreprod58.4.988. [DOI] [PubMed] [Google Scholar]
- 61.Sharpe KL, Vernon MW. Polypeptides synthesized and released by rat ectopic uterine implants differ from those of the uterus in culture. Biol Reprod. 1993;48:1334–40. doi: 10.1095/biolreprod48.6.1334. [DOI] [PubMed] [Google Scholar]
- 62.Du P, Kibbe WA, Lin SM. lumiMouseIDMapping: Illumina Identifier mapping for Mouse. R package [Google Scholar]
- 63.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–8. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlson M, Falcon S, Pages H, Li N. GO.db: A set of annotation maps describing the entire Gene Ontology. R package [Google Scholar]
- 65.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–8. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 66.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–7. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 67.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Altered gene expression by microarray in the endometriotic lesion at 3 days post-induction.
Supplementary Table 2: Altered gene expression by microarray in the endometriotic lesion at 29 days post-induction.
Supplementary Table 3: Gene ontology terms overrepresented at 3 days post-induction.
Supplementary Table 4: Gene ontology terms overrepresented at 29 days post-induction.