Abstract
Dynamic treatment regime is a decision rule in which the choice of the treatment of an individual at any given time can depend on the known past history of that individual, including baseline covariates, earlier treatments, and their measured responses. In this paper we argue that finding an optimal regime can, at least in moderately simple cases, be accomplished by a straightforward application of nonparametric Bayesian modeling and predictive inference. As an illustration we consider an inference problem in a subset of the Multicenter AIDS Cohort Study (MACS) data set, studying the effect of AZT initiation on future CD4-cell counts during a 12-month follow-up.
Keywords: Bayesian nonparametric regression, causal inference, dynamic programming, monotonicity, optimal dynamic regimes
1. Introduction
A dynamic (sequential) treatment regime is a decision rule which determines the treatment decisions to be taken over time, and it can in general depend on both the patient’s earlier known history and the anticipated future consequences of the decision. The definition of optimality of such a rule can involve formal consideration of loss or utility functions; here, however, we take optimality to simply mean the treatment regime which produces the highest expected value for a response variable of interest at the end of the study period.
Important contributions to the study of optimal treatment regimes have been provided, in particular, by Robins (1986, 1994, 2004) and Murphy (2003). The respective approaches of the two authors are summarized and compared by Moodie et al. (2007). Murphy (2003) proposed methodology for estimating optimal dynamic regimes from observational data. As discussant of this paper, Arjas (2003) suggested to solve the inferential problem using simulation from the predictive distributions of potential outcomes of the response variable, given alternative treatment regimes (see also Arjas and Parner, 2004). In contrast, Murphy parameterizes the problem directly in terms of differences of mean response values of the different treatment regimes to the mean of the optimal regime, calling these functions regrets. Basically, any unbiased estimation of ‘causal effects’ from observational data requires an assumption of no unmeasured confounders, meaning that the treatment decisions made at each point in time are based only on observed data that are available also in the statistical analysis. Rosthøj et al. (2006) present an application of Murphy’s method in a situation where the treatment variable (dose of anticoagulation medication) is continuous, concentrating on parametric regret functions. In addition to the regret functions, the estimation procedure requires the determination of probability distributions for the treatment actions.
Determination of optimal decisions in more conventional likelihood-based analysis requires the modeling of the longitudinal distribution of the data, combined with backward induction (also known as dynamic programming) in time. This approach requires working out all the future optimal decisions, backwards from the last observation point to the first. One way to achieve this, at least in the case of discrete decision alternatives, is to draw samples from the predictive distributions estimated from the observed data, for each of the alternative treatment options. The expectations are then evaluated using Monte Carlo integration.
Carlin et al. (1998) describe this approach in the context of optimal stopping rules in clinical trials, where the decisions to be made are binary (stop the trial if a large enough positive or negative treatment effect has been observed, or continue the trial to collect more data). If determination of one expectation using Monte Carlo integration requires a sample of size m, using nested simulation rounds through k backward steps would require a total of m(mk − 1)/(m − 1) simulations. For large values of k this leads to computational problems, and therefore Carlin et al. describe also a forward sampling algorithm applicable in certain special cases of likelihood functions. Here the nested integration can be replaced with an optimization task over 2k − 1 decision rule boundaries. Also in the context of sequential clinical trials, Wathen and Thall (2008) revamp this algorithm by assuming more restrictive parametric forms for the boundaries. To account for uncertainty in the model definition, they incorporate a model selection based on approximate Bayes factors over a preselected set of candidate models.
Moodie et al. (2007) demonstrate optimal dynamic regime analysis based on modeling of regret or blip (Robins, 2004) functions. They argue that likelihood-based modeling of longitudinal data is sensitive to model misspecification. However, it is unclear whether misspecification of the regret functions is any less serious of a problem. In this paper we aim to wipe the dust off the combination of likelihood and backward induction in optimal dynamic regime analysis and claim that it is still a viable alternative for such purpose. Probability-based data analysis carries some obvious advantages over alternative methods, including the ability to handle missing data and providing a natural quantification of uncertainties at different levels of model specification. In order to avoid problems associated with too rigid model assumptions we apply Bayesian nonparametric monotonic regression, where a monotonicity assumption is used in place of more restrictive parametric or structural assumptions. In contrast to Wathen and Thall (2008), in our approach model selection is built into a part of the probability model and statistical inference is based on model averaged results over random realizations drawn from the space of possible models. We argue that, at least in situations where the decision space is small, the optimal regimes can be worked out using standard MCMC techniques, with the optimal decisions obtained as a byproduct of a single sequence of MCMC samples. We illustrate this approach by using the same example as in Moodie et al. (2007), based on data from Multicenter AIDS Cohort (MACS) study (Kaslow et al., 1987).
The plan of the paper is as follows: In the next section we briefly reintroduce the data and the notation of Moodie et al. (2007), and then present our probability model for the data. In Section 3 we discuss probability-based inference of the optimal treatment regime and the computational issues involved. We also pay attention to the problems caused by missing data and censoring to causal inference in such observational settings. The paper concludes with a discussion in Section 4.
2. Statistical model
Multicenter AIDS Cohort study (MACS) was a longitudinal observational study starting from 1984 and following up HIV-positive homosexual and bisexual men recruited from four U.S. cities. As in Moodie et al. (2007), we restrict our attention to a small subset of this dataset, consisting of the first three consecutive follow-up visits (separated by approximately six months each) after antiviral drug azidothymidine (AZT) became available. We included HIV-positive individuals who did not have (self-reported) AIDS defining illness at the first eligible visit. The interest is in studying the effect of AZT treatment initiated before the 6 or 12 month follow-up visits to the response variable, which is the count of CD4-positive helper cells measured at the 12 month visit. First suppressing an index i referring to an individual, let
X1 = CD4-cell count at the beginning of the study,
A1 = 1 if AZT treatment initiated at 0 < t < 6, and A1 = 0 otherwise;
X2 = CD4-cell count at t = 6 months;
A2 = 1 if AZT treatment initiated at 6 < t < 12 (only possible if A1 = 0), and A2 = 0 otherwise;
Y = CD4-cell count at t = 12 months.
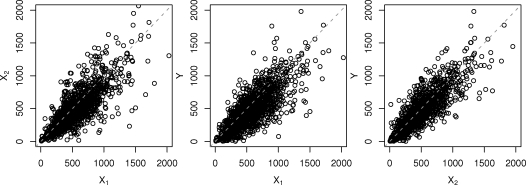
Further, let = {i : A1i = 1}, = {i : A1i = 0}, = {i : A1i = 0, A2i = 1}, and = {i : A1i = 0, A2i = 0} denote the sets of individuals in the data observed in the different treatment alternatives. The observed counts in the dataset were | | = 132, | | = 1295, | | = 131 and | | = 1164. Figure 1 displays the scatterplots of the three CD4-cell measurements, showing that there are strong serial correlations between the consecutive cell counts, and also that there is a visible decreasing trend in the individual CD4-levels as a function of time.
Figure 1:
Scatterplots for three CD4-level measurements
We assume that treatment assignment after the baseline visit can depend on the measured covariate value X1, but that it otherwise satisfies the condition of no unmeasured confounders. The MACS data include many possibly relevant measured covariates, both questionnaire based and laboratory results. Here, however, we take this condition to mean simply that, given the CD4-count X1i of individual i at time t = 0, there are no additional known individual characteristics of i, represented in the model by (latent) random variables, that would predict the assignment of i to either A1i = 0 or A1i = 1, nor does such a prediction of assignment depend on the model parameters (considered in detail below) which we use for describing the development of the CD4-count levels Yi and X2i. For more formal definition of no unmeasured confounders, see e.g. Arjas and Parner (2004). In similar fashion, when considering the assignment of treatment alternatives A2i after time t = 6, we assume that they can depend on the, then known, covariate readings X2i and X1i, but that they otherwise satisfy a similar no unmeasured confounders postulate.
The statistical model is now specified in such a way that it is conditional on the observed values of X1i, i = 1, …, n, in the data. Moreover, as our statistical inferences will be based on forming the likelihood of the data, the above no unmeasured confounders implies that the likelihood contributions arising from treatment assignments can be treated as constant proportionality factors, and therefore have no influence on the inferences based on such likelihoods. Thus, under the no unmeasured confounders postulate we can specify our statistical model in such a way that it is at t = 6 conditioned directly on the observed values of both X1i and A1i. The model is specified as a modular structure, where each module corresponds to a possible transition from one ‘state’ to the next ‘state’. In the following, we use the notation [.] to refer to both probability density and probability mass functions, depending on whether the random variate is continuous or discrete.
Module 1: Model structure for treatment branch A1 = 1:
Consider first a nonparametric Bayesian model for ‘the systematic part’ of predicting response Yi of individual i, given covariate value X1i at baseline t = 0. Denote this systematic part by f1(X1i). Since the function f1, which is assumed to be common to all individuals, is unknown, we view it to be random and will estimate it from the combined data. In order to facilitate this, we make the following assumption: f1 is (that is, all realizations of f1 are) monotonically increasing. Considering that both Yi and X1i are CD4-counts measured from the same individual i, this assumption would seem to be well justified.
Modeling individual variation: It is obvious from Figure 1 that the variability of the CD4-levels over time depends on the absolute CD4-level. To take this into account, conditionally on f1 and X1i, and independently across individuals, we draw a random variable, say Z1i, from a gamma distribution: [Z1i | A1i = 1, X1i, f1, α1] = Gamma(α1f1(X1i), α1). Here α1 is a hyperparameter, which can be either specified separately or estimated from the data; here we choose the latter option and therefore specify a prior [α1] for it. This parameter controls the variance of the gamma distribution. Note that E(Z1i | A1i = 1, X1i, f1, α1) = f1(X1i).
Modeling the response Yi: Conditionally on (A1i = 1, X1i, f1, Z1i), suppose that [Yi | A1i = 1, X1i, f1, Z1i] = Poisson(Z1i). Note that then E(Yi | A1i = 1, X1i, f1, α1) = f1(X1i).
In summary, we have postulated the following hierarchical model structure for the individuals i ∈ in branch A1 = 1, conditionally on the treatment assignments and the corresponding X1i,
| (1) |
Module 2: The model for X2i in the treatment branch A1 = 0 is specified exactly as the model for Yi above, with a different nonparametrically specified and monotonically increasing function f0 and the related parameterization, resulting in a joint distribution of the form
| (2) |
The next step in the model construction is to consider the progression in branch A1 = 0 from time t = 6 onwards, up to Y measured at t = 12, separately for the alternatives A2 = 1 and A2 = 0. The main difference to the above two constructions is that now there are two covariate measurements, X1 and X2, in the history that should be accounted for when predicting Y. Before continuing the model specification, we return briefly to the issue of treatment assignment, now at time 6 < t < 12. As stated earlier, we assume that A2 can depend on, then known, covariate readings X1 and X2 (as well as, in obvious way, on A1), but that they otherwise satisfy the no unmeasured confounders postulate. As a consequence, the likelihood contribution arising from observing the value of A2 for an individual in the data can be treated as a constant, and therefore need not be considered when drawing statistical inferences, unless it is specifically of interest (in Section 3.2 we will note one possible situation where this might be the case).
Module 3: Considering first the case A1 = 0 and A2 = 1, we introduce a nonparametrically defined function f01(X1, X2), and assume that it is monotonically increasing in X1 and X2 − X1. The motivation behind this choice (instead of using the two measured levels directly as covariates) was to separate the baseline level and the longitudinal trend, thus resulting in two less correlated variables, both of which are seemingly compatible with the monotonicity postulate. We postulate a hierarchical model structure
| (3) |
where
and [Yi | Z01i] = Poisson(Z01i). Note that
Module 4: Finally, considering the combination A1 = 0 and A2 = 0, we introduce a random function f00(X1, X2), similarly monotonically increasing in X1 and X2 − X1, and postulate the joint distribution
| (4) |
where the distributions for Z00i and Yi are defined analogously to above.
3. Statistical inference
3.1. Determining the optimal treatment assignment
Given the observed data , the model parameters f0, f1, f00, f01, α0, α1, α00, and α01 can now be estimated. The nonparametric random functions are defined and estimated following Saarela and Arjas (2009). Our approach to the inferential problem is Bayesian, with the consequence that we obtain the joint posterior distribution of these parameters, and can then consider the corresponding posterior predictive distribution of how a generic individual drawn from the same population as the actual individuals in the data (that is, we are using the same model for both) would respond to different treatment alternatives. Denote the ‘potentially observed’ characteristics of such an individual by , and Y*. Note that the choice of the values of and can be viewed as representing ‘do’-conditioning (Pearl, 2000). (That we can switch from observed treatment values in the data to ‘optional’ or ‘forced’ in the predictions is again consequence of the no unmeasured confounders postulate.) It is now straightforward to compute, for example, the posterior predictive expectation of the response Y* at time t = 12, as a function of the CD4-count when , that is, if the AZT treatment is initiated after the baseline visit. We get
where the outer expectation in the second line is with respect to the unknown parameters f1 and α1. Similarly,
and
The computation of these expectations is carried out in practice most easily alongside the MCMC sampling which is used for estimating the model parameters, applying data augmentation. A more detailed description of the computational algorithm is given in the Appendix.
To solve the main problem, that is, finding the optimal treatment regime, we need to consider how the choice between the three alternatives ( ), ( , ) and ( , ) should be made in a situation in which the measurement is available at t = 0, and, if is chosen, also is measured at t = 6 in order to facilitate the choice between and . Following the principles of dynamic programming, we consider first how this latter choice after t = 6 should be made, conditioning on the observed past history ( , , ) of a generic individual as described above, and then moving backwards in time in order to make a choice between branch ( ) and branch ( ).
After t = 6, the key comparison to be made is between the posterior expectations and , both of which were obtained above. We now define the optimal treatment choice after t = 6 to be the one under which, for given values of and , the expected value of Y* is larger, denoted by indicator
After t = 0, we then make a comparison between and
where the outer expectation is with respect to the posterior predictive distribution of , and let
The optimal dynamic treatment regime Aopt will thereby be defined as follows: For 0 < t < 6 choose and for 6 < t < 12, if , choose .
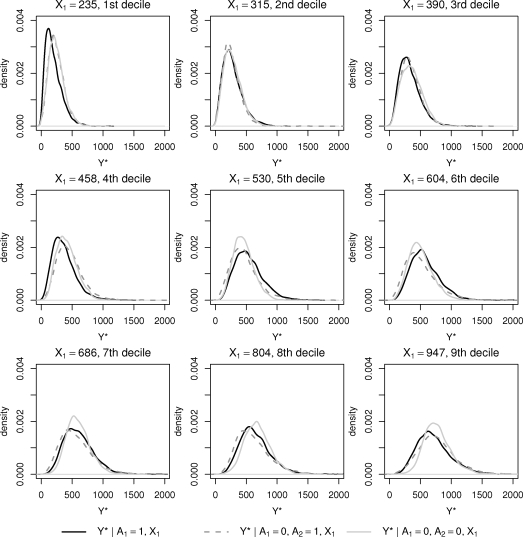
It is of some interest to compare results obtained when following the three deterministic regimes ( ), ( ) and ( ) to each other, and all these to Aopt. The comparison becomes most clear and intuitive if we present it in terms of the posterior predictive distributions of the responses Y*, and since these predictions will depend on the value of covariate determined at the baseline, we should study the predictive distributions as functions of . In each case, producing the predictive distributions can be realized with relative ease by data augmentation within the MCMC sampling scheme, by augmenting the hierarchical models (1), (2), (3) and (4) with the potential outcomes, with the inferential model, and then considering marginals of Y*. In Figure 2 we show the predictive distributions of Y* for the three different regimes given observed decile points of X1 in the data. The three distributions are often overlapping, and even if some systematic differences between the groups could be discerned, they appear to be quite small.
Figure 2:
Predictive distributions for three alternative treatment regimes
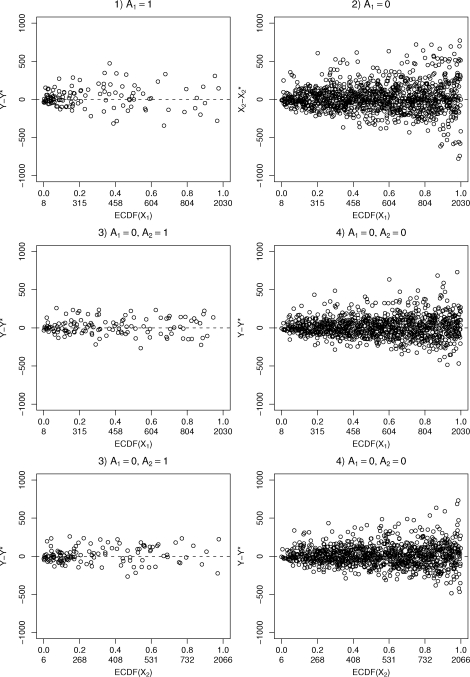
Model fit can be evaluated by comparing the predictive distributions, drawn given the individual covariate levels, to the actual observed outcome values. Figure 3 shows the medians of the posterior distributions of residuals from the four modules. For modules 3 and 4 these are plotted with respect to both X1 and X2. Only the observations which contribute to the likelihood in the specific module are shown. The residual plots seem to indicate reasonably good model fit.
Figure 3:
Posterior median residuals
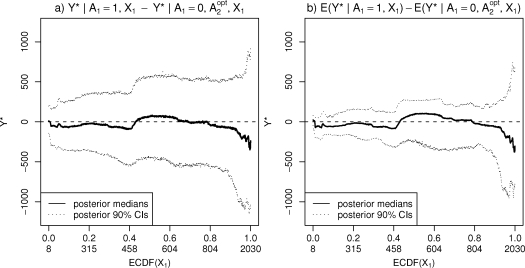
Producing predictive distributions corresponding to the optimal regime Aopt ( ) is slightly more involved since then the choice of what particular module, of the four possibilities, is used in the simulation will depend on the current values of and , the former being chosen in advance for each prediction, and the latter being obtained in the course of the simulation if (see Appendix). We evaluated the optimal regime for each observed X1-value present in the data; in Figure 4 the results are presented as differences between the decision to initiate the treatment after the baseline visit, and the decision not to initiate, given that the later decision after t = 6 is optimal. In panel a) the medians and credible intervals obtained from the posterior predictive distributions are presented, as functions of X1, for the response itself, while panel b) shows these statistics for the corresponding means, represented in the model by the random functions f. The credible intervals indicate no real differences between the groups based on these data.
Figure 4:
Comparison between two alternative treatment decisions after the baseline visit
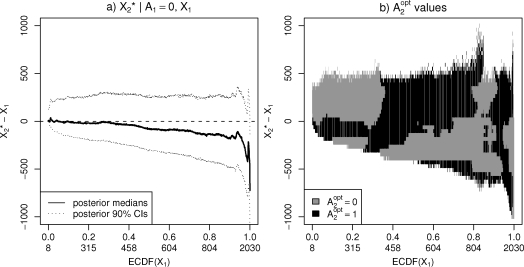
When two CD4-measurements have been recorded, finding the optimal decision will be based on both of these. Figure 5 a) shows the predictive distributions of the 6 months change in the CD4-level as a function of the baseline value. A decreasing trend is clearly visible here. Panel b) shows the values as a function of the baseline CD4-level and of the 6 months change. There is no clear pattern separating the alternative decisions, indicating again that the differences between the groups in these data are small.
Figure 5:
Treatment decision based on two covariate measurements
3.2. Considerations due to missing data and censoring
The probability models defined in Section 2 were written for fully observed measurement data and follow-up histories. In reality the data described were subject to various kinds of incomplete observation. Least problematic of these are ordinary missing data in the CD4 laboratory measurements. There are only a small number of these in the data, and due to the full probability model they can be handled easily by using Bayesian data augmentation.
Incompletely observed treatment histories due to missed follow-up visits are a more problematic issue. Consider first the individuals who dropped out from the follow-up after the first or second visit. (It is possible that the same individuals have reappeared for later MACS visits, but since we consider only follow-up of 12 months we do not utilize this information here.) The data included 200 individuals who were censored from the follow-up after the baseline visit and another 245 who were censored after the intermediate visit. These we omitted from the analysis. Effects of the drop-out for the statistical inference depend on the (typically unknown) censoring mechanism (Little, 1995). For example, here missing at random would refer to the assumption that the censoring after the baseline visit would depend only on the observed baseline CD4 measurement X1, while the censoring after the intermediate visit would depend only on observed values of (X1, X2, A1). This assumption is in many ways analogous to the no unmeasured confounders postulate, although the latter may be easier to understand, since it means that that the clinician making the treatment decisions and the statistician analyzing the data would be working on the basis of the same observed variables. In contrast, missing at random means assuming that confounding variables really do not exist. If the assumption is valid, censored individuals are similar to individuals who remained in the study with similar covariate histories and the inference is unbiased. A nonignorable censoring mechanism would mean that the drop-out depends on the unobserved response values themselves, or on some latent characteristics related to them. Correcting the resulting bias with likelihood-based inference would require also modeling of the censoring process (including its dependence on the outcome). However, in practice there may be little or no information available for such modeling.
If only the intermediate visit of the three is missed, and since the drug usage items in the MACS questionnaire concern medications taken after the previous follow-up visit, it is possible to infer the corresponding treatment branch and the corresponding likelihood contribution for those who missed the intermediate visit and in the 12 months visit did not report having received AZT treatment (there were 119 such individuals in our dataset). Assuming missing at random, we can include them in the branch (A1 = 0, A2 = 0), with missing X2 measurements to be handled as ordinary missing data, allowing also dependence on the observed values of Y. However, those who in their 12 months visit reported having taken AZT may have initiated the treatment either before or after 6 months. To utilize these individuals in the inference, we would also have to define a probability model for A1 in order to augment the missing treatment combination. Since there were only 25 such individuals in the data, we did not pursue this here. However, in case of more complicated designs with many different treatment alternatives possibly only partially observed, and in order to utilize as much of the data as possible, it would be necessary to model also the probabilities of different treatment decisions and their dependence on the covariates.
Rubin (2006) discusses causal inference in situations where follow-up histories are subject to censoring due to death and the response variable reading is thus unobserved for the censored individuals. Here the membership in a ‘principal stratum’ is an unobservable individual characteristic which is a source of nonignorable censoring. For example, the MACS data could include individuals who would have survived with AZT treatment, but who did not receive the treatment and were then censored due to death before 12 months (Rubin’s principal stratum ‘LD’). If treated, an individual belonging to this principal stratum survives, but may end up with a lower CD4-count than those who would survive through the follow-up period irrespective of the treatment decision (principal stratum ‘LL’), thus confounding causal inference. In the MACS study, a more common situation than censoring due to death may well be that an individual simply feels too sick to go to a follow-up visit. This may be even more problematic than censoring due to death since here knowing the dates and the causes of death of the individuals in the study would not help the modeling task. For example, if we knew who has died of AIDS during the 12 month period, we could try to include these individuals in the analysis using zero as the outcome CD4-level. This would not help in cases where the cause of censoring is also unknown.
If the missing at random assumption is unverifiable in practice, how should one then interpret the results obtained from such an observational setting? At least, the results should always be presented conditionally on the baseline measurement of CD4-level (as we have done throughout). As discussed by Rubin (2006), such a covariate may be predictive of both the final and intermediate outcomes, including possible censoring, and thus may shed some light on the unobservable principal strata. For example, in the MACS study we could speculate that those with normal levels of CD4-cells (e.g. > 500) would be primarily individuals who would survive through the follow-up period irrespective of the treatment decision, and thus the results for this group would be less affected by a possible bias due to censoring. Correspondingly, the results may be more uncertain for those whose disease has already progressed to a more advanced stage at the baseline.
4. Discussion
In this paper we have presented a brief illustration of how methods belonging to Bayesian nonparametric inference and dynamic programming can be used in establishing an optimal treatment regime. Although our example was simplified, essentially the same methods can be applied directly to more complicated designs as long as the treatment decisions are discrete and there are sufficiently many observations in each branch of the tree of alternative treatment histories. In practice the modeling would likely involve more covariates than what we considered here. Since completely nonparametric inference of high dimensional problems is usually not feasible, dimension reducing modeling assumptions will be required. For higher dimensional problems we propose Bayesian hierarchical parameterization and “packaging” of covariates (see e.g. Arjas and Liu, 1996), where parametric assumptions are relaxed in the most critical parts of the model. Further work is required with models which could accommodate continuous treatment decisions.
A main challenge is to collect enough covariate information, in order to convince oneself (and others) about the adequacy of the no unmeasured confounders postulate in the considered context, and then model such processes in a way that would lead to plausible predictive distributions. However, this approach alone does not provide a complete solution in problems involving incomplete observation, which cannot be fitted into the standard probability-based missing data framework. As was evident in the present example, the no unmeasured confounders postulate, even if true, will not ensure unbiased inference if the longitudinal design is subject to self-selection due to censoring caused by a missed follow-up visit. At least, attempts should be made to gather information on the causes of such censoring (Little, 1995). Also, the assumed conditions for non-confounded statistical inference should be made explicit in an intuitively understandable way. Here the concept of principal stratification (Frangakis and Rubin, 2002; Rubin, 2006) might be helpful.
In reporting the results from an empirical study, there is often considerable emphasis on statistical tests, or estimates, relating to the parameters of simple (often simplistic) statistical models. Given that sufficient amounts of data are available, nonparametric Bayesian modeling, combined with MCMC methodology, offers an attractive and flexible alternative for statistical inference. This task is to some extent facilitated by the use of constrained (for example, monotone or U-shaped) multivariate nonparametric Bayesian models. Our preference would be to report the results from an empirical study in the form of predictive distributions of the response of interest, with each such prediction corresponding to a specific (sequence of) intervention(s) or choice(s) of control variables. In studies involving real data the computational challenge can become formidable, and even exceed what is feasible in practice. Nevertheless, we would view the relative simplicity of the present conceptual, entirely probabilistic, framework to be a valuable asset in such an enterprise.
Appendix: Computational details
We outline an algorithm for the sequential inference problem described in Section 3, which requires only a single MCMC sample of size m from the joint distribution of all model parameters and potential outcomes. This differs from the sampling scheme described by Carlin et al. (1998), which utilized nested rounds of simulations in calculating the integrals involved. Let
| (5) |
and equivalently,
| (6) |
Note here that the posterior distributions of the random functions f01 and f00 do not depend on the pair ( , ), since these potential outcomes are not ‘data’ and thus do not affect the estimates of the unknown parameters in the model. With fixed ( , ), the above integrals can be evaluated using Monte Carlo integration, by sampling from the posterior distributions of f01 and f00. To determine , we have to evaluate the expectation
| (7) |
Again, some obvious conditional independencies have been applied in the last expression. The split of the joint posterior distribution into the conditional distributions shown above suggests how these will be used in the MCMC simulation. The algorithm to evaluate (7) now goes as follows. For k = 1, …, m, given a fixed value of , do:
Draw parameter values ( , ) from the posterior .
Draw the latent variable from .
Draw the corresponding value from .
Draw function realizations and from and , respectively.
Optionally, if the predictive distributions of the response variable are of interest, draw a value of Yk given both ( ) and ( ), similarly as in steps 2.-3.
Simultaneously, an MCMC sample of parameter values and potential outcomes is also produced from model (1). Now using the obtained samples ( , ), k = 1, …, m, of the function realizations, the expectations (5) and (6) are evaluated at each point ( , ), k = 1, …, m, to determine the values of the indicators . Now, with the values of known, expectation (7) can also be evaluated using Monte Carlo integration, using, depending on the value of at the point ( , ), the realization or in place of . Note again that the posterior distributions of the random functions do not depend on the simulated potential outcomes and thus the evaluation of the function level at any given point ( , ) is purely deterministic, given the current realization of the random function. Now (7) can be compared to , evaluated using the sample from model (1), to determine the indicator .
The MCMC sampler described here was ran for 25000 iterations after a 15000 round burn-in, saving every 5th state of the chain. Gamma(0.1, 0.1) prior was used for the α parameters. One iteration took 2.7 seconds on a standard desktop computer. To put this into context, each of these iterations involved 100 proposals to update each the four random functions f. The computational time needed in our approach would generally depend roughly linearly on the numbers of knots (likelihood modules) in the tree representing the different treatment alternatives.
References
- Arjas E, Jennison C, Dawid AP, Cox DR, Senn S, Cowell RG, Didelez V, Gill RD, Kadane JB, Robins JM, Murphy SA. Optimal dynamic treatment regimes - Discussion on the paper by Murphy. Journal of the Royal Statistical Society, Series B. 2003;65:355–366. [Google Scholar]
- Arjas E, Liu L. Non-parametric Bayesian approach to hazard regression: a case-study with a large number of missing covariate values. Statistics in Medicine. 1996;15:1757–1770. doi: 10.1002/(SICI)1097-0258(19960830)15:16<1757::AID-SIM336>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Arjas E, Parner J. Causal reasoning from longitudinal data. Scandinavian Journal of Statistics. 2004;31:171–187. doi: 10.1111/j.1467-9469.2004.02-134.x. [DOI] [Google Scholar]
- Carlin BP, Kadane JB, Gelfand AE. Approaches for optimal sequential decision analysis in clinical trials. Biometrics. 1998;54:964–975. doi: 10.2307/2533849. [DOI] [PubMed] [Google Scholar]
- Frangakis CE, Rubin DB. Principal stratification in causal inference. Biometrics. 2002;58:21–29. doi: 10.1111/j.0006-341X.2002.00021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo CR., Jr The Multicenter AIDS Cohort study: rationale, organization, and selected characteristics of the participants. American Journal of Epidemiology. 1987;126:310–318. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- Little RJA. Modeling the drop-out mechanism in repeated-measures studies. Journal of the American Statistical Association. 1995;90:1112–1121. doi: 10.2307/2291350. [DOI] [Google Scholar]
- Moodie EEM, Richardson TS, Stephens DA. Demystifying optimal dynamic treatment regimes. Biometrics. 2007;63:447–455. doi: 10.1111/j.1541-0420.2006.00686.x. [DOI] [PubMed] [Google Scholar]
- Murphy SA. Optimal dynamic treatment regimes. Journal of the Royal Statistical Society, Series B. 2003;65:331–355. doi: 10.1111/1467-9868.00389. [DOI] [Google Scholar]
- Pearl J. Causality: models, reasoning and inference. Cambridge: Cambridge University Press; 2000. [Google Scholar]
- Robins JM. A new approach to causal inference in mortality studies with a sustained exposure period - application to control of the healthy worker survivor effect. Mathematical Modelling. 1986;7:1393–1512. doi: 10.1016/0270-0255(86)90088-6. [DOI] [Google Scholar]
- Robins JM. Correcting for non-compliance in randomized trials using structural nested mean models. Communications in Statistics. 1994;23:2379–2412. doi: 10.1080/03610929408831393. [DOI] [Google Scholar]
- Robins JM. Optimal structural nested models for optimal sequential decisions. In: Lin DY, Heagerty P, editors. Proceedings of the second Seattle symposium in biostatistics. New York: Springer; 2004. pp. 189–326. [Google Scholar]
- Rosthøj S, Fullwood C, Henderson R, Stewart S. Estimation of optimal dynamic anticoagulation regimes from observational data: a regret-based approach. Statistics in Medicine. 2006;25:4197–4215. doi: 10.1002/sim.2694. [DOI] [PubMed] [Google Scholar]
- Rubin DB. Causal inference through potential outcomes and principal stratification: application to studies with “censoring” due to death. Statistical Science. 2006;21:299–309. doi: 10.1214/088342306000000114. [DOI] [Google Scholar]
- Saarela O, Arjas E.2009Postulating monotonicity in nonparametric Bayesian regressionSubmitted [Google Scholar]
- Wathen JK, Thall PF. Bayesian adaptive model selection for optimizing group sequential clinical trials. Statistics in Medicine. 2008;27:5586–5604. doi: 10.1002/sim.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]