Abstract
Proteolysis by the ubiquitin-proteasome pathway (UPP) has emerged as a new molecular mechanism that controls wide-ranging functions in the nervous system, including fine-tuning of synaptic connections during development and synaptic plasticity in the adult organism. In the UPP, attachment of a small protein, ubiquitin, tags the substrates for degradation by a multisubunit complex called the proteasome. Linkage of ubiquitin to protein substrates is highly specific and occurs through a series of well-orchestrated enzymatic steps. The UPP regulates neurotransmitter receptors, protein kinases, synaptic proteins, transcription factors, and other molecules critical for synaptic plasticity. Accumulating evidence indicates that the operation of the UPP in neurons is not homogeneous and is subject to tightly managed local regulation in different neuronal subcompartments. Investigations on both invertebrate and vertebrate model systems have revealed local roles for enzymes that attach ubiquitin to substrate proteins, as well as for enzymes that remove ubiquitin from substrates. The proteasome also has been shown to possess disparate functions in different parts of the neuron. Here I give a broad overview of the role of the UPP in synaptic plasticity and highlight the local roles and regulation of the proteolytic pathway in neurons.
The ability of the nervous system to change in response to environmental stimuli allows organisms to adapt, to survive, and to propagate. The property of the nervous system to change itself has fascinated scientists for many years and has been studied in great depth and breadth at molecular, cellular, and behavioral levels. These investigations have revealed a role for phosphorylation of pre-existing substrates by protein kinases such as calcium-calmodulin-dependent protein kinase II (CaMKII), cAMP-dependent protein kinase (PKA), mitogen-activated protein (MAP) kinase, and protein kinase C (PKC) in regulating short-term and intermediate-term plasticity (Lisman 1994; Reissner et al. 2006; Sossin 2007; Abel and Nguyen 2008). The past research has also determined that the signaling from neurotransmitter receptors to the nucleus, mediated by protein kinases, controls gene transcription and protein synthesis required for long-term synaptic plasticity (Chen and Tonegawa 1997; Kandel 2001). In the recent past, a new molecular mechanism that modulates both short-term and long-term synaptic plasticity has emerged: protein degradation through the ubiquitin-proteasome pathway (UPP).
The ubiquitin-proteasome pathway (UPP): Basics
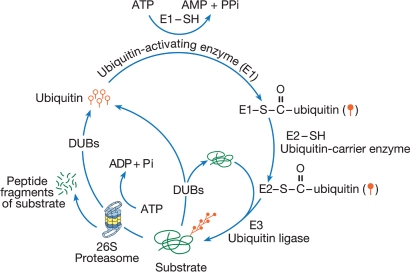
In the UPP, the protein substrates are marked by a covalent attachment of ubiquitin for degradation by a huge proteolytic complex called the proteasome. Ubiquitin is a small protein of 76 amino acids that is highly conserved through evolution. Linkage of ubiquitin (ubiquitination) occurs through action of three classes of enzymes termed E1, E2, and E3 (Fig. 1). Previously it was believed that there is only one E1 gene, although it was known that the E1 mRNA can generate two El isoforms because of alternative translation initiation sites (Shang et al. 2001). Recently, a second E1 gene, which functions with specific E2s, has been discovered (Chiu et al. 2007; Jin et al. 2007; Pelzer et al. 2007). There are multiple genes encoding E2s and E3s. The ubiquitination process begins with activation of ubiquitin by E1. Activated ubiquitin is then passed onto E2s. E2s transfer ubiquitin to the substrates directly or, in the case of some ligases, through generation of E3–ubiquitin thioester intermediates. The substrate specificity of ubiquitin ligation is essentially determined by E3s. Sometimes an E3 might prefer a particular E2 by binding to a specific noncatalytic amino or carboxyl terminal extension in an E2. In such cases only specific E2–E3 pairs can ligate ubiquitin to substrates. For example, UbcH8, a human E2, can interact with parkin and E6-AP, whereas a similar E2 UbcH5 functions with Rsp5 and BRCA1-BARD1 (Kumar et al. 1997; Zhang et al. 2000; Brzovic et al. 2003). During ubiquitination, after the first ubiquitin is attached to the substrate, another ubiquitin is attached to an internal Lys residue in the first ubiquitin; thus a polyubiquitin chain grows. The polyubiquitinated substrate is then recognized by the proteasome and is degraded to small peptides and amino acids. The polyubiquitin chains are not degraded but disassembled by deubiquitinating enzymes (DUBs). There are two classes of DUBs: Low molecular weight DUBs are called ubiquitin C-terminal hydrolases (UCHs) and high molecular weight DUBs are called ubiquitin-specific proteases (UBPs or USPs). UCHs and UBPs differ with respect to substrate preference. The proteasome that degrades polyubiquitinated proteins is referred to as the 26S proteasome, which has a 20S catalytic core and two 19S regulatory particles (RP) attached to either end of the cylindrical 20S core. In eukaryotes, the 20S core is made up of two outer rings with seven α subunits (α1–α7) in each ring and two inner rings consisting of seven β subunits (β1–β7). The catalytic activity of the proteasome is provided by three of the seven β subunits (β1, β2, and β5). The catalytic sites in these β subunits are located at their N-termini, which are situated inside the catalytic chamber with an opening of 13 Å in diameter (Cheng 2009). Thus, only an unfolded substrate can pass through this aperture. The unfolding activity is believed to be provided by the ATPases that are present in the base of the 19S RP, which contains six ATPase subunits Rpt1–Rpt6 (Regulatory particle ATPase 1–6) and four non-ATPase subunits Rpn1, Rpn2, Rpn10, and Rpn13 (Regulatory particle non-ATPases 1, 2, 10, and 13). The other part of the 19S RP is called the “lid,” which comprises only non-ATPase subunits (Rpn3, Rpn5, Rpn6-9, Rpn11, Rpn12, and Rpn15) (Marques et al. 2009; Hegde 2010). Among the Rpn subunits, Rpn11 (also called Poh1) and Rpn13 (also called Uch37) are DUBs that are integral parts of the 19S RP that assist in deubiquitination of the substrate as it is unfolded and threaded into the catalytic chamber of the 20S core. Another DUB called Usp14 (also known as Ubp6) reversibly associates with the Rpn1 and stimulates substrate degradation through deubiquitination (Leggett et al. 2002; Peth et al. 2009).
Figure 1.
The ubiquitin-proteasome pathway (UPP). In this proteolytic pathway, ubiquitin (single ubiquitin molecules are represented by open circles with straight tails) is selectively and covalently linked to the substrate. The enzymatic process of attaching ubiquitin to substrates is called ubiquitination or ubiquitin conjugation and depends on the action of three different classes of enzymes, E1, E2, and E3. First, ubiquitin is activated by E1 to form a ubiquitin-AMP intermediate. Activated ubiquitin (closed circles with straight tails) is passed on to E2 (ubiquitin carrier enzymes). E2s transfers ubiquitin to an E3 (ubiquitin ligase), which ligates the activated ubiquitin to the substrate. To the ubiquitin, which is attached to a substrate, another ubiquitin is attached and thus, through successive linkages of ubiquitin, a polyubiquitin chain forms. Polyubiquitinated substrates are degraded by a proteolytic complex called the 26S proteasome in an ATP-dependent reaction. Ubiquitin is not degraded but the polyubiquitin chain is disassembled and ubiquitin is recycled by deubiquitinating enzymes (DUBs). Before being committed to be degraded by the proteasome, ubiquitination is reversible. DUBs can disassemble the polyubiquitin chain if a substrate is ubiquitinated erroneously and prevent the degradation of the substrate. (Figure modified from Hegde 2004 and reprinted with permission from Elsevier © 2004.)
The 20S proteasome can exist not only as a core of 26S, but also as a separate population that cannot degrade ubiquitinated proteins (Rechsteiner et al. 1993). The 20S proteasome by itself has chymotrypsin-like, trypsin-like, and post-glutamyl peptidase activities, which cleave after hydrophobic, basic, and acidic residues, respectively. The 19S RP recognizes the polyubiquitinated substrate, and channels the substrate into the catalytic 20S core of the proteasome. The catalytic core is the part that ultimately cleaves the ubiquitinated protein into small peptides anywhere from three amino acids to 32 amino acids long. The peptides generated are likely to be further hydrolyzed to generate free amino acids by other proteases and amino peptidases (Marques et al. 2009; Hegde 2010).
Ubiquitin ligases (E3s)
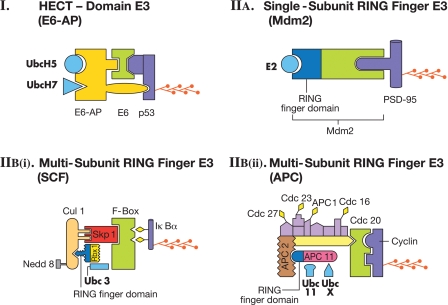
E3s are the enzymes that attach ubiquitin to specific substrates. E3s can be single proteins or complexes of proteins (Fig. 2). Some E3s can accept ubiquitin in a thioester linkage from E2s (see Fig. 1) and ligate ubiquitin to the substrate, while other E3s bring the E2s and the substrates together and facilitate the transfer of ubiquitin to substrates. There are two major classes of E3s: (1) HECT (homologous to E6-AP carboxyl-terminus) domain E3s and (2) RING (really interesting new gene) finger E3s.
Figure 2.
Classes of ubiquitin ligases (E3s). (I) HECT-domain E3. E6-AP ubiquitin ligase in combination with E6 protein and one of the two E2s (UbcH5 or UbcH7) ligates ubiquitin to the p53 tumor suppressor protein. (IIA) A single-subunit RING finger E3. Mdm2 ligates ubiquitin to PSD-95 with the help of an E2 enzyme. (IIBi) A multisubunit RING finger E3. SCF ligases contain the substrate recognition site on an F-box protein. Skp1 is an adaptor that joins the F-box protein to Cul1. RING finger domain is on Rbx 1. The E2 is Ubc3. Cul 1 is modified by Nedd8, a ubiquitin-like protein leading to an increase in the activity of the ligase complex. The substrate is phosphorylated (diamonds) IκBα. (IIBii) A multisubunit RING finger E3. APC is a more complex example of multisubunit RING finger E3s and has a subunit composition distinct from that of SCF. Cdc20 protein in APC has the substrate (Cyclin) recognition site. The RING finger domain is on APC11. The E2s Ubc 11 or UbcX can function with the APC ligase. In addition, several adaptor proteins, some labeled (Cdc27, Cdc23, APC 1, Cdc16) and some unlabeled, interact with Cdc20 and APC11. Diamonds on the adaptor subunits indicate phosphorylation. The polyubiquitin chain is shown on the substrates in each panel. In all panels, E2s are light blue, RING finger domains are dark blue, and the substrates are purple in color. (Figure modified from Hegde 2004 and reprinted with permission from Elsevier © 2004.)
HECT Domain E3s
The first E3 discovered in this class is called E6-AP, which ligates ubiquitin to the tumor suppressor protein p53 (Fig. 2I) in human papilloma virus (HPV) infected cells (Beer-Romero et al. 1997). E6 is a protein encoded by oncogenic strains of HPV. E6 associates with a cellular protein called E6-associated protein (E6-AP). The C-terminal region of E6-AP contains the catalytic domain of the ubiquitin ligase (Huibregtse et al. 1993). Subsequent studies found that a family of proteins ubiquitin ligases with homology to the catalytic domain of E6-AP exists. These ubiquitin ligases came to be called homologous to E6-AP carboxyl-terminus (HECT) domain E3s (Huibregtse et al. 1995). A characteristic feature of HECT ligases is that they accept activated ubiquitin from an E2 and form an E3–ubiquitin thioester intermediate (Scheffner et al. 1995). The human E6-AP is now referred to as UBE3A and has been found to be defective in patients suffering from a disorder called Angelman syndrome characterized by mental retardation (Kishino et al. 1997; Matsuura et al. 1997). A mouse model of Angelman syndrome exhibits defects in long-term potentiation (LTP) and impairment in contextual learning (Jiang et al. 1998). A recent study suggests that UBE3A regulates excitatory synapse development by targeting Arc, which is known to play a role in synaptic plasticity (Greer et al. 2010).
RING finger E3s
These E3s are called RING finger E3s because they contain a RING finger domain, which consists of seven cysteine residues and one histidine residue forming a single-folded domain binding two zinc ions. The RING finger motif was originally discovered by sequence database searches using the N-terminal sequence of a protein encoded by a new gene called really interesting new gene 1 (RING1). During the past few years several ubiquitin ligases were found to contain the RING finger. Unlike the HECT ligases, the RING finger ligases do not form a thioester intermediate with ubiquitin. Rather, these E3s bind both the substrate and the E2 and mediate the transfer of ubiquitin from the E2 to the substrate. It is now generally believed that the RING finger motif plays a critical role in ubiquitin ligation to substrates or to RING finger proteins themselves (Glickman and Ciechanover 2002; Hegde 2004, 2010; Deshaies and Joazeiro 2009).
The RING finger category of E3s can be subdivided into RING finger E3s with a single subunit or RING finger E3s with multiple subunits.
Single-subunit RING finger E3s
Single-subunit RING finger E3s contain the RING finger domain and the substrate recognition site in the same protein. A ubiquitin ligase called Mdm2 is such an enzyme. Mdm2 has been shown to ubiquitinate PSD-95 (postsynaptic density protein of 95 kDa) (Fig. 2IIA; Colledge et al. 2003). PSD-95 provides an anchor for a type of glutamate receptor called the AMPA (α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid) receptor and degradation of PSD-95 reduces the AMPA receptor number on the plasma membrane.
Multisubunit RING finger E3s
Two well-characterized multisubunit RING finger E3s are SCF (SKP1-cullin-F-Box protein) and APC (Anaphase promoting complex).
SCF
Central to the functioning of the SCF complex is the RING finger domain-containing protein Rbx1 (Fig. 2IIBi). The SCF-type ligases have another invariant protein called cullin. The theme appears to be that the cullins interact with adaptor proteins, such as Skp1 to recruit substrate-interacting proteins. In many SCFs the substrate binding subunits are the F-Box proteins. In some cullins, such as Cullin3, the adaptor and substrate-binding functions are fused in a single polypeptide containing a BTB (Broad-complex/Tramtrack/Bric-a-brac) domain. There are at least five different cullins in mammals (Petroski and Deshaies 2005). There are several F-Box proteins as well. The human genome contains 68 genes with F-box motifs and the mouse genome has 74 genes with identifiable F-boxes (Jin et al. 2004). Therefore, just with the cullin F-Box combination alone, it would be possible to generate hundreds of E3s with differing specificities. SCF ligases recognize phosphorylated substrates. For example, when a protein called IκBα is phosphorylated on two closely spaced serine residues, it is recognized by the F-box protein β-TrCP and is ubiquitinated by an SCF containing Rbx1, a protein with a RING finger domain (Winston et al. 1999; Tanaka et al. 2001).
SCF ligases have been found to control several critical substrates in neurons. For example, an SCF ligase that contains the F-box protein β-TrCP has been shown to target SPAR (spine-associated Rap GTPase activating protein) for ubiquitination and degradation (Ang et al. 2008). SPAR is a postsynaptic density protein that regulates spine morphogenesis (Pak and Sheng 2003).
APC
The APC (also known as cyclosome; hence often referred to as APC/C) ligases, although they are multisubunit RING finger E3s like SCF ligases and contain a subunit (APC2) with a cullin-homology region, they are distinct from the SCF ligase in overall subunit combination (van Leuken et al. 2008; Simpson-Lavy et al. 2010). For example, instead of one adaptor found in SCF ligases (such as Skp1), APC has multiple subunits that serve as adaptors (Fig. 2IIBii). Also, unlike SCF ligases, substrate phosphorylation is not an important determinant for specific substrate recognition by the APC ligase. Rather, substrate specificity of APC ligases appears to be modulated by incorporation of “specificity factors” into the ligase complex. Two such factors, Cdc20 and Cdh1, are substrate-specific activators of APC. Cdc20 enables APC to degrade substrates at the onset of anaphase such as the anaphase inhibitor Pds1p, whereas it does not promote degradation of other substrates, such as Clb2 and Ase1. By contrast, Cdh1 incorporation into APC promotes degradation of Clb2 and Ase1 but not that of Pds1 (Visintin et al. 1997). APC acts together with an E2 Ubc11 or UbcX. One of the most studied substrates of APC is mitotic cyclin. This substrate has a short stretch of nine amino acids called the “destruction box,” which is critical for recognition by the APC ubiquitin ligase (King et al. 1996).
Several studies have indicated that APC plays a critical role in post-mitotic neurons. For example, APC is critical for determining synaptic size and number in Drosophila neuromuscular junctions (van Roessel et al. 2004). A Cdh1-containing APC is critical for axonal growth in the cerebellum (Stegmuller et al. 2006). A Cdc-20 containing APC plays a role in presynaptic differentiation, as well as dendritic morphogenesis (Kim et al. 2009).
E4s
An additional class of proteins called E4s that elongates the polyubiquitin chain has been discovered. A protein product of a gene previously known as ubiquitin fusion degradation protein 2 (UFD2) in yeast was found to catalyze the ubiquitin chain assembly along with E1, E2, and E3 and was named E4 (Koegl et al. 1999). The hallmark of E4s is that these enzymes contain a conserved motif originally found in UFD2 and consequently named the U-box. Biochemical studies showed that U-box proteins have ubiquitin ligase activity that is dependent on E1 and E2, but independent of E3 (Hatakeyama et al. 2001). Therefore, some E4s might be ubiquitin ligases. In support of this idea, U-boxes are considered modified RING finger domains. Moreover, comparative nuclear magnetic resonance studies of the U-box and the RING finger domain revealed that the two domains are structurally similar (Ohi et al. 2003). If E4s are indeed ubiquitin ligases, they might represent a subfamily of E3s. Since there have not been many studies on E4s and the first E4 discovered functions as a cofactor for an E3, it might be premature to conclude that E4s belong to a special class of E3s. U-box proteins/E4s are much fewer in number compared to E3s. For example, the human genome contains an estimated 19 U-box proteins (Patterson 2002).
Although E4s are less well studied in the nervous system compared to other ligases, there is evidence that E4s have a role in the nervous system. For example, a protein called WldS, which protects against Wallerian degeneration of axons, contains the ubiquitination factor E4B (also known as UFD2) fused to nicotinamide mononucleotide adenyltransferase (Nmnat) (Hoopfer et al. 2006). This chimeric gene is present in a natural mouse mutant. The E4B/UFD2 part contains only 70 of the 1173 amino acids of the mouse Ufd2a gene and hence it is not clear whether this part plays a role in assembly of polyubiquitin chains. Ufd2a protein is abundantly expressed in neurons of adult mice. Heterozygous Ufd2a mutants (Ufd2a+/−) exhibit axonal dystrophy, Purkinje cell degeneration, and severe motor deficits (Kaneko-Oshikawa et al. 2005).
Achieving specificity of ubiquitin conjugation: Combinatorial coding by E2s and E3s
Thus far, the evidence available indicates that the conjugation of ubiquitin to a substrate is a highly specific reaction. How is this specificity achieved? As explained above, ubiquitin conjugation requires three enzymes, E1, E2, and E3. Two E1s activate ubiquitin and each E1 transfers the activated ubiquitin to specific E2s. Thus some degree of specificity is achieved at this step. There is an additional degree of specificity at the E2 step, which mainly comes from specific E2–E3 interactions. E2s interact with E3s through two loops (named L1 and L2) and an N-terminal α-helix 1 in the three-dimensional structure of E2s. Small amino acid sequence variations in these structural elements contribute to the specificity of E2s binding to E3s (Ye and Rape 2009). In addition to the structural elements mentioned above, unique parts of some E2s contribute to the specificity of E3 binding as well. For example, the C-terminal tail of a yeast E2 called Cdc34, which is dissimilar to that of a closely related E2 Ubc4, confers specificity of Cdc34 binding to an SCF ligase (Kolman et al. 1992; Silver et al. 1992). The E3s are the most specific to a given substrate, however. Initially it was thought that there is a specific E3 for each substrate. Having a dedicated E3 for each substrate would be untenable because of the coding burden it places on the genome. Rather, the specificity is derived from a combination of recognition modules as shown in Figure 3. Interaction between a given E3 and its substrate is believed to be specific. In some instances an E3 ligates ubiquitin to only one substrate. In other cases, an E3 ligates ubiquitin to more than one substrate. In the latter case specificity of E3–substrate interaction might be determined by other factors such as post-translational modification of the substrate (Hegde 2010). A genome-wide study estimated that the human genome contains 617 genes encoding putative E3s (Li et al. 2008). The diversity of E3 is further increased by the fact that some E3s, such as SCFs and APCs, are modular and by mixing and matching subunits additional unique E3s could be generated. The estimate for the number of genes coding for E2s is around 25–30. Considering that there are about 25,000 genes in the human genome (Stein 2004), E2s and E3s together potentially could generate a unique combination for every gene. Besides the unique E2–E3 combinations, specificity can be generated by the state of the substrate (vulnerable or resistant to degradation), as well as regulation of E3s through post-translational modification such as phosphorylation (Hegde and DiAntonio 2002; Hegde 2010). Thus, the ubiquitin conjugation machinery can be highly specific to a given substrate.
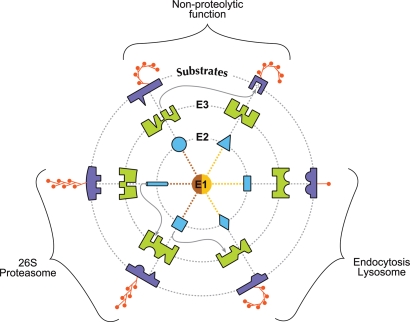
Figure 3.
Combinatorial coding of specificity in ubiquitin conjugation. Two E1s (indicated by two colors in the center) provide some degree of specificity to an ubiquitination reaction. E2s preferentially interact with some E3s but not others. An E3, which may be a single molecule or a complex of molecules, is believed to be specific for each substrate. E2s, E3s, and substrates generate a large number of combinations to “code” for the specificity of an ubiquitination reaction. The shapes on each concentric circle (from inside to outside) represent specific E2s, E3s, and substrates. The notches or projections in each shape represent specific domains in the enzymes or substrates. Radiating dotted lines indicate shapes that fit into each other, indicating specific interactions. Occasionally, an E2 can interact with more than one E3 and a given E3 can ubiquitinate more than one substrate (wavy arrows between circles). These interactions still are specific because they likely occur through different recognition domains in these molecules. The ubiquitinated proteins can undergo degradation by the proteasome or endocytosis or could have a nonproteolytic role. The polyubiquitin chains that are targeted to the 26S proteasome are Lys-48 linked chains, whereas those that subserve nonproteolytic functions and endocytosis are Lys-63 linked (curvy) chains. Monoubiquitin chains also participate in marking some substrates for endocytosis. (Figure modified from Hegde 2004 and reprinted with permission from Elsevier © 2004.)
Another factor that adds to the combinatorial coding capacity of the ubiquitin conjugation reaction is that the type of ubiquitin linkage to a given substrate determines its fate. Ubiquitin is attached to the side chain (called the ϵ-amino group) of Lys residues in the substrate. The first ubiquitin is attached to the substrate through a covalent linkage via the C-terminus of ubiquitin. Attachment of a single ubiquitin at one site in the substrate (monoubiquitination) or at multiple sites (multi-monoubiquitination) generally determines binding of the ubiquitinated substrate to other proteins and is utilized for functions such as endocytosis and modulation of protein activity. Attachment of many ubiquitin molecules to the substrate (polyubiquitination) serves diverse purposes, the main one of which is marking the protein for degradation by the proteasome. Polyubiquitin chains are built through successive addition of single-ubiquitin molecules to an internal lysine residue in the previously attached ubiquitin. Ubiquitin attachment to other ubiquitins could occur through any of the seven lysine residues in the ubiquitin molecules. For marking the substrate for ubiquitin-proteasome-mediated degradation, additional ubiquitin are attached to the first ubiquitin at its 11th or 48th Lys residue. Lys-63 linked polyubiquitin chains modulate protein function such as NFκ-B activation (Deng et al. 2000). Although polyubiquitin chains that are formed through second ubiquitin linkage to Lys-6, Lys-27, Lys-29, and Lys-33 of the first ubiquitin attached to the substrate are known to occur, the functions of such chains are not understood (Komander 2009; Ye and Rape 2009).
The UPP and long-term synaptic plasticity
Although ubiquitin was used as a marker for brain pathology, such as neurofibrillary tangles in Alzheimer's disease and Lewy bodies in Parkinson's disease (Mori et al. 1987; Lowe et al. 1988), no physiological or pathological role for ubiquitin in the nervous system was found until about a decade and a half ago. The first discovery of ubiquitin-proteasome-mediated degradation of a substrate relevant to synaptic plasticity in the nervous system was that of R subunits of PKA (Hegde et al. 1993). Since then several substrates of the UPP in the nervous system have been identified (Hegde 2010).
Degradation of R subunits of PKA and proteolytic removal of a CREB repressor
A role for the UPP in synaptic plasticity was discovered during the investigation of persistent activation of PKA. Studies on the biochemical mechanism of long-term facilitation (LTF) (Greenberg et al. 1987) in Aplysia indicated that PKA was persistently activated in the absence of elevated cAMP. LTF underlies behavioral sensitization of defensive reflexes in Aplysia, which is a simple form of memory (Abrams 1985). How is PKA activated in the absence of sustained increase in cAMP? It was found that the R subunits of PKA were decreased without any change in the catalytic (C) subunit during induction of LTF. Since there was no change in mRNA for either the R subunit or the C subunit, it was concluded that R subunits were diminished perhaps through proteolysis. What is the mechanism of R subunit degradation? Hegde et al. (1993) found through a series of biochemical experiments that R subunits were substrates for ubiquitination and proteasome-mediated degradation. Moreover, a UCH ([Ap-uch] Aplysia ubiquitin C-terminal hydrolase ) that interacts with the proteasome was found to be induced by serotonin, the neurotransmitter that induces LTF. Ap-uch was found to be critical for induction of LTF (Hegde et al. 1997). Subsequently, Chain et al. (1999) showed that at sensory-motor neuron synapses, injection of lactacystin, a specific proteasome inhibitor blocked induction of LTF. Since R subunits inhibit the activity of C subunits of PKA, the results suggested that the UPP operates to remove inhibitory constraints on the formation of long-term memory. This has been corroborated by work carried out on the rat hippocampus. Lopez-Salon and co-workers (2001) demonstrated that bilateral infusion of lactacystin to the CA1 region of the rat hippocampus caused total retrograde amnesia for a one-trial avoidance learning. They also showed that total ubiquitination increases in the hippocampus 4 h after the training. These results are consistent with the idea that a decrease in some critical inhibitory proteins during long-term memory formation (Abel et al. 1998) is mediated by the UPP.
Additional evidence that the UPP might function to degrade proteins that normally inhibit long-term synaptic plasticity has also been obtained using the Aplysia model. Stimulation protocols that induce LTF in Aplysia, cause ubiquitination and degradation of a CREB repressor called CREB1b. Both ubiquitination and degradation of CREB1b are increased by protein kinase C. It remains to be seen whether CREB1b or the ligase that targets CREB1b for ubiquitination is modulated by PKC (Upadhya et al. 2004).
Recent studies on vertebrates suggest that the UPP may have much broader and more complex roles than just degrading the inhibitory constraints on long-term synaptic plasticity and memory, such as R subunits and the CREB repressor. For example, infusion of the proteasome inhibitor β-lactone into the CA1 region of the hippocampus prevents extinction of contextual fear memory (Lee et al. 2008). Also, in using infusion of lactacystin into the CA3 region of the hippocampus it was shown that protein degradation is important for consolidation as well as reconsolidation of spatial memory (Artinian et al. 2008). The mechanistic details as to how the UPP contributes to the extinction of fear memory or reconsolidation of spatial memory are not clearly understood.
Modulation and essential function of a DUB in long-term synaptic plasticity
Subsequent to the finding on ubiquitin-proteasome-mediated degradation of R subunits of PKA, a crucial role in LTF for a neuronal specific Ap-uch was discovered. Ap-uch is the homolog of human UCH-L1 and is induced by stimuli that produce LTF, but not stimuli that lead to short-term facilitation. Injection of antibodies or antisense oligonucleotides specific to Ap-uch into sensory neurons synapsing onto motor neurons in culture blocked induction of LTF (Hegde et al. 1997). Investigation on biochemical functions of Ap-uch indicated that Ap-uch is capable of cleaving small attachments to linearly attached ubiquitin molecules such as ubiquitin–ubiquitin–cysteine, but not large attachments like Glutathione S-transferase (GST) in substrates like ubiquitin–GST. Interestingly, additional biochemical analyses showed that Ap-uch associates with the proteasome. The association of Ap-uch increases the rate of degradation by the proteasome. For example, addition of recombinant Ap-uch to in vitro degradation systems showed that there was approximately a two-fold increase in degradation of the R subunit of PKA. Since persistent activation of PKA has been shown to be critical for induction of LTF and the R subunits of PKA were found to be substrates for the UPP, the experiments on Ap-uch provided some molecular explanation for the role of regulated proteolysis in long-term facilitation (LTF) (Hegde et al. 1997). Computational modeling has provided support for the idea that persistently active PKA induces Ap-uch, which in turn provides a positive feedback loop for increasing the PKA activity through the enhancement of the R subunit degradation (Song et al. 2006).
How does Ap-uch increase the rate of degradation by the proteasome? Using recombinant ubiquitin with its Lysine-48 mutated to Arg, which cannot support a Lys-48 type of polyubiquitin linkage to a protein substrate, it was shown that Ap-uch stimulates the release of ubiquitin from substrates in the presence of the proteasome (Hegde et al. 1997). Ubiquitin with Arg-48 can form single- or multiple-monoubiquitin linkages on the substrate. Therefore, it can be inferred that Ap-uch perhaps cleaves the first ubiquitin in the polyubiquitin chain attached to the peptide remnant of the substrate. Such a function of Ap-uch has to occur after the DUBs that are tightly associated with the proteasome finish bulk of the polyubiquitin chain disassembly as the unfolding of the substrate and its degradation progresses. The function of UCHs in synaptic plasticity appears to be evolutionarily conserved. The mammalian homolog of Ap-uch, UCH-L1, has been shown to play a role in long-term memory in mice (Gong et al. 2006). Also, the Gong et al. (2006) study showed a link between UCH-L1 and the R subunit degradation in the mouse hippocampus.
Recent studies have expanded the role of Ap-uch and the proteasome in Aplysia to long-term depression (LTD). In Aplysia, sensory-motor neuron synapses undergo transcription-dependent LTD in response to treatment with the neuropeptide Phe-Met-Arg-Phe-NH2 (FMRFa). Application of the proteasome inhibitor lactacystin blocked FMRFa-induced LTD. Also, FMRFa was found to up-regulate Ap-uch mRNA (Fioravante et al. 2008). Thus, Ap-uch could have a role in LTD in Aplysia perhaps through its action on a different set of downstream targets compared to those affected by Ap-uch during LTF.
Presynaptic and postsynaptic roles of the UPP in short-term plasticity and synaptic transmission
Ubiquitin-proteasome-mediated proteolysis regulates key proteins at the synaptic terminals (presynaptic) as well as in the postsynaptic compartment. These roles of the UPP contribute to the regulation of synaptic transmission as well as short-term synaptic plasticity.
Presynaptic roles of the UPP
The UPP, in addition to regulating molecules such as PKA, which are critical for long-term synaptic plasticity, also acutely modulates proteins, thus affecting synaptic transmission and short-term synaptic plasticity. For instance, the protein Dunc-13, which is critical in priming the synaptic vesicles, is ubiquitinated and degraded by the proteasome in Drosophila neuromuscular synapse. Application of proteasome inhibitors and the dominant-negative mutation in a core subunit (β6) of the Drosophila proteasome both lead to an increase in the quantity of Dunc-13 protein in presynaptic terminals. Also, the application of the proteasome inhibitors lactacystin and epoxomycin cause an increase in the excitatory junctional current suggesting that stabilization of Dunc-13 and the resultant increase in the net Dunc-13 quantity leads to increased synaptic transmission (Speese et al. 2003).
Does the UPP have a wider role in controlling short-term synaptic plasticity? If so, regulated proteolysis might control the amounts of other presynaptic proteins as well. Other synaptic vesicle proteins such as syntaxin 1 and RIM1α (Rab3-interacting molecule 1α) have been shown to be substrates for ubiquitin-proteasome-mediated degradation. Syntaxin 1 is a presynaptic protein that has a role in synaptic vesicle exocytosis. Evidence for ubiquitin-proteasome-mediated degradation of syntaxin 1 was obtained through identification of an ubiquitin ligase called staring (syntaxin 1-interacting RING finger protein) using the yeast two-hybrid system (Chin et al. 2002). Coexpression of staring with syntaxin 1 in HeLa cells increases the degradation of syntaxin 1, which can be inhibited by the proteasome inhibitor MG132. The physiological effect of ubiquitin-proteasome-mediated degradation of syntaxin 1 remains to be determined. RIM1α functions to form a presynaptic scaffold that links synaptic vesicles with fusion machinery. A ubiquitin ligase named SCRAPPER (an acronym whose derivation is not clearly defined) has been shown to regulate the amount of RIM1α. A series of experiments using miniature postsynaptic current (mEPSC) measurements established that SCRAPPER regulates synaptic transmission. It was also found that in mice lacking SCRAPPER short-term synaptic plasticity was impaired (Yao et al. 2007).
A role for the proteasome has been found in recycling of synaptic vesicles in hippocampal neurons in primary culture. Proteasome inhibition causes an increase in the size of the recycling pool of vesicles. A blockade of neuronal activity significantly reduces the effect of proteasome inhibition, decreasing vesicle numbers. Inhibition of the proteasome, however, does not increase the transmitter release probability. It appears that in vertebrate neurons, the proteasome functions to maintain vesicle homeostasis (Willeumier et al. 2006). Recent data add another layer to the complexity of UPP function in neurons. In cultured mammalian hippocampal neurons, applications of proteasome inhibitors increase mEPSC frequency without any effect on the amplitude, indicating a presynaptic role for the UPP. Contrary to expectations, stabilization of the presynaptic proteins (RIM1 or Munc13) was not observed (Rinetti and Schweizer 2010). Another study, however, found a decrease in Rim 1 and Munc 13 during persistent presynaptic silencing induced by depolarization (Jiang et al. 2010). The results from these two studies seem to be at odds with each other even though both used postnatal rat hippocampal neurons in culture and antibodies against Rim 1 and Munc 13 from the same commercial sources. Perhaps the discrepancy was due to the fact that the study by Jiang et al. (2010) measured Rim 1 and Munc 13 after K+-induced depolarization, whereas the study by Rinetti and Schweizer (2010) tested Rim 1 and Munc 13 levels in relation to changes in mEPSCs and spontaneous EPSCs. Therefore, it is likely that the degradation of Rim 1 and Munc 13 is triggered by neuronal depolarization rather than baseline activity.
Postsynaptic roles of the UPP
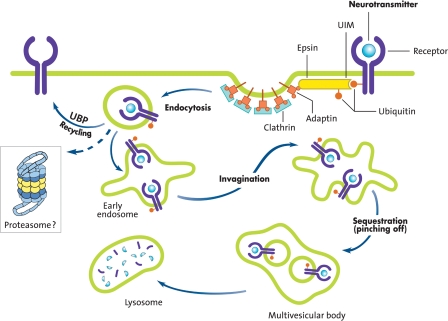
Much of the evidence for postsynaptic roles of the UPP in synaptic plasticity is indirect. Many studies have indicated that the UPP modulates neurotransmitter receptors, structural proteins, and regulatory molecules in the postsynaptic compartment. Regulation of the neurotransmitter receptors mainly occurs through ubiquitination that marks proteins for endocytosis as shown in Figure 4. Endocytosis is mainly mediated by the attachment of a single-ubiquitin (monoubiquitination) or a Lys-63-linked polyubiquitin chain. The ubiquitinated protein that is endocytosed may be recycled back to the plasma membrane if the ubiquitin is removed by DUBs or targeted to the lysosome via the multivesicular body. Some membrane proteins, upon endocytosis are degraded by the proteasome instead of being routed to the lysosome for degradation (Fig. 4).
Figure 4.
Ubiquitin and endocytosis. Receptors on the plasma membrane undergo monoubiquitination as a result of ligand (e.g., neurotransmitter) binding to them. Ubiquitinated receptors bind to proteins called epsins through a ubiquitin-interacting motif (UIM). The epsins in turn interact with adaptor proteins (adaptin) bound to clathrin-coated pits. Ubiquitination also functions to sort the internalized membrane protein into early endosomes, which directs them to degradation by lysosome via the multivesicular body. If ubiquitin from the endocytosed receptors is removed by a UBP, the receptor recycles back to the membrane. Proteasome inhibitors block endocytotic degradation of some proteins, such as glutamate receptor subunits, indicating a possible role for the proteasome. In several cases, a Lys-63-linked polyubiquitin chain attachment (instead of monoubiquitination) plays a role in endocytosis (not depicted). (Figure modified from Hegde 2004 and reprinted with permission from Elsevier © 2004.)
Studies on Caenorhabditis elegans showed a role for ubiquitin in endocytosis of a GLR-1 type of glutamate receptor (Burbea et al. 2002). In mammalian hippocampal neurons, treatment with the proteasome inhibitor MG132 blocks agonist-induced endocytosis of AMPA-type glutamate receptors (Patrick et al. 2003). Also, NMDA-induced AMPA receptor internalization is prevented by the application of the proteasome inhibitor. The exact mechanism of AMPA receptor endocytosis in mammals is not clear. It is possible that the AMPA receptor itself is not the target of proteasome action, but a protein(s) that interacts with AMPA receptor is degraded by the proteasome. Given that both NMDA and AMPA agonists stimulate internalization that is inhibited by the proteasome, it is likely that this is a case of passive internalization and not an instance of regulated AMPA receptor endocytosis. In support of this idea, a postsynaptic density protein PSD-95 was shown to be regulated by ubiquitin-proteasome-mediated degradation (Colledge et al. 2003). PSD-95 is a major component of the postsynaptic scaffold, which through interaction with another protein called stargazin provides a docking site for AMPA receptors (Schnell et al. 2002). Degradation of PSD-95 leads to AMPA receptor internalization, and mutations that block PSD-95 ubiquitination block NMDA-induced AMPA receptor endocytosis (Colledge et al. 2003). Furthermore, application of the proteasome inhibitor MG132 to hippocampal slices reduces the magnitude of hippocampal LTD (Colledge et al. 2003). Because the transient, protein-synthesis-independent LTD (Sajikumar and Frey 2003) requires a net reduction in synaptic AMPA receptors (Malenka and Bear 2004), these data further support a role for the proteasome in decreasing the AMPA receptor amount at synaptic sites. A direct role for ubiquitin in regulated endocytotic degradation of AMPA receptors is still an open question, although it is likely that targeting of AMPA receptors for degradation after internalization depends on ubiquitination, which might play a role in short-term plasticity such as the transient form of LTD mentioned above.
A role for the proteasome in short-term synaptic plasticity has also been obtained through LTP experiments. A type of LTP called early-phase LTP (E-LTP), which is independent of protein synthesis, is enhanced by pre-incubation of hippocampal slices with the proteasome inhibitor β-lactone (Dong et al. 2008). Although the mechanisms by which E-LTP is enhanced by proteasome inhibition have not been elucidated, it is likely that the AMPA receptor stabilization and the consequent increase in the AMPA receptor number at postsynaptic sites might contribute to the increase in E-LTP.
The UPP likely plays a broad role in regulating the neurotransmitter receptors. The NMDA receptors are retrotranslocated and degraded by the UPP in an activity-dependent fashion. An F-box protein called Fbx2 is critical for this process (Kato et al. 2005), suggesting that an SCF-type ligase targets the NMDA receptors for ubiquitination. Endocytosis of other neurotransmitter receptors might be regulated by ubiquitination. A glycine receptor has been shown to be internalized upon ubiquitination (Buttner et al. 2001). A protein associated with GABAA receptors, Plic-1, indirectly controls the removal of GABAA through endocytosis (Bedford et al. 2001). It was found that proteasome inhibitors prevent degradation of internalized GABAA receptors. Subsequent studies showed that GABAA receptor ubiquitination is controlled by neuronal activity. A chronic blockade of neuronal activity by tetrodotoxin increases the level of GABAA receptor ubiquitination, and an increase in neuronal activity decreases the GABAA receptor ubiquitination and improves insertion of these receptors into the plasma membrane (Saliba et al. 2007). The GABAA receptors are heteropentameric proteins typically consisting of two α subunits, two β subunits, and one γ subunit. In the brain, the β subunits of the GABAA receptors are either β2 or β3 (Rudolph and Mohler 2006). The site of ubiquitination is the β3 subunit of the receptor. Activity blockade reduces the insertion of a β3-containing GABAA wild-type receptor, but not of a receptor containing mutant β3 that cannot be ubiquitinated (Saliba et al. 2007).
Ubiquitin-proteasome-mediated proteolysis appears to regulate several other proteins in addition to PSD-95 in the postsynaptic density including several structural proteins. For example, Shank, GKAP, and AKAP79/150 are degraded through the UPP. Unlike for the degradation of PSD-95, physiological relevance of the proteolytic removal of Shank, GKAP, and AKAP79/150 is not clear because the studies were correlative and a direct link between ubiquitin-proteasome-mediated degradation of the postsynaptic density (PSD) proteins and the structural remodeling was not established (Ehlers 2003).
There is also evidence that the UPP controls a protein that regulates spine shape. SPAR controls dendritic spine shape by reorganizing the actin cytoskeleton. During activity-dependent remodeling of synapses, SPAR was found to be degraded by the UPP. Degradation of SPAR is stimulated by serum inducible kinase (SNK). Activity induces SNK mRNA in the cell body and the induced SNK is targeted to the dendritic spines. Because of the time required for SNK mRNA to travel to the spines, one speculation is that SPAR may function to oppose synaptic remodeling after elevated activity (Pak and Sheng 2003).
Local proteolysis and synaptic plasticity: Roles for spatial control of ubiquitination and proteasome-mediated degradation
A role for local ubiquitin-proteasome-mediated degradation in synaptic plasticity was previously proposed (Hegde 2004). Many studies carried out since then provide support to this idea. Others working in this field are embracing the idea of local degradation as well (Segref and Hoppe 2009). It appears that local, regulated degradation of substrate proteins plays an important role in synaptic plasticity, as well as many other aspects of the nervous system such as development and fine-tuning of synaptic connections. Spatially restricted degradation can achieve synapse-specific effects. Cell-wide degradation would have consequences on all synapses made by a given neuron (Hegde 2004).
How might local protein degradation be achieved in neurons? A simple way would be to restrict the substrate to a subcellular location. For example, proteins whose expression is largely restricted to the synapses could be locally degraded because all the requisite UPP components are present at the synapse. Also substrates can be made vulnerable (or resistant) to ubiquitination by phosphorylation, which can be locally controlled in neurons. Similarly, activation (or inactivation) of ubiquitin ligases by phosphorylation or other post-translational modifications (such as the attachment of ubiquitin-like protein Nedd8 to Cul1 that activates SCF ligases; see Fig. 2IIBi) can be locally controlled as well. Moreover, specific E3 ligases can also be sequestered to specific cellular compartments. Experimental evidence has been obtained for some of these possibilities. Accumulating evidence indicates that proteasome activity is also differentially regulated in different neuronal compartments. A few examples of the local roles of ubiquitination and the local roles of the proteasome in neuronal compartments are discussed below.
Local roles of ubiquitination
As described previously, the specificity of ubiquitination is largely controlled at the level of E3 ubiquitin ligases. The specificity of ubiquitination could also be regulated at the level of E2s because of diversity in E2s, as well as availability of unique E2–E3 combinations (see Fig. 3). Recent data show evidence for the local roles of E2s and E3s, as well as for DUBs during the development of the synaptic connections.
E2s
In Drosophila, an E2 called ubcD1 controls dendritic pruning where local degradation appears to be critical. In this insect, most of the larval neurons die during metamorphosis but a small population of neurons, including a group of peripheral sensory neurons called the class IV dendritic arborization (C4da) neurons, survive to adulthood (Kuo et al. 2005). These neurons extensively remodel their dendrites by completely degrading the old arborizations and by growing a new elaborate set of dendrites. During the remodeling of dendrites, axons are kept intact. Hence, the molecular processes have to be spatially restricted. It was found that disruptions of the UPP by overexpression of an exogenous DUB called UBP2 from yeast, or mutations in E1 or a 19S proteasome subunit all disrupted dendritic pruning. Subsequent studies identified the essential role of ubcD1 in this process (Kuo et al. 2006). Mutations in ubcD1 led to a blockade of dendritic pruning and retention of larval dendrites in C4da neurons. Based on additional experiments it was inferred that ubcD1 targets DIAP1 (Drosophila inhibitor of apoptosis 1), an E3 ubiquitin ligase. DIAP1 is required for degradation of a caspase called Dronc. Therefore, degradation of DIAP1 allows for a local activation of the Dronc caspase in dendrites. Because the Dronc caspase is critical for severing dendrites of C4da neurons, restricted dendritic activation of this caspase allows the preservation of C4da neurons while removing their dendrites (Kuo et al. 2005, 2006).
E3s
Investigation of the Drosophila neuromuscular junction, using a genetic screen for identifying the mutants that enhance synaptic growth, found that a loss-of-function in a gene called highwire (hiw) causes a significant increase in the number of synapses. The hiw gene encodes a huge protein with 5233 amino acids. The highwire protein contains a RING finger domain, which is a characteristic feature of some ubiquitin ligases (Wan et al. 2000).
Subsequent work carried out on the C. elegans homolog of the hiw gene called RPM-1 showed that the ligase functions to regulate presynaptic differentiation. The RPM-1 protein is localized to the periactive zone, a presynaptic region excluded from the active zone and synaptic vesicles. RPM-1 combines with an F-box protein called FSN-1 and the C. elegans homologs of SKP1 and Cullin to form an SCF-like ubiquitin ligase complex. The localized function of this ubiquitin ligase in the periactive zone appears to be critical for presynaptic differentiation in C. elegans (Liao et al. 2004). The downstream target of RPM-1 in C. elegans is a MAP kinase kinase kinase (MAPKKK) called DLK-1 (Delta-like homolog 1), which is also localized to the periactive zone like RPM-1. Inactivation of the DLK-1 pathway suppresses RPM-1 loss of function phenotypes, whereas overexpression of DLK-1 causes synaptic aberrations resembling RPM-1 mutations (Nakata et al. 2005). In Drosophila, the downstream target of highwire is a MAPKKK encoded by a gene called wallenda (Collins et al. 2006). Although the downstream effectors of DLK-1 and the wallenda proteins are different, attenuation of the signaling mediated by these proteins inhibits synaptic growth in similar ways (Fulga and Van Vactor 2008).
The function of the highwire protein in the development of neuronal connections seems to be evolutionarily conserved. During an effort to discover new genes critical for axon navigation in mice, a mutation in Phr1, a gene encoding a ubiquitin ligase, which is a vertebrate homolog of highwire, was found. Studies using mice with a mutation in the Phr1 gene (a mutation called Magellan), which lacks the C-terminal ligase domain, revealed that the Phr1 protein is localized to an axon shaft and is largely excluded from growth cones and distal processes. The substrate of Phr1 is most likely DLK in mice as well. Distribution of DLK is nonoverlapping with that of Phr1; DLK is present in growth cones with only low levels in the axon shaft (Lewcock et al. 2007).
Recent data suggest that RPM-1 may have additional roles in the postsynaptic compartment. Park et al. (2009) found that RPM-1 regulates AMPA receptor trafficking in interneurons in C. elegans. RPM-1 works by negatively regulating the levels of DLK-1 (MAPKKK) (Park et al. 2009).
Local regulation of other E3 ligases has also been reported. For example, in hermaphrodite-specific motor neurons of C. elegans, an SCF ligase containing the protein SKR-1 and an F-box protein called SEL-10 mediates developmental elimination of synapses. A synaptic adhesion molecule called SYG-1 binds to SKR-1 and inhibits the assembly of the SCF complex, which protects the nearby synapses (Ding et al. 2007).
DUBs
The fate of a substrate marked for degradation by ubiquitination can be reversed by removal of the attached ubiquitin molecules by DUB. Thus, DUBs provide important negative regulation of protein degradation. Like ligases DUBs can act locally to reverse ubiquitination. A search for molecules that regulate the size and strength of synapses in Drosophila found that a DUB encoded by the fat facets (faf) gene functions in synapse formation. Overexpression of faf in the developing Drosophila nervous system causes synaptic overgrowth and perturbs synaptic transmission. A similar phenotype is observed when a yeast DUB is expressed in the fruit fly CNS (DiAntonio et al. 2001).
In mammals also DUBs have a local synaptic role. A DUB called Usp14 is essential for synaptic development and function in mouse neuromuscular junctions. The role for Usp14 in the nervous system was originally discovered through studies on mice with the ataxia (axj) mutation, a recessive mutation characterized by severe tremors, hind limb paralysis, and postnatal lethality (Wilson et al. 2002). The axj gene encodes Usp14, the protein product of which associates with the proteasome and is believed to help disassemble polyubiquitin chains and recycle ubiquitin (see Fig. 1), thus, maintaining ubiquitin levels in the cell. Accordingly, the loss of Usp14 results in reduced ubiquitin levels in many tissues of the axj mice including the brain (Anderson et al. 2005). The motor defects of the axj mice could be rescued and viability restored with transgenic Usp14 suggesting that Usp14 deficiency is the cause of neurological defects in these mice (Crimmins et al. 2006). Subsequent studies demonstrated that in Usp14-deficient axj mice ubiquitin loss occurred in the spinal cord and the sciatic nerve. Biochemical experiments showed that the highest loss of ubiquitin occurred in synaptosomal fractions, suggesting Usp14 at synaptic sites was critical. Loss of Usp14 caused presynaptic defects, such as nerve terminal sprouting and poor arborization of motor nerve terminals, and transgenic expression of Usp14 rescued these defects. Thus, it appears that local Usp14 function is critical for maintaining the ubiquitin levels and hence protein degradation at the synapse (Chen et al. 2009).
Does the proteasome have local roles in synaptic plasticity?
Modulation of the proteasome adds another level to regulation of proteolysis by the UPP. Even though it was not previously appreciated, the recent data indicate that proteasome is not homogeneous throughout the neuron. The impetus for closely looking at the regulation of the neuronal proteasome came from conflicting results obtained with proteasome inhibitors on LTF in Aplysia. Originally, it was found that proteasome inhibitors block the induction of LTF (Chain et al. 1999). Later studies on LTF, however, showed that bath application of the active form of lactacystin, clasto-lactacystin β-lactone, to sensory–motor neuron synapses resulted in enhanced LTF and an increase in neurite outgrowth in isolated sensory neuron (Zhao et al. 2003). The increase in neurite elongation is consistent with results obtained in PC12 and Neuro2A cells in which lactacystin induces neurite outgrowth (Fenteany et al. 1994). Both sets of results can be reconciled if one postulates that proteasome has different roles in different cellular compartments (Hegde 2004). In the same neuron, the proteasome is likely to carry out different tasks in different subcellular compartments, resulting in different physiological consequences at different loci. Therefore, blocking different roles of the proteasome during the induction of memory would lead to distinct, and even opposite, effects on synaptic strength. For example, the proteasome is known to degrade transcription repressors. Degradation of transcription repressors should allow transcription activators to induce gene expression, which in turn leads to the development of LTF. If the proteasome is inhibited only in the nucleus before the repressors are degraded, gene expression and hence induction of LTF should be blocked. Degradation of the CREB repressor CREB1b by the UPP in response to LTF-inducing protocols (Upadhya et al. 2004) supports this idea. On the other hand, if the degradation of proteins needed at the synapse for developing LTF is inhibited by the proteasome, LTF should be enhanced. As previously proposed, the main purpose of transcription during induction of LTF or other forms of long-term memory is to provide mRNAs for the synthesis of “rapidly turning over proteins” needed for memory formation (Hegde 2004). If the degradation of these proteins is prevented, then long-term memory formation becomes independent of transcription. In support of this idea, Zhao et al. (2003) found that proteasome inhibitor-induced synaptic strengthening depends on translation but not transcription.
The biochemical experiments on the proteasome also support the differential function of the proteasome in different neuronal compartments. The results of these experiments showed that both in the Aplysia nervous system and the mouse brain, proteasome activity in the synaptic terminals is significantly higher than that of the nuclear proteasome. Moreover, the proteasome activity in the two compartments is differentially regulated by protein kinases relevant to synaptic plasticity, such as PKA, PKC, and MAP kinase (Upadhya et al. 2006). Recently, others have found that CaMKII can stimulate proteasome activity in cultured hippocampal neurons (Djakovic et al. 2009).
As discussed above, differential activity of the proteasome in Aplysia might explain conflicting results obtained in different studies. Does differential proteasomal activity affect synaptic plasticity differentially in vertebrates? It has been found that the proteasome has differential roles during induction and maintenance phases of the late phase of long-term potentiation (L-LTP) (Dong et al. 2008), which is discussed in detail in the next section.
Evidence from other studies using cultured rat hippocampal neurons showed dynamic local regulation of the proteasome at the dendrites. It was found that proteasome is redistributed from dendritic shafts to synaptic spines in an NMDA receptor-dependent manner. How does the redistribution of the proteasome occur? The experiments showed that activity only modestly increased the entry of the proteasome into dendritic shafts, but significantly reduced their exit. Furthermore, the results suggested that the proteasome was sequestered persistently in the spines through association with cytoskeleton (Bingol and Schuman 2006). Subsequent studies showed that a protein called NAC1, which is induced by psychostimulants, modulates the recruitment of the proteasome into the dendritic spines (Shen et al. 2007). Since the bulk of the evidence in this study is for the catalytic 20S core of the proteasome, it remains to be seen whether the recruitment of the full proteasome complex (26S) that degrades polyubiquitinated proteins is also regulated by NAC1. A recent study has suggested that the CaMKIIα subunit acts as a scaffold for the proteasome (Bingol et al. 2010). It is not clear how, or if, the functions of NAC1 and CaMKIIα relate to each other in sequestering the proteasome.
Proteasome might function to locally regulate other processes required for synaptic plasticity such as translation of mRNA. For example, the fragile X mental retardation protein (FMRP), which is thought to regulate translation of a subset of mRNAs in dendrites, is regulated by the proteasome. Furthermore, regulation of FMRP by the proteasome appears to be critical for metabotropic glutamate receptor-dependent LTD (Hou et al. 2006).
Differential local roles of the proteasome in induction and maintenance of L-LTP: Evidence for opposing dendritic and nuclear functions
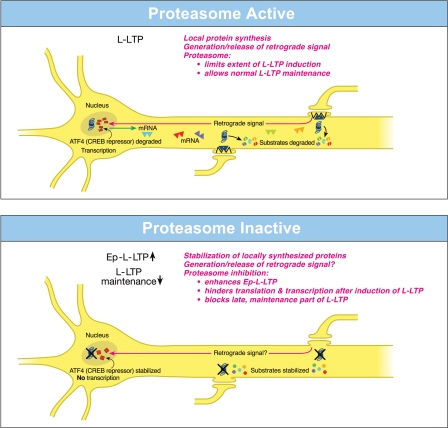
Evidence for functional significance of local roles of the proteasome came from studies on the hippocampal late-phase LTP (L-LTP). Recent investigations showed that the proteasome inhibitor application to the hippocampal slices prior to the induction of L-LTP caused an increase in the magnitude of the early, induction phase but an inhibition of the late, maintenance phase (Dong et al. 2008). What is the basis of these differential effects of the proteasome on phases of L-LTP? The enhancement of the early induction phase (this early phase of L-LTP is referred to as Ep-L-LTP for convenience) by the proteasome inhibitor β-lactone is blocked by a prior application of the translation inhibitor anisomycin but not by a transcription inhibitor actinomycin D. The increase in Ep-L-LTP caused by β-lactone is also prevented by prior application of rapamycin, which blocks signaling that controls translation of a subset of mRNAs (Gingras et al. 2001). Moreover, Ep-L-LTP is augmented in dendrites isolated from the cell body by means of a surgical cut. These lines of evidence suggest that proteasome inhibition enhances Ep-L-LTP by stabilizing proteins locally translated from pre-existing mRNAs (Fig. 5, top; Dong et al. 2008).
Figure 5.
Opposing local roles of the proteasome in dendrites and in the nucleus during L-LTP. (Top) Proteasome active: When the proteasome in the dendrites is highly active, protein substrates that positively regulate L-LTP are degraded (broken spheres), and therefore the extent of L-LTP is limited and only normal L-LTP ensues. A retrograde signal is likely transmitted to the nucleus. Proteasome aids the transcription of genes by degrading the CREB repressor ATF4 (broken squares in the nucleus) thus allowing for normal L-LTP maintenance. Transcribed mRNAs (triangles) travel to activated synapses. (Bottom) Proteasome inactive: When the proteasome is inhibited (indicated by crosses on the proteasome), newly synthesized proteins in the dendrites are stabilized (intact spheres) and the L-LTP-inducing stimulation protocols dramatically increase (upward arrow) the early part of L-LTP (Ep-L-LTP). The proteasome inhibition obstructs CREB-mediated transcription by preventing the degradation of transcription repressor ATF4 (intact squares in the nucleus). Proteasome inhibition could also inhibit the generation of the retrograde signal. Therefore, L-LTP is not maintained but decays (downward arrow). It is likely that proteasome inhibition also causes failure of sustained translation because of the stabilization of the translation repressors, which accumulate after the induction of L-LTP, thus contributing to the blockade of the L-LTP maintenance.
How does proteasome inhibition block maintenance of L-LTP? The proteasome inhibitor β-lactone blocks maintenance of L-LTP only if applied prior to the induction of L-LTP, but not if applied 2 h after the induction of L-LTP. Previous studies have established that the critical time window for the transcription required for the maintenance of L-LTP is 2 h (Nguyen et al. 1994). These results suggest that proteasome inhibition blocks maintenance of L-LTP by inhibiting transcription. Additional molecular evidence supports this notion. The application of β-lactone to the hippocampal slices significantly reduced induction of BDNF (brain-derived neurotrophic factor) mRNA by chemically induced LTP (cLTP) or L-LTP induced by a theta-burst protocol (Dong et al. 2008). BDNF is a CREB-inducible gene linked to maintenance of L-LTP (Barco et al. 2005).
What is the mechanism of transcription blockade caused by inhibition of the proteasome? One possibility is that normally the UPP aids the degradation of transcription repressors. Hence, proteasome inhibition would result in accumulation of these repressors, thus blocking transcription. In support of this idea, it was found that a CREB repressor ATF4 is degraded by the UPP during cLTP and the β-lactone application to the hippocampal slices prevents degradation of ATF4. Furthermore, ATF4-ubiquitin conjugates accumulate during cLTP when proteasome is inhibited (Fig. 5, bottom; Dong et al. 2008).
These studies have also revealed the changing role of the proteasome even in dendrites through progression of L-LTP. The application of β-lactone to the isolated dendrites also results in the blockade of the dendritic L-LTP (Dong et al. 2008). Under these conditions, there is no supply of newly transcribed mRNA from the cell body. Thus, a blockade of transcription by proteasome inhibition does not explain this phenomenon. The most likely possibility is that proteasome inhibition leads to a slow accumulation of translation repressors in dendrites. The buildup of translation repressors would also occur in the cell body, which would hinder translation of newly transcribed mRNAs. Thus, late stages of translation in both dendrites and the cell body would be blocked by stabilization of translation repressors by proteasome inhibition. One line of experimental evidence supports this idea. If the translation inhibitor anisomycin is applied after the application of β-lactone to the hippocampal slices, then the inhibition of the maintenance phase is rescued (Dong et al. 2008). Recent work on the rat hippocampal neuronal culture renders further support to the idea of accumulation of translation repressors. In hippocampal neurons a negative regulator of translation called Mov10 is degraded by the proteasome in NMDA- and activity-dependent manners (Banerjee et al. 2009). Mov10 is the mammalian homolog of the Drosophila Armitage. Previous work has shown that Armitage, which is a component of the RNA-induced silencing complex (RISC), regulates the synaptic protein synthesis required for long-lasting odor memory. Armitage is also degraded by the proteasome and the degradation occurs in response to neuronal activity (Ashraf et al. 2006). In cultured hippocampal neurons Mov10 inhibits the translation of key plasticity-related mRNAs, such as that of CaMKIIα (Banerjee et al. 2009). Thus, prolonged proteasome inhibition is likely to cause buildup of Mov10 and block the synthesis of key proteins required for long-lasting plasticity.
Other studies have investigated the effect of proteasome inhibition on LTP. These studies failed to discover differential functions of proteasome in LTP because one study used MG-132 (Karpova et al. 2006), which is not a highly specific proteasome inhibitor (Chain et al. 1999; Tang and Leppla 1999), and the other used proteasome inhibitors lactacystin and epoxomycin at nanomolar concentration (Fonseca et al. 2006), which is significantly lower than the effective concentration (micromolar) required to block proteasome activity.
The UPP and synaptic malfunction
In addition to the myriad of roles in a normal synaptic function, the UPP has also been linked to synaptic malfunction observed in many diseases and disorders of the brain. Defects in the UPP are believed to play some role in the development of Alzheimer disease (AD) (de Vrij et al. 2004), Parkinson's disease (PD) (Upadhya and Hegde 2005), and Huntington's disease (HD) (Rubinsztein 2006). Given its role in synaptic plasticity the UPP may also play a role in synaptic defects underlying cognitive impairment observed in these diseases.
Impairment in synaptic plasticity and its connection to the UPP is better understood in AD compared to PD and HD. Cognitive defects that occur early in AD likely occur because of synaptic malfunction (Selkoe 2002). In mouse models of AD, deficits in LTP and memory are known to occur and have been shown to correlate well with the accumulation of Aβ (Hsiao et al. 1996). Ubiquitin immunoreactivity is found in plaques and tangles of AD brains. A blockade of the UPP in the neurons of AD brains might be responsible for accumulation of ubiquitinated proteins (Upadhya and Hegde 2005, 2007). Although, how the UPP connects to AD pathology and cognitive impairment is not understood some hints regarding the role of the UPP in AD have been discovered. For example, the application of oligomeric Aβ inhibits LTP, which can be rescued by treatment with exogenous UCHL1 (mammalian homolog of Ap-uch). In AD model mice carrying amyloid precursor protein and presenilin1 transgenes, deficits in LTP and memory can also be rescued by treatment with exogenous UCHL1 (Gong et al. 2006).
A direct link between the UPP and pathogenesis of AD was provided by a finding that the brains of some AD patients contained an aberrant form of ubiquitin that has 20 additional amino acids at its C-terminus (UBB+1) (van Leeuwen et al. 1998). Transgenic mice that postnatally express UBB+1 in neurons show proteasome dysfunction and deficits in contextual memory (Fischer et al. 2009). Considering that proteasome inhibition throughout the neuron blocks maintenance of L-LTP because it hinders transcription and sustained translation (Dong et al. 2008), it is interesting to speculate that the memory deficits in the UBB+1 mice result from impaired synaptic plasticity owing to neuron-wide proteasome dysfunction.
A new kind of connection of the UPP to neurodegneration has recently emerged. It has been found that another cellular degradative process, autophagy, utilizes an enzymatic pathway similar to ubiquitin conjugation, which attaches ubiquitin-like proteins such as Atg12 to some of the proteins that regulate the autophagic process (Nakatogawa et al. 2009). Autophagy is the mechanism by which a double membrane vesicle (called the autophagosome) engulfs parts of the cytoplasm or organelles and delivers it to the lysosome. A direct connection between ubiquitination and autophagy has also been found. In SH-SY5Y cells, Lys-63-linked polyubiquitination promotes inclusion bodies, which are cleared by autophagy (Tan et al. 2008). Another study found that parkin, which is an E3 ligase, promotes Lys-63-linked polyubiquitin chain attachment to misfolded proteins. The Lys-63-polyubiquitin chain appears to serve as a signal to couple the misfolded proteins to the dynein motor complex through histone deacetylase 6 (which serves as an adaptor), and thus aiding in sequestration of misfolded proteins into specialized inclusion bodies called aggresomes, which are cleared by autophagy (Olzmann and Chin 2008). A recent study has found that autophagy promotes synapse development in Drosophila (Shen and Ganetzky 2009). It remains to be seen whether autophagy is connected to synaptic malfunction observed in neurodegenerative diseases.
Unanswered questions and future directions
There are many unanswered questions with respect to the role of the UPP in developmental synaptic plasticity as well as plasticity in the fully developed brain. Because the levels of many proteins in neurons are likely regulated by protein degradation, we have probably only scratched the surface as far as identifying the substrates of proteolysis that play a role in synaptic plasticity. Also, there are many gaps in our knowledge regarding the already identified substrates and the enzymes of the UPP. For example, APC appears to have nuclear as well as synaptic roles, but it is not clear how this ligase is locally regulated in different compartments. Although there is evidence to indicate differential regulation of the proteasome in different parts of neurons the mechanisms of such regulation have not been identified. In the coming years, we are likely to see many new discoveries on the roles of the UPP in synaptic plasticity and other functions of the nervous system. In the future, we might also expect breakthroughs in linking UPP defects to synaptic dysfunction observed in neurodegenerative diseases, because of the intimate connection between impairment of the UPP and these diseases.
Acknowledgments
I thank Dr. Sudarshan Upadhya for critical comments on the manuscript. This research is supported by grants from the National Institutes of Health (MH060225 and NS066583).
References
- Abel T, Nguyen PV 2008. Regulation of hippocampus-dependent memory by cyclic AMP-dependent protein kinase. Prog Brain Res 169: 97–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel T, Martin KC, Bartsch D, Kandel ER 1998. Memory suppressor genes: Inhibitory constraints on the storage of long-term memory. Science 279: 338–341 [DOI] [PubMed] [Google Scholar]
- Abrams TW 1985. Activity-dependent presynaptic facilitation: An associative mechanism in Aplysia. Cell Mol Neurobiol 5: 123–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, Wilson SM 2005. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem 95: 724–731 [DOI] [PubMed] [Google Scholar]
- Ang XL, Seeburg DP, Sheng M, Harper JW 2008. Regulation of postsynaptic RapGAP SPAR by Polo-like kinase 2 and the SCFβ-TRCP ubiquitin ligase in hippocampal neurons. J Biol Chem 283: 29424–29432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artinian J, McGauran AM, De JX, Mouledous L, Frances B, Roullet P 2008. Protein degradation, as with protein synthesis, is required during not only long-term spatial memory consolidation but also reconsolidation. Eur J Neurosci 27: 3009–3019 [DOI] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S 2006. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124: 191–205 [DOI] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS 2009. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron 64: 871–884 [DOI] [PubMed] [Google Scholar]
- Barco A, Patterson S, Alarcon JM, Gromova P, Mata-Roig M, Morozov A, Kandel ER 2005. Gene expression profiling of facilitated L-LTP in VP16-CREB mice reveals that BDNF is critical for the maintenance of LTP and its synaptic capture. Neuron 48: 123–137 [DOI] [PubMed] [Google Scholar]
- Bedford FK, Kittler JT, Muller E, Thomas P, Uren JM, Merlo D, Wisden W, Triller A, Smart TG, Moss SJ 2001. GABAA receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat Neurosci 4: 908–916 [DOI] [PubMed] [Google Scholar]
- Beer-Romero P, Glass S, Rolfe M 1997. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene 14: 595–602 [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM 2006. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature 441: 1144–1148 [DOI] [PubMed] [Google Scholar]
- Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M 2010. Autophosphorylated CaMKIIα acts as a scaffold to recruit proteasomes to dendritic spines. Cell 140: 567–578 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D III, Fukuda M, Ohta T, Klevit R 2003. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA 100: 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbea M, Dreier L, Dittman JS, Grunwald ME, Kaplan JM 2002. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron 35: 107–120 [DOI] [PubMed] [Google Scholar]
- Buttner C, Sadtler S, Leyendecker A, Laube B, Griffon N, Betz H, Schmalzing G 2001. Ubiquitination precedes internalization and proteolytic cleavage of plasma membrane-bound glycine receptors. J Biol Chem 276: 42978–42985 [DOI] [PubMed] [Google Scholar]
- Chain DG, Casadio A, Schacher S, Hegde AN, Valbrun M, Yamamoto N, Goldberg AL, Bartsch D, Kandel ER, Schwartz JH 1999. Mechanisms for generating the autonomous cAMP-dependent protein kinase required for long-term facilitation in Aplysia. Neuron 22: 147–156 [DOI] [PubMed] [Google Scholar]
- Chen C, Tonegawa S 1997. Molecular genetic analysis of synaptic plasticity activity-dependent neural development learning and memory in the mammalian brain. Annu Rev Neurosci 20: 157–184 [DOI] [PubMed] [Google Scholar]
- Chen PC, Qin LN, Li XM, Walters BJ, Wilson JA, Mei L, Wilson SM 2009. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J Neurosci 29: 10909–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y 2009. Toward an atomic model of the 26S proteasome. Curr Opin Struct Biol 19: 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin LS, Vavalle JP, Li L 2002. Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J Biol Chem 277: 35071–35079 [DOI] [PubMed] [Google Scholar]
- Chiu YH, Sun Q, Chen ZJ 2007. E1-L2 activates both ubiquitin and FAT10. Mol Cell 27: 1014–1023 [DOI] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD 2003. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 40: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Wairkar YP, Johnson SL, DiAntonio A 2006. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron 51: 57–69 [DOI] [PubMed] [Google Scholar]
- Crimmins S, Jin Y, Wheeler C, Huffman AK, Chapman C, Dobrunz LE, Levey A, Roth KA, Wilson JA, Wilson SM 2006. Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J Neurosci 26: 11423–11431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ 2000. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- de Vrij FM, Fischer DF, van Leeuwen FW, Hol EM 2004. Protein quality control in Alzheimer's disease by the ubiquitin proteasome system. Prog Neurobiol 74: 249–270 [DOI] [PubMed] [Google Scholar]
- DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, Goodman CS 2001. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature 412: 449–452 [DOI] [PubMed] [Google Scholar]
- Ding M, Chao D, Wang G, Shen K 2007. Spatial regulation of an E3 ubiquitin ligase directs selective synapse elimination. Science 317: 947–951 [DOI] [PubMed] [Google Scholar]
- Djakovic SN, Schwarz LA, Barylko B, DeMartino GN, Patrick GN 2009. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. J Biol Chem 284: 26655–26665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Upadhya SC, Ding L, Smith TK, Hegde AN 2008. Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learn Mem 15: 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD 2003. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci 6: 231–242 [DOI] [PubMed] [Google Scholar]
- Fenteany G, Standaert RF, Reichard GA, Corey EJ, Schreiber SL 1994. A β-lactone related to lactacystin induces neurite outgrowth in a neuroblastoma cell line and inhibits cell cycle progression in an osteosarcoma cell line. Proc Natl Acad Sci USA 91: 3358–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravante D, Liu RY, Byrne JH 2008. The ubiquitin-proteasome system is necessary for long-term synaptic depression in Aplysia. J Neurosci 28: 10245–10256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer DF, van Dijk R, van Tijn P, Hobo B, Verhage MC, van der Schors RC, Li KW, van Minnen J, Hol EM, van Leeuwen FW 2009. Long-term proteasome dysfunction in the mouse brain by expression of aberrant ubiquitin. Neurobiol Aging 30: 847–863 [DOI] [PubMed] [Google Scholar]
- Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nagerl UV 2006. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron 52: 239–245 [DOI] [PubMed] [Google Scholar]
- Fulga TA, Van Vactor D 2008. Synapses and growth cones on two sides of a highwire. Neuron 57: 339–344 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev 15: 807–826 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A 2002. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A, Moolman D, Zhang H, Shelanski M, Arancio O 2006. Ubiquitin hydrolase Uch-L1 rescues β-amyloid-induced decreases in synaptic function and contextual memory. Cell 126: 775–788 [DOI] [PubMed] [Google Scholar]
- Greenberg SM, Castellucci VF, Bayley H, Schwartz JH 1987. A molecular mechanism for long-term sensitization in Aplysia. Nature 329: 62–65 [DOI] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, et al. 2010. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 140: 704–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI 2001. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276: 33111–33120 [DOI] [PubMed] [Google Scholar]
- Hegde AN 2004. Ubiquitin-proteasome-mediated local protein degradation and synaptic plasticity. Prog Neurobiol 73: 311–357 [DOI] [PubMed] [Google Scholar]
- Hegde AN 2010. Ubiquitin-dependent protein degradation. In Comprehensive natural products II chemistry and biology, Vol. 5 (ed. Mander L, Lui H-W), pp. 699–752 Elsevier, Oxford, UK [Google Scholar]
- Hegde AN, DiAntonio A 2002. Ubiquitin and the synapse. Nat Rev Neurosci 3: 854–861 [DOI] [PubMed] [Google Scholar]
- Hegde AN, Goldberg AL, Schwartz JH 1993. Regulatory subunits of cAMP-dependent protein kinases are degraded after conjugation to ubiquitin: A molecular mechanism underlying long-term synaptic plasticity. Proc Natl Acad Sci USA 90: 7436–7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, Martin KC, Kandel ER, Schwartz JH 1997. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell 89: 115–126 [DOI] [PubMed] [Google Scholar]
- Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O'Leary DD, Luo L 2006. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron 50: 883–895 [DOI] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E 2006. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron 51: 441–454 [DOI] [PubMed] [Google Scholar]
- Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G 1996. Correlative memory deficits, Aβ elevation, and amyloid plaques in transgenic mice. Science 274: 99–102 [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM 1993. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol 13: 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA 92: 2563–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL 1998. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 21: 799–811 [DOI] [PubMed] [Google Scholar]
- Jiang X, Litkowski PE, Taylor AA, Lin Y, Snider BJ, Moulder KL 2010. A role for the ubiquitin-proteasome system in activity-dependent presynaptic silencing. J Neurosci 30: 1798–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW 2004. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 18: 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Li X, Gygi SP, Harper JW 2007. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447: 1135–1138 [DOI] [PubMed] [Google Scholar]
- Kandel ER 2001. The molecular biology of memory storage: A dialogue between genes and synapses. Science 294: 1030–1038 [DOI] [PubMed] [Google Scholar]
- Kaneko-Oshikawa C, Nakagawa T, Yamada M, Yoshikawa H, Matsumoto M, Yada M, Hatakeyama S, Nakayama K, Nakayama KI 2005. Mammalian E4 is required for cardiac development and maintenance of the nervous system. Mol Cell Biol 25: 10953–10964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova A, Mikhaylova M, Thomas U, Knopfel T, Behnisch T 2006. Involvement of protein synthesis and degradation in long-term potentiation of Schaffer collateral CA1 synapses. J Neurosci 26: 4949–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Rouach N, Nicoll RA, Bredt DS 2005. Activity-dependent NMDA receptor degradation mediated by retrotranslocation and ubiquitination. Proc Natl Acad Sci USA 102: 5600–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M, Rowitch D, Bonni A 2009. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell 136: 322–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW 1996. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell 7: 1343–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J 1997. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet 15: 70–73 [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96: 635–644 [DOI] [PubMed] [Google Scholar]
- Kolman CJ, Toth J, Gonda DK 1992. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J 11: 3081–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D 2009. The emerging complexity of protein ubiquitination. Biochem Soc Trans 37: 937–953 [DOI] [PubMed] [Google Scholar]
- Kumar S, Kao WH, Howley PM 1997. Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J Biol Chem 272: 13548–13554 [DOI] [PubMed] [Google Scholar]
- Kuo CT, Jan LY, Jan YN 2005. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci USA 102: 15230–15235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN 2006. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 51: 283–290 [DOI] [PubMed] [Google Scholar]
- Lee SH, Choi JH, Lee N, Lee HR, Kim JI, Yu NK, Choi SL, Lee SH, Kim H, Kaang BK 2008. Synaptic protein degradation underlies destabilization of retrieved fear memory. Science 319: 1253–1256 [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D 2002. Multiple associated proteins regulate proteasome structure and function. Mol Cell 10: 495–507 [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Genoud N, Lettieri K, Pfaff SL 2007. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron 56: 604–620 [DOI] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA 2008. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One 3: 1487 doi: 10.1371/journal.pone.0001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao EH, Hung W, Abrams B, Zhen M 2004. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature 430: 345–350 [DOI] [PubMed] [Google Scholar]
- Lisman J 1994. The CaM kinase II hypothesis for the storage of synaptic memory. Trends Neurosci 17: 406–412 [DOI] [PubMed] [Google Scholar]
- Lopez-Salon M, Alonso M, Vianna MR, Viola H, Mello e Souza Izquierdo I, Pasquini JM, Medina JH 2001. The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci 14: 1820–1826 [DOI] [PubMed] [Google Scholar]
- Lowe J, Blanchard A, Morrell K, Lennox G, Reynolds L, Billett M, Landon M, Mayer RJ 1988. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson's disease, Pick's disease, and Alzheimer's disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J Pathol 155: 9–15 [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF 2004. LTP and LTD: An embarrassment of riches. Neuron 44: 5–21 [DOI] [PubMed] [Google Scholar]
- Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ 2009. Catalytic mechanism and assembly of the proteasome. Chem Rev 109: 1509–1536 [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL 1997. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet 15: 74–77 [DOI] [PubMed] [Google Scholar]
- Mori H, Kondo J, Ihara Y 1987. Ubiquitin is a component of paired helical filaments in Alzheimer's disease. Science 235: 1641–1644 [DOI] [PubMed] [Google Scholar]
- Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, Jin Y 2005. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell 120: 407–420 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y 2009. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Nguyen PV, Abel T, Kandel ER 1994. Requirement of a critical period of transcription for induction of a late phase of LTP. Science 265: 1104–1107 [DOI] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL 2003. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol 10: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Chin LS 2008. Parkin-mediated K63-linked polyubiquitination: A signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy 4: 85–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Sheng M 2003. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science 302: 1368–1373 [DOI] [PubMed] [Google Scholar]
- Park EC, Glodowski DR, Rongo C 2009. The ubiquitin ligase RPM-1 and the p38 MAPK PMK-3 regulate AMPA receptor trafficking. PLoS One 4: 4284 doi: 10.1371/journal.pone.0004284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick GN, Bingol B, Weld HA, Schuman EM 2003. Ubiquitin-mediated proteasome activity is required for agonist-induced endocytosis of GluRs. Curr Biol 13: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Patterson C 2002. A new gun in town: The U box is a ubiquitin ligase domain. Sci STKE 2002: pe4 doi: 10.1126/stke.2002.116.pe4 [DOI] [PubMed] [Google Scholar]
- Pelzer C, Kassner I, Matentzoglu K, Singh RK, Wollscheid HP, Scheffner M, Schmidtke G, Groettrup M 2007. UBE1L2, a novel E1 enzyme specific for ubiquitin. J Biol Chem 282: 23010–23014 [DOI] [PubMed] [Google Scholar]
- Peth A, Besche HC, Goldberg AL 2009. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell 36: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20 [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Hoffman L, Dubiel W 1993. The multicatalytic and 26 S proteases. J Biol Chem 268: 6065–6068 [PubMed] [Google Scholar]
- Reissner KJ, Shobe JL, Carew TJ 2006. Molecular nodes in memory processing: Insights from Aplysia. Cell Mol Life Sci 63: 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinetti GV, Schweizer FE 2010. Ubiquitination acutely regulates presynaptic neurotransmitter release in mammalian neurons. J Neurosci 30: 3157–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC 2006. The roles of intracellular protein–degradation pathways in neurodegeneration. Nature 443: 780–786 [DOI] [PubMed] [Google Scholar]
- Rudolph U, Mohler H 2006. GABA-based therapeutic approaches: GABAA receptor subtype functions. Curr Opin Pharmacol 6: 18–23 [DOI] [PubMed] [Google Scholar]
- Sajikumar S, Frey JU 2003. Anisomycin inhibits the late maintenance of long-term depression in rat hippocampal slices in vitro. Neurosci Lett 338: 147–150 [DOI] [PubMed] [Google Scholar]
- Saliba RS, Michels G, Jacob TC, Pangalos MN, Moss SJ 2007. Activity-dependent ubiquitination of GABAA receptors regulates their accumulation at synaptic sites. J Neurosci 27: 13341–13351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM 1995. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373: 81–83 [DOI] [PubMed] [Google Scholar]
- Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA 2002. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci USA 99: 13902–13907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Hoppe T 2009. Think locally: Control of ubiquitin-dependent protein degradation in neurons. EMBO Rep 10: 44–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ 2002. Alzheimer's disease is a synaptic failure. Science 298: 789–791 [DOI] [PubMed] [Google Scholar]
- Shang F, Deng G, Obin M, Wu CC, Gong X, Smith D, Laursen RA, Andley UP, Reddan JR, Taylor A 2001. Ubiquitin-activating enzyme (E1) isoforms in lens epithelial cells: Origin of translation E2 specificity and cellular localization determined with novel site-specific antibodies. Exp Eye Res 73: 827–836 [DOI] [PubMed] [Google Scholar]
- Shen W, Ganetzky B 2009. Autophagy promotes synapse development in Drosophila. J Cell Biol 187: 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Korutla L, Champtiaux N, Toda S, LaLumiere R, Vallone J, Klugmann M, Blendy JA, Mackler SA, Kalivas PW 2007. NAC1 regulates the recruitment of the proteasome complex into dendritic spines. J Neurosci 27: 8903–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver ET, Gwozd TJ, Ptak C, Goebl M, Ellison MJ 1992. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): Implications for the structure, function and evolution of the E2s. EMBO J 11: 3091–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Lavy KJ, Oren YS, Feine O, Sajman J, Listovsky T, Brandeis M 2010. Fifteen years of APC/cyclosome: A short and impressive biography. Biochem Soc Trans 38: 78–82 [DOI] [PubMed] [Google Scholar]
- Song H, Smolen P, Av-Ron E, Baxter DA, Byrne JH 2006. Bifurcation and singularity analysis of a molecular network for the induction of long-term memory. Biophys J 90: 2309–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossin WS 2007. Isoform specificity of protein kinase Cs in synaptic plasticity. Learn Mem 14: 236–246 [DOI] [PubMed] [Google Scholar]
- Speese SD, Trotta N, Rodesch CK, Aravamudan B, Broadie K 2003. The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr Biol 13: 899–910 [DOI] [PubMed] [Google Scholar]
- Stegmuller J, Konishi Y, Huynh MA, Yuan Z, Dibacco S, Bonni A 2006. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron 50: 389–400 [DOI] [PubMed] [Google Scholar]
- Stein LD 2004. Human genome: End of the beginning. Nature 431: 915–916 [DOI] [PubMed] [Google Scholar]
- Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, Ho MW, Troncoso J, Gygi SP, Lee MK, et al. 2008. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet 17: 431–439 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kawakami T, Tateishi K, Yashiroda H, Chiba T 2001. Control of IκBα proteolysis by the ubiquitin-proteasome pathway. Biochimie 83: 351–356 [DOI] [PubMed] [Google Scholar]
- Tang G, Leppla SH 1999. Proteasome activity is required for anthrax lethal toxin to kill macrophages. Infect Immun 67: 3055–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya SC, Hegde AN 2005. Ubiquitin-proteasome pathway components as therapeutic targets for CNS maladies. Curr Pharm Des 11: 3807–3828 [DOI] [PubMed] [Google Scholar]
- Upadhya SC, Hegde AN 2007. Role of the ubiquitin proteasome system in Alzheimer's disease. BMC Biochem 8 (Suppl 1): S12 doi: 10.1186/1471-2091-8-S1-S12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya SC, Smith TK, Hegde AN 2004. Ubiquitin-proteasome-mediated CREB repressor degradation during induction of long-term facilitation. J Neurochem 91: 210–219 [DOI] [PubMed] [Google Scholar]
- Upadhya SC, Ding L, Smith TK, Hegde AN 2006. Differential regulation of proteasome activity in the nucleus and the synaptic terminals. Neurochem Int 48: 296–305 [DOI] [PubMed] [Google Scholar]
- van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Koycu S, Ramdjielal RD, Salehi A, Martens GJ, et al. 1998. Frameshift mutants of β amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science 279: 242–247 [DOI] [PubMed] [Google Scholar]
- van Leuken R, Clijsters L, Wolthuis R 2008. To cell cycle, swing the APC/C. Biochim Biophys Acta 1786: 49–59 [DOI] [PubMed] [Google Scholar]
- van Roessel P, Elliott DA, Robinson IM, Prokop A, Brand AH 2004. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell 119: 707–718 [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A 1997. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science 278: 460–463 [DOI] [PubMed] [Google Scholar]
- Wan HI, DiAntonio A, Fetter RD, Bergstrom K, Strauss R, Goodman CS 2000. Highwire regulates synaptic growth in Drosophila. Neuron 26: 313–329 [DOI] [PubMed] [Google Scholar]
- Willeumier K, Pulst SM, Schweizer FE 2006. Proteasome inhibition triggers activity-dependent increase in the size of the recycling vesicle pool in cultured hippocampal neurons. J Neurosci. 26: 11333–11341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, Householder DB, Fletcher CF, Miller RJ, Copeland NG, Jenkins NA 2002. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet 32: 420–425 [DOI] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW 1999. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev 13: 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao I, Takagi H, Ageta H, Kahyo T, Sato S, Hatanaka K, Fukuda Y, Chiba T, Morone N, Yuasa S, et al. 2007. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell 130: 943–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M 2009. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM 2000. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein CDCrel-1. Proc Natl Acad Sci USA 97: 13354–13359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hegde AN, Martin KC 2003. The ubiquitin proteasome system functions as an inhibitory constraint on synaptic strengthening. Curr Biol 13: 887–898 [DOI] [PubMed] [Google Scholar]