Abstract
Multidrug resistance protein 4 (MRP4; ABCC4) is an ATP binding cassette transporter that facilitates the excretion of bile salt conjugates and other conjugated steroids in hepatocytes and renal proximal tubule epithelium. MRP4/Mrp4 undergoes adaptive upregulation in response to oxidative and cholestatic liver injury in human and animal models of cholestasis. However, the molecular mechanism of this regulation remains to be determined. The aryl hydrocarbon receptor (AhR) and NF-E2-related factor 2 (Nrf2) play important roles in protecting cells from oxidative stress. Here we examine the role of these two nuclear factors in the regulation of the expression of human MRP4. HepG2 cells and human hepatocytes were treated with the AhR and Nrf2 activators, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), 3-methylcholanthrene (3-MC), or oltipraz and other nuclear receptor agonists. TCDD, 3-MC, and oltipraz significantly increased MRP4 expression at mRNA and protein levels. Computer program analysis revealed three Xenobiotic response element (XRE) and one Maf response element sites within the first 500 bp of the MRP4 proximal promoter. Luciferase reporter assay detected strong promoter activity (53-fold higher than vector control) in this region. TCDD and 3-MC also induced promoter activity in the reporter assays. Mutation of any of these XRE sites significantly decreased MRP4 promoter activity in reporter assays, although XRE2 demonstrated the strongest effects on both basal and TCDD-inducible activity. EMSA and chromatin immunoprecipitation assays further confirmed that both AhR and Nrf2 bind to the proximal promoter of MRP4. Our findings indicate that AhR and Nrf2 play important roles in regulating MRP4 expression and suggest that agents that activate their activity may be of therapeutic benefit for cholestasis.
Keywords: ABC transporter, xenobiotic receptor, transcription factor, gene regulation, cholestasis, drug metabolism
the multidrug resistance-associated protein 4 (MRP4, ABCC4) is a member of the ATP-binding cassette transporters and is capable of extruding a large variety of endogenous and foreign compounds from cells including bile acid sulfate conjugates and conjugated steroids (28, 29, 44). MRP4 is localized to the basolateral membrane of hepatocytes and the apical membrane of renal proximal tubule epithelium (6). Under normal physiological conditions, MRP4 is expressed at very low levels in hepatocytes since bile salts and other foreign compounds and toxicants can be excreted into bile by canalicular apical ABC transporters including bile salt export pump (BSEP; ABCB11), MDR1 (ABCB1), MRP2 (ABCC2), and breast cancer resistance protein (BCRP; ABCG2). In contrast, MRP4/Mrp4 expression is significantly upregulated in the liver and kidney in cholestatic conditions in patients with primary biliary cirrhosis (47), in children with Progressive Familial Intrahepatic Cholestasis (15), and in rodents following bile duct ligation (21). Although the expression of human MRP4 is highly variable as determined in an analysis of a large group of human liver samples, both MRP4 mRNA and protein expression in cholestatic livers are significantly higher than in “normal” liver (12).
Several other hepatic basolateral transporters are also upregulated during the adaptive response to cholestatic liver injury, including both human and rodent organic solute transporter alpha and beta (Ostα and Ostβ) and rodent Mrp3 (Abcc3) (7, 14, 32). The relative functional importance of MRP4/Mrp4 in this adaptive protective response is further emphasized from studies in bile duct-ligated Mrp4−/−, Mrp3−/−, and Ostα−/− mice. In these studies, hepatic injury was more severe in Mrp4−/− mice (21), not changed in Mrp3−/− mice (5, 45), and paradoxically protected in Ostα knockout mice (33). These findings strongly suggest that MRP4/Mrp4 is the principal mechanism for the basolateral extrusion of bile acids during the adaptive responses to cholestasis and that successful upregulation of the expression of hMRP4 might represent a potential therapeutic target for patients with cholestatic liver diseases. However, to pursue this goal, it will be necessary to have a clear understanding of the molecular mechanisms of the transcriptional regulation of hMRP4 in the hepatocyte.
Recent studies suggest that the nuclear receptor constitutive androstane receptor (Car, Nr1i3) and the nuclear factor E2-related factor 2 (Nrf2) are involved in transcriptional regulation of Mrp4 in rodents (3, 11, 20). In these studies, oltipraz (OPZ), an activator of Nrf2, increased hepatic Mrp4 expression in mice (2, 34). Nrf2 is a transcription factor that is a crucial regulator of cellular redox homeostasis (16, 24). Nrf2 is retained in the cytoplasm by associating with the actin-kelch-like-ECH-associated protein 1 (Keap1). Under conditions of oxidative stress, the disulfide bonds between Nrf2 and Keap1 are cleaved. Subsequently, Nrf2 translocates to the nucleus and forms a heterodimer with a partner protein, usually small Maf proteins. The Nrf2-Maf heterodimer then binds to antioxidant-responsive elements (AREs) in the promoter of its target genes, thereby activating transcription of these targets. Nrf2 directly upregulates the expression of a number of phase II enzymes, including NAD(P)H quinone oxidoreductase 1 (Nqo1) (2), the glutathione (GSH) synthesis enzyme glutamate cysteine ligase (Gclc) (39), and UDP-glucuronosyltransferase 1A1 (UGT1A1) (42). Cholestasis can result in the formation of reactive oxygen species in the liver and lead to oxidative stress (31, 35). The importance of Nrf2 to liver toxicity is further demonstrated by studies in Nrf2 knockout mice that show marked increases in sensitivity to acetaminophen-induced liver toxicity (9) and ethanol-induced liver injury (18).
The aryl hydrocarbon receptor (AhR) is another crucial transcription factor that plays a central role in protection of dioxin hepatotoxicity and the regulation of xenobiotic metabolism. However, AhR activation can also initiate defense mechanisms for oxidative stress (25). AhR is a ligand-activated transcription factor that belongs to the basic helix-loop-helix/PER-aryl hydrocarbon receptor nuclear translocator (ARNT)-SIM (PAS) family (8). Upon activation, AhR translocates to the nucleus where it forms a heterodimer with ARNT and binds to xenobiotic responsive elements (XRE) in the promoter of its target genes and initiates transcription. Many phase I and phase II enzymes are AhR targets, including CYP1A1 (10), CYP1A2(46), CYP1B1 (36), NQO1, UGT1A (43), UGT1A6, and GST2A. In addition, there is a reciprocal relationship between AhR and Nrf2 in regulating their targets as activation of either receptor leads to increase expression of the other one (23, 30). This interaction represents a bidirectional “cross talk” of AhR and Nrf2 signal pathways in cellular detoxification.
In this report, we have assessed the role of AhR and Nrf2 in the regulation of MRP4. We found that both Nrf2 and AhR activators increased MRP4 mRNA and protein expression in HepG2 cells and primary human hepatocytes. The MRP4 proximal promoter was characterized, and AhR and Nrf2 response elements were identified in this region. These findings provide initial insights that might lead to new therapeutic strategies for patients with cholestatic liver disorders.
MATERIALS AND METHODS
Materials.
Chemicals were purchased from Sigma (St. Louis, MO), except when the source is specified. OPZ was obtained from Thermo Fisher (Pittsburgh, PA). The human hepatoma HepG2 cell line and culture medium MEM were obtained from the American Type Culture Collection (ATCC; Manassas, VA). Heat-inactivated fetal bovine serum (FBS), penicillin and streptomycin, trypsin-EDTA, and PBS were purchased from Invitrogen (Carlsbad, CA). Hepatocyte maintenance medium (HMM) and penicillin-streptomycin-amphotericin B were purchased from Lonza (Walkersville, MD). DNA and RNA oligos were synthesized by Integrated DNA Technologies (IDT; Coralville, IA) and DNA sequencing was provided by the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University. TaqMan primer/probes for real-time PCR were purchased from Applied Biosystems (Foster City, CA). Luciferase reporter assay, DIG-Gel Shift, and chromatin immunoprecipitation (ChIP) assay kits were purchased from Promega (Madison, WI), Roche (Indianapolis, IN), and Millipore (Temecula, CA), respectively. Antibodies against AhR, ARNT, and Nrf2 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). MRP4 antibody was purchased from Everest Biotech (Oxfordshire, UK).
Plasmid constructs.
Human AhR and ARNT expression plasmids (pSport-AhR and pSport-ARNT) were kindly provided by Dr. Christopher Bradfield (University of Wisconsin-Madison Medical School), and mouse Nrf2/pEF1α was a gift from Dr. Jefferson Chan (University of California-Irvine). The 5′-flanking regions of the MRP4 gene were amplified by PCR using a BAC clone (RP11-789G22 provided by the Sanger Institute, Hinxton, UK) as template. Three constructs containing −238 to −72, −761 to −72, and −2034 to −72 fragments (positions relative to translation initiation site) were inserted into a pGL3-basic vector (Promega), respectively. The specific oligonucleotides are listed in Table 1. Mutant reporter constructs were prepared by using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA), and the specific primers are also listed in Table 1.
Table 1.
Oligonucleotides for reporter gene cloning, mutagenesis, EMSA, ChIP assay, and ribonucleotide sequences for siRNA targeting of human AhR
| Name | Sequence 5′ → 3′ |
|---|---|
| −2.3kb-KpnI-forward | GGTACCGCTGGGATTATGCGCTTGTAC |
| −761bp-XhoI-forward | CATACTCTCGAGTTACCCGGCTTTC |
| −238bp-SmaI-forward | GCTCCCGGGCTCCAGGCGGCGGCGC |
| −72bp-HindIII-reverse | TCCAAGCTTACCTCAAGCAGGGATGCTG |
| XRE1 wt | −119GAGCGGACGGCGTGGCGGCCGGAG- |
| XRE1 mut | −119GAGCGGACGGCTATGCGGCCGGAG−9 |
| XRE2 wt | −181CGGGGACGGGGCGTGCCGCCGGCG−157 |
| XRE2 mut | −181CGGGGACGGGGCTATCCGCCGGCG−157 |
| XRE3 wt | −264GCCGCGCTCGCACGCGGTGCGCAGC−240 |
| XRE3 mut | −264GCCGCGCTCGCAAACGGTGCGCAGC−240 |
| MARE wt | −144AGCGCTGCTTCACAGGCTCC−124 |
| MARE deletion | −144AGCG - -TTCACAGGCTCC−124 |
| Primers for ChIP assay | |
| Forward | CTGCTACCCTCTCCATCCAG |
| Reverse | ACCTCAAGCCAGGGATGCTG |
| Sequence of AhR siRNA | |
| 1Sense | GUGGAAGUAUUGAUCCAUCUUUCCCU |
| Antisense | CACCUUCAUAACUAGGUAGAAAGGG |
| 2Sense | AAUGCAACAUCAAAGAAGCUCUUGGCU |
| Antisense | TTACCGUUGUAGUUUCUUCGAGAACC |
| 3Sense | AUAAAUAGACUCAUCUUGUUGCAUCAU |
| Antisense | TAUUUAUCUGAGUAGAACAACGUAG |
Antisenses were made at the same length and complementary to the sense sequence. ChIP, chromatin immunoprecipitation; siRNA, small interfering RNA; AhR, aryl hydrocarbon receptor; wt, wild type; mut, mutant.
Quantitative real-time PCR analysis.
Total RNA was extracted from HepG2 cells or human hepatocytes by using Trizol reagent according to the manufacturer's instructions. Two micrograms of total RNA were converted to cDNA by reverse transcription using AffinityScript cDNA synthesis kit (Stratagene). TaqMan real-time PCR was performed in 20 μl/well triplicates in an ABI7500 Sequence Detection System. Primer-probe sequences for Q-PCR are ABCC4 ID:Hs00195260_m1, AhR ID:Hs00169233_ml, NQO1 ID:Hs00168547_ml and GAPDH ID:Hs99999905_m1, respectively (Applied Biosystems). All mRNA levels were normalized to GADPH. Data presented are averages from at least three independent experiments.
Cell culture, transfection, and reporter gene assays.
Human hepatocytes were obtained from the Liver Tissue Procurement and Distribution System of the National Institutes of Health (NIH) (Dr. Stephen Strom, University of Pittsburgh) and maintained on collagen-coated plates by use of HMM medium containing 1% penicillin-streptomycin-amphotericin B and overlaid with Matrigel (BD Bioscience, Bedford, MA) in 37°C, 5% CO2 atmosphere incubator. Briefly, cells were plated in 6-well or 12-well plates for at least 20 h before shipping. Upon arrival, the cells remained in shipping medium for 2–3 h in a 37°C incubator, followed by Matrigel overlay in HMM medium with 0.1 μM dexamethasone, 0.1 μM insulin, and 1% penicillin-streptomycin-amphotericin B. The treatments were started 24 h after arrival. All cells were harvested within a week from the time of the hepatocyte isolation procedure. HepG2 cells were routinely maintained in MEM supplemented with 10% FBS and 1% penicillin G-streptomycin in 5% CO2 atmosphere incubator. For transfection, cells were seeded in 24-well plates at a density of 1 × 105 cells per well and grown in complete medium for 24 h. Cells were then transfected with 0.3 μg plasmid DNA and 5 ng phRL-CMV in complex with 0.6 μl of FuGENE HD reagent (Roche) in Opti-MEM (Invitrogen) per well. At 24 h after transfection, cells were treated with the indicated chemicals for the designated time points. Passive lysis buffer was applied to cells for the dual- luciferase reporter assay (Promega). Promoter activities were normalized to the control Renilla activity and were calculated as the fold induction of promoter construct over control. Each transfection was performed in triplicate. Data presented are averages from at least three independent experiments.
Western blots.
For total protein, cells were lysed with RIPA buffer (50 mM Tris·HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing Halt protease inhibitor cocktail (Pierce, Rockford, IL). Cell surface protein expression of MRP4 was determined using a membrane impermeant biotin reagent, sulfo-NHS-LC-biotin (Pierce), and the biotinylated proteins were precipitated with ImmunoPure immobilized streptavidin beads (Pierce). The biotinylated proteins were then eluted and separated by SDS-PAGE using a 7.5% Tricine gel, transferred to nitrocellulose membranes. The molecular mass was estimated by comparison with chemiluminescent protein size standards (MagicMark Western XP; Invitrogen). The intensity of protein bands was analyzed by use of FOTODYNE Imaging and TL-100 image analysis software.
Immunofluorescence and confocal microscopy.
Primary human hepatocytes were grown on glass coverslips and treated with DMSO, OPZ (50 μM), 4-PBA (1 mM), or 3-MC (1 μM) for 48 h. Cells were then fixed with methanol for 10 min at −20°C and incubated with anti-MRP4 polyclonal antibody for 2 h at room temperature. Alexa488 anti-rabbit immunoglobulin G (Molecular Probes; Eugene, OR) was used as the secondary antibody. The fluorescent image was visualized with a Zeiss LSM510 confocal scanning microscope (Carl Zeiss; Thornwood, NY), and images were processed with Adobe PhotoshopCS (Moutainview, CA).
Gene expression knockdown with siRNA.
Three sets of small interfering RNA (siRNA) oligonucleotides that targeted human AhR were synthesized by IDT. The sequences are listed in Table 1. To knock down AhR expression in HepG2 cells, the cells were reverse transfected with either AhR siRNA duplexes (30 nM) or control siRNA duplex (Ambion, Austin, TX) by use of siPORT NeoFX Transfection Agent (Ambion) for 24 h. After transfection, the cells were maintained in normal culture conditions for an additional 24 or 48 h before harvesting.
EMSA and supershift assay.
Nuclear protein extract from HepG2 cells was isolated by using NE-PER kit (Pierce). Oligonucleotides used for gel shift analysis are listed in Table 1. EMSA and supershift assays were performed by using the DIG-Gel kit with digoxigenin-labeled probes by following manufacturer's instructions, where 5 μg of nuclear protein extract was used in each reaction. In competition experiments, unlabeled oligonucleotide was incubated with nuclear extract for 10 min before the addition of the labeled probe. In supershift studies, 1 μg of the relevant rabbit polyclonal antibody was incubated with nuclear extract on ice for 30 min before addition of the labeled probe. The total binding reaction (20 μl) was resolved on a 5% native polyacrylamide gel in 0.25 × Tris borate-EDTA buffer and then transferred to a positively charged nylon membrane (Amersham, Piscataway, NJ).
ChIP assay.
HepG2 cells were grown on 10-cm dishes. After reaching 80% confluence, the cells were cross-linked by adding formaldehyde directly into the media to a final concentration of 1%. Cross-linking was stopped by adding glycine to a final concentration of 125 mM. After cross-linking, chromatin DNA was sheared into 200- to 1,000-bp fragments by sonication using a Bioruptor (Diagenode SA, Liege, Belgium). The sheared chromatin was incubated with AhR or Nrf2 rabbit polyclonal antibodies or rabbit IgG (as a negative control) at 4°C overnight. The immunocomplexes were precipitated with Agarose-Protein G beads. After reversing the cross-link, the precipitated DNA was analyzed by PCR using the primer pairs listed in Table 1. The amplicon encompasses the three potential binding sites for AhR (at −109, −171, and −254 bp) and one for Nrf2 (at −141 bp).
Statistical analysis.
Data are expressed as means ± SD. Differences between experimental groups were assessed by the two-tail paired Student's t-test. Differences with P < 0.05 were considered statistically significant.
RESULTS
TCDD and OPZ induced MRP4 mRNA and protein expression in HepG2 cells and primary human hepatocytes.
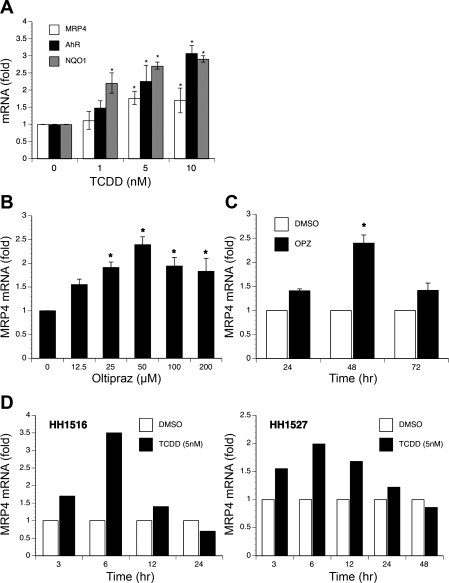
To search for transcription factors that might regulate MRP4 expression, we treated HepG2 cells with various compounds, including OPZ (an Nrf2 and AhR activator), t-BHQ (an Nrf2 activator), 4-PBA (a low-molecular-weight fatty acid that acts as a chemical chaperone), phenobarbital and bilirubin (CAR activators), CDCA (a farnesoid X receptor ligand), rifampicin [a pregnane X receptor (PXR) ligand], clofibrate (a peroxisome proliferator-activated receptor-α ligand), and TCDD (an AhR activator). Quantitative real-time PCR analysis demonstrated that only TCDD, OPZ, and 4-PBA significantly induced MRP4 mRNA levels. Treatment for 24 h with TCDD significantly stimulated MRP4, NQO1 (a classic target of AhR), and AhR mRNA expression in a dose-dependent manner. Five nanomolar TCDD stimulated MRP4, AhR, and NQO1 expression by 1.8, 2.3, and 2.7-fold, respectively, in HepG2 cells (Fig. 1A). OPZ also induced MRP4 mRNA expression in dose- and time-dependent manners in HepG2 cells, where 50 μM OPZ increased MRP4 mRNA expression by 2.3-fold after 48 h treatment (Fig. 1, B and C). In addition, we confirmed that TCDD (5 nM) and OPZ (50 μM) can also stimulate MRP4 mRNA expression approximately two- to threefold in human hepatocytes from two donors, HH1516 and HH1527, with maximum induction at 6 h (Fig. 1D).
Fig. 1.
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) and oltipraz (OPZ) induce the expression of multidrug resistance-associated protein 4 (MRP4) mRNA in HepG2 cells and primary human hepatocytes. A: dose-dependent induction by TCDD in HepG2 cells after 24-h treatment. *P < 0.05, n = 3. B: OPZ upregulated MRP4 mRNA levels in a dose-dependent manner in HepG2 cells (48-h treatment). C: OPZ induction of MRP4 mRNA is time dependent. HepG2 cells were treated with 50 μM OPZ for different time points. All data were normalized to GAPDH level. (n = 3). D: TCDD induced MRP4 mRNA expression in primary human hepatocytes. Human hepatocytes from 2 individuals were exposed to DMSO or 5 nM TCDD for indicated times. The data presented are averages from triplicate measurements.
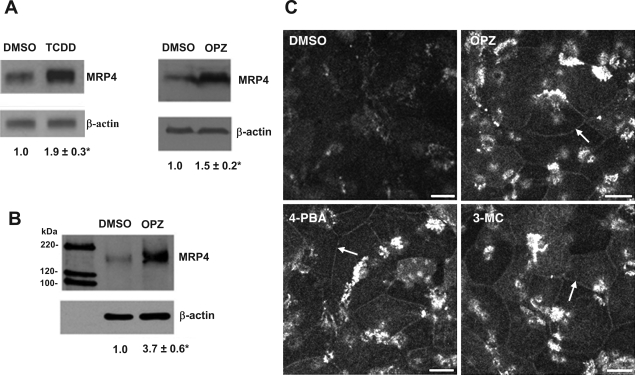
The induction of MRP4 mRNA expression by TCDD and OPZ were accompanied by significant increases in MRP4 protein in HepG2 cells compared with the solvent control (Fig. 2A). Furthermore, biotin conjugation of cell surface protein detected a 3.7-fold increase in MRP4 membrane expression (Fig. 2B). As shown in Fig. 2C, immunofluorescent labeling of human hepatocytes also indicated that MRP4 expression was increased on the basolateral membrane of OPZ-, 3-MC-, and 4-PBA-treated cells. Taken together, these results indicate that OPZ and TCDD can induce MRP4 expression in both HepG2 cells and human hepatocytes by transcriptional as well as posttranscriptional mechanisms. OPZ has been previously reported to be a bifunctional activator for both Nrf2 and AhR (22). Therefore, we speculated that both AhR and Nrf2 might be involved in regulating MRP4 expression.
Fig. 2.
TCDD and OPZ upregulate MRP4 protein expression. Western blots analysis of total protein expression (A) and cell surface protein expression (B) in HepG2 cells 48 h after 50 μM OPZ treatment. C: immunofluorescence staining of MRP4 in primary human hepatocytes. Human hepatocytes were treated with DMSO, OPZ (50 μM), 3-methylcholanthrene (3-MC; 1 μM), or 4-PBA (1 mM) for 48 h. Bar represents 10 μm. Arrows point to the increase in plasma membrane staining of MRP4.
AhR knockdown resulted in reduction of MRP4 expression in HepG2 cells.
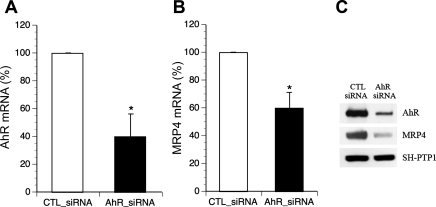
To verify the functional role of AhR on MRP4 expression, we knocked down AhR expression in HepG2 cells using siRNA transfection. As illustrated in Fig. 3, reduction of AhR mRNA and protein (Fig. 3, A and C) resulted in a significant decrease in MRP4 expression at both mRNA (Fig. 3B) and protein levels (Fig. 3C). This result indicates that AhR is a positive regulator for MRP4 expression and suggests that there may be an XRE in the MRP4 promoter.
Fig. 3.
Endogenous MRP4 expression is decreased in aryl hydrocarbon receptor (AhR) knockdown HepG2 cells. Reduced AhR expression (A) led to diminished MRP4 mRNA (B) and protein (C) expression. CTL, control; siRNA, small interfering RNA. *P < 0.01, n = 3.
Putative AhR and Nrf2 response elements are identified in MRP4 gene proximal promoter.
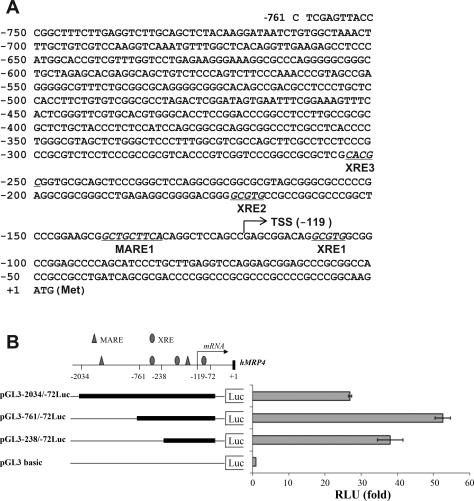
To identify whether there are XREs and AREs in the MRP4 promoter, we analyzed a 761-bp sequence of the hMRP4 gene 5′-flanking region using MatInspector software (Genomatrix, Munich, Germany). Three putative XRE sites were identified that were located at −109 bp, −171 bp, and −254 bp upstream from the MRP4 translational start site, termed XRE1, 2, and 3, respectively. In addition, one potential Nrf2 binding site was also detected at −141 bp, termed Maf response element (MARE; Fig. 4A). To assess whether these predicted response elements play a role in the basal expression of MRP4, we first cloned a 2,034-bp fragment from the MRP4 gene 5′-flanking region into a pGL3-basic vector (named pGL3-2034/-72Luc). A dual-luciferase assay demonstrated that this construct had 26-fold higher promoter activity than the empty vector control in HepG2 cells (Fig. 4B). To further characterize the proximal promoter region, we generated two more reporter constructs of hMRP4 gene 5′-flanking regions, pGL3–761/-72Luc and pGL3–238/-72Luc. As shown in Fig. 4B, both pGL3–761/-72Luc and pGL3–238/-72Luc demonstrated even higher promoter activity than pGL3-2034/-72Luc, increasing 53- and 38-fold, respectively, compared with the vector control. These results indicate that the basal promoter of the MRP4 gene is located to its proximal 5′-flanking region.
Fig. 4.
A: sequence of the proximal promoter of the MRP4 gene. The numbered positions refer to the translational initiation codon (ATG). →, Transcription start site at −119 bp. Potential XRE and MARE-like regulatory elements are underlined. B: reporter assays demonstrate that the highest promoter activity is within the first 700-bp 5′-flank region of MRP4 gene. The regulatory DNA elements are indicated by the symbols: triangle, MARE; oval, XRE. The promoter activity of each reporter constructs is presented as fold to pGL3-basic vector.
Functional XREs are identified in the MRP4 promoter.
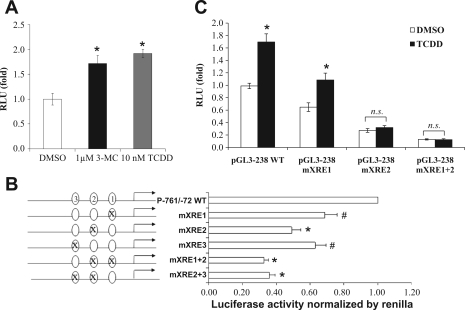
To determine whether the predicted XREs in the proximal region of MRP4 gene were involved in regulation of MRP4 expression, HepG2 cells were transfected with pGL3–761/-72Luc construct and treated with two prototypical AhR ligands, TCDD or 3-MC. As shown in Fig. 5A, 5 nM TCDD or 1 μM 3-MC significantly induced MRP4 promoter activity by 1.7-fold and 2-fold, respectively, indicating that the proximal region of the MRP4 gene contains functional AhR-XRE sites. These findings suggest that TCDD induction of endogenous expression of MRP4 in HepG2 cells and human hepatocytes results from activation of its promoter (Fig. 1, A and B).
Fig. 5.
A: 3-MC or TCDD increased MRP4 promoter activity in reporter assays. The pGL3–761/-72Luc reporter construct was transfected in HepG2 cells and treated for 24 h; *P < 0.05, n = 3. B: mutations of XREs reduced MRP4 promoter activity in reporter assays. Ovals on the left of the bars represent XREs. *P < 0.005, #P < 0.05, n = 3. C: the XRE2 mutation prevents TCDD induction of the MRP4 promoter activity in reporter assay. HepG2 cells were transfected for 24 h and treated with 10 nM TCDD for additional 24 h. Data were normalized to Renilla luciferase activity. *P < 0.05, n = 3. n. s., Not significant.
To verify the role of these predicted XREs in stimulating MRP4 promoter activity, we selectively mutated these XRE sites in pGL3–761/-72Luc. Reporter assays demonstrated that mutations of XRE1, 2, and 3 reduced MRP4 promoter activities by 25, 45, and 32%, respectively, compared with the wild-type control (Fig. 5B). Double mutations of XRE1 and 2 or XRE2 and 3 further reduced the promoter activities more than any single XRE mutation. Because the XRE2 mutant exhibited the greatest reduction of these three mutations, we then determined whether the XRE2 site was indeed responsible for TCDD- and 3-MC-induced MRP4 promoter activity. HepG2 cells transfected with the XRE1 mutant, XRE2 mutant, or XRE1+2 double mutant were treated with 5 nM TCDD for 24 h. As shown in Fig. 5C, the mutation of XRE2 greatly reduced not only MRP4 promoter basal activity but also its inducibility by TCDD, whereas mutations in XRE1 or XRE3 (data not shown) did not significantly change TCDD inducibility. Mutations in XRE2 also diminished 3-MC (1 μM) inducibility in this reporter assay (data not shown). Taken together, these findings indicate that MRP4 upregulation is AhR-XRE dependent and suggest that the XRE2 site may be the most important for AhR-mediated MRP4 expression.
Nrf2 activates the MRP4 promoter.
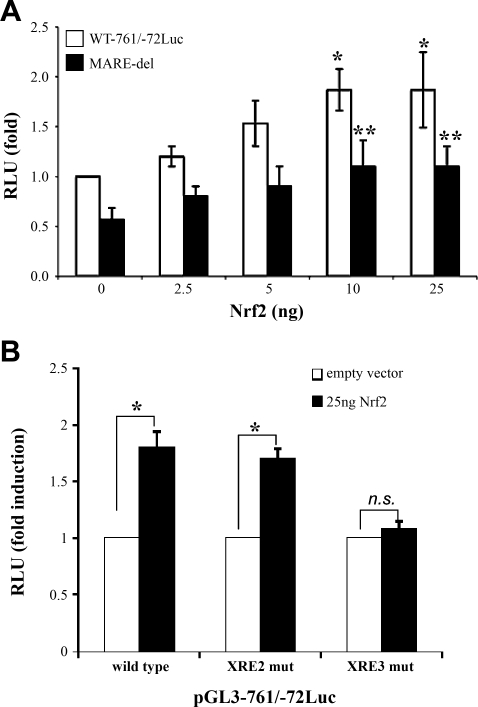
Since OPZ is also an Nrf2 activator and the computer program analysis also predicted one MARE site (at −141 bp) within the proximal promoter of the MRP4 gene, we hypothesized that this MARE-like site might mediate Nrf2 regulation of MRP4 expression. To assess this possibility, we cotransfected the pGL3–761/-72Luc construct with an Nrf2 expression plasmid into HepG2 cells. As demonstrated in Fig. 6A, cotransfection of Nrf2 increased MRP4 promoter activity in a dose-dependent manner. Disruption of the putative MARE (at −141 bp) by deletion of the core sequence, CTGC, reduced the basal activity of MRP4 promoter by 35%, but did not abolish the Nrf2 induction. These findings suggest that there may be an additional mechanism mediating Nrf2 regulation of MRP4 expression besides binding to its DNA response element. To assess whether Nrf2 could mediate its effect through the AhR-XRE pathway, we cotransfected a Nrf2 expression plasmid with pGL3–761/−72Luc wild type or its XRE2 and XRE3 mutant constructs. As shown in Fig. 6B, the mutation of XRE3, but not XRE2, greatly reduced the induction by Nrf2.
Fig. 6.
A: cotransfection of NF-E2-related factor 2 (Nrf2) increases MRP4 promoter activity and deletion of MARE core sequence reduces the promoter activity in reporter assay. HepG2 cells were cotransfected with MRP4 promoter constructs of either the wild-type or the deletion of MARE core motifs for 48 h; n = 3 experiments. *P < 0.05, pGL3–761/-72Luc plus Nrf2 expression plasmid DNA vs. pGL3–761/-72Luc plus empty vector, **P < 0.05, the wild-type vs. the deletion of MARE, n = 3. B: mutation of XRE3 prevented the induction by Nrf2. HepG2 cells were cotransfected with pGL3–761/-72Luc wild type, XRE2 mutant or XRE3 mutant construct with either empty vector or Nrf2 expression plasmid (25 ng). The results are represented as fold induction compared with its control vector. *P < 0.05, n = 3.
EMSA and ChIP assays confirm the role of AhR and Nrf2 in MRP4 expression.
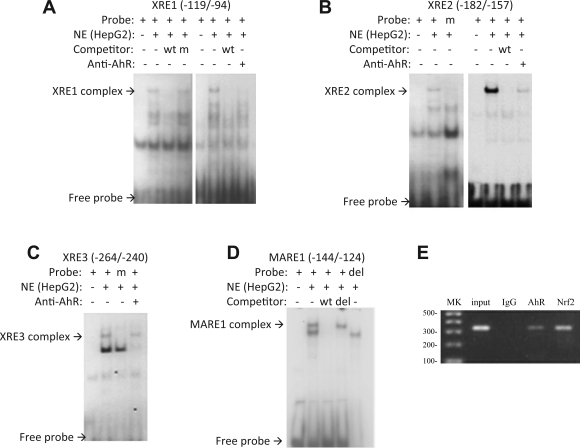
Because the XRE2 and MARE sites appeared to be important for the expression of MRP4 in HepG2 cells, we tested the specificity of the identified XRE2 and MARE sites for AhR and Nrf2 by performing EMSA using nuclear extract from HepG2 cells. As shown in Fig. 7, A, B, and C, labeled oligonucleotides containing XRE1, 2 and 3, respectively, were specifically shifted after incubation with HepG2 nuclear extract. Mutation of the corresponding XRE site and/or competition using unlabeled oligo abolished the binding. When 1 μg of rabbit polyclonal AhR antibody was incubated with each of these reaction mixtures, the specifically shifted bands were diminished, although no supershifted bands were seen. For the Nrf2-MARE site, similar results were obtained. Specifically, the wild-type oligo formed a complex with proteins in the nuclear extract. The shifted band was competed by unlabeled wild-type oligonucleotide, but not by mutated unlabeled oligonucleotide. The labeled mutant oligo also did not yield a band shift (Fig. 7D). A supershift band was also not seen when Nrf2 antibody was added to the mixture (data not shown), suggesting that the specific epitopes may be sterically hindered.
Fig. 7.
A–C: EMSA demonstrate XRE1, XRE2, or XRE3 forms a complex (retarded band) with extracts from HepG2 cells. wt, Wild type; m, mutated; NE, nuclear extract. D: the specificity of MARE complex (retarded band) was confirmed by competition with an unlabeled oligo (wt) or the unlabeled deleted oligo (del). Loss of the complex occurred when the reaction mix was incubated with the labeled 4-bp deleted oligos. E: chromatin immunoprecipitation assay (ChIP) confirms that AhR and Nrf2 bind to the proximal promoter region of MRP4. MK, 100-bp DNA marker.
To further confirm the binding of AhR and Nrf2 to the MRP4 promoter, we performed ChIP assays in HepG2 cells. In this experiment, a specific DNA fragment targeting the proximal promoter of MRP4 was detected by PCR only when AhR or Nrf2 specific antibodies were used for immunoprecipitation, but not when a nonspecific IgG control antibody was used (Fig. 7E). When the cells were treated with 5 nM TCDD, the intensity of the specific band increased (data not shown), suggesting that TCDD stimulates AhR binding to the MRP4 promoter. Together, these results indicate that both AhR and Nrf2 bind to the proximal promoter of MRP4 and participate in MRP4 gene regulation.
DISCUSSION
Both AhR and Nrf2 are transcription factors that respond to oxidative stress and mediate transcriptional events that facilitate protective responses in animal models of cholestasis (26, 37). Previous studies by us and others have indicated that MRP4/Mrp4 may play a protective role in cholestasis (12, 21, 47). Therefore, we examined the role of AhR and Nrf2 in the regulation of hMRP4 expression.
Our findings reported here indicate that AhR and Nrf2 are two major transcription factors that regulate hMRP4 expression. This conclusion is based on the following observations: 1) activators of AhR and Nrf2, TCDD and OPZ, respectively, induced both mRNA and protein expression of MRP4 in HepG2 cells and primary human hepatocytes (Fig. 1 and Fig. 2); 2) knockdown of AhR resulted in reduced MRP4 expression in HepG2 cells (Fig. 3); 3) both AhR and Nrf2 increased MRP4 promoter activity in luciferase reporter assays; 4) mutations or deletion of putative XRE and MARE sites reduced MRP4 promoter activity and inducibility in reporter assays; and, finally, 5) both AhR and Nrf2 can bind to the MRP4 promoter as demonstrated by EMSA and ChIP assays (Fig. 7).
Of 14 compounds tested in initial screening studies, only OPZ, TCDD or 3-MC, and 4-PBA significantly induced MRP4 mRNA and protein levels in both human hepatocytes and HepG2 cells. OPZ and TCDD are well-known inducers of phase I and II enzymes by activating Nrf2 and AhR in the liver. Furthermore, recent studies also indicate that there is signaling cross talk between AhR and Nrf2 (13, 19) so that OPZ and TCDD are now defined as bifunctional inducers for both Nrf2 and AhR (4, 19, 22). As we have shown here, MRP4 expression is regulated by both AhR and Nrf2 and thus should be added to the “TCDD-inducible AhR-Nrf2 gene battery” (41).
Because MRP4 transports xenobiotics as well as conjugated bile acids, its expression may also be regulated by xenobiotic activated transcription factors that would function coordinately to protect the liver from accumulation of foreign and endogenous toxins. Indeed, Mrp4 expression is reduced in Car knockout mice, suggesting that Car (a major xenobiotic activated nuclear receptor) can also regulate Mrp4 expression in the mouse although a Car response element has not been identified in its promoter (3). When we exposed HepG2 cells and human hepatocytes to phenobarbital, a human CAR activator, MRP4 mRNA expression was significantly increased ∼2.1-fold (P < 0.005, n = 3) in primary human hepatocytes (unpublished data), suggesting that CAR may also regulate MRP4 expression but by as-yet-unidentified mechanisms. As in mice, CAR response elements have not been identified in the MRP4 promoter. However, since recent studies indicate that AhR is involved in extensive cross talk with other transcription factors, including the estrogen receptor (1) and PXR/CAR (17), and that AhR activation can regulate CAR levels in murine and human liver (27), we cannot exclude that MRP4 regulation by AhR might also involve cross talk with CAR.
Other studies have previously reported that Mrp4 expression is reduced in Nrf2−/−-null mice, suggesting that Nrf2 plays a role in regulating Mrp4 expression in the mouse (20). These authors identified an ARE site 3.5 kb upstream of the mouse Mrp4 transcription initiation site. However, it remains to be verified whether this ARE site plays a functional role in mouse Mrp4 promoter activity. In the present report, we identified a MARE site at the proximal promoter region of the MRP4 gene where ChIP and reporter assays demonstrated that Nrf2 may bind. Deletion of the core sequence in this MARE reduced, but did not abolish, Nrf2 induction in MRP4 promoter activity in reporter assays, suggesting that there are other binding sites for Nrf2 in its proximal promoter region that may regulate MRP4 expression. In fact, mutation of XRE3, an AhR binding site, greatly reduced Nrf2 induction in the reporter assay (Fig. 6B). This result suggests that there may be an Nrf2 binding site that overlaps with XRE3. However, we cannot exclude that AhR and Nrf2 may regulate MRP4 through cross talk as discussed above, although the molecular mechanism remains to be elucidated.
We have observed a discrepancy in the time of MRP4 mRNA induction by TCDD and OPZ between human hepatocytes and HepG2 cells since the peak elevations occurred more rapidly in human hepatocytes (∼6 h) than in HepG2 cells (24–48 h) (Fig. 1D). We have also observed similar time discrepancies for induction of BSEP (ABCB11) by TCDD and OPZ between human hepatocytes and HepG2 cells (38). Possibly, these transcription factors may be activated by metabolites of TCDD and OPZ because the expression level of drug metabolism enzymes is generally much higher in primary hepatocytes than in differentiated hepatoma HepG2 cells (40).
The present study focuses on mechanisms of transcriptional regulation of MRP4. However, our studies in human hepatocytes indicate that posttranscriptional events are also significant since treatment of human hepatocytes with activators of Nrf2 and AhR resulted in clear increases in expression of MRP4 on the plasma membrane by immunofluorescence assays and a 3.7-fold increase of protein in HepG2 cells (Fig. 2). It is therefore of interest that MRP4 protein levels were induced threefold in patients with primary biliary cirrhosis, whereas MRP4 mRNA levels remained unchanged (47). Future studies will need to examine the mechanism of posttranscriptional regulation of MRP4 in the liver in more detail.
In summary, we have characterized the 5′-flanking region of the MRP4 gene and demonstrated that xenobiotic induction of the MRP4 gene is regulated through the AhR-XRE and Nrf2-ARE cellular detoxification pathways. Coordinated regulation of the MRP4 may be mediated by AhR and Nrf2 to facilitate the role of this gene in drug metabolism and disposition and in its response to cholestatic liver injury.
GRANTS
This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK34989 and DK25636. Normal human hepatocytes were obtained through the Liver Tissue Cell Distribution System, Pittsburgh, PA, which was funded by NIH Contract no. N01-DK-7-0004/HHSN 267200700004C.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1.Ahmed S, Valen E, Sandelin A, Matthews J. Dioxin increases the interaction between aryl hydrocarbon receptor and estrogen receptor alpha at human promoters. Toxicol Sci 111: 254–266, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aleksunes LM, Slitt AL, Maher JM, Dieter MZ, Knight TR, Goedken M, Cherrington NJ, Chan JY, Klaassen CD, Manautou JE. Nuclear factor-E2-related factor 2 expression in liver is critical for induction of NAD(P)H:quinone oxidoreductase 1 during cholestasis. Cell Stress Chaperones 11: 356–363, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assem M, Schuetz EG, Leggas M, Sun D, Yasuda K, Reid G, Zelcer N, Adachi M, Strom S, Evans RM, Moore DD, Borst P, Schuetz JD. Interactions between hepatic Mrp4 and Sult2a as revealed by the constitutive androstane receptor and Mrp4 knockout mice. J Biol Chem 279: 22250–22257, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Auyeung DJ, Kessler FK, Ritter JK. Mechanism of rat UDP-glucuronosyltransferase 1A6 induction by oltipraz: evidence for a contribution of the Aryl hydrocarbon receptor pathway. Mol Pharmacol 63: 119–127, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Belinsky MG, Dawson PA, Shchaveleva I, Bain LJ, Wang R, Ling V, Chen ZS, Grinberg A, Westphal H, Klein-Szanto A, Lerro A, Kruh GD. Analysis of the in vivo functions of Mrp3. Mol Pharmacol 68: 160–168, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Borst P, de Wolf C, van de Wetering K. Multidrug resistance-associated proteins 3, 4, and 5. Pflügers Arch 453: 661–673, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Boyer J, Trauner M, Mennone A, Soroka CJ, Cai SY, Moustafa T, Zollner G, Lee JY, Ballatori N. Upregulation of a basolateral FXR-dependent bile acid efflux transporter Ostα/Ostβ in cholestasis in humans and rodents. Am J Physiol Gastrointest Liver Physiol 290: G1124–G1130, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Burbach KM, Poland A, Bradfield CA. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci USA 89: 8185–8189, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci USA 98: 4611–4616, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong L, Ma Q, Whitlock JP., Jr DNA binding by the heterodimeric Ah receptor. Relationship to dioxin-induced CYP1A1 transcription in vivo. J Biol Chem 271: 7942–7948, 1996 [DOI] [PubMed] [Google Scholar]
- 11.Geier A, Wagner M, Dietrich CG, Trauner M. Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochim Biophys Acta 1773: 283–308, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Gradhand U. Variability in human hepatic MRP4 expression: influence of cholestasis and genotype. Pharmacogenomics J 8: 42–52, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Hayes JD. Toxicological highlight cross-talk between transcription factor AhR and Nrf2: lessons for caner chemoprevention from dioxin. Toxicol Sci 111: 199–201, 2009 [DOI] [PubMed] [Google Scholar]
- 14.Hirohashi T, Suzuki H, Sugiyama Y. Characterization of the transport properties of cloned rat multidrug resistance-associated protein 3 (MRP3). J Biol Chem 274: 15181–15185, 1999 [DOI] [PubMed] [Google Scholar]
- 15.Keitel V, Burdelski M, Warskulat U, Kuhlkamp T, Keppler D, Haussinger D, Kubitz R. Expression and localization of hepatobiliary transport proteins in progressive familial intrahepatic cholestasis.[see comment][erratum appears in Hepatology 42: 242, 2005]. Hepatology 41: 1160–1172, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47: 89–116, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Kohle C, Bock KW. Coordinate regulation of human drug-metabolizing enzymes, and conjugate transporters by the Ah receptor, pregnane X receptor and constitutive androstane receptor. Biochem Pharmacol 77: 689–699, 2009 [DOI] [PubMed] [Google Scholar]
- 18.Lamle J, Marhenke S, Borlak J, von Wasielewski R, Eriksson CJP, Geffers R, Manns MP, Yamamoto M, Vogel A. Nuclear factor-eythroid 2-related factor 2 prevents alcohol-induced fulminant liver injury. Gastroenterology 134: 1159–1168, 2008 [DOI] [PubMed] [Google Scholar]
- 19.Ma Q, Kinneer K, Bi Y, Chan JY, Kan YW. Induction of murine NAD(P)H:quinone oxidoreductase by 2,3,7,8-tetrachlorodibenzo-p-dioxin requires the CNC (cap ‘n’ collar) basic leucine zipper transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2): cross-interaction between AhR (aryl hydrocarbon receptor) and Nrf2 signal transduction. Biochem J 377: 205–213, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, Dalton TP, Yamamoto M, Klaassen CD. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology 46: 1597–1610, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Mennone A, Soroka CJ, Cai SY, Harry K, Adachi M, Hagey L, Schuetz JD, Boyer JL. Mrp4−/− mice have an impaired cytoprotective response in obstructive cholestasis. Hepatology 43: 1013–1021, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Miao W, Hu L, Kandouz M, Batist G. Oltipraz is a bifunctional inducer activating both phase I and phase II drug-metabolizing enzymes via the xenobiotic responsive element. Mol Pharmacol 64: 346–354, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Miao W, Hu L, Scrivens PJ, Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem 280: 20340–20348, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci USA 91: 9926–9930, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem Pharmacol 59: 65–85, 2000 [DOI] [PubMed] [Google Scholar]
- 26.Okada K, Shoda J, Taguchi K, Maher JM, Ishizaki K, Inoue Y, Ohtsuki M, Goto N, Sugimoto H, Utsunomiya H, Oda K, Warabi E, Ishii T, Yamamoto M. Nrf2 counteracts cholestatic liver injury via stimulation of hepatic defense systems. Biochem Biophys Res Commun 389: 431–436, 2009 [DOI] [PubMed] [Google Scholar]
- 27.Perdew G. Aryl-hydrocarbon receptor activation regulates constitutive androstane receptor levels in murine and human liver. Hepatology 46: 209–218, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rius M, Hummel-Eisenbeiss J, Hofmann AF, Keppler D. Substrate specificity of human ABCC4 (MRP4)-mediated cotransport of bile acids and reduced glutathione. Am J Physiol Gastrointest Liver Physiol 290: G640–G649, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Russel FGM, Koenderink JB, Masereeuw R. Multidrug resistance protein 4 (MRP4/ABCC4): a versatile efflux transporter for drugs and signalling molecules. Trends Pharmacol Sci 29: 200–207, 2008 [DOI] [PubMed] [Google Scholar]
- 30.Shin S, Wakabayashi N, Misra V, Biswal S, Lee GH, Agoston ES, Yamamoto M, Kensler TW. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol 27: 7188–7197, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokol RJ, Winklhofer-Roob BM, Devereaux MW, McKim JM., Jr Generation of hydroperoxides in isolated rat hepatocytes and hepatic mitochondria exposed to hydrophobic bile acids. Gastroenterology 109: 1249–1256, 1995 [DOI] [PubMed] [Google Scholar]
- 32.Soroka CJ, Lee JM, Azzaroli F, Boyer JL. Cellular localization and up-regulation of multidrug resistance-associated protein 3 in hepatocytes and cholangiocytes during obstructive cholestasis in rat liver. Hepatology 33: 783–791, 2001 [DOI] [PubMed] [Google Scholar]
- 33.Soroka CJ, Mennone A, Hagey LR, Ballatori N, Boyer JL. Mouse organic solute transporter alpha deficiency enhances renal excretion of bile acids and attenuates cholestasis. Hepatology 51: 181–190, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka Y, Aleksunes LM, Cui YJ, Klaassen CD. ANIT-induced intrahepatic cholestasis alters hepatobiliary transporter expression via Nrf2-dependent and independent signaling. Toxicol Sci 108: 247–257, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai LY, Lee KT, Liu TZ. Evidence for accelerated generation of hydroxyl radicals in experimental obstructive jaundice of rats. Free Radic Biol Med 24: 732–737, 1998 [DOI] [PubMed] [Google Scholar]
- 36.Tsuchiya Y, Nakajima M, Yokoi T. Critical enhancer region to which AhR/ARNT and Sp1 bind in the human CYP1B1 gene. J Biochem (Tokyo) 133: 583–592, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Volz DC, Kullman SW, Howarth DL, Hardman RC, Hinton DE. Protective response of the Ah receptor to ANIT-induced biliary epithelial cell toxicity in see-through medaka. Toxicol Sci 102: 262–277, 2008 [DOI] [PubMed] [Google Scholar]
- 38.Weerachayaphorn J, Cai SY, Soroka CJ, Boyer JL. Nuclear factor erythroid 2-related factor 2 is a positive regulator of human bile salt export pump expression. Hepatology 50: 1588–1596, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wild AC, Moinova HR, Mulcahy RT. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J Biol Chem 274: 33627–33636, 1999 [DOI] [PubMed] [Google Scholar]
- 40.Wilkening S, Bader A. Influence of culture time on the expression of drug-metabolizing enzymes in primary human hepatocytes and hepatoma cell line HepG2. J Biochem Mol Toxicol 17: 207–213, 2003 [DOI] [PubMed] [Google Scholar]
- 41.Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Introducing the “TCDD-inducible AhR-Nrf2 gene battery.” Toxicol Sci 111: 238–246, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yueh MF, Tukey RH. Nrf2-Keap1 signaling pathway regulates human UGT1A1 expression in vitro and in transgenic UGT1 mice. J Biol Chem 282: 8749–8758, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Yueh MF, Huang YH, Hiller A, Chen S, Nguyen N, Tukey RH. Involvement of the xenobiotic response element (XRE) in Ah receptor-mediated induction of human UDP-glucuronosyltransferase 1A1. J Biol Chem 278: 15001–15006, 2003 [DOI] [PubMed] [Google Scholar]
- 44.Zelcer N, Reid G, Wielinga P, Kuil A, van der Heijden I, Schuetz JD, Borst P. Steroid and bile acid conjugates are substrates of human multidrug-resistance protein (MRP) 4 (ATP-binding cassette C4). Biochem J 371: 361–367, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zelcer N, van de Wetering K, de Waart R, Scheffer GL, Marschall HU, Wielinga PR, Kuil A, Kunne C, Smith A, van der Valk M, Wijnholds J, Elferink RO, Borst P. Mice lacking Mrp3 (Abcc3) have normal bile salt transport, but altered hepatic transport of endogenous glucuronides. J Hepatol 44: 768–775, 2006 [DOI] [PubMed] [Google Scholar]
- 46.Zhou SF, Chan E, Zhou ZW, Xue CC, Lai X, Duan W. Insights into the structure, function, and regulation of human cytochrome P450 1A2. Curr Drug Metab In Press [DOI] [PubMed] [Google Scholar]
- 47.Zollner G, Wagner M, Fickert P, Silbert D, Gumhold J, Zatloukal K, Denk H, Trauner M. Expression of bile acid synthesis and detoxification enzymes and the alternative bile acid efflux pump MRP4 in patients with primary biliary cirrhosis. Liver Int 27: 920–929, 2007 [DOI] [PubMed] [Google Scholar]