Abstract
The guanidinium alkaloids tetrodotoxin (TTX) and saxitoxin (STX) are classic ligands of voltage-gated sodium channels (VGSCs). Like TTX and STX, μ-conotoxin peptides are pore blockers but with greater VGSC subtype selectivity. μ-Conotoxin KIIIA blocks the neuronal subtype NaV1.2 with nanomolar affinity and we recently discovered that KIIIA and its mutant with one fewer positive charge, KIIIA[K7A], could act synergistically with TTX in a ternary peptide·TTX·NaV complex. In the complex, the peptide appeared to trap TTX in its normal binding site such that TTX could not readily dissociate from the channel until the peptide had done so; in turn, the presence of TTX accelerated the rate at which peptide dissociated from the channel. In the present study we examined the inhibition of NaV1.2, exogenously expressed in Xenopus oocytes, by STX (a divalent cation) and its sulfated congener GTX2/3 (with a net +1 charge). Each could form a ternary complex with KIIIA and NaV1.2, as previously found with TTX (a monovalent cation), but only when STX or GTX2/3 was added before KIIIA. The KIIIA·alkaloid·NaV complex was considerably less stable with STX than with either GTX2/3 or TTX. In contrast, ternary KIIIA[K7A]·alkaloid·NaV complexes could be formed with either order of ligand addition and were about equally stable with STX, GTX2/3, or TTX. The most parsimonious interpretation of the overall results is that the alkaloid and peptide are closely apposed in the ternary complex. The demonstration that two interacting ligands (“syntoxins”) occupy adjacent sites raises the possibility of evolving a much more sophisticated neuropharmacology of VGSCs.
INTRODUCTION
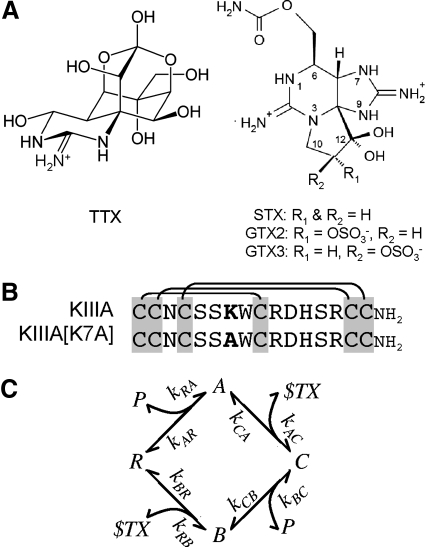
Voltage-gated sodium channels (VGSCs) are responsible for the upstroke of action potentials in excitable cells. Tetrodotoxin (TTX) and saxitoxin (STX) are guanidinium alkaloids (Fig. 1A) that block six of the nine isoforms of the pore-bearing α-subunit of mammalian VGSCs with Kd values in the nanomolar range (Catterall et al. 2005). μ-Conotoxins, which also block VGSCs, are peptides of 16 to 25 residues, including six cysteines that form a characteristic disulfide framework (see Fig. 1B) (Terlau and Olivera 2004). In contrast to the promiscuity of TTX and STX, GIIIA, the best-studied μ-conotoxin, is significantly more potent against the skeletal muscle subtype of channel NaV1.4 (half-maximal inhibitory concentration [IC50] = 19 nM), than other TTX-sensitive subtypes, such as brain NaV1.2 (IC50 ≈ 2.7 μM) (Safo et al. 2000). TTX, STX, and GIIIA competitively bind to site 1 of NaV1.4, located at the outer vestibule of the channel's pore (Al-Sabi et al. 2006; Cestele and Catterall 2000; French and Terlau 2004; Moczydlowski et al. 1986).
Fig. 1.
Structures of guanidinium alkaloids and peptides whose activities were examined in this study. A: tetrodotoxin (TTX, left) and saxitoxin (STX) and GTX2/3 (STX congener with a net +1 charge; right). At neutral pH, TTX has a single positive charge, whereas STX has 2. GTX2/3 is a mixture of epimers GTX2 and GTX3 and differs from STX by having a sulfate group, so its net charge is +1 (Hall et al. 1990). B: aligned sequences of μ-conotoxin KIIIA and its point mutant KIIIA[K7A]. These peptides are identical except for bold residue, which is Lys7 in KIIIA and Ala7 in KIIIA[K7A]. Three disulfide bridges characteristic of all μ-conotoxins are indicated by lines linking Cys residues. Carboxyl terminals of both peptides are amidated. C: proposed reaction scheme for the syntoxin interactions between alkaloid and peptide when they bind to NaV1.2. The channel (i.e., receptor) is represented by R and its ligands by P for μ-conopeptide and $TX for alkaloid (TTX or STX). A is the binary peptide·channel complex, B is the binary alkaloid·channel complex, and C is the ternary peptide·alkaloid·channel complex. The formation of the same ternary complex, independent of order of ligand addition, was inferred from the interactions of TTX and KIIIA or KIIIA[K7A] with NaV1.2 (Zhang et al. 2009) and is also consistent with the results in the present experiments with STX and GTX2/3. The notation for the rate constants correspond to that in the formulae, used to numerically simulate the kinetics of the reactions, in methods.
We recently discovered μ-conotoxins KIIIA and SIIIA, which block NaV1.2 with higher affinity than NaV1.4 (Bulaj et al. 2005; Yao et al. 2008; Zhang et al. 2007). Single-channel analyses of brain VGSCs in planar lipid bilayers showed that KIIIA produced only a partial block (Zhang et al. 2007). In saturating concentrations of KIIIA or its mutant KIIIA[K7A], a residual sodium current (rINa) could be macroscopically recorded in voltage-clamped Xenopus oocytes expressing NaV1.2 (Zhang et al. 2009). Remarkably, TTX blocked the rINa with either peptide, albeit 102- to 104-fold more slowly than control INa. When TTX was bound to the channel first, only a modest nearly 25% decrease in the rate of peptide binding was observed. The rates of recovery from block of the ternary complexes (koff values) were the same regardless of the order of ligand addition. The koff values for the bimolecular complex KIIIA[K7A]·NaV and the ternary complexes KIIIA·TTX·NaV and KIIIA[K7A]·TTX·NaV were similar; in contrast, the koff value for KIIIA·NaV was 10-fold smaller, indicating that the presence of TTX decreased the stability of KIIIA·TTX·NaV. We hypothesized that TTX and peptide bound abutting microsites (Olivera et al. 1991), within the greater confines of a macrosite synonymous with neurotoxin receptor site 1 designated by Catterall (Catterall 1980; Cestele and Catterall 2000).
In this report we tested STX, which has two positive charges (Bordner et al. 1975; Schantz et al. 1975), in contrast to TTX's single charge (Hille 1975), and GTX2/3, a naturally occurring STX congener with a net +1 charge (Fig. 1A). In view of their rather different structures, it is not surprising that STX and TTX do not bind in an identical fashion to VGSCs. For example, when Phe 385 near the selectivity filter of NaV1.2 is mutated to Cys, the channel's affinity for TTX is reduced 3,000-fold (Zhang et al. 2009), whereas that for STX is reduced (only) 340-fold (Heinemann et al. 1992). Herein, we show that STX and GTX2/3 behave differently from each other as well as from TTX in the formation and dissociation of complexes of peptide and alkaloid with NaV1.2. Nevertheless, the overall results are explicable in terms of the reaction scheme in Fig. 1C and support the hypothesis that in the ternary peptide·alkaloid·NaV complex, KIIIA or KIIIA[K7A] cooccupies site 1 with alkaloid, irrespective of whether it is STX, GTX2/3, or TTX.
METHODS
Oocyte electrophysiology
Oocytes expressing rat NaV1.2 were prepared and two-electrode voltage clamped essentially as previously described (Fiedler et al. 2008; Zhang et al. 2007). Oocytes were harvested from Xenopus frogs, following a protocol approved by the University of Utah's Institutional Animal Care and Use Committee that conforms to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Briefly, oocytes were bathed in ND96 and sodium currents (INa) were recorded by holding the membrane potential at −80 mV and stepping to −10 mV for 50 ms every 20 s. The oocytes were exposed to toxin in a small (30 μl) static bath to conserve material. Recordings were done at room temperature (∼21°C).
Toxins
μ-Conopeptides were synthesized as previously described (Zhang et al. 2007). TTX was obtained from Alomone Labs (Jerusalem, Israel), STX from either Calbiochem (San Diego, CA) or the National Research Council Canada (Halifax, Nova Scotia, Canada), and GTX2/3 from the National Research Council Canada. In experiments involving high concentrations, STX and GTX2/3 were lyophilized then dissolved in ND96. All stocks were stored at −20°C in a nonfrost-free freezer until use.
Data analysis
Off-rate constants (koff values) were determined from single-exponential fits of peak INa following toxin washout; observed rate constants (kobs values) were obtained by single exponential fits of INa following toxin addition and on-rate constants (kon values) were obtained from slopes of kobs versus [toxin], all as reported previously (West et al. 2002). In particular instances (see the text immediately following), the time course of the recovery of INa following toxin washout was biphasic and the recovery was fit to a double-exponential curve as previously reported (Zhang et al. 2009).
Numerical simulation of the reaction scheme in Fig. 1C
To model the time-dependent changes in the concentrations of the various components, the reaction scheme in Fig. 1C was simulated by reiteration of the following formulas in a for-loop using the graphical programming language LabVIEW (National Instruments, Austin, TX)
Pseudofirst-order kinetics was assumed; i.e., the concentration of alkaloid, denoted by [$] (representing STX, GTX2/3, or TTX), and that of μ-conopeptide, denoted by [P], remained constant. When concentrations were expressed in micromoles (μM) and time in minutes, a time increment Δt of 10−4 proved sufficiently small to accommodate the concentrations and rate constants used for the simulations used in this study. The rate constants are as denoted in Fig. 1C and their values were obtained by measuring the kinetics of block and recovery of INa following exposure to, and washout of, alkaloid and peptide, each alone or in combinations, as follows: kRA and kAR were the kon and koff values, respectively, of the reaction of NaV with peptide alone; kBR and kRB were the kon and koff values, respectively, of the reaction of NaV with alkaloid alone; and kAC was determined from the kon value for the block of residual current of the peptide·NaV complex at saturating [peptide]. There was no direct means to determine kCA, but its value could be calculated, since the product of the rate constants for the four clockwise steps equals that for the four counterclockwise steps; that is, kCA = [kRA*kAC*kCB*kBR]/[kRB*kBC*kAR]. When all of the NaV was converted into a ternary complex by use of saturating concentrations of toxins, the time course for functional recovery after toxin washout could be fit by a single-exponential curve with a rate constant koff, which was smaller than kBR (and larger than kAR), indicating that kCB was rate-liming during toxin washout; therefore kCB could be approximated by koff. A more accurate value of kCB could be estimated from the experimentally determined values for six of the rate constants, kRA, kAR, kAC, kRB, kBR, and kBC (see next paragraph for experimental determination of kBC), and calculating the seventh, kCA, as described earlier, while the value of kCB was varied until the mean squared error between the simulated and experimental time courses of recovery from block reached a minimum. Values of kCA and kCB determined in this fashion are presented in Table 4.
Table 4.
Equilibrium constants (nM) of reactions in Fig. 1C
|
A. Kdvalues of peptides for channel alone versus channel saturated with peptide | |||
|---|---|---|---|
| Channel Alone |
Channel Saturated With Alkaloid |
||
| Peptide | None Kd (kBR/kRB) | KIIIA Kd (kCA/kAC) | KIIIA[K7A] Kd (kCA/kAC) |
| Alkaloid | |||
| TTX | 38 ± 6 | 730 ± 800 | 45 ± 22 |
| STX | 8 ± 2 | — | 6 ± 3 |
| GTX2/3 | 5 ± 1 | — | 7 ± 4 |
|
B. Kdvalues of peptides for channel alone versus channel saturated with alkaloid | ||||
|---|---|---|---|---|
| Channel Alone |
Channel Saturated With Alkaloid |
|||
| Alkaloid | None Kd (kAR/kRA) | TTX Kd (kCB/kBC) | STX Kd (kCB/kBC) | GTX2/3 Kd (kCB/kBC) |
| Peptide | ||||
| KIIIA | 5 ± 5 | 105 ± 25 | 4 (1–12) × 103† | 290 ± 30 |
| KIIIA[K7A] | 115 ± 40 | 138 ± 30 | 87 ± 18 | 156 ± 62 |
Equilibrium Kd values, mean ± SD, were calculated from koff/kon ratios (notations of corresponding rate constants in Fig. 1C are indicated in parentheses); values for these rate constants were obtained from Tables 1–3, except for those of kCB and kCA, which were obtained by numerical simulation, as described in methods. Values of kCB, in min−1, were (for indicated peptide–alkaloid combinations): 0.021 (KIIIA–TTX); 0.35 (KIIIA–STX); 0.029 (KIIIA–GTX2/3); 0.022 (KIIIA[K7A]–TTX); 0.013 (KIIIA[K7A]–STX); and 0.014 (KIIIA[K7A]–GTX2/3); SDs of koff values for corresponding ternary complexes in Table 2 served as proxies for SDs for kCB values and these were used in calculating SDs for corresponding kCA and Kd values. Mean values of kCA ± SD, in min−1, were (for indicated peptide–alkaloid combinations): 0.0022 ± 0.0024 (KIIIA–TTX); 0.025 ± 0.012 (KIIIA[K7A]–TTX); 0.0038 ± 0.0018 (KIIIA[K7A]–STX); and 0.004 ± 0.0021 (KIIIA[K7A]–GTX2/3).
Values in parentheses indicate range of uncertainty of predicted Kd, from limits of uncertainty for corresponding kBC value in Table 3.
The value of kBC was estimated from experiments in which alkaloid was added before peptide and exploiting the large disparity between kCB and kBR (except in one instance, involving KIIIA and STX; see following paragraph), which produced a double-exponential time course for the recovery of INa following toxin washout. The coefficients of the double-exponential fits provided the koff values and spans of the fast and slow phases. In every case, the fast koff had a value close (within a factor of ∼2) to kBR (i.e., the rate constant for the recovery from block of the binary alkaloid·NaV complex) and the slow koff had a value close (within a factor of ∼2) to kCB (i.e., the rate constant for recovery from block of the ternary complex formed with saturating concentrations of both alkaoid and peptide applied for sufficient time to convert all of the NaV into a ternary complex). These results supported the notion that the span of the slow phase was likely to provide a good estimate of the fraction of the channels that were in the ternary peptide·alkaloid·NaV complex at the start of toxin washout (Zhang et al. 2009). It was assumed that preceding toxin washout, the fraction of ternary complex formed (FTCF) as a function of time followed a single-exponential time course and therefore the estimated value for the observe rate constant kobs was calculated from the equation kobs = [ln (FTCF)]/time. For the sake of simplicity, data were obtained only at a single time point. This kobs was used with the equation kobs = kon*[peptide] + koff and, since koff (i.e., ∼kCB) was invariably about an order of magnitude smaller than kobs, the apparent kon (i.e., kBC) was calculated from kobs/[peptide].
A procedure different from that described earlier had to be used to estimate kBC in the case of STX and KIIIA because the difference between kCB and kBR was only fourfold and the experimental time course of recovery of INa following toxin washout could not be fit by a double-exponential curve better than a single-exponential one. Instead, the experiment was simulated using different values for kBC and the value producing simulated results that corresponded best with the experimental data was used to approximate kBC. The remaining rate constants used in this approximation are specified in results.
Molecular modeling and docking of NaV1.2, μ-conotoxin KIIIA, and the alkaloids TTX and STX
A model of rat NaV1.2 was constructed using as a template the model of rat NaV1.4 proposed by Lipkind and Fozzard (2000), which was composed of 12 parts from the four domains. Each domain was represented by three structural fragments: S5-segment, P-loop, and S6-segment. Sequence analysis of these 12 parts from NaV1.2 and NaV1.4 shows 91.0% identity (data not shown). We found every structural fragment for each domain of NaV1.2 that corresponded to Lipkind and Fozzard's model of NaV1.4. Our model of NaV1.2 was built by introducing modifications of specific residues using PyMOL (DeLano Scientific, Palo Alto, CA); the residues of the NaV1.4 model were replaced by corresponding backbone-dependent rotamer residues of the NaV1.2 using the PyMOL mutagenesis function. The conopeptide KIIIA was constructed with coordinates from Khoo et al. (2009) and the coordinates of TTX and STX were from Tikhonov and Zhorov (2005).
Automated docking of TTX, STX, and KIIIA to the model of NaV1.2 was performed using the AutoDock4.2 software package (Morris et al. 2009). Flexible torsions of ligands were assigned using AUTOTORS and a grid box defined by 60 × 60 × 60 grid points and spacing of 0.375 Å with AUTOGRID. Docking simulations were performed using a Lamarckian genetic algorithm (Morris et al. 1998), with the following parameters: population size = 300, mutation rate = 0.02, crossover rate = 0.8. The numbers of dockings per simulation were 50 and 256 for conopeptide and alkaloid, respectively. The number of dockings per simulation for conopeptide was lower than that for alkaloid due to the larger values of flexible torsions and corresponding longer simulation times. The maximum number of energy evaluations was set to 250,000 and the maximum number of generations was 27,000. One cluster-representative structure was selected from each of the docking runs. Clusters were examined for quantity of conformations and those with the greatest number were chosen for further analysis. A secondary criterion for selecting a cluster-representative structure was the value of binding energy.
RESULTS
Residual Na current (rINa) with KIIIA[K7A] can be blocked by both STX and GTX2/3; however, block of the rINa with KIIIA by either of these alkaloids could not be demonstrated
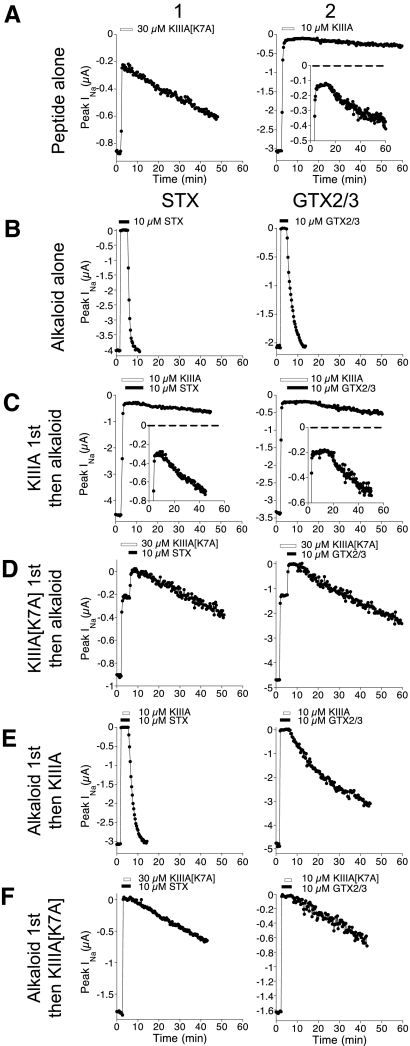
As we previously showed (Zhang et al. 2009), when oocytes expressing NaV1.2 were voltage clamped and exposed to saturating concentrations of either KIIIA or KIIIA[K7A], a residual sodium current (rINa) of 5 and 23% of control INa, respectively, persisted (Fig. 2A). In contrast, saturating concentrations of the alkaloids STX or GTX2/3 produced essentially a complete block (>99%), which was much more quickly reversible than the block by either conopeptide alone (Fig. 2, A and B).
Fig. 2.
Block of NaV1.2 by KIIIA, KIIIA[K7A], STX, or GTX2/3, each alone or in combination. Voltage-gated sodium currents (INa) of voltage-clamped oocytes expressing NaV1.2 were measured as described in methods. Plotted are peak INa as a function of time during exposure to, and subsequent washout of, peptide alone (row A) or alkaloid alone (row B) or both (rows C–F). Bars above plots indicate when peptide (white bar) or alkaloid (black bar) were present. Except for row A, 1st column represents STX and 2nd column, GTX2/3. A: block by saturating concentration of either KIIIA[K7A] or KIIIA alone (30 or 10 μM, respectively). Inset in KIIIA plot shows curve with INa-axis expanded. Note residual current (rINa) persisting in the presence of each peptide. B: block by saturating concentration of STX or GTX2/3 alone (each 10 μM). C: rINa with KIIIA was blocked by neither STX nor GTX2/3. Insets show plots with INa-axis expanded; note rate of recovery following toxin washout was unaffected (cf. inset of A2). D: in contrast, STX and GTX2/3 each blocked rINa with KIIIA[K7A]. E and F: recovery from block during simultaneous washout of both alkaloid and peptide following exposure to alkaloid first, then KIIIA (E) or KIIIA[K7A] (F). E: with KIIIA, washout rates were much faster than those following exposure to KIIIA alone (A2) but slower than those following exposure to alkaloid alone (B), consistent with KIIIA retarding the washout of alkaloid and alkaloid reciprocating by accelerating the washout of KIIIA. F: with KIIIA[K7A], washout rates were similar to those following exposure to peptide alone (A1) and slower than those following exposure alkaloid alone (B), consistent with peptide retarding alkaloid washout. Time bases (abscissa) of all plots have the same scale. Representative experiments are shown; for quantitative summaries see Tables 1 and 2.
The rINa with KIIIA was not blocked by either STX or GTX2/3 at the highest concentrations tested (10 μM); i.e., ≲10%, if any, of the rINa was blocked (Fig. 2C). In contrast, the rINa with KIIIA[K7A] could be readily blocked by STX and GTX2/3 (Fig. 2D). With each alkaloid, complete block was achieved with the formation of a ternary KIIIA[K7A]·alkaloid·NaV complex, as evident from the rate of recovery from block after toxin washout, which occurred in a single phase (Fig. 2D) without an initial fast phase that would indicate the recovery from block by alkaloid alone (also see following text).
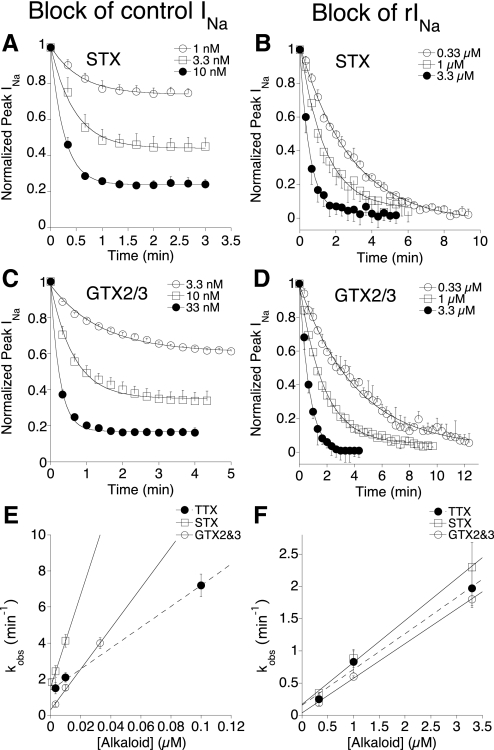
The time course of the onset of block by STX and GTX2/3 of control INa and rINa with KIIIA[K7A] could be fit with single-exponential curves (Fig. 3,A–D), and the resulting observed rate constants (kobs values) were linearly dependent on the concentration of alkaloid (Fig. 3, E and F). For comparison, also illustrated in Fig. 3, E and F are previously acquired results with TTX (Zhang et al. 2009). The slopes of the lines yielded apparent kon values, which are tabulated in Table 1. The presence of KIIIA[K7A] decreased the on rates of all three alkaloids by at least two orders of magnitude.
Fig. 3.
Rates of block of control INa and rINa with KIIIA[K7A] by different concentrations of STX and GTX2/3. Experiments were done as in Fig. 2, B and D. Time courses of block of control INa (A, C; cf. Fig. 2B) and rINa (B, D; cf. Fig. 2D) by various alkaloid concentrations were each fit to a single exponential (solid curve) to obtain kobs. Dependence of kobs on alkaloid concentration for block of control INa (E) or rINa with 30 μM KIIIA[K7A] (F) (STX, open squares; GTX2/3, open circles; or TTX, closed circles). Data for TTX are from Zhang et al. (2009) and are included for comparison. Straight lines represent linear regression slopes whose values (i.e., kon values) are given in Table 1. Rates of block of rINa were significantly slower than those of control INa (note ∼30-fold difference in x-axis scale between E and F). Steady-state block of control INa at different concentrations of STX (A) and GTX2/3 (C) yielded half-maximal inhibitory concentration (IC50) values of 3 ± 0.3 and 4.6 ± 1.0 nM, respectively, which are close to (within a factor of 3 of) their respective Kd values of 7.9 ± 1.7 and 5.3 ± 0.8 nM calculated from koff and kon values (Tables 1 and 2). In contrast, steady-state block of rINa is essentially complete, even at the lowest [alkaloid] tested (B, D); using lower concentrations takes too long to reach steady state, precluding determination of IC50 values for this reaction. Data points represent mean ± SD (n ≥ 3 oocytes).
Table 1.
Rates of block of rINa and control INa by the various guanidinium alkaloids, corresponding to kAC and kRB of Fig. 1C, respectively
| Apparent kon, μM−1·min−1† |
||||
|---|---|---|---|---|
| Alkaloid | rINa With KIIIA | rINa With KIIIA [K7A] | Control INa | Reference |
| TTX | 0.003 ± 0.0004 | 0.56 ± 0.08 | 58 ± 5.3 | Zhang et al. (2009) |
| STX | ∼0‡ | 0.65 ± 0.10 | 254 ± 41.0 | Thisstudy |
| GTX2/3 | ∼0‡ | 0.55 ± 0.03 | 113 ± 8.7 | Thisstudy |
Ternary complexes of KIIIA·alkaloid·NaV and KIIIA[K7A]·alkaloid·NaV can be formed by adding STX or GTX2/3 first, followed by peptide
Instead of adding peptide first, then alkaloid, experiments were also performed in which the order of ligand addition was reversed. Formation of a ternary complex was evident from the single-phase nature of the recovery of activity when both toxins were washed out (Fig. 2, E and F). The rates of functional recovery during dissociation of the ternary KIIIA[K7A]·alkaloid·NaV complex after toxin washout were similar for both STX and GTX2/3. This was so regardless of whether alkaloid or peptide was added first and the off rates were essentially the same as those for the recovery from block of the binary KIIIA[K7A]·NaV complex (Fig. 2, A, D, and F, Table 2).
Table 2.
Rates of recovery from block during washout of ligand(s) from binary and ternary complexes, corresponding to kBR and kAR and approximation of kCB of Fig. 1C, respectively
|
koff of complex, min−1† |
||||
|---|---|---|---|---|
| Alkaloid | KIIIA·X·NaV | KIIIA[K7A]·X·NaV | X·NaV | Reference |
| None | 0.0016 ± 0.0016 | 0.015 ± 0.005 | — | Zhang et al. (2009) |
| TTX | 0.020 ± 0.005 (0.017 ± 0.002) | 0.024 ± 0.004 (0.025 ± 0.003) | 2.2 ± 0.3 | Zhang et al. (2009) |
| STX | 0.350 ± 0.039 (NA)‡ | 0.014 ± 0.002 (0.015 ± 0.007) | 2.0 ± 0.3 | This study |
| GTX2/3 | 0.029 ± 0.001 (NA)‡ | 0.014 ± 0.003 (0.018 ± 0.005) | 0.6 ± 0.08 | This study |
Values are means ± SD, n ≥3 oocytes.
Rate constants of recovery from block during washout of toxin(s) following exposure to: each peptide alone (top data row), or each alkaloid alone (right data column), or both alkaloid and peptide (remaining data entries). Ternary complexes were formed by exposure first to alkaloid as in Fig. 2, E and F (or exposure first to peptide as in Fig. 2D), followed by mixture of alkaloid plus peptide. Values of koff were obtained by fitting the time course of recovery of activity after toxin(s) were washed out to a single-exponential curve, values without parentheses are those of the complex formed by exposure to alkaloid first, and values in parentheses are those of the complex formed by exposure to peptide first.
NA, not available because ternary complex was not readily formed by adding peptide first.
Although we were unable to show that the rINa with KIIIA could be blocked by STX or GTX2/3, formation of a ternary KIIIA·alkaloid·NaV complex could be readily demonstrated by adding STX or GTX2/3 first, followed by KIIIA: on toxin washout, the respective rates of recovery of block were >100- and >10-fold faster than that after exposure to KIIIA alone and 6- and 20-fold slower than that after exposure to the respective alkaloid alone (Fig. 2E, Table 2).
Rates of formation of ternary complexes when peptide was added after alkaloid
In view of the dramatic reduction by the presence of peptide in the rate of binding of alkaloid to the channel, it was of interest to see how the presence of alkaloid affected the rate of binding of peptide to the channel. After exposure to a saturating concentration of alkaloid to form the fully blocked binary alkaloid·NaV complex, the amount of the ternary complex formed following addition of peptide was determined by analyzing the recovery from block following toxin washout as described in methods. For all but the KIIIA·STX·NaV complex, after exposure to both alkaloid and peptide using a protocol similar to that used in Fig. 2, E and F but with tenfold lower peptide concentration, the time course of recovery of activity following toxin washout had two distinct phases that could be fit by a double-exponential curve (Fig. 4, A and B): an early fast phase attributed to the off rate of the binary alkaloid·NaV complex and a slower phase attributed to the dissociation of the ternary peptide·alkaloid·NaV complex—these attributions were inferred because the respective koff values were within a factor of 2 of the expected values in Table 2. As elaborated in methods, the span of the slow component yields the fraction of ternary complex formed (FTCF) from which the apparent kon value for the reaction peptide + alkaloid·NaV → peptide·alkaloid·NaV, provided in Table 3, was calculated. The approach used here was a simplified version of the protocol used to determine the apparent kon of KIIIA and KIIIA[K7A] binding to the channel preoccupied with TTX, where a fixed concentration of peptide was used and the peptide exposure time was varied (Zhang et al. 2009); in those experiments the apparent kon value for the reaction peptide + TTX·NaV → peptide·TTX·NaV was 0.13 ± 0.006 and 0.20 ± 0.008 μM−1·min−1 with KIIIA and KIIIA[K7A], respectively. These values are close to the corresponding kon values obtained in the present experiments with these two peptides and TTX (Table 3) and suggest that good estimates of kon were obtained by the simpler protocol used here.
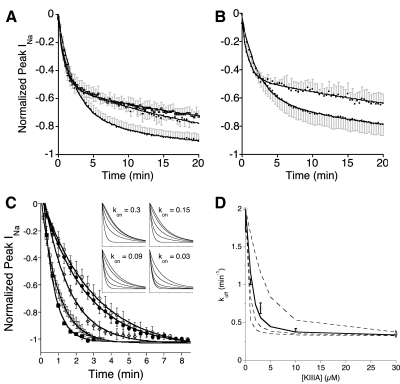
Fig. 4.
Time course of recovery from block when toxins are washed out before all of the channels have been converted into a ternary complex. Experiments were done essentially as in Fig. 2, E and F, except a lower concentration of peptides was used and results were processed as described in methods. A and B: oocytes were exposed first to a saturating concentration (10 μM) of alkaloid for 2 min, followed by 10 μM alkaloid + 1 μM peptide for 4 min before toxin washout. Plotted are recoveries of INa following toxin washout (normalized mean ± SD, n = 3 oocytes). Solid lines represent data fit to double-exponential curves; in each case, fast and slow components had rate constants (see following text) within a factor of 2 of the koff values of the corresponding alkaloid·NaV or peptide·alkaloid·NaV complex, respectively (Table 2). Spans of slow components (fractions of ternary complex formed [FTCFs]) are given in Table 3. A: recovery from block by KIIIA[K7A] and either GTX2/3, TTX, or STX (bottom, middle, and top traces at 20 min, respectively); fast rate constants were 0.45 ± 0.02, 1.9 ± 0.3, and 1.0 ± 0.1 min−1, respectively, and slow constants were 0.025 ± 0.004, 0.04 ± 0.002, and 0.027 ± 0.002 min−1, respectively. B: recovery from block by KIIIA and either GTX2/3 or TTX (bottom and top traces at 20 min, respectively); fast rate constants were 0.43 ± 0.02 and 1.1 ± 0.1 min−1, respectively; slow constants were 0.031 ± 0.004 and 0.02 ± 0.002 min−1, respectively. C: recovery from block following washout after oocytes were exposed to 10 μM STX for 2 min, then for 4 min to 10 μM STX + x μM KIIIA, where x was 0, 1, 3, 10, and 30 (briefest to longest-lasting curves, respectively). Data points represent normalized mean peak INa ± SD (n = 3 oocytes) and solid lines are data fit to single-exponential curves. Insets: simulated curves, obtained as described in methods, when kon values for formation of ternary complex were (left to right, top to bottom) 0.3, 0.15, 0.09, and 0.03 μM−1·min−1 and where for each plot [KIIIA] = 0, 1, 3, 10, and 30 μM (briefest to longest-lasting curves, respectively). Ranges of x and y axes are the same as those of the main experimental plot. Note that of the simulated plots, the pattern of curves in the bottom left, where kon was 0.09 μM−1·min−1, most closely resembles that of the experimental plot. D: decay constants of experimental curves in C, koff (mean ± SD), plotted as a function of KIIIA concentration. Lines plot decay constants of simulated curves in inset of C, where (top to bottom) kon = 0.03 (dashed), 0.09 (solid), 0.15 (dashed), and 0.3 (dashed) μM−1·min−1, with additional decay constants representing those with [KIIIA] of 2 and 5 μM. Correspondence of simulated and experimental data suggests a kon of 0.09 μM−1·min−1, with a proposed uncertainty ranging from 0.03 to 0.3 μM−1·min−1.
Table 3.
Fraction of ternary complex formed (FTCF), apparent kon for the reaction peptide + alkaloid·NaV → peptide·alkaloid·NaV, and kon for the reaction peptide + NaV → peptide·NaV, corresponding to kBC and kRA of Fig. 1C, respectively
| Peptide | Alkaloid | FTCF† | Apparent kon, min−1·μM−1† | kon, min−1·μM−1‡ |
|---|---|---|---|---|
| KIIIA[K7A] | TTX | 0.48 ± 0.08 | 0.16 ± 0.02 | — |
| KIIIA[K7A] | STX | 0.46 ± 0.07 | 0.15 ± 0.02 | — |
| KIIIA[K7A] | GTX2/3 | 0.30 ± 0.07 | 0.09 ± 0.03 | — |
| KIIIA[K7A] | None | — | — | 0.13 ± 0.006 |
| KIIIA | TTX | 0.56 ± 0.03 | 0.20 ± 0.01 | — |
| KIIIA | STX | — | 0.09 (0.03–0.3)§ | — |
| KIIIA | GTX2/3 | 0.31 ± 0.03 | 0.10 ± 0.01 | — |
| KIIIA | None | — | — | 0.30 ± 0.030 |
Values are means ± SD, n ≥3 oocytes, except for the following.
Values in parentheses, which span a decade, indicate proposed limits of uncertainty for kon; see Fig. 4, C and D.
From results for formation of ternary complex illustrated in Fig. 4, A and B, processed as described in methods.
From slope of kobs versus [peptide] for formation of binary peptide·NaV complex, from Zhang et al. (2009).
As noted previously, with KIIIA and STX, the off rate for the recovery from block of the ternary complex was only sixfold slower than that of the binary STX·NaV complex (Table 2). Consequently, when the experimental protocol used earlier was used with these ligands, the washout data were not fit by a double- any better than by a single-exponential curve, presumably in part because of experimental noise. Nevertheless, the formation of the ternary KIIIA·STX·NaV complex could be discerned from the observation that the rate constant of the fit to a single-exponential curve decreased as the concentration of KIIIA increased (Fig. 4, C and D). The reaction scheme in Fig. 1C was numerically simulated for KIIIA and STX as ligands using the algorithm described in methods. The following rate constants were used in the simulation: kRA and kAR, 0.3 μM−1·min−1 and 0.0016 min−1, respectively (Zhang et al. 2009); kRB, 254 μM−1·min−1 (Table 1); kBR, 2.0 min−1 (Table 2); kAC and kCA, both 0 (since STX was unable to block rINa with KIIIA, as described earlier); kCB, 0.35 min−1 (from the limiting koff value in Fig. 4D at high [KIIIA]); and, finally, kBC (i.e., kon for the reaction KIIIA + STX·NaV → KIIIA·STX·NaV) was varied over a tenfold range from 0.03 to 0.3 μM−1·min−1, where the latter value is that of kRA (i.e., the kon for the reaction KIIIA + NaV → KIIIA·NaV) (Zhang et al. 2009). Simulated decay curves for the recovery from block are illustrated in the inset of Fig. 4C. It is apparent by visual inspection that the pattern of the simulated curves with the kon of 0.09 μM−1·min−1 most closely resembles that of the experimental curves. On fitting each of the experimental and simulated decay curves in Fig. 4C to a single-exponential time course and plotting the resulting koff values as a function of [KIIIA], the formation of the ternary complex was reflected by the decreasing koff value as [KIIIA] increased, until a limiting value of koff was achieved at presumably saturating levels [KIIIA] (30 μM) (Fig. 4D). In the figure, the congruence between the experimental koff values (circles) and those obtained by simulation (solid line) suggests that the kon for the reaction KIIIA + STX·NaV → KIIIA·STX·NaV was about 0.09 μM−1·min−1.
Table 4 shows the equilibrium binding constants (i.e., koff/kon ratios or Kd values) for each of the four limbs of the reaction scheme in Fig. 1C. The table also indicates the predicted rate constants obtained by numerical simulation that were used, along with rate constants presented in previous tables, to derive the Kd values. It might be noted that direct determinations of steady-state IC50 values for alkaloids, corresponding to the Kd values in saturating [peptide], were not obtained because the time it would have taken to reach steady-state block of rINa, using alkaloid concentrations near the anticipated IC50 values (≈Kd values), was hours and outside the roughly 1-h time window of our experiments. Corresponding IC50 values for peptides in saturating [alkaloid] were not obtained because, of course, no sodium currents are detectable under those circumstances.
DISCUSSION
In this study, we used two μ-conotoxins and two alkaloids and tested all four peptide–alkaloid combinations on NaV1.2. The results, together with those obtained previously with TTX (Zhang et al. 2009), support the idea that bound conopeptide not only increases the access resistance to sodium ions, as evident by the diminutive rINa, but moreover also restricts the alkaloid's access to its binding site on the channel. The results also support the notion that in the ternary peptide·alkaloid·NaV complex, the peptide traps the alkaloid at the latter's binding site and the alkaloid does not dissociate from the channel until the peptide has dissociated.
Implications of the rates of functional recovery following toxin washout of ternary complexes
A ternary complex could be formed by either order of toxin addition in four peptide–alkaloid pairs: one pair with KIIIA (in which the alkaloid was TTX) and three pairs with KIIIA[K7A] (in which the alkaloid was, respectively, TTX, STX, and GTX2/3). In each instance, essentially the same koff value was obtained for a given pair (Table 2), suggesting that for each pair the same ternary complex was formed regardless of order of toxin addition. Thus all totaled, it appears that six unique ternary (peptide·alkaloid·NaV) complexes can be formed with the two peptides and three alkaloids tested thus far.
Comparison of the koff values of the six ternary peptide·alkaloid·NaV complexes indicates that by far the least stable ternary complex was KIIIA·STX·NaV (Table 2). The other ternary complexes had remarkably similar koff values, as if TTX and GTX2/3 can shield Lys7 of KIIIA from the same site(s) on the channel. The presence of alkaloid destabilized the KIIIA[K7A]·alkaloid·NaV complex minimally with TTX and hardly at all with STX and GTX2/3. Compared with TTX and GTX2/3, STX has an extra positive charge, as does KIIIA compared with KIIIA[K7A]; thus we hypothesize that charge repulsion accounts for the relatively high instability of the ternary KIIIA·STX·NaV complex.
Implications of the on rates for the formation of the ternary complex
TTX blocked the rINa with KIIIA at a rate 104-fold slower than it blocked control INa (Zhang et al. 2009) and the block of rINa by STX and GTX2/3 were not observed even at the highest concentrations tested (10 μM) (Table 1). Thus KIIIA bound to the channel severely restricted the access of TTX and that of STX and GTX2/3 even more so. In striking contrast, for the block of the channel preoccupied by KIIIA[K7A], the kon values of all three alkaloids were more modestly reduced and, furthermore, to essentially the same value (Fig. 3F, Table 1). A possible mechanism for this is that the binary peptide·NaV complex flickers between two states: a permissive state wherein an alkaloid can “sneak” by the peptide and a state in which the alkaloid cannot. The rate at which an alkaloid sneaks by would be dictated by the fraction of time the complex dwells in the permissive state and how readily the alkaloid can permeate that state. The permissive state with KIIIA may be permeable to TTX but not to STX or GTX2/3, whereas that with KIIIA[K7A] may be equally permeable to all three alkaloids. This would explain why the alkaloids have similar on rates in blocking the rINa with KIIIA[K7A], even though they block the control INa at very different rates; i.e., the rate-limiting factor for rINa block is the fractional dwell time of the permissive state, which is intrinsic to the peptide and independent of the specific alkaloid.
Examination of the rates of recovery from block allowed the apparent kon for the reaction
to be estimated (Fig. 4). When alkaloid was already bound to the channel, the rate at which peptide could subsequently bind to the channel appeared to be only modestly impeded, if at all. With KIIIA[K7A] and STX, GTX2/3, or TTX (Fig. 4A) and with KIIIA and GTX2/3 or TTX (Fig. 4B), the apparent kon values were within a factor of 2 of each other (Table 3). This was consistent with previous results with KIIIA and KIIIA[K7A], where the presence of TTX on the channel was observed to reduce the apparent kon of the peptide by only about 25% by Zhang et al. (2009). STX bound to the channel reduced the apparent kon of KIIIA by about threefold, from 0.3 μM−1·min−1 under control conditions (Zhang et al. 2009) to about 0.1 μM−1·min−1 (Fig. 4, C and D).
Equilibrium constants derived for the proposed reaction scheme in Fig. 1C
The upper and lower limbs on the left side of the reaction scheme in Fig. 1C represent simply the binding of peptide and alkaloid, respectively, to the channel. The upper and lower limbs on the right represent binding of alkaloid and peptide, respectively, to the channel preoccupied with the opposite ligand; their convergence to form a single species of ternary complex for a given peptide/alkaloid pair, although hypothetical, is compatible with the results of our experiments. Table 4 lists the apparent binding constants of the reactions in all four limbs of the scheme. The Kd of TTX for the channel saturated with KIIIA was 20-fold larger than that without the peptide, where the presence of KIIIA decreased TTX's kon (i.e., block of control INa vs. rINa) about 20,000-fold (Table 1) and koff [i.e., koff of binary (Table 2) vs. ternary complex (predicted kCA, footnote Table 4)] about 1,000-fold. (Note that the SD of the predicted kCA is responsible for the large SD in Kd here.) In contrast, the presence of KIIIA[K7A] only modestly altered the Kd values of TTX, STX, and GTX—the apparent kon (Table 1) and predicted koff (kCA, Table 4) values for a given alkaloid were both decreased by two orders of magnitude or more. This raises the possibility that the binding site of the alkaloids per se may not have been strongly affected by KIIIA[K7A]; instead, access to and egress from the binding site was retarded by the presence of the peptide. This conjecture is compatible with the flicker mechanism for the binary peptide·NaV complex suggested in the preceding section.
For each ternary complex, the predicted kCB value (Table 4) was close to the measured koff value for the recovery of INa during toxin washout (Table 2), consistent with the idea that the rate-limiting step in disassembly of the ternary complex is the dissociation of peptide. Likewise, kCB was always greater than kCA except for the KIIIA[K7A]·TTX·NaV complex, in which the two rate constants were predicted to be similar (Table 4).
The Kd of KIIIA[K7A] binding was approximately the same irrespective of whether the channel was preoccupied with alkaloid, regardless of the alkaloid species; here, neither kon nor predicted koff was affected much by the presence of alkaloid (Table 4B). In stark contrast, the Kd of KIIIA was increased close to 20-, 800-, and 60-fold by TTX, STX, and GTX2/3, respectively; in each case, an increased koff value was responsible for most of the elevation in Kd (compare kCB values in Table 4 with the koff, i.e., kAR, value in Table 2).
Our overall results support the conclusion that the presence of alkaloid minimally interfered with the accessibility of peptide, whereas the presence of peptide severely occluded the accessibility of alkaloid. This striking asymmetry is consistent with the picture of the ternary complex wherein the alkaloid is sandwiched between the peptide and selectivity filter of the channel (Zhang et al. 2009). We explored the feasibility of this “sandwich” model in silico, as described in the immediately following text.
Structural models of alkaloid and peptide docked at the outer vestibule of the channel
Following the advent of the cloning of VGSCs, experiments to pinpoint the binding site for TTX and STX have been carried out by several investigators; the general consensus is that the alkaloids bind at a site deep in the outer vestibule of the channel to cork its pore (Choudhary et al. 2002, 2003, 2007; Lipkind and Fozzard 1994, 2000; Penzotti et al. 1998, 2001; Santarelli et al. 2007; Terlau et al. 1991; Tikhonov and Zhorov 2005). On the other hand, studies with GIIIA and NaV1.4 indicate that the conopeptide binds at a more superficially located site (Chang et al. 1998; Choudhary et al. 2007; Dudley Jr et al. 1995, 2000; Hui et al. 2002; Li et al. 2001; Xue et al. 2003).
Given this background work by others, it behooved us to simply see whether both KIIIA and TTX or STX would fit in the vestibule. We present here our initial attempts to examine this by docking alkaloid and peptide in silico, as described in methods. These docking experiments are based on the sodium channel model proposed by Lipkind and Fozzard (2000). The model, which consists of the S5- and S6-fragments and P-loop present in each of the four domains of the channel, was built starting with the structural framework of the KcsA potassium channel (Doyle et al. 1998). The approach of using such models to analyze the results of electrophysiological measurements of sodium channel mutants has proven instructive (see references published after 2000 cited in preceding paragraph).
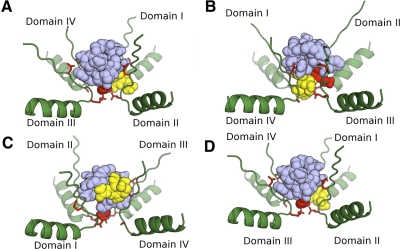
Illustrated in Fig. 5 are the pore loops at the outer vestibule of NaV1.2, including the DEKA “ring” forming the ion selectivity filter (Hille 2001) and the more superficial EEDD ring of negative charges that are common to all mammalian NaVs (except NaV1.9, where the latter ring is EEED). STX and TTX were each docked before KIIIA (Fig. 5, A and B, respectively) as well as after KIIIA (Fig. 5, C and D, respectively). Indeed, both KIIIA and alkaloid can simultaneously fit in the vestibule. Furthermore, when KIIIA was docked first, TTX could also dock close to the selectivity filter (Fig. 5D); however, this was not the case with STX (Fig. 5C). Thus these results are, to a first approximation, consistent with our experimental observations. It would be interesting to examine the docking behaviors of the alkaloids when KIIIA[K7A] is docked first, as well as the docking of other peptides and alkaloids that have yet to be tested experimentally (see following text).
Fig. 5.
Possible arrangements of KIIIA (lavender) and STX or TTX (yellow) nestled in outer vestibule of NaV1.2 (green). Only the 4 pore loops of the channel are illustrated: backbone ribbons are alpha-helices and beta-strands, with associated red sticks representing side chains of EEDD ring at the top and DEKA ring of selectivity filter at the bottom (only 3 of the 4 side chains of residues in each ring are visible in each panel). Models were constructed using AutoDock4, as described in methods where: STX was docked before KIIIA (A); TTX was docked before KIIIA (B); KIIIA was docked before STX (C); and KIIIA was docked before TTX (D). Each panel shows the ternary complex from a perspective that provided good visibility of alkaloid, peptide, and DEKA ring. Note, alkaloid managed to dock close to DEKA ring in all panels except C.
Aside from GTX2/3, 15 other naturally occurring congeners of STX have been identified (Yasumoto and Murata 1993). Experiments with: 1) these congeners as well as possible synthetic derivatives of both STX and TTX (good progress in guanidinium alkaloid syntheses is promising in this regard; Andresen and Du Bois 2009; Mulcahy and Du Bois 2008; Nishikawa et al. 2004; Sato et al. 2008); 2) close relatives of KIIIA, such as the μ-conotoxins SIIIA and SmIIIA (Bulaj et al. 2005; West et al. 2002); and 3) mutants of the channel, such as NaV1.2[F385C] (Zhang et al. 2009), should advance our understanding of the mechanism of action of guanidinium alkaloids and μ-conopeptides that target site 1 of VGSCs. Furthermore, if active, covalently linked alkaloid–peptide adducts can be synthesized, these should provide insights into the arrangement of the units when bound to site 1, as well as also serve as second-generation ligands for sodium channels.
We introduced the descriptor syntoxin to denote the phenomenon wherein toxins such as TTX and KIIIA can act in concert (Zhang et al. 2009). From a broader perspective, investigation of the syntoxin interactions of guanidinium alkaloids and conopeptides with VGSCs may contribute to more than just our understanding of the structure and function of sodium channels per se. For example, STX is a major active factor in paralytic shellfish poisoning, a serious public health hazard (Al-Sabi et al. 2006; Bricelj et al. 2005). KIIIA is a lead compound in the development of an antidote to STX in the form of a contratoxin; i.e., a ligand that shields the target receptor against a toxin (Zhang et al. 2009). KIIIA's ability to prevent STX binding augurs well for its development into a contratoxin, although it remains to be seen whether KIIIA can be sculpted to minimize its efficacy in interfering with sodium conductance (that is, engineered to increase its rINa) without excessively compromising both its affinity for the channel and its ability to interfere with STX binding. It also remains to be shown how conserved are the properties of the toxins when tested against sodium channels expressed in mammalian cells (as opposed to Xenopus oocytes). In any event, the exploration of the syntoxin and contratoxin properties of μ-conotoxins and their derivatives opens a new window through which to view the pharmacology of sodium channels.
GRANTS
This work was supported by National Institutes of Health Grants GM-48677 to G. Bulaj, B. M. Olivera, and D. Yoshikami and R21 NS-055845 to G. Bulaj and D. Yoshikami.
DISCLOSURES
G. Bulaj is a cofounder of NeuroAdjuvants, Inc. B. M. Olivera is a cofounder of Cognetix, Inc.
ACKNOWLEDGMENTS
We thank A. L. Goldin for the NaV1.2 clone, L. Azam for cRNA, and R. J. French and J. R. McArthur for providing the structural coordinates for NaV1.4.
Present address of A. Walewska: Faculty of Chemistry, University of Gdansk, 80-952 Gdansk, Poland.
REFERENCES
- Al-Sabi et al., 2006.Al-Sabi A, McArthur J, Ostroumov V, French RJ. Marine toxins that target voltage-gated sodium channels. Mar Drugs 4: 157–192, 2006 [Google Scholar]
- Andresen and Du Bois, 2009.Andresen BM, Du Bois J. De novo synthesis of modified saxitoxins for sodium ion channel study. J Am Chem Soc 131: 12524–12525, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordner et al., 1975.Bordner J, Thiessen WE, Bates HA, Rapoport H. The structure of a crystalline derivative of saxitoxin. The structure of saxitoxin. J Am Chem Soc 97: 6008–6012, 1975 [DOI] [PubMed] [Google Scholar]
- Bricelj et al., 2005.Bricelj VM, Connell L, Konoki K, Macquarrie SP, Scheuer T, Catterall WA, Trainer VL. Sodium channel mutation leading to saxitoxin resistance in clams increases risk of PSP. Nature 434: 763–767, 2005 [DOI] [PubMed] [Google Scholar]
- Bulaj et al., 2005.Bulaj G, West PJ, Garrett JE, Watkins M, Zhang MM, Norton RS, Smith BJ, Yoshikami D, Olivera BM. Novel conotoxins from Conus striatus and Conus kinoshitai selectively block TTX-resistant sodium channels. Biochemistry 44: 7259–7265, 2005 [DOI] [PubMed] [Google Scholar]
- Catterall, 1980.Catterall WA. Neurotoxins that act on voltage-sensitive sodium channels in excitable membranes. Annu Rev Pharmacol Toxicol 20: 15–43, 1980 [DOI] [PubMed] [Google Scholar]
- Catterall et al., 2005.Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure–function relationships of voltage-gated sodium channels. Pharmacol Rev 57: 397–409, 2005 [DOI] [PubMed] [Google Scholar]
- Cestele and Catterall, 2000.Cestele S, Catterall WA. Molecular mechanisms of neurotoxin action on voltage-gated sodium channels. Biochimie 82: 883–892, 2000 [DOI] [PubMed] [Google Scholar]
- Chang et al., 1998.Chang NS, French RJ, Lipkind GM, Fozzard HA, Dudley SC., Jr Predominant interactions between mu-conotoxin Arg-13 and the skeletal muscle Na+ channel localized by mutant cycle analysis. Biochemistry 37: 4407–4419, 1998 [DOI] [PubMed] [Google Scholar]
- Choudhary et al., 2007.Choudhary G, Aliste MP, Tieleman DP, French RJ, Dudley SC., Jr Docking of mu-conotoxin GIIIA in the sodium channel outer vestibule. Channels 1: 344–352, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary et al., 2002.Choudhary G, Shang L, Li X, Dudley SC., Jr Energetic localization of saxitoxin in its channel binding site. Biophys J 83: 912–919, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary et al., 2003.Choudhary G, Yotsu-Yamashita M, Shang L, Yasumoto T, Dudley SC., Jr Interactions of the C-11 hydroxyl of tetrodotoxin with the sodium channel outer vestibule. Biophys J 84: 287–294, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle et al., 1998.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280: 69–77, 1998 [DOI] [PubMed] [Google Scholar]
- Dudley et al., 2000.Dudley SC, Jr, Chang N, Hall J, Lipkind G, Fozzard HA, French RJ. mu-Conotoxin GIIIA interactions with the voltage-gated Na(+) channel predict a clockwise arrangement of the domains. J Gen Physiol 116: 679–690, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley et al., 1995.Dudley SC, Jr, Todt H, Lipkind G, Fozzard HA. A mu-conotoxin-insensitive Na+ channel mutant: possible localization of a binding site at the outer vestibule. Biophys J 69: 1657–1665, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler et al., 2008.Fiedler B, Zhang MM, Buczek O, Azam L, Bulaj G, Norton RS, Olivera BM, Yoshikami D. Specificity, affinity and efficacy of iota-conotoxin RXIA, an agonist of voltage-gated sodium channels Na(V)1.2, 1.6 and 1.7. Biochem Pharmacol 75: 2334–2344, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French and Terlau, 2004.French RJ, Terlau H. Sodium channel toxins: receptor targeting and therapeutic potential. Curr Med Chem 11: 3053–3064, 2004 [DOI] [PubMed] [Google Scholar]
- Hall et al., 1990.Hall S, Strichartz GR, Moczydlowski E, Ravindran A, Reichardt PB. The saxitoxins: sources, chemistry, and pharmacology. In: Marine Toxins (ACS Symposium Series 418), edited by Hall S, Strichartz GR. Washington, DC: American Chemical Society, 1990, p. 29–65 [Google Scholar]
- Heinemann et al., 1992.Heinemann SH, Terlau H, Imoto K. Molecular basis for pharmacological differences between brain and cardiac sodium channels. Pflügers Arch 422: 90–92, 1992 [DOI] [PubMed] [Google Scholar]
- Hille, 1975.Hille B. The receptor for tetrodotoxin and saxitoxin. A structural hypothesis. Biophys J 15: 615–619, 1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, 2001.Hille B. Ion Channels of Excitable Membranes (3rd ed.). Sunderland, MA: Sinauer; 2001 [Google Scholar]
- Hui et al., 2002.Hui K, Lipkind G, Fozzard HA, French RJ. Electrostatic and steric contributions to block of the skeletal muscle sodium channel by mu-conotoxin. J Gen Physiol 119: 45–54, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo et al., 2009.Khoo KK, Feng ZP, Smith BJ, Zhang MM, Yoshikami D, Olivera BM, Bulaj G, Norton RS. Structure of the analgesic mu-conotoxin KIIIA and effects on the structure and function of disulfide deletion. Biochemistry 48: 1210–1219, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al., 2001.Li RA, Ennis IL, French RJ, Dudley SC, Jr, Tomaselli GF, Marban E. Clockwise domain arrangement of the sodium channel revealed by (mu)-conotoxin (GIIIA) docking orientation. J Biol Chem 276: 11072–11077, 2001 [DOI] [PubMed] [Google Scholar]
- Lipkind and Fozzard, 1994.Lipkind GM, Fozzard HA. A structural model of the tetrodotoxin and saxitoxin binding site of the Na+ channel. Biophys J 66: 1–13, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkind and Fozzard, 2000.Lipkind GM, Fozzard HA. KcsA crystal structure as framework for a molecular model of the Na(+) channel pore. Biochemistry 39: 8161–8170, 2000 [DOI] [PubMed] [Google Scholar]
- Moczydlowski et al., 1986.Moczydlowski E, Olivera BM, Gray WR, Strichartz GR. Discrimination of muscle and neuronal Na-channel subtypes by binding competition between [3H]saxitoxin and mu-conotoxins. Proc Natl Acad Sci USA 83: 5321–5325, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris et al., 1998.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19: 1639–1662, 1998 [Google Scholar]
- Morris et al., 2009.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2583–2791, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy and Du Bois, 2008.Mulcahy JV, Du Bois J. A stereoselective synthesis of (+)-gonyautoxin 3. J Am Chem Soc 130: 12630–12631, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa et al., 2004.Nishikawa T, Urabe D, Isobe M. An efficient total synthesis of optically active tetrodotoxin. Angew Chem Int Ed Engl 43: 4782–4785, 2004 [DOI] [PubMed] [Google Scholar]
- Penzotti et al., 1998.Penzotti JL, Fozzard HA, Lipkind GM, Dudley SC., Jr Differences in saxitoxin and tetrodotoxin binding revealed by mutagenesis of the Na+ channel outer vestibule. Biophys J 75: 2647–2657, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzotti et al., 2001.Penzotti JL, Lipkind G, Fozzard HA, Dudley SC., Jr Specific neosaxitoxin interactions with the Na+ channel outer vestibule determined by mutant cycle analysis. Biophys J 80: 698–706, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safo et al., 2000.Safo P, Rosenbaum T, Shcherbatko A, Choi DY, Han E, Toledo-Aral JJ, Olivera BM, Brehm P, Mandel G. Distinction among neuronal subtypes of voltage-activated sodium channels by mu-conotoxin PIIIA. J Neurosci 20: 76–80, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli et al., 2007.Santarelli VP, Eastwood AL, Dougherty DA, Horn R, Ahern CA. A cation-pi interaction discriminates among sodium channels that are either sensitive or resistant to tetrodotoxin block. J Biol Chem 282: 8044–8051, 2007 [DOI] [PubMed] [Google Scholar]
- Sato et al., 2008.Sato K, Akai S, Shoji H, Sugita N, Yoshida S, Nagai Y, Suzuki K, Nakamura Y, Kajihara Y, Funabashi M, Yoshimura J. Stereoselective and efficient total synthesis of optically active tetrodotoxin from D-glucose. J Org Chem 73: 1234–1242, 2008 [DOI] [PubMed] [Google Scholar]
- Schantz et al., 1975.Schantz EJ, Ghazarossian VE, Schnoes HK, Strong FM, Springer JP, Pezzanite JO, Clardy J. The structure of saxitoxin (Letter). J Am Chem Soc 97: 1238, 1975 [DOI] [PubMed] [Google Scholar]
- Terlau et al., 1991.Terlau H, Heinemann SH, Stuhmer W, Pusch M, Conti F, Imoto K, Numa S. Mapping the site of block by tetrodotoxin and saxitoxin of sodium channel II. FEBS Lett 293: 93–96, 1991 [DOI] [PubMed] [Google Scholar]
- Terlau and Olivera, 2004.Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev 84: 41–68, 2004 [DOI] [PubMed] [Google Scholar]
- Tikhonov and Zhorov, 2005.Tikhonov DB, Zhorov BS. Modeling P-loops domain of sodium channel: homology with potassium channels and interaction with ligands. Biophys J 88: 184–197, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West et al., 2002.West PJ, Bulaj G, Garrett JE, Olivera BM, Yoshikami D. mu-Conotoxin SmIIIA, a potent inhibitor of tetrodotoxin-resistant sodium channels in amphibian sympathetic and sensory neurons. Biochemistry 41: 15388–15393, 2002 [DOI] [PubMed] [Google Scholar]
- Xue et al., 2003.Xue T, Ennis IL, Sato K, French RJ, Li RA. Novel interactions identified between micro-conotoxin and the Na+ channel domain I P-loop: implications for toxin-pore binding geometry. Biophys J 85: 2299–2310, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao et al., 2008.Yao S, Zhang MM, Yoshikami D, Azam L, Olivera BM, Bulaj G, Norton RS. Structure, dynamics, and selectivity of the sodium channel blocker mu-conotoxin SIIIA. Biochemistry 47: 10940–10949, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto and Murata, 1993.Yasumoto T, Murata M. Marine toxins. Chem Rev 93: 1897–1909, 1993 [Google Scholar]
- Zhang et al., 2007.Zhang MM, Green BR, Catlin P, Fiedler B, Azam L, Chadwick A, Terlau H, McArthur JR, French RJ, Gulyas J, Rivier JE, Smith BJ, Norton RS, Olivera BM, Yoshikami D, Bulaj G. Structure/function characterization of micro-conotoxin KIIIA, an analgesic, nearly irreversible blocker of mammalian neuronal sodium channels. J Biol Chem 282: 30699–30706, 2007 [DOI] [PubMed] [Google Scholar]
- Zhang et al., 2009.Zhang MM, McArthur JR, Azam L, Bulaj G, Olivera BM, French RJ, Yoshikami D. Synergistic and antagonistic interactions between tetrodotoxin and mu-conotoxin in blocking voltage-gated sodium channels. Channels 3: 32–38, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]