SUMMARY
RUNX1/AML1 is required for definitive hematopoiesis and is frequently targeted by chromosomal translocation in acute myeloid leukemias (AML). The t(8;21) related AML1-ETO fusion protein blocks differentiation of myeloid progenitors. Here, we show by immunofluorescence microscopy that during interphase, endogenous AML1-ETO localizes to nuclear microenvironments distinct from those containing native RUNX1/AML1 protein. At mitosis, we clearly detect binding of AML1-ETO to nucleolar organizing regions (NORs) in AML derived Kasumi-1 cells and binding of RUNX1/AML1 to NORs in Jurkat cells. Both RUNX1/AML1 and AML1-ETO occupy ribosomal DNA repeats during interphase, as well as interact with the endogenous RNA Pol I transcription factor UBF-1. Promoter cytosine methylation analysis indicates that RUNX1/AML1 binds to rDNA repeats that are more highly CpG methylated than those bound by AML1-ETO. Down-regulation by RNA interference reveals that RUNX1/AML1 negatively regulates rDNA transcription, while AML1-ETO is a positive regulator in Kasumi-1 cells. Taken together, our findings identify a novel role for the leukemia-related AML1-ETO protein in epigenetic control of cell growth through upregulation of RNA Pol I-mediated ribosomal gene transcription, consistent with the hyper-proliferative phenotype of myeloid cells in AML patients.
Keywords: acute myelogenous leukemia, Runx1, ribosomal DNA transcription, RNA polymerase I, UBF1, nucleolar organizing region
INTRODUCTION
The most frequent target of chromosomal translocations in acute myeloid leukemia (AML) is the Runt-related transcription factor RUNX1/AML1, a key regulator of hematopoiesis (Setoguchi et al., 2008; Pabst and Mueller, 2007; Growney et al., 2005; Ito, 2004; Nucifora and Rowley, 1995; Romana et al., 1995). RUNX1/AML1 directly regulates multiple distinct myeloid and lymphoid genes that are involved in hematopoietic lineage commitment (Huang et al., 2007; Otto et al., 2003; Frank et al., 1995; Nuchprayoon et al., 1994). The protein contains an N-terminal DNA-binding domain (runt homology domain) and a C-terminal regulatory domain that contains a nuclear matrix targeting signal (NMTS) and several context-dependent transcriptional activation or repression domains (Wheeler et al., 2000; Meyers and Hiebert, 2000; Stein et al., 1999).
The 8;21 leukemic translocation fuses the RUNX1/AML1 gene to MTG8/ETO coding sequences resulting in the AML1-ETO fusion protein (Miyoshi et al., 1993; Erickson et al., 1992; Licht, 2001; Davis et al., 2003; Peterson et al., 2007a). AML1-ETO retains the DNA binding function of the RUNX1/AML1 protein but does not contain the transactivation domain or the nuclear matrix targeting signal (NMTS) of RUNX1/AML1 (Zeng et al., 1997; McNeil et al., 1999). Previous studies have shown that exogenously expressed RUNX1/AML1 and AML1-ETO exhibit differential subnuclear targeting which may be responsible in part for the aberrant function of the fusion protein (McNeil et al., 1999; Barseguian et al., 2002). Altered subnuclear targeting of AML1 in patients with the 8;21 translocation may contribute to the pathology of AML, because RUNX1/AML1 mutations that alter subnuclear routing and fidelity of transcriptional control result in a differentiation block and increase proliferation of myeloid progenitors (Vradii et al., 2005; Zaidi et al., 2007; Zaidi et al., 2005). In addition, a large number of co-factors interact with gene regulatory domains of RUNX1/AML1, including the C-terminus that is removed in AML1-ETO (Wotton et al., 1994; Giese et al., 1995; Hiebert et al., 1996; Rhoades et al., 1996; Petrovick et al., 1998; Rubnitz and Look, 1998; Osato et al., 1999). Through the recruitment of unique co-regulators that interact with the ETO moiety in lieu of the AML1 C-terminus, the AML1-ETO fusion protein antagonizes the transcriptional function of native RUNX1/AML1 (Hiebert et al., 2001). Thus, there are several plausible mechanisms by which the pathological formation of the AML1-ETO protein may block differentiation of myeloid progenitors and promote leukemia.
In addition to regulating hematopoiesis-specific genes, RUNX1/AML1 is also implicated in the regulation of cell-cycle genes, including p21WAF1/CIP1, which encodes a cyclin-dependent kinase inhibitor important for checkpoint control and terminal differentiation (Lutterbach et al., 2000; Peterson et al., 2007b). RUNX1/AML1 controls cell cycle progression by shortening the G1/S phase in hematopoietic cells and is negatively regulated by cyclin D3 (Strom et al., 2000; Peterson et al., 2005). Levels of RUNX1/AML1 increase as cells progress into S phase and are downregulated at the G2/M transition (Bernardin-Fried et al., 2004; Biggs et al., 2006). The closely related osteoblastic transcription factor RUNX2 also controls proliferation and is cell cycle regulated with maximal levels in G1 (Pratap et al., 2003; Galindo et al., 2005). Furthermore, the presence of RUNX2 in osteogenic mesenchymal cells during mitosis may reinforce cell fate through an epigenetic mechanism that retains phenotypic gene expression patterns after cell division (Young et al., 2007b; Young et al., 2007a). Because RUNX1/AML1 has also been detected during mitosis (Zaidi et al., 2003), it may perform an analogous function in hematopoietic cells.
In this study, we examined the biological functions of Runx1/AML1 and AML1-ETO during mitosis and interphase in relation to their subcellular localization in hematopoietic cells. Among the main findings from our study is the observation that both proteins can associate with mitotic chromosomes and regulate transcription of ribosomal RNA genes, a fundamental process that supports the growth of cells and is tightly coupled with cell differentiation. Our results indicate that RUNX1/AML1 mediates epigenetic mechanisms that convey regulatory information to progeny cells and that these mechanisms are perturbed by the leukemia related AML1-ETO fusion protein.
RESULTS
During interphase RUNX/AML proteins localize in nuclear micro-environments where the transcriptional machinery is organized. As cells emerge from mitosis there is a stringent requirement for expression of RUNX1/AML1 responsive genes. While RUNX1/AML1 is destabilized during the G2/M transition (Bernardin-Fried et al., 2004), at least a subset of RUNX1/AML1 remains associated with mitotic chromosomes (Zaidi et al., 2003). However, many AML patients express the AML1-ETO fusion protein and the important question arises whether this leukemia associated protein is similarly capable of interacting with mitotic chromosomes.
Endogenous RUNX1/AML1 and AML1-ETO are targeted to distinct nuclear microenvironments
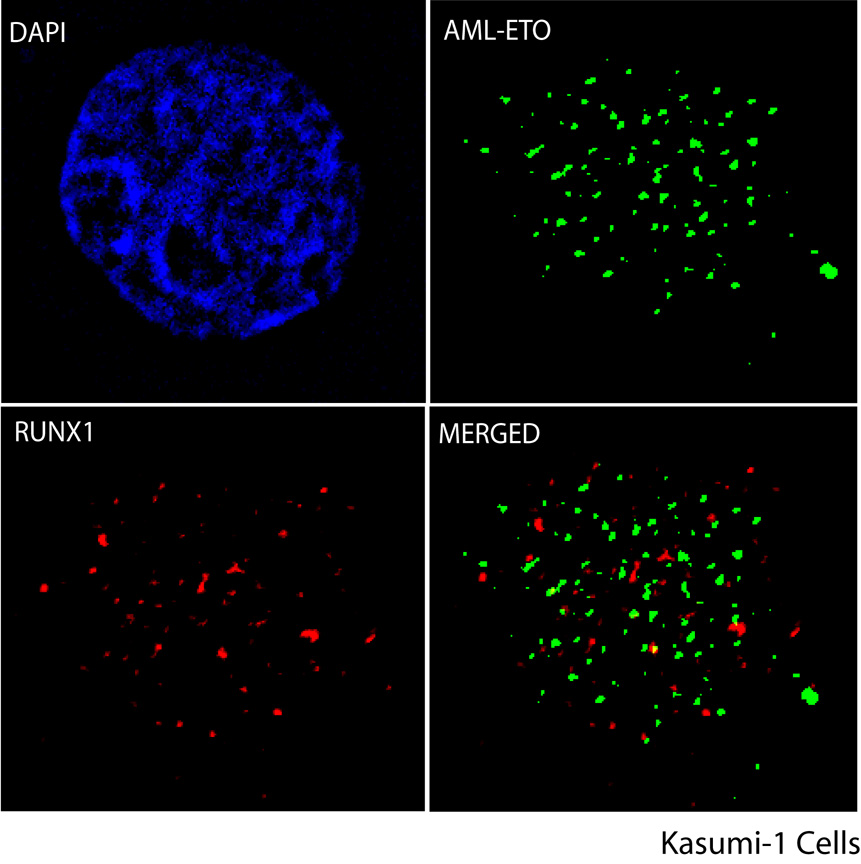
To begin examining the localization of AML1-ETO during interphase and mitosis, we used immunofluorescence microscopy to monitor the subcellular distribution of endogenously expressed RUNX1/AML1 and AML1-ETO in asynchronously growing Kasumi-1 cells that were derived from an AML patient with an 8;21 chromosomal translocation (Fig. 1). Double label-comparisons of immunofluorescence signals revealed absence of significant overlap in the distribution patterns of RUNX1/AML1 and AML1-ETO and most sites were detected as distinct green or red immunofluorescence signals (Fig. 1). Biochemical fractionation of Kasumi-1 cells followed by immunoblotting validated the nuclear distribution of RUNX1/AML1 and AML1-ETO (data not shown). Hence, both RUNX1/AML1 and the AML1-ETO fusion protein are predominantly present in the nucleus, but are each directed to distinct subnuclear compartments. The distinct locations may reflect differences in the regulatory activities of these factors in leukemic cells.
Figure 1. RUNX1/AML1 and AML1-ETO are targeted to distinct subnuclear locations.
Immunofluorescence microscopy for endogenous RUNX1/AML1 (red) and AML1-ETO (green) with DAPI staining (blue) in Kasumi-1 cells as well as merged images are shown. RUNX1/AML1 and AML1-ETO fusion protein are predominantly present in the nucleus but do not co-localize in the Kasumi-1 cells. An antibody detecting ETO was used to visualize the AML1-ETO fusion protein, whereas an antibody recognizing the C-terminal domain of RUNX1/AML1 was used to detect endogenous RUNX1/AML1.
RUNX1/AML1 and AML1-ETO are specifically associated with metaphase chromosomes
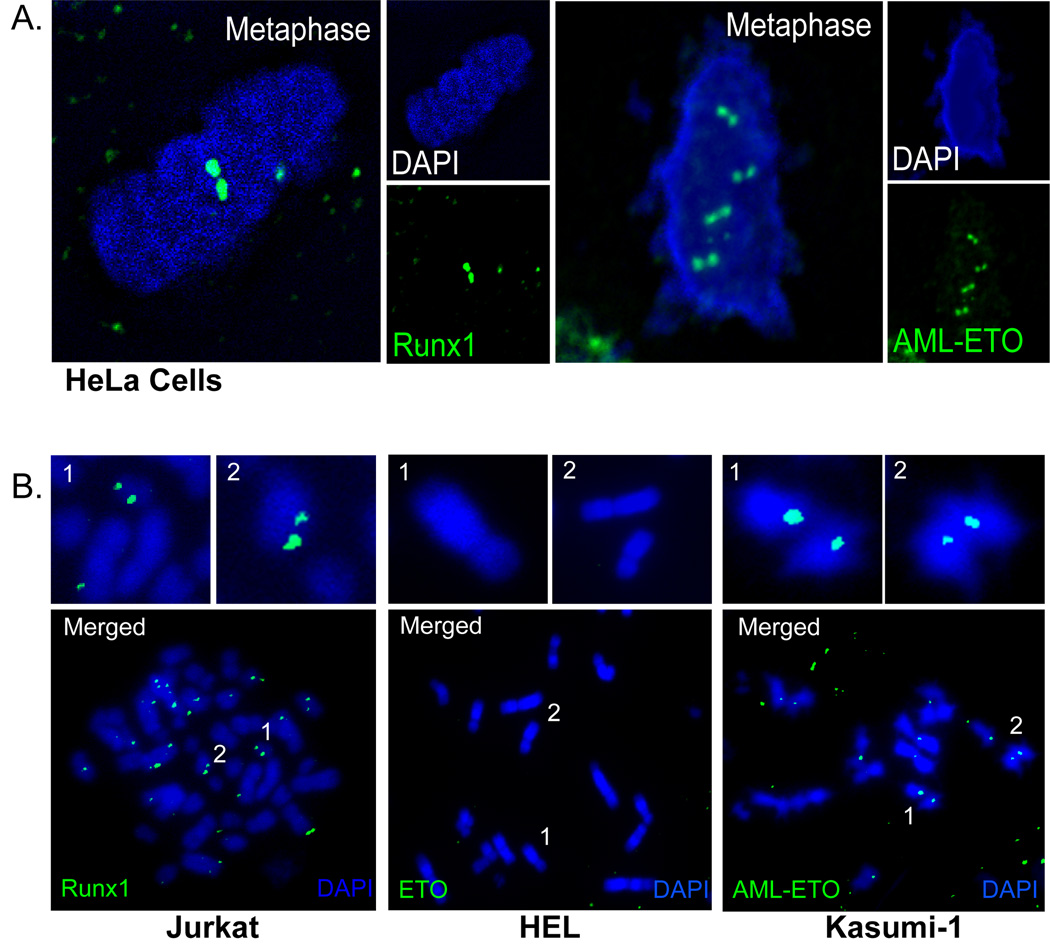
To assess whether the leukemia-associated AML1-ETO fusion protein interacts with mitotic chromosomes, we investigated its presence in metaphase chromosomes by immunofluorescence microscopy (Fig. 2). We first examined the association of RUNX1/AML1 and AML1-ETO with mitotic chromosomes by transfecting actively growing HeLa cells with epitope tagged expression constructs. Immunofluorescence microscopy reveals that ectopically expressed RUNX1/AML1 and AML1-ETO each associate with chromosomes in mitotic cells and are detected as paired foci (Fig. 2A). The pairwise organization of these foci is similar to that reported for Runx2 during mitosis (Young et al., 2007a).
Figure 2. RUNX1/AML1 and AML1-ETO associate with metaphase chromosomes in pairs.
A) Epitope-tagged RUNX1/AML1 and AML1-ETO were examined by in situ immunofluorescence in HeLa cells and metaphase cells were visually selected. RUNX1/AML1 and AML1-ETO foci (green) are associated with chromosomes (blue) in pairs (shown in deconvoluted images). B) Mitotic chromosome spreads were prepared for human Jurkat, HEL, and Kasumi-1 cells and processed for immunofluorescence microscopy using antibodies directed against the RUNX1/AML1, ETO and AML1-ETO proteins. RUNX1/AML1 and AML1-ETO show distinct foci on the mitotic chromosome spreads, while ETO is not detected on mitotic chromosomes. Panels on the top of each image show enlarged areas marked by numbers in the image.
To determine the endogenous cellular organization of RUNX1/AML1 and AML1-ETO proteins during mitosis, we prepared metaphase chromosomal spreads for human Jurkat cells endogenously expressing RUNX1/AML1, HEL cells exhibiting physiological expression of ETO, and Kasumi-1 cells expressing the AML1-ETO translocation fusion protein. Endogenous RUNX1/AML1 (in Jurkat cells) and AML1-ETO (in Kasumi-1 cells) each show distinct paired foci on metaphase chromosome spreads, while ETO (in HEL cells) is not detected (Fig. 2B). Interestingly, RUNX1/AML1 cannot be detected on mitotic chromosomes in Kasumi-1 cells (data not shown), suggesting that AML1-ETO may exclude RUNX1/AML1 from its sites at mitosis (see below). Regardless of this possibility, our data clearly show that AML1-ETO specifically associates with mitotic chromatin in human leukemia cells, and thus provide the first indication that AML1-ETO may interfere with the normal function of RUNX1/AML1 at mitosis.
RUNX1/AML1 and AML1-ETO each bind to human ribosomal DNA
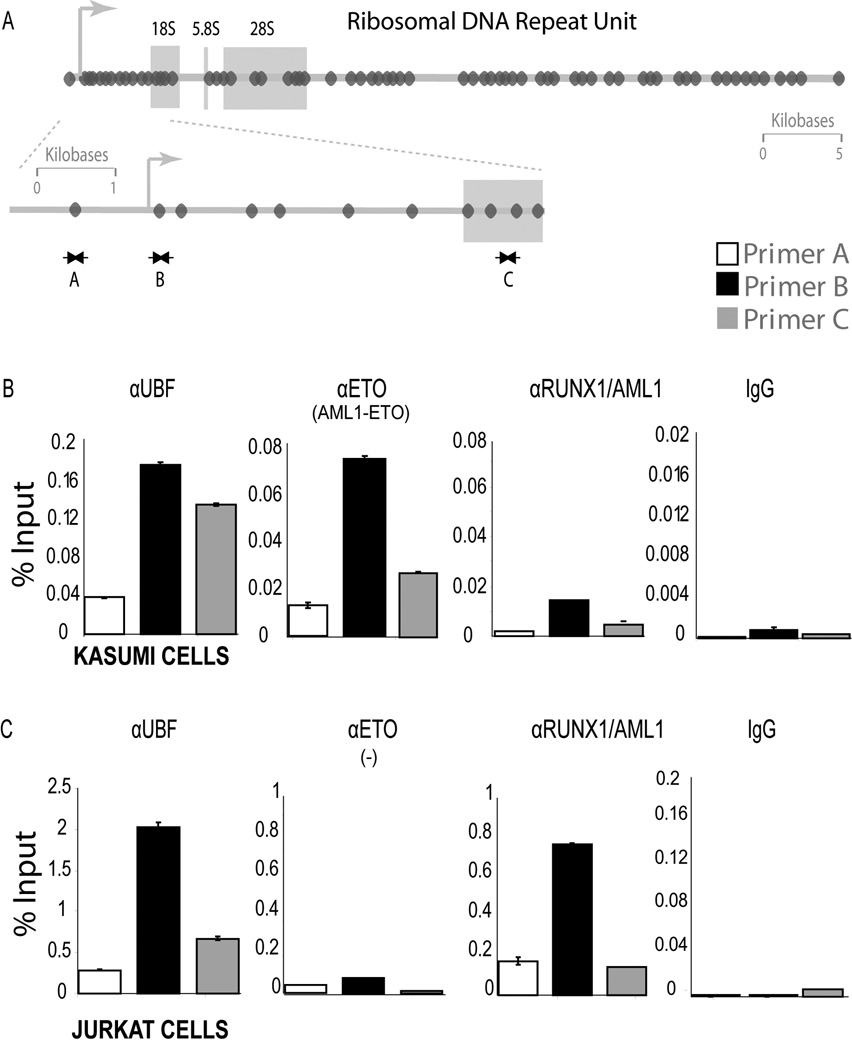
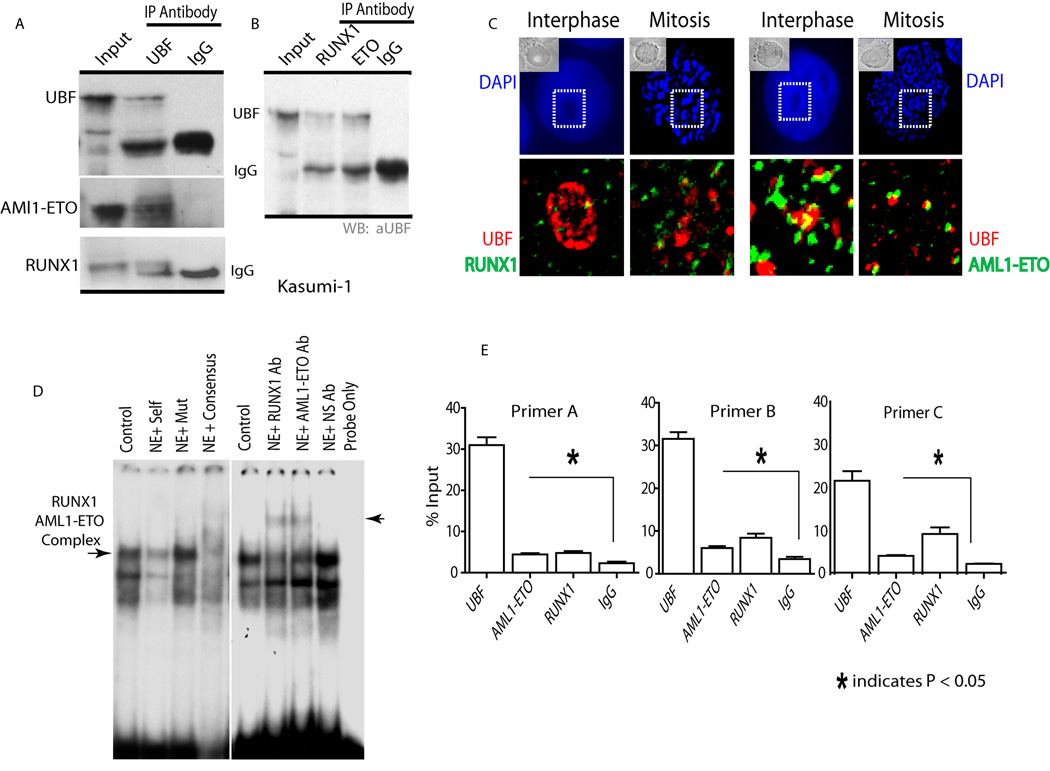
The symmetrical pairing of large foci containing RUNX1/AML1 and AML1-ETO on mitotic chromosomes is reminiscent of previous results with osteoblasts showing mitotic association of Runx2 with Nucleolar Organizing Regions (NORs) that represent sites containing tandemly repeated ribosomal DNA (rDNA) genes (Young et al., 2007a). Therefore, we postulated that RUNX1/AML1 and AML1-ETO may also bind to NORs and ribosomal repeats during mitosis in hematopoietic cells. We performed chromatin immunoprecipitation (ChIP) assays with primers spanning rDNA sequences to determine in vivo occupancy of rDNA repeats by RUNX1/AML1 and AML1-ETO in Kasumi-1 and Jurkat cells (Fig. 3A). The RNA Pol I regulatory protein Upstream Binding Factor 1 (UBF1), which binds directly to rDNA and to mitotic NORs, was used as a positive control for ChIP assays. Quantitative PCR data show that AML1-ETO occupies rDNA repeats during mitosis in Kasumi-1 cells. However, association of RUNX1/AML1 to the rDNA genes is barely detectable (Fig. 3B), consistent with the immunofluorescence data (see results above). For comparison, Jurkat cells that have high levels of RUNX1/ AML1 but do not express the AML1-ETO fusion protein show rDNA occupancy by RUNX1/AML1 but not AML1-ETO, as expected (Fig. 3C). Thus, these biochemical data establish that RUNX1/AML1 (in Jurkat cells) and the AML1-ETO fusion protein (in Kasumi-1 cells) are each associated with rDNA repeats during mitosis that are visualized as NORs by immunofluorescence microscopy (see Fig. 2). Clear detection of the binding of AML1-ETO but not RUNX1/AML1 to mitotic chromosomes in Kasumi-1 cells indicates that AML1-ETO may perturb the normal function of RUNX1/AML1 during mitosis.
Figure 3. AML1-ETO shows enhanced occupancy of rDNA repeats during mitosis.
A) The panel shows a schematic of the Runx consensus elements (dark ovals) in the human rDNA repeats and depicts the locations of ChIP primers. B and C) ChIPs were done with antibodies for RUNX1/AML1, ETO and UBF1, as well as non immune IgG in Kasumi-1 and Jurkat cells blocked in mitosis. An antibody detecting ETO was used to immunoprecipitate the AML1-ETO fusion protein, whereas an antibody recognizing the C-terminal domain of RUNX1/AML1 was used to pull down endogenous RUNX1/AML1. Quantitative PCR data are normalized to genomic DNA and denoted as percent input.
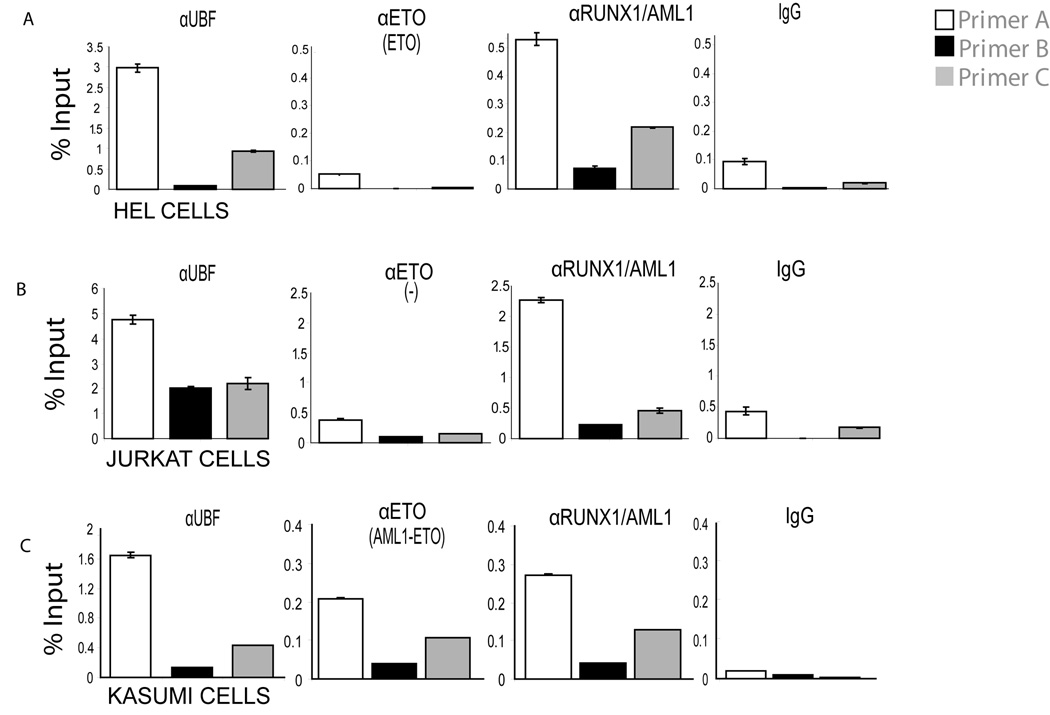
Because RUNX1/AML1 and AML1-ETO bind to rDNA repeats during mitosis, we used ChIP assays to examine whether these proteins are still capable of association during interphase in Kasumi-1, HEL and Jurkat cells. As positive controls, we show that the RNA Pol I factor UBF1 can interact with rDNA repeats in all three cell types (Fig. 4). The native ETO protein endogenously expressed in HEL cells is not associated with the rDNA repeats, nor is ETO detected on rDNA repeats in Jurkat cells that express neither ETO nor the AML1-ETO fusion protein (Fig. 4A and 4B). Of note we observe that both RUNX1/AML1 and AML1-ETO occupy rDNA repeats in Kasumi-1 cells during interphase (Fig. 4C). The latter finding suggests that RUNX1/AML1 and AML1-ETO may be concurrently bound to rDNA genes during interphase.
Figure 4. Both RUNX1/AML1 and AML1-ETO occupy rDNA repeats in interphase Kasumi-1 cells.
Chromatin immunoprecipitation was done in asynchronously growing HEL, Jurkat and Kasumi-1 cells using RUNX1/AML1, ETO, UBF1 and IgG antibodies. Three different PCR primer sets spanning Runx consensus elements were used (see Figure 3). A and B) An antibody detecting ETO was used to immunoprecipitate the AML1-ETO fusion protein, whereas an antibody recognizing the C-terminal domain of RUNX1/AML1 was used to pull down endogenous RUNX1/AML1. Quantitative PCR data show that endogenous ETO does not bind to rDNA repeats in HEL cells nor in Jurkat cells (where ETO is not expressed), while UBF1 occupies the rDNA repeats in vivo in all the three cell lines tested. C) RUNX1/AML1 and AML1-ETO both occupy rDNA repeats in Kasumi-1 cells. Quantitative PCR data are normalized to genomic DNA and denoted as percent input.
RUNX1/AML1 and AML1-ETO associate with UBF1 on rDNA repeats in vivo
Our finding that RUNX1/AML1, AML1-ETO and UBF1 each associate with rDNA repeats suggests that RUNX1/AML1 and AML1-ETO may interact with UBF1 in vivo. We carried out co-immunoprecipitation assays to examine directly whether UBF-1 is present in a protein complex with RUNX1/AML1 or AML1-ETO. AML1-ETO and RUNX1/AML1 are detected in immunoprecipitates obtained with a UBF1 antibody (Fig. 5A). To validate these results, reciprocal immunoprecipitation assays with antibodies against RUNX1/AML1 and AML1-ETO were performed. Indeed, UBF1 precipitated when immuno-complexes were prepared using either RUNX1/AML1 or ETO antibodies (Fig. 5B). We also observe co-localization, albeit limited, of both RUNX1/AML1 and AML1-ETO with nucleolar UBF1 during interphase using immunofluorescence microscopy (Fig. 5C). Thus, RUNX1/AML1 and AML1-ETO each associate with UBF-1, consistent with the interaction of these proteins with the regulatory sequences of rDNA repeats.
Figure 5. Endogenous RUNX1/AML1 and AML1-ETO interact with UBF1 on the rDNA repeats.
A) Immunoprecipitation analysis was carried out with an antibody for UBF1 followed by western blotting with AML1-ETO and RUNX1/AML1 specific antibodies. Both RUNX1 and AML1-ETO are detected in western blot analysis, however AML1-ETO shows greater interaction with UBF1 when compared to RUNX1/AML1. B) Endogenous RUNX1/AML1 and AML1-ETO were immunoprecipitated from Kasumi-1 cells using rabbit polyclonal antibodies that specifically recognize the C-terminus of RUNX1/AML1 or the ETO moiety. A mouse monoclonal antibody was used to detect endogenous UBF1 by immunoblotting. UBF1 and IgG heavy chain are indicated. C) Immunofluorescence microscopy was performed in Kasumi-1 cells to detect endogenous RUNX1/AML1 (green), UBF1 (red) or AML1-ETO (green) with DAPI staining (blue). There is co-localization, albeit limited, of both RUNX1/AML1 and AML1-ETO with nucleolar UBF1 during interphase. D) Electrophoretic mobility shift assays were performed with a human rDNA probe spanning a Runx binding element and nuclear extracts from Kasumi-1 cells. Competition assays with 100-fold molar excess of unlabeled wild type, mutant or Runx consensus oligonucleotide were performed to establish the specific protein-DNA complex (lanes 1–4) as indicated. Super-shift immunoassays were performed by incubating binding reactions with the indicated antibodies (Lanes 5–8). Normal IgG was used as negative control. The arrow at the right marks the supershift band. E) ChIP-reChIP assays with endogenous proteins in interphase Kasumi-1 cells using UBF1 antibody (primary ChIP) and second immunoprecipitation (reChIP) with antibodies directed against UBF1, RUNX1/AML1, AML1-ETO or IgG. The re-ChIP data are plotted as a percentage immuno-precipitation of the primary ChIP (set as 100%). Each of the regions was immunoprecipitated with similar efficiency in the primary ChIP. These results show that RUNX1/AML1 and AML1-ETO each can co-occupy rDNA repeats with UBF1.
To demonstrate that both RUNX1/AML1 and AML1-ETO bind directly to the rDNA repeats, we conducted electrophoretic mobility shift assays with a human rDNA probe spanning a Runx binding element and nuclear extracts from Kasumi-1 cells. Competition assays with 100-fold molar excess of unlabeled wild type, mutant or Runx consensus oligonucleotide were performed to establish the specific protein-DNA complex (Fig. 5D). Addition of antibodies against RUNX1 or AML1-ETO resulted in retarded mobility (supershift) of the complex (Fig. 5D). These results indicate that RUNX1 and AML1-ETO can interact with Runx elements in the rDNA repeat.
To assess whether RUNX1/AML1 or AML1-ETO co-occupy rDNA repeats with the RNA Pol I transcription factor UBF-1, ChIP-reChIP assays were performed in asynchronous Kasumi-1 cells. UBF1 antibody was used for the primary ChIP, followed by a second immunoprecipitation (reChIP) with either non-immune IgG (as negative control) or antibodies directed against UBF1, RUNX1/AML1 or AML1-ETO. Quantitative PCR data show that at least some UBF1 bound rDNA fragments are also occupied by RUNX1/AML1 or AML1-ETO proteins (Fig. 5E). These results indicate that RUNX1/AML1 and AML1-ETO have limited but significant presence on ribosomal DNA repeats that interact with UBF1 in vivo.
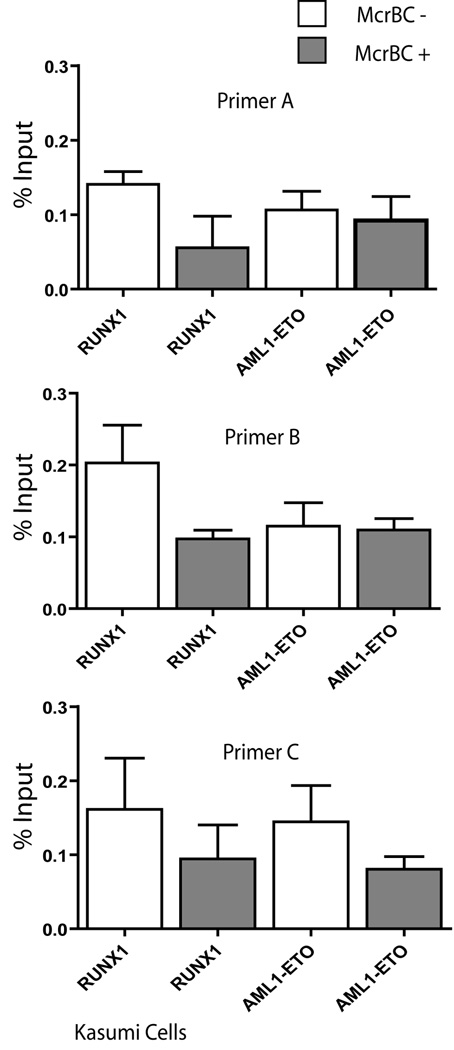
AML1-ETO occupancy of rDNA repeats is correlated with DNA hypomethylation and altered histone H3 methylation
It is well established that only a subset of rRNA genes are transcriptionally active at any one time (Grummt and Pikaard, 2003). We therefore investigated whether genomic occupancy of rDNA repeats by RUNX1/AML1 or AML1-ETO is linked to epigenetic chromatin modifications. To monitor the association of RUNX1/AML1 and AML1-ETO at methylated and unmethylated rRNA genes, we used the ChIP-CHOP assay (Lawrence et al., 2004). Chromatin from Kasumi cells was immunoprecipitated with antibodies against RUNX1 or AML1-ETO (ChIP) and the resulting DNA was digested with McrBC enzyme (CHOP) prior to qPCR using rDNA primers. Only DNA that is methylated at two or more cytosines (within 55–3000 bp) is digested by McrBC. We find that rDNA regulatory regions associated with RUNX1/AML1 are sensitive to digestion by the McrBC enzyme (Figure 6, primers A and B), while those associated with AML1-ETO are not. Interestingly, the 18s rRNA coding region detected by primer C was equally sensitive to McrBC in both RUNX1/AML1 and AML1-ETO bound fractions. Taken together, our results indicate that rDNA repeats bound by RUNX1/AML1 are hyper-methylated relative to those that are bound by AML1-ETO (Figure 6).
Figure 6. RUNX1/AML1 associates with hypermethylated rDNA repeats.
Chromatin from Kasumi cells was immunoprecipitated with antibodies against RUNX1/AML1 or AML1-ETO and the resulting DNA was digested with McrBC enzyme prior to qPCR using indicated rDNA primers. An antibody detecting ETO was used to immunoprecipitate the AML1-ETO fusion protein, whereas an antibody recognizing the C-terminal domain of RUNX1/AML1 was used to pull down endogenous RUNX1/AML1. Quantitative PCR data are normalized to genomic DNA and denoted as percent input. These results indicate that rDNA repeats bound by RUNX1/AML1 are hyper-methylated relative to those that are bound by AML1-ETO.
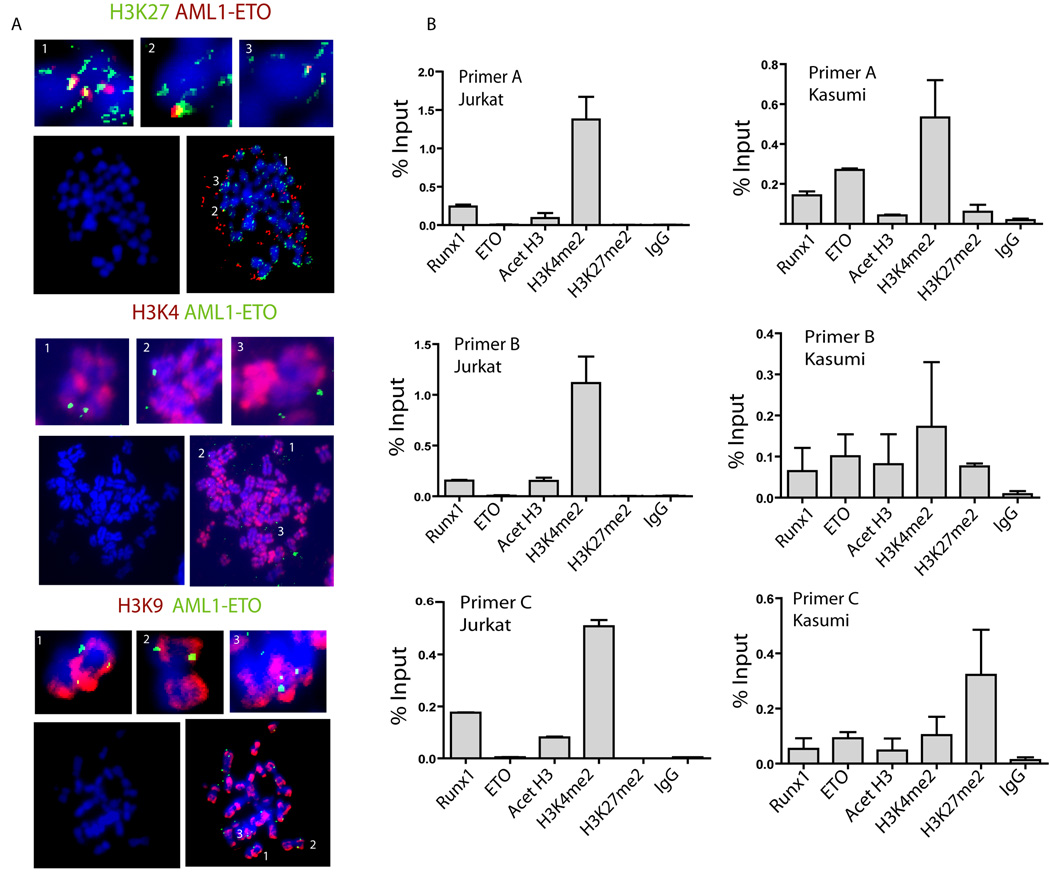
We next investigated the presence of AML1-ETO relative to post-translational modifications of nucleosomal histones. By immunofluorescence microscopy of Kasumi cells we observed significant co-localization of histone H3 dimethyl lysine 27 (H3K27me2) with AML1-ETO foci on mitotic chromosomes (Figure 7A). There was limited co-localization with H3K9me2 and none with H3K4me2. We also performed ChIP assays to monitor modified histones on rDNA repeats in Kasumi-1 cells that express AML1-ETO and RUNX1/AML1 or in Jurkat cells that express only RUNX1/AML1. We find that methylation of H3K4 is higher for rDNA repeats in Jurkat cells while the presence of AML1-ETO in Kasumi cells correlates with histone H3K27 methylation on the ribosomal genes (Fig. 7B). The function of H3K27 methylation in Pol I transcription remains to be established. Taken together, our results suggest that the presence of AML1-ETO at ribosomal genes results in epigenetic alterations that may reflect the pathological consequences of the 8;21 translocation.
Figure 7. AML1-ETO presence on rDNA repeats correlates with altered histone H3 methylation.
A).AML1-ETO colocalizes with histone H3 dimethyl lysine 27 (H3K27) on mitotic chromosomes. Immunofluorescence microscopy with antibodies raised against post-translational modifications of nucleosomal histones in metaphase spreads prepared from Kasumi-1 cells. The antibodies used to detect histone modifications were as follows: H3K9me2, H3K4me2, and H3K27me2. Each group of five panels shows merged images co-stained with antibodies against AML1-ETO. The left lower panels in each group show DAPI staining only. The top three panels in each group (indicated by numbers) show enlargements of the areas marked in the lower right of each group. B) AML1-ETO occupancy of rDNA repeats is associated with histone H3 lysine 27 methylation (H3K27me2). ChIPs were performed with RUNX1/AML1, AML1-ETO, IgG and histone modification antibodies were done in Jurkat (left panels) and Kasumi-1 cells (right panels). The antibodies used to detect histone modifications were as follows: Acetylated histone H3 (Acet-H3), H3K4me2 and H3K27me2. Presence of AML1-ETO in Kasumi-1 cells correlates with elevated histone H3K27 methylation on the rDNA repeats in comparison to Jurkat cells which express only RUNX1/AML1.
RUNX1/AML1 and AML1-ETO have opposing effects on ribosomal biogenesis
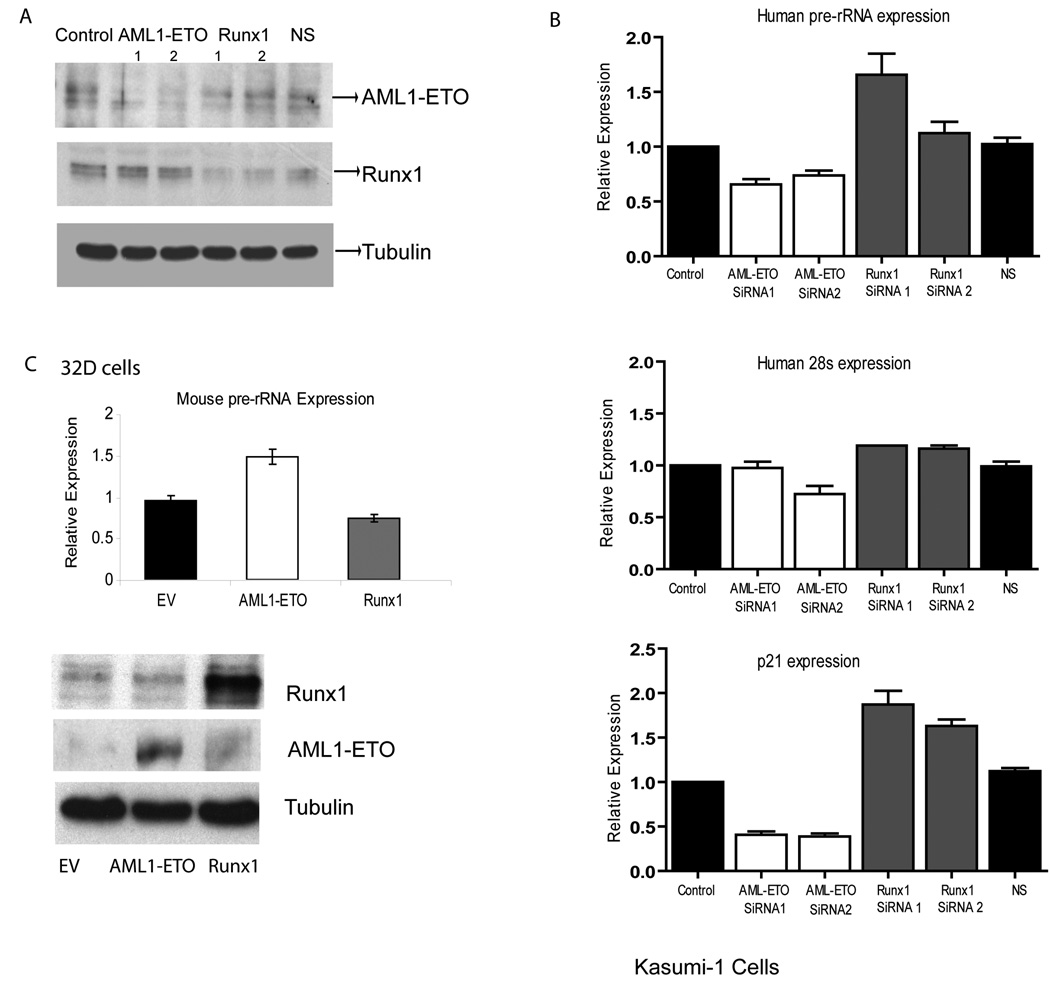
We addressed the functional relevance of rDNA occupancy by RUNX1/AML1 and AML1-ETO using RNA interference. Small interfering RNAs (siRNAs) specifically targeted against the mRNA junction sequences of the AML1-ETO fusion protein (Heidenreich et al., 2003) selectively reduced AML1-ETO protein levels by 50%–80% without affecting RUNX1/AML1 levels in Kasumi-1 cells (Fig. 8). Conversely, RUNX1/AML1 siRNAs specifically down-regulated RUNX1/AML1 protein levels by 50%–80% without any effect on AML1-ETO (Fig. 8A). Neither RUNX1 nor AML1-ETO protein was affected by control siRNAs. Quantitative analysis reveals that pre-rRNA synthesis is decreased significantly by depletion of AML1-ETO in Kasumi-1 cells. In contrast, down-regulation of RUNX1/AML1 protein significantly increases pre-rRNA synthesis (Fig. 8B). In neither case did we observe significant changes in the large pre-existing pools of total 28s rRNA. As a positive control, we assessed mRNA levels of the CDK inhibitor p21, which is upregulated by AML1-ETO (Peterson et al., 2007b), while RUNX1/AML1 represses p21 gene transcription (Lutterbach et al., 2000). Consistent with prior findings, our qPCR analysis indicates that p21 levels are down-regulated upon depletion of AML1-ETO, while RUNX1/AML1 siRNA treatment increases p21 mRNA levels. To rule out an indirect cell cycle effect resulting from changes in p21 levels, we directly depleted p21 mRNA by RNA interference and found no effect on pre-rRNA expression (data not shown and (Budde and Grummt, 1999)). Thus, both RUNX1/AML1 and AML1/ETO levels control pre-rRNA synthesis, but have opposing effects.
Figure 8. RUNX1/AML1 or AML1-ETO deficiency alters rRNA synthesis.
Kasumi-1 cells were transfected with two independent RUNX1/AML1 or AML1-ETO siRNAs or non-silencing (NS) siRNA. A) To check the efficiency of knockdown, protein expression of RUNX1/AML1, AML1-ETO and α-tubulin was examined by western blot analysis. B) Expression of unprocessed rRNA (pre-rRNA synthesis), 28s RNA and p21 was examined by RT-PCR analysis. Bars represent expression levels relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (±SEM) from three independent experiments performed in triplicate. C) Epitope tagged RUNX1/AML1 and AML1-ETO were each ectopically expressed in mouse 32D myeloid progenitor cells. Pre-rRNA synthesis was measured by RT-qPCR relative to GAPDH in equal numbers of cells (top). Expression of the exogenous proteins was analyzed by western blot analysis (bottom).
To further examine the role of RUNX1/AML1 and AML1-ETO in regulation of ribosomal genes, we electroporated epitope tagged RUNX1/AML1 and AML1-ETO into mouse 32D myeloid progenitor cells. The expression of the exogenous proteins was confirmed by immunoblotting with specific antibodies (Fig. 8C). Total cellular RNA from transfected cells was isolated and quantified by RT-qPCR. The results reveal that pre-rRNA synthesis is significantly increased by exogenous expression of AML1-ETO, but forced expression of RUNX1/AML1 diminishes pre-rRNA levels (Fig. 8C). Together, the RNA interference and forced expression studies demonstrate that RUNX1/AML1 negatively regulates rDNA transcription, while AML1-ETO is a positive regulator.
DISCUSSION
Using immunofluorescence microscopy and chromatin immunoprecipitations, we have shown here that the leukemia-related AML1-ETO fusion protein and the native RUNX1/AML1 factor associate with ribosomal gene loci on mitotic chromosomes at nuclear organizing regions, the precursors to interphase nucleoli. This principal finding provides the first indication that these regulatory proteins contribute to epigenetic control of ribosomal gene expression in pre-leukemic and leukemic cells in part by ‘book-marking’ these genes during mitosis. Decreased interaction of RUNX1/AML1 with mitotic chromosomes in AML derived Kasumi-1 cells suggests that AML1-ETO may interfere with RUNX1/AML1 function not only during interphase (Hiebert et al., 2001) but also during mitosis. Because regulation of ribosomal RNA genes is a fundamental process that supports the growth of cells and is tightly coupled with cell differentiation, our findings have immediate ramifications for the deregulation of growth control that is characteristic of cancer cells.
Transcription by RNA polymerase I regulates the rate of ribosome biogenesis and the biosynthetic potential of the cell (Moss, 2004; White, 2005). RNA Pol I activity is also tightly linked to the signals that control cell growth, and a number of physiological and pathological stimuli affect the rate of RNA Pol I transcription (Russell and Zomerdijk, 2006). It is clear that cancer involves significant changes to transcription factors, such as p53, that interact with the Pol I complex (Grummt, 1999). The fact that elevated rRNA synthesis has recently been shown to accelerate proliferation of transformed cells (Zhao et al., 2003) provides further reason to believe that ribosomal biogenesis has a profound impact on cancer biology.
We evaluated the functional roles for RUNX1/AML1 and the leukemia related AML1-ETO in rDNA transcription. Although immunoprecipitation and immunofluorescence microscopy results indicate that both proteins interact with UBF-1 in interphase cells, the combined results from our studies suggest that AML1-ETO and RUNX1/AML1 may perform opposing activities in control of ribosomal gene transcription. Indeed, down-regulation of RUNX1/AML1 or AML1-ETO by RNA interference in Kasumi-1 cells reveals that RUNX1/AML1 negatively regulates rDNA transcription, while AML1-ETO is a positive regulator. Consistent with a negative role for RUNX1/AML1 in ribosomal RNA synthesis we find that it binds to highly methylated rDNA regulatory sequences. Additionally, Kasumi-1 cells expressing the AML1-ETO fusion protein and Jurkat cells that express only AML1 differ in post-translational epigenetic marks of histone proteins at the rDNA repeats. Hence, our findings suggest a novel pathological role for the leukemogenic AML1-ETO protein in epigenetic regulation of cell growth through control of RNA Pol I-mediated ribosomal gene transcription.
We also have observed that endogenous RUNX1/AML1, ETO and AML1-ETO proteins are directed to distinct subcellular compartments. In interphase cells, the AML1-ETO fusion protein is localized in the nucleus but targeted to nuclear microenvironments distinct from those containing endogenous RUNX1/AML1 protein. Thus, as we previously reported for ectopically expressed RUNX1/AML1 and AML1-ETO proteins (McNeil et al., 1999; Barseguian et al., 2002), localization of the leukemia-related fusion protein is deregulated both during interphase and mitosis. The altered subnuclear location of AML1-ETO is a direct consequence of the elimination of the RUNX1/AML1 targeting signal and the addition of specific determinants residing in ETO that are fused to AML1 during the t(8;21) chromosomal rearrangement. In contrast, unlike the AML1-ETO protein, native ETO endogenously expressed in HEL cells is excluded from mitotic chromosomes. Thus, the runt-homology DNA binding domain that is fused to ETO in the chimeric AML1-ETO protein supports the recruitment of the ETO moiety to the rDNA repeats to deregulate rDNA transcription.
RUNX1/AML1 is a scaffolding protein that recruits many co-regulatory transcription factors to focally organized nuclear microenvironments (Zaidi et al., 2007; Lian et al., 2004). Many of these cofactors interact with the C-terminus of RUNX1/AML1 and can support either repression or activation by RUNX1/AML1 (e.g., TLE/Groucho, LEF1,CBP, p300 and MOZ) (reviewed in (Durst and Hiebert, 2004)). The loss of the RUNX1/AML1 C-terminus in the AML1-ETO fusion protein thus precludes the recruitment of a large group of possible co-factors to rDNA repeats and other RUNX1/AML1 target genes. Furthermore, the acquisition of ETO-related protein coding sequences in AML-ETO expressing cells will mediate the interactions of a distinct group of co-factors during interphase and mitosis. For example, the AML1-ETO fusion protein has been shown to aberrantly recruit co-repressor complexes (e.g., N-CoR/Sin3/HDAC1) to actively shut down transcription from RUNX1/AML1 target genes important for normal hematopoiesis (Hiebert et al., 2001). However, AML1-ETO does not always function as a transcriptional repressor. For example, expression of AML1-ETO has been shown to transactivate the BCL-2 and MDR1 promoters in reporter gene assays, although binding to a RUNX element may not be necessary (Klampfer et al., 1996; Hines et al., 2007; Burel et al., 2001). Additionally, AML1-ETO up-regulates C/EBPε to induce the expression of the G-CSF receptor and synergistically activates the M-CSF receptor promoter in combination with AML1 (Rhoades et al., 1996; Shimizu et al., 2000). We propose that modified association of co-factors with rDNA genes activates rDNA transcription in leukemia cells expressing the 8;21 fusion protein. However, the actual mechanism by which AML1-ETO compromises fidelity of rDNA transcription may be complex. An indirect mechanism that is not ruled out by our data is that AML1-ETO binds to the regulatory element of another gene that is responsible for activation of rDNA transcription.
In conclusion, there are two major implications of the results presented in our study. First, the AML1-ETO fusion protein not only affects RNA polymerase II gene regulation but also RNA polymerase I mediated ribosomal RNA transcription during interphase. Second, AML1-ETO perturbs normal functions of RUNX1/AML1 in mitotic cells to alter hematopoietic lineage-specific control of ribosomal RNA genes in progeny cells. The current findings suggest that AML1-ETO deregulates multiple gene regulatory pathways that control growth, proliferation and lineage identity. The coordinated deregulation of these intricate and biologically linked processes may clarify the potent properties of AML1-ETO in altering normal hematopoiesis and promoting development of leukemia.
MATERIALS AND METHODS
Cell culture
The human erythroleukemia (HEL) cells and the t(8;21) carrying cell line Kasumi-1 were cultured in RPMI supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and 100 U/mL penicillin and 100 µg/ml streptomycin. Cells were maintained at 37°C in a humidified atmosphere with 95% air, 5% CO2 at a concentration between 0.5 and 1 × 106 cells/ml.
Plasmid constructs and transfection experiments
Constructs pCMV5-HA-AML1/ETO and pCMV5-HA-RUNX1/AML1 were used in this study (Barseguian et al., 2002). HeLa cells were seeded in 6-well culture plates at a density of 1.5–2.0 × 105 cells/well, and transient transfections were performed 24 h later using FuGENE 6 according to the manufacturer's instructions (Roche, Indianapolis, IN). Cells were harvested 24 h after transfection for immunofluorescence microscopy. Amaxa nucleofection of HEL and Kasumi-1 cells was performed according to the protocol suggested by the manufacturer (Amaxa, Gaithersburg, MD). The immunostaining was performed using anti HA antibody.
Metaphase preparation
For metaphase preparation, cells were incubated with colcemid (Invitrogen Corporation, Carlsbad, CA) to a final concentration of 0.05 µg/ml at 37°C for 3–4 h. Chromosome spreads were generated by incubating mitotic cells in 0.075M KCl solution for 20 min at 37°C, fixed with methanol to acetic acid (3:1 vol/vol), and dropped onto frosted glass microscope slides, and air-dried.
Immunofluorescence microscopy
Cells were grown in regular growth medium for 1 to 2 days and then processed for the in situ immunofluorescence. Five hundred µl of cell suspension were deposited onto glass slides in a Shandon Cytospin 2 centrifuge. Cells were rinsed with ice-cold phosphate buffered saline (PBS) and fixed in 3.7% formaldehyde in PBS for 10 min on ice. After rinsing once with PBS, the cells were permeabilized in 0.25% Triton X-100 in PBS, rinsed twice in PBSA (0.5% bovine serum albumin (BSA) in PBS) and stained with antibodies.
The following primary antibodies and dilutions were used: UBF rabbit polyclonal (H-300), UBF mouse monoclonal (both 1:100; Santa Cruz Biotechnology); AML1 rabbit polyclonal (1:100; Active Motif), AML1 mouse monoclonal (1:100; 2B5 generous gift from Yoshiaki Ito, National University of Singapore, Singapore); ETO rabbit polyclonal (1:100; Calbiochem, San Diego, CA). For localization of antigen/antibody complexes, we used the following complementary fluorescent secondary antibodies: Alexa-488 goat anti-rabbit IgG, Alexa-488 goat anti-mouse IgG, and Alexa-594 goat anti-mouse IgG (1:800; Molecular Probes/Invitrogen).
Staining of cell preparations and chromosome spreads was recorded with a CCD camera attached to an epifluorescence Zeiss Axioplan 2 (Zeiss Inc., Thorwood, NY) microscope. For interphase studies single image planes were acquired and deconvoluted using the Metamorph Imaging Software (Universal Imaging, Dowingtown, PA). For metaphase spreads Z-series image stacks were acquired at 0.25 µm intervals with 67 nm/pixel (xy). Restoration of images was carried out by 3D deconvolution using a measured point-spread function as described previously (Carrington et al., 1995).
Chromatin immunoprecipitation and analysis
Chromatin immunoprecipitation assays (ChIPs) were performed by crosslinking asynchronously growing cells with 1% formaldehyde in RPMI for 10 min at room temperature. Crosslinking was quenched by adding glycine to a final concentration of 250 mM for 10 min. Cells were collected and washed twice with PBS. Cell pellets were resuspended in 2.5 ml of lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0, 1% NP-40, 25 µM MG-132, and 1X Complete® Protease inhibitor cocktail (Roche). After 10 min on ice, cells were sonicated to obtain DNA fragments of ~500 bp as determined by agarose gel electrophoresis with ethidium bromide staining. Protein-DNA complexes were isolated by centrifugation at 15,000 rpm for 20 min. Supernatants with protein-DNA complexes were incubated for 16 h with 3 µg Rabbit polyclonal antibody directed against each protein. The following primary antibodies were used: UBF rabbit polyclonal, ETO rabbit polyclonal (Santa Cruz Biotechnology) and AML1 rabbit polyclonal (Active Motif). The antibodies used to detect histone modifications were as follows: Acetylated histone H3, dimethyl H3K4 and dimethyl H3K27 (Upstate Biotechnology, Lake Placid, NY). Antibody-Protein-DNA complexes were further incubated with 50–60 µl 30% protein A/G beads (Santa Cruz Biotechnology) to isolate antibody bound fractions of chromatin. Immuno-complexes were washed with the following buffers: low salt (20 mM Tris-Cl, pH 8.1, 150 mM NaCl, 1% Triton X-100, 2 mM EDTA, 1X complete protease inhibitor), high salt (20 mM Tris-Cl, pH 8.1, 500 mM NaCl, 1% Triton X-100, 2 mM EDTA), LiCl (10 mM Tris-Cl, pH 8.1, 250 mM LiCl, 1% deoxycholate, 1% NP-40, 1 mM EDTA) and twice in TE (10 mM Tris-Cl, pH 8.1, 1 mM EDTA). Protein-DNA complexes were eluted in 1% SDS and 100 mM NaHCO3. Crosslinks of pulldown fractions and inputs (2% of total IP fraction) were reversed by incubation overnight in elution buffer and 0.2M NaCl. DNA then was extracted, purified, precipitated, and resuspended in TE for qPCR. ChIP-reChIP experiments were carried out essentially as described. Briefly, UBF1 ChIP complexes were eluted in 10 mM DTT buffer for 30 min at 37°C, diluted 1:40 in ChIP lysis buffer and subjected to a second immunoprecipitation (i.e., re-ChIP) as described above. Quantitative PCR was done to quantify the immunoprecipitated DNA as described previously (Frank et al., 2001). For ChIP-CHOP-PCR (Lawrence et al., 2004), 10% of the immunoprecipitated DNA was digested with 10 units of McrBC (New England Biolabs) in reaction buffer (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol, 100 µg/ml bovine serum albumin, 1 mM GTP [pH 7.9]) at 37°C for 2–3 hr. Quantitative PCR was done to quantify the immunoprecipitated DNA as described previously. All ChIP and ChIP-CHOP-PCR experiments were repeated at least twice.
Co-immunoprecipitation analysis
Kasumi-1 cells (50–70% confluent) were used for co-immunoprecipitation studies as described previously (Hassan et al., 2004). Equal amounts of cell lysate were immunoprecipitated with antibodies for UBF1 (F-9 or H-300), ETO (H-54, Santa Cruz Biotechnology) and AML1 (39000, Active Motif), overnight in phosphate buffered saline with 5 mM EDTA. After a 2 h incubation with protein A/G beads followed by 3 washes with PBS, the immunocomplexes were separated in 10% SDS-PAGE and western blotted with the indicated antibodies.
Electrophoretic mobility shift Assay (EMSA)
Asynchronously growing Kasumi cells were harvested in ice-cold PBS buffer, cell pellets were lysed, and nuclear extracts prepared. The following oligonucleotides (double-stranded) representing wild-type (WT) and mutant (MT) Runx binding elements of the human rDNA promoter were synthesized: WT, GGCTATCTATTTTGTGGTTAGAATAAAGTT; and MT, GGCTATCTATTTTGTAC-TTAGAATAAAGTT. The probes were end labeled with [γ-32P] ATP by using T4 poly-nucleotide kinase (New England Biolabs, MA). Consensus and mutant oligonucleotides were used as competitors. Nuclear protein extracts (5 µg) were incubated for 30 min at room temperature with 1 µg of nonspecific competitor DNA poly (dI·dC) (Pharmacia, Piscataway, NJ) and 80,000 cpm of labeled oligonucleotides. Competition assays were performed by mixing 100-fold molar excess of unlabeled oligonucleotides (wild type or mutant) with nuclear extracts before addition of probes. RUNX1 and ETO antibodies (2 µg each; Santa Cruz Biologicals) were used for super shift experiments. Normal rabbit IgG was used as a nonspecific control. Protein–DNA complexes were visualized by autoradiography of a 5% polyacrylamide gel.
RNA interference
Exponentially growing Kasumi-1 cells were electroporated using Amaxa Nucleofector (Amaxa Biosystems) according to the manufacturer’s protocol, with siRNAs against RUNX1/AML1 (Dharmacon, Lafayette, CO) or AML1-ETO as described previously (Heidenreich et al., 2003). A non-silencing siRNA (Qiagen) were used as a negative control. Total RNA and protein were isolated for further analysis.
Quantitative Reverse Transcription-PCR (qRT-PCR)
RNA was extracted from all samples using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Purified total RNA was subjected to DNase I digestion, followed by column purification using the DNA Free RNA Kit (Zymo Research, Orange, CA). Eluted total DNA-free RNA was quantitated by spectophotometry, and 1 µg was added to a reverse transcription reaction using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA) with a mixture of random hexamers and oligo(dT) primers. Relative quantitation was determined using the ABI PRISM 7000 sequence detection system (Applied Biosystems, Foster City, CA) measuring real-time SYBR Green supermix fluorescence. The relative level of each mRNA was determined using the comparative CT method for relative quantitation using GAPDH or mCox as an endogenous reference. Primer sets used here have been described previously (Young et al., 2007a).
Acknowledgements
We thank the members of our laboratory, especially Shirwin Pockwinse, Prachi Ghule, Jitesh Pratap, Syed Ali and Klaus Becker, for stimulating discussions and/or reagents. We thank Judy Rask for expert editorial assistance with the preparation of the manuscript. We also thank Dr. Jeffrey Nickerson and Jean Underwood for assistance with confocal microscopy. This work was supported in part by NIH grant CA082834. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- Barseguian K, Lutterbach B, Hiebert SW, Nickerson J, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Multiple subnuclear targeting signals of the leukemia-related AML1/ETO and ETO repressor proteins. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15434–15439. doi: 10.1073/pnas.242588499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardin-Fried F, Kummalue T, Leijen S, Collector MI, Ravid K, Friedman AD. AML1/RUNX1 increases during G1 to S cell cycle progression independent of cytokine-dependent phosphorylation and induces cyclin D3 gene expression. J. Biol. Chem. 2004;279:15678–15687. doi: 10.1074/jbc.M310023200. [DOI] [PubMed] [Google Scholar]
- Biggs JR, Peterson LF, Zhang Y, Kraft AS, Zhang DE. AML1/RUNX1 phosphorylation by cyclin-dependent kinases regulates the degradation of AML1/RUNX1 by the anaphase-promoting complex. Mol. Cell Biol. 2006;26:7420–7429. doi: 10.1128/MCB.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- Burel SA, Harakawa N, Zhou L, Pabst T, Tenen DG, Zhang DE. Dichotomy of AML1-ETO functions: growth arrest versus block of differentiation. Mol. Cell Biol. 2001;21:5577–5590. doi: 10.1128/MCB.21.16.5577-5590.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington WA, Lynch RM, Moore ED, Isenberg G, Fogarty KE, Fay FS. Superresolution three-dimensional images of fluorescence in cells with minimal light exposure. Science. 1995;268:1483–1487. doi: 10.1126/science.7770772. [DOI] [PubMed] [Google Scholar]
- Davis JN, McGhee L, Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10. doi: 10.1016/s0378-1119(02)01172-1. [DOI] [PubMed] [Google Scholar]
- Durst KL, Hiebert SW. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 2004;23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- Frank R, Zhang J, Uchida H, Meyers S, Hiebert SW, Nimer SD. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. The bone-specific expression of RUNX2 oscillates during the cell cycle to support a G1 related anti-proliferative function in osteoblasts. J. Biol. Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, Speck NA, Gilliland DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res Mol. Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol. Cell. Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich O, Krauter J, Riehle H, Hadwiger P, John M, Heil G, Vornlocher HP, Nordheim A. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood. 2003;101:3157–3163. doi: 10.1182/blood-2002-05-1589. [DOI] [PubMed] [Google Scholar]
- Hiebert SW, Lutterbach B, Amann J. Role of co-repressors in transcriptional repression mediated by the t(8;21), t(16;21), t(12;21), and inv(16) fusion proteins. Curr. Opin. Hematol. 2001;8:197–200. doi: 10.1097/00062752-200107000-00003. [DOI] [PubMed] [Google Scholar]
- Hiebert SW, Sun W, Davis JN, Golub T, Shurtleff S, Buijs A, Downing JR, Grosveld G, Roussell MF, Gilliland DG, Lenny N, Meyers S. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol. Cell. Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines R, Boyapati A, Zhang DE. Cell type dependent regulation of multidrug resistance-1 gene expression by AML1-ETO. Blood Cells Mol. Dis. 2007;39:297–306. doi: 10.1016/j.bcmd.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, Shivdasani RA, Gilliland DG, Speck NA, Nimer SD, Tenen DG. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat. Genet. 2007;40:51–60. doi: 10.1038/ng.2007.7. Erratum in: Nat Genet. 2008 Feb;40(2):255. [DOI] [PubMed] [Google Scholar]
- Ito Y. Oncogenic potential of the RUNX gene family: 'overview'. Oncogene. 2004;23:4198–4208. doi: 10.1038/sj.onc.1207755. [DOI] [PubMed] [Google Scholar]
- Klampfer L, Zhang J, Zelenetz AO, Uchida H, Nimer SD. The AML1/ETO fusion protein activates transcription of BCL-2. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14059–14064. doi: 10.1073/pnas.93.24.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev. Eukaryot. Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- Licht JD. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene. 2001;20:5660–5679. doi: 10.1038/sj.onc.1204593. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Westendorf JJ, Linggi B, Isaac S, Seto E, Hiebert SW. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J Biol. Chem. 2000;275:651–656. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- McNeil S, Zeng C, Harrington KS, Hiebert S, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The t(8;21) chromosomal translocation in acute myelogenous leukemia modifies intranuclear targeting of the AML1/CBFalpha2 transcription factor. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14882–14887. doi: 10.1073/pnas.96.26.14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers S, Hiebert SW. Alterations in subnuclear trafficking of nuclear regulatory factors in acute leukemia. J Cell Biochem. 2000;35:93–98. doi: 10.1002/1097-4644(2000)79:35+<93::aid-jcb1131>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. At the crossroads of growth control; making ribosomal RNA. Curr. Opin. Genet. Dev. 2004;14:210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Nuchprayoon I, Meyers S, Scott LM, Suzow J, Hiebert SW, Friedman AD. PEBP2/CBF, the murine homolog of the human myeloid AML1 and PEBP2β/CBFβ proto-oncoproteins, regulates the murine myeloperoxidase and neutrophil elastase genes in immature myeloid cells. Mol. Cell. Biol. 1994;14:5558–5568. doi: 10.1128/mcb.14.8.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora G, Rowley JD. AML1 and the 8;21 and 3;21 translocations in acute and chronic myeloid leukemia. Blood. 1995;86:1–14. [PubMed] [Google Scholar]
- Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, Suzushima H, Takatsuki K, Kanno T, Shigesada K, Ito Y. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2alphaB gene associated with myeloblastic leukemias. Blood. 1999;93:1817–1824. [PubMed] [Google Scholar]
- Otto F, Lubbert M, Stock M. Upstream and downstream targets of RUNX proteins. J Cell Biochem. 2003;89:9–18. doi: 10.1002/jcb.10491. [DOI] [PubMed] [Google Scholar]
- Pabst T, Mueller BU. Transcriptional dysregulation during myeloid transformation in AML. Oncogene. 2007;26:6829–6837. doi: 10.1038/sj.onc.1210765. [DOI] [PubMed] [Google Scholar]
- Peterson LF, Boyapati A, Ahn EY, Biggs JR, Okumura AJ, Lo MC, Yan M, Zhang DE. Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts. Blood. 2007a;110:799–805. doi: 10.1182/blood-2006-11-019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LF, Boyapati A, Ranganathan V, Iwama A, Tenen DG, Tsai S, Zhang DE. The hematopoietic transcription factor AML1 (RUNX1) is negatively regulated by the cell cycle protein cyclin D3. Mol. Cell Biol. 2005;25:10205–10219. doi: 10.1128/MCB.25.23.10205-10219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LF, Yan M, Zhang DE. The p21Waf1 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO. Blood. 2007b;109:4392–4398. doi: 10.1182/blood-2006-03-012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovick MS, Hiebert SW, Friedman AD, Hetherington CJ, Tenen DG, Zhang DE. Multiple functional domains of AML1: PU.1 and C/EBPalpha synergize with different regions of AML1. Mol. Cell Biol. 1998;18:3915–3925. doi: 10.1128/mcb.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi J-Y, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Cell growth regulatory role of Runx2 during proliferative expansion of pre-osteoblasts. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- Rhoades KL, Hetherington CJ, Rowley JD, Hiebert SW, Nucifora G, Tenen DG, Zhang DE. Synergistic up-regulation of the myeloid-specific promoter for the macrophage colony-stimulating factor receptor by AML1 and the t(8;21) fusion protein may contribute to leukemogenesis. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11895–11900. doi: 10.1073/pnas.93.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romana SP, Poirel H, Leconiat M, Flexor M-A, Mauchauffe M, Jonveaux P, Macintyre EA, Berger R, Bernard OA. High frequency of t(12;21) in childhood B lineage acute lymphoblastic leukemia. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- Rubnitz JE, Look AT. Molecular basis of leukemogenesis. Curr. Opin. Hematol. 1998;5:264–270. doi: 10.1097/00062752-199807000-00006. [DOI] [PubMed] [Google Scholar]
- Russell J, Zomerdijk JC. The RNA polymerase I transcription machinery. Biochem. Soc. Symp. 2006:203–216. doi: 10.1042/bss0730203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Kitabayashi I, Kamada N, Abe T, Maseki N, Suzukawa K, Ohki M. AML1-MTG8 leukemic protein induces the expression of granulocyte colony-stimulating factor (G-CSF) receptor through the up-regulation of CCAAT/enhancer binding protein epsilon. Blood. 2000;96:288–296. [PubMed] [Google Scholar]
- Stein GS, van Wijnen AJ, Stein JL, Lian JB, Javed A, McNeil S, Pockwinse SM. Insight into regulatory factor targeting to transcriptionally active subnuclear sites. Exp. Cell Res. 1999;253:110–116. doi: 10.1006/excr.1999.4680. [DOI] [PubMed] [Google Scholar]
- Strom DK, Nip J, Westendorf JJ, Linggi B, Lutterbach B, Downing JR, Lenny N, Hiebert SW. Expression of the AML-1 oncogene shortens the G(1) phase of the cell cycle. J. Biol. Chem. 2000;275:3438–3445. doi: 10.1074/jbc.275.5.3438. [DOI] [PubMed] [Google Scholar]
- Vradii D, Zaidi SK, Lian JB, van Wijnen AJ, Stein JL, Stein GS. A point mutation in AML1 disrupts subnuclear targeting, prevents myeloid differentiation, and results in a transformation-like phenotype. Proc. Natl. Acad. Sci., USA. 2005;102:7174–7179. doi: 10.1073/pnas.0502130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JC, Shigesada K, Gergen JP, Ito Y. Mechanisms of transcriptional regulation by Runt domain proteins. Semin. Cell Dev. Biol. 2000;11:369–375. doi: 10.1006/scdb.2000.0184. [DOI] [PubMed] [Google Scholar]
- White RJ. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- Wotton D, Ghysdael J, Wang S, Speck NA, Owen MJ. Cooperative binding of Ets-1 and core binding factor to DNA. Mol. Cell. Biol. 1994;14:840–850. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang X-Q, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc. Natl. Acad. Sci. USA. 2007b;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Mntecino M, Stein JL, van Wijnen AJ, Lian JB, Stein GS. The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep. 2005;6:128–133. doi: 10.1038/sj.embor.7400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van WA, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nat. Rev. Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Pockwinse SH, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic partitioning and selective reorganization of tissue specific transcription factors in progeny cells. Proc. Natl. Acad. Sci. USA. 2003;100:14852–14857. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, van Wijnen AJ, Stein JL, Meyers S, Sun W, Shopland L, Lawrence JB, Penman S, Lian JB, Stein GS, Hiebert SW. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBFα transcription factors. Proc. Natl. Acad. Sci. USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yuan X, Frodin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell. 2003;11:405–413. doi: 10.1016/s1097-2765(03)00036-4. [DOI] [PubMed] [Google Scholar]