Abstract
Chromosomal translocations are common in leukemia, but little is known about their mechanism. Metnase (also termed SETMAR) is a fusion of a histone methylase and transposase protein that arose specifically in primates. Transposases were thought to be extinct in primates because they would mediate deleterious DNA movement. In primates, Metnase interacts with DNA Ligase IV (Lig IV), and promotes non-homologous end joining (NHEJ) DNA repair. We show here that the primate-specific protein Metnase can also enhance NHEJ in murine cells, and can also interact with murine Lig IV, indicating that it integrated into the pre-existing NHEJ pathway after its development in primates. Significantly, expressing Metnase in murine cells significantly reduces chromosomal translocations. We propose that the fusion of the histone methylase SET domain and the transposase domain in the anthropoid lineage to form primate Metnase promotes accurate intra-chromosomal NHEJ, and thereby suppresses inter-chromosomal translocations. Thus, Metnase may have been selected for because it has a function opposing transposases, and may thus play a key role in suppressing translocations that underlie oncogenicity.
Keywords: Metnase, Translocations, Transposase, DNA repair, Evolution, Genomic Stability
INTRODUCTION
Discovering that specific chromosomal translocations can be used to classify leukemia was a seminal discovery in cancer biology [1, 2]. Leukemic translocations have been widely studied, but their molecular mechanisms are poorly understood. Syndromes associated with faulty DNA repair predispose to leukemia, such as Ataxia Telangiectasia, and several studies have addressed this issue in the specific context of translocations [2, 3]. It appears that malfunctions in DNA double-strand break (DSB) repair pathways are indeed the primary culprit. Two DSB pathways are known to produce balanced translocations: non-homologous end joining (NHEJ) and single strand annealing (SSA). However, the best evidence is that most oncogenic chromosomal translocations result from NHEJ as the primary mechanism [4].
Transposons are ancient mobile DNA elements that encode the enzymatic machinery for their own mobility, termed transposases, and are found in genomes of species from bacteria to mammals [5–9]. These mobile elements move within genomes by two major mechanisms: 1) an excision and ligation strategy utilized by DNA transposons, and 2) by forming RNA intermediates in the case of retrotransposons. Because transposases mediate DNA mobility, they are candidates for mediating oncogenic chromosomal translocations [5–9]. However, transposase activity was postulated to be extinct in primates, perhaps because unregreulated DNA mobility would be deleterious to a long lived organism. Nonetheless, we identified a translated protein, which we termed Metnase, that has a transposase domain [10].
Metnase (also called SETMAR) arose in primates through a fusion of SET (protein methylase) and Mariner transposase/nuclease domains which are both present separately in the mouse genome [8]. Metnase methylates histone 3, enhances NHEJ DNA double strand break repair, improves retroviral genomic integration, enhances chromosome decatenation by enhancing Topoisomerase IIα and increasing resistance to etoposide and adriamycin, enhances cell survival after ionizing radiation, and protects DNA ends during NHEJ [5, 10–16]. Metnase’s role in NHEJ likely depends on its interaction with human DNA ligase IV (Lig IV) [12]. Thus, Metnase improved DNA repair, as opposed to the more common characteristic of transposases generally, increasing DNA mobility.
These findings raised the question of the role of Metnase in chromosomal translocations, which we investigated in this study. We show here that Metnase promotes NHEJ and DNA integration in murine cells, and interacts with murine Lig IV. Interestingly, Metnase expression in murine embryonic stem cells (mES) suppressed I-SceI-induced chromosomal translocations, although it did not alter the accuracy of the translocation joints. Furthermore, expression of full-length Metnase with point mutations that inactivate either the SET or nuclease domains fails to suppress translocations, indicating that both domains play a role in preventing inappropriate NHEJ-mediated translocations. Finally, expression of the SET domain alone increased translocations by 3 fold, whereas the nuclease domain had no effect. Altogether, we propose a model in which fusion of two genomic elements in lower mammals resulted in a genome stabilizing phenomenon.
METHODS
Plasmids, cell lines and immunoprecipitation
Wild-type Metnase, point mutants, domains and I-SceI were transiently expressed from the pCAGGS vector as described [4, 10]. Translocation reporter p5rE and r15 mES cells were cultured and transfected as described [4]. Mouse Embryonic Fibroblasts were cultured as recommended by the ATCC. Immunoprecipitation of V5-tagged Metnase and mouse Lig IV was performed as described [12].
NHEJ-integration assay
NIH-3T3 and r15 cells expressing pCAGGS control or various Metnase proteins were analyzed for NHEJ-integration as described [10, 12]. NIH-3T3 and r15 cells were co-transfected with pCAGGS control, wild-type Metnase, SET-defective, or nuclease-defective Metnase expression vectors and HindIII-linearized U6 plasmid as described [12]. Briefly, cells were transfected with a U6 plasmid that is cut between the U6 promoter and hygromycin gene. To show measurable resistance to hygromycin, the cell must accurately rejoin the plasmid intracellularly and then properly integrate it into their genome, both steps in which Metnase has been previously shown to enhance in primate cells. Cells were selected with hygromycin-B (hyg) (NIH-3T3, 300 μg/mL; r15, 100 μg/mL) and colonies were scored after 10–14 days. All experiments were performed in triplicate and NHEJ-integration frequencies were calculated as the number of hyg-resistant colonies per viable cell determined by parallel plating in drug-free medium.
Chromosome translocation assay
V5-tagged Metnase, point mutants or domains were analyzed for chromosomal translocations as described except PCR primers for der(17) were modified (primers- 5′: tctctactaaaaatacaaaaatgagctgag and 3′: cacattgttttaatttctttaactcacagt) [4, 10, 12]. V5-tagged Metnase, specific point mutants or domains were co-transfected with an I-SceI expression vector into p5rE mES cells as described [4, 10, 12]. . After 24 hr, 300,000–500,000 cells were seeded in 10 cm dishes, allowed to recover and then 200 μg/mL G418 was added. Translocation frequencies were calculated as the number of G418-resistant colonies per viable cell as above. To determine the accuracy of NHEJ at translocation junctions, G418-resistant colonies were expanded, genomic DNA was harvested using the Archive Pure DNA kit (5 Prime, Gaithersburg, MD), amplified by PCR (primers- 5′: tctctactaaaaatacaaaaatgagctgag and 3′: cacattgttttaatttctttaactcacagt), and sequenced as described [12].
RESULTS
Metnase enhances NHEJ and interacts with Lig IV in mouse cells
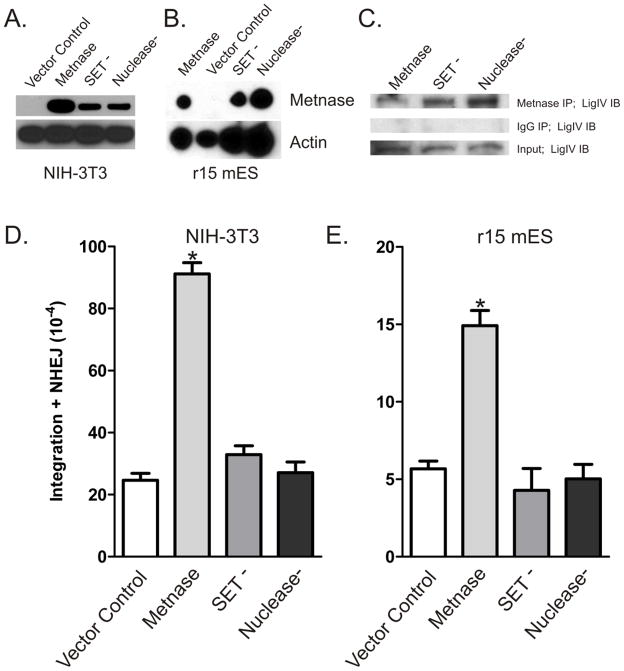
Metnase arose late in primate evolution so it is not present in mice. However, the fact that Metnase functions in ancient processes including NHEJ, viral DNA integration, and chromosome decatenation [10–12, 14–16] suggests that Metnase may function when inserted into lower mammals. We therefore investigated whether expression of Metnase in mouse NIH-3T3 or mES cells would enhance NHEJ, similar to human cells, and prove to be evolutionarily advantageous [6–8]. In Fig. 1, expression of Metnase enhanced two-step NHEJ-integration of linear plasmid DNA in NIH-3T3 (Fig. 1A, D) and mES (Fig. 1B, E) cells. We next evaluated whether Metnase interacts with mouse Lig IV. We expressed Metnase in mES cells and found that it interacted with mouse Lig IV by co-immunoprecipitation (Fig. 1C). These data indicate that although Metnase is not expressed in mouse cells, this human protein functionally interacts with the essential conserved mouse NHEJ machinery.
Figure 1. Human Metnase interacts with and functionally enhances NHEJ in mouse cells.
(A) V5-tagged Metnase or specific point mutants inactivating either the SET (SET−) or Nuclease (Nuclease−) domains were expressed in NIH 3T3 (A) or ES (B) mouse cells. (C) V5-tagged wild-type or mutant Metnase was expressed in mouse ES cells was immunoprecipitated with anti-V5 antibodies (top) or non-specific IgG (middle). These samples and input control were analyzed by western blot with anti-LigIV antibodies. (D) NIH-3T3 or (E) r15 ES were co-transfected with a linearized U6 (hyg) plasmid and vectors expressing wild-type Metnase, SET- or nuclease-defective Metnase, or empty control vector. Two-step NHEJ/integration frequencies are averages (+ SEM) for 3 determinations. * indicates P ≤ 0.0011 (t test).
Metnase suppresses chromosomal translocations
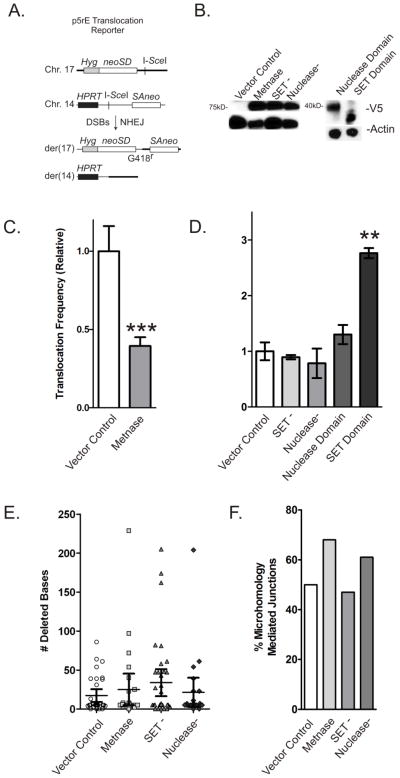
Although translocations can be initiated by DSBs and require joining of broken ends, the end-joining machinery normally suppresses translocations as seen by the increase in translocations after the removal of the NHEJ protein Ku70/80 [17]. Thus, defects in NHEJ prevent rapid re-joining of broken ends to their proximal partner, and they are free to migrate to other nuclear regions and join inappropriately [18]. Since expression of Metnase in mouse cells enhances NHEJ, we investigated whether Metnase could regulate translocations in p5rE mES cells using a previously described model (Figure 2A) [4]. As shown in Fig. 2C, translocations induced by I-SceI DSBs in chromosomes 14 and 17 were significantly suppressed by 2.5-fold in mES cells expressing wild-type Metnase (Fig. 2B). This is suggests that Metnase functionally interacts with the mouse NHEJ machinery and implicates Metnase as a regulator of translocations in cells experiencing multiple DSBs. The stimulation of NHEJ and suppression of translocations by Metnase is analogous to the effects of the Ku NHEJ protein on these endpoints [17]. Altogether, these results indicate that while Metnase arose late in the evolutionary process, its function may still be critical to maintain the complexity of the primate genome in the face of genomic damage including DNA DSBs via the NHEJ pathway.
Figure 2. Metnase suppresses chromosomal translocations.
(A) Dual DSB translocation assay in p5rE mES cells as described by Weinstock et al. [4]. I-SceI induces DSBs near splice-acceptor and -donor (SA, SD) sequences attached to truncated neo genes on chromosomes 14 and 17 that become linked by translocation producing a functional neo gene. (B) Expression of V5-tagged Metnase and Metnase mutants and domains (top row) and actin control (bottom row) in p5rE mES cells. (C) Translocation frequencies in mES cells expressing wild-type Metnase are shown relative to cells with empty vector control. Values are averages (+ SEM) for 3 determinations. *** indicates P < 0.0005 (t test). (D) Similar to C, but cells expressing mutant Metnase or domains as labeled. Values are averages (+ SEM) for 3 determinations. * indicates P < 0.05, ** indicates P < 0.01 (t test). (E) Average deletion size among 23–36 products per cell type; no statistically significant differences. (F) Percentage of products with microhomology mediated re-joining in each cell type; no statistically significant differences.
To gain further insight into how Metnase regulates translocations, we examined translocations in cells expressing SET (SET−) or nuclease (Nuclease−) defective mutants of Metnase (Fig. 2B). Previously and verified here, both of these mutants have been shown to be defective in NHEJ [12]. As shown in Fig. 2D, neither mutant had a significant effect on translocation efficiency, indicating that functional SET and nuclease domains are required to suppress translocations, which is consistent with our previous finding that both domains are required for Metnase stimulation of NHEJ.
Finally, to determine if the evolutionarily conserved portions of Metnase play a role in translocation formation, we expressed the SET and nuclease domains in p5rE cells (Fig. 2B). The SET domain shows a three fold increase in translocations and the nuclease domain is null (Fig. 2D), indicating that the possibly expressed SET protein in mice induces genome instability and the combination with a transposase was a genome stabilizing event.
Metnase does not affect sequences at translocation junctions
We previously showed that over-expressing Metnase increases NHEJ and decreases long deletions during DSB repair by NHEJ in plasmid end-joining assays [4]. Since all detectable translocations in p5rE cells are produced by NHEJ, we investigated whether Metnase expression would affect the extent of deletions during chromosome translocations. As shown in Fig. 2E–F and Table 1, over-expression of wild-type or mutant Metnase had no discernible effect on the average length of deletions, microhomology mediated joining, or their distribution. Incomplete Metnase expression within the transfected cell population could also account for this lack of significance or that the construct evaluated here is integrated into the genome and the previous end-joining assays were performed on transfected plasmids. Overall, this finding suggests that Metnase decreases the likelihood, but not the final outcome, of chromosome translocations.
Table 1. Translocation products.
All sequenced translocation ends are shown. The error free product is shown first for comparison and then products arising in cells with pCAGGS, wild-type Metnase, or Metnase SET-deficient or nuclease-deficient expression vectors are listed. The 14 bp sequences surrounding the translocation junctions are shown for both 5′ and 3′ directions along with the number of deleted bases outside of the AATA I-SceI overhangs. Products arising more than once are shown with a single sequence and with clone numbers listed. Micro-homology mediated repair products are represented by *.
| Error-free translocation product: | |||
| der(17) | …ttcaaaaaaaaTAGGGATAA | CAGGGTAATgcatgca… | |
| sequence | …aagttttttttATCCC | TATTGTCCCATTAcgtacgt… | |
| pCAGGS: | |||
| 18,24,26,31 | …aaaaaaaaataggg | 0 del | cagggtaatgcatg… |
| 1 | …acagagcaagactc | 54 del | tttgtgatgcaacc… |
| 2 | …gggaggttgcagtg | 86 del | cttggctccgaaat… |
| 3,30 | …caaaaaaaataggg | 4 del | gtaatgcatgcaag… |
| 4,8,10,12,14,16,17,19,20,22,27 | …caaaaaaaataggg | 5* del | taatgcatgcaagc… |
| 5 | …tccagcctgggtga | 61 del | atgatttttgtgat… |
| 6 | …acagagcaagactc | 39 del | tggctccgaaatga… |
| 7 | …agcaagactccgtc | 38 del | ctccgaaatgattt… |
| 9 | …gacagagcaagact | 40 del | tggctccgaaatga… |
| 11,21,25 | …cgtctcaaaaaaaa | 5* del | cagggtaatgcatg… |
| 13 | …tctcaaaaaaaata | 8 del | taatgcatgcaagc… |
| 15 | …gcctgggtgacaga | 31 del | ggtaatgcatgcaa… |
| 23 | …ccagcctgggtgac | 64 del | atttttgtgatgca… |
| 28 | …tgacagagcaagac | 39 del | cttggctccgaaat… |
| 29 | …gtctcaaaaaaaat | 13 del | gcatgcaagcttgg… |
| 32 | …ctcaaaaaaaatag | 2* del | cagggtaatgcatg… |
| Metnase WT: | |||
| 19 | …aaaaaaaaataggg | 0 del | cagggtaatgcatg… |
| 1 | …aaaaaaaaataggg | 7* del | atgcatgcaagctt… |
| 2 | …tctcaaaaaaaata | 3* del | cagggtaatgcatg… |
| 3 | …ctcaaaaaaaatag | 97 del | gaaatgcgagctaa… |
| 4 | …gactccgtctcaaa | 40 del | atgatttttgtgat… |
| 5 | …caaaaaaaataggg | 4 del | gtaatgcatgcaag… |
| 6,9,10,11,14–17,20,21 | …caaaaaaaataggg | 5* del | taatgcatgcaagc… |
| 7 | …tgacagagcaagac | 54 del | atttttgtgatgca… |
| 8 | …ggtgacagagcaag | 23 del | cagggtaatgcatg… |
| 12 | …aaaaaaaaataggg | 8* del | tgcatgcaacgttg… |
| 13 | …ctccgtctcaaaaa | 8* del | cagggtaatgcatg… |
| 18 | …ctcaaaaaaaatag | 2* del | cagggtaatgcatg… |
| 22 | …acagagcaagactc | 72 del | attttaggggacct… |
| 23 | …agggcctctagtct | 229* del | agctaagtagacac… |
| 24 | …aaaaaaaaataggg | 6* del | aatgcatgcaagct… |
| 25 | …caaaaaaaaatagg | 25 del | tccgaaatgatttt… |
| Set Domain Point Mutant: | |||
| 15,20,24,31,33 | …aaaaaaaaataggg | 0 del | cagggtaatgcatg… |
| 1 | …tctcaaaaaaaata | 3* del | cagggtaatgcatg… |
| 2,4,7–9,13,14,16,19,25,35 | …caaaaaaaataggg | 5* del | taatgcatgcaagc… |
| 3 | …caaaaaaaataggg | 7* del | atgcatgcaagctt… |
| 5,21 | …cgtctcaaaaaaaa | 5* del | cagggtaatgcatg… |
| 6 | …ccgtctcaaaaaaa | 58 del | attttaggggacct… |
| 10 | …gagctgagatcgcg | 81 del | cgaaatgatttttg… |
| 11 | …gtgagctgagatcg | 82 del | ccgaaatgattttt… |
| 12,17,27 | …tcaaaaaaaatagg | 47 del | aaatgatttttgtg… |
| 18 | …gtgacagagcaaga | 162 del | ctaagtagacacaa… |
| 22 | …caagactccgtctc | 48 del | ttggctccgaaatg… |
| 23 | …aacctgcccaacat | 205 del | catgcaagcttggc… |
| 26 | …acaagggcctctag | 174 del | atgcaaccctattt… |
| 28 | …tgggtgacagagca | 43 del | ttggctccgaaatg… |
| 29 | …ctgcactccagcct | 61 del | tccgaaatgatttt… |
| 30 | …tgggtgacagagca | 27 del | cagggtaatgcatg… |
| 32 | …aaaaaaaaataggg | 22 del | gctccgaaatgatt… |
| 34 | …caaaaaaaaatagg | 6* del | taatgcatgcaagc… |
| 36 | …aaaaaaaaataggg | 30* del | atgatttttgtgat… |
| Nuclease Domain Point Mutant: | |||
| 14,18 | …aaaaaaaaataggg | 0 del | cagggtaatgcatg… |
| 1 | …aaaaaaaaataggg | 6* del | aatgcatgcaagct… |
| 2,5,8–10,13,19,20,23 | …caaaaaaaataggg | 5* del | taatgcatgcaagc… |
| 3 | …ccactgcactccag | 61 del | tggctccgaaatga… |
| 4 | …acctgcccaacatg | 204 del | catgcaagcttggc… |
| 6,16,21 | …caaaaaaaataggg | 7* del | atgcatgcaagctt… |
| 7 | …tggtgacaagggcc | 54 del | gtaatgcatgcaag… |
| 11 | …actccgtctcaaaa | 11 del | gaaatgcgagctaa… |
| 12 | …cgtctcaaaaaaaa | 5* del | cagggtaatgcatg… |
| 15 | …ctgggtgacagagc | 39 del | gcAAGCTTggctcc… |
| 17 | …ggtgacagagcaag | 23 del | cagggtaatgcatg… |
| 22 | …gagcaagactccgt | 23 del | atgcatgcaagctt… |
Micro-homology mediated repair product
CONCLUSIONS
Our results provide evidence of a novel role for Metnase in suppressing chromosomal translocations. Metnase increases the rate of NHEJ, and increases the rate that the two broken ends of a single DSB will rejoin rapidly [12]. This reduces the chance that ends will migrate to other nuclear domains and rejoin improperly with other broken ends to produce a translocation, similar to the mechanism proposed for Ku mediated suppression of translocations [17]. In this scenario, when translocations occur, it may reflect a transient deficiency of Metnase in the vicinity of the DSBs, slowing NHEJ reaction kinetics and allowing more time for errors to occur [12, 13, 19, 20]. The fact that translocation junctions were unaffected by Metnase expression is consistent with this model because Metnase activity is currently thought to be restricted to an early stage in NHEJ and will thus prevent translocation frequency, but not sequence fidelity.
Metnase therefore represents a domesticated transposase: its transposase biochemical activities that might cause genome rearrangements and predispose to malignancy have been subverted by primates to repress DNA movement by tethering with a SET domain. It is ironic that Metnase, instead of mobilizing DNA fragments and moving them to alternative locations, promotes repair of free DNA ends. One of the most intriguing features of Metnase is its complete presence solely in primates. Genome stability is certainly important for higher organisms with longer life spans, and Metnase may have arisen late in evolution in response to increasing threats posed by invading transposons or simply to the increase in total genomic complexity which was a result of higher evolutionary processes. There are certainly other possible explanations for its late arrival. For example, there is a mental retardation syndrome that has a deletion at 3p26 near Metnase, and one could speculate that Metnase may play a role in neuronal stability [21]. There are other interesting questions that the function of Metnase raises. For example, how can a protein that arose so recently interact with proteins involved in critical aspects of DNA dynamics and regulate so many important cellular functions? One answer could be that the Metnase’s SET and transposase domains were each present long before they fused in primates [8]. Thus, these distinct domains probably evolved as interaction partners independently, long before they fused into a single gene. In this view, this gene fusion brought together distinct domains, which yielded novel beneficial biochemical properties. This is unlikely, though, because the SET domain alone induces a 3 fold increase in translocations. Since Metnase’s SET domain was genomically present long before its fusion to the transposase domain, it is possible that losing the individual SET activity might beneficially alter transcriptional regulation of gene expression during primate evolution in some manner. It could be that the addition of the transposase domain yielded some extended specificity to the SET domain, altering its methylated targets. Similarly, this fusion clearly altered the classic biochemical activities of its transposase domain, which may have improved the genomic stability of primates simply by decreasing genomic instability. Thus, the loss of specific biochemical activities, including methylation of multiple non-specific targets or the transposase DNA cleaving function, by this gene fusion event may have also played an important yet undiscovered role in primate development.
In summary, we show in this study that Metnase expression significantly increases the efficiency of the mouse NHEJ machinery and reduces the frequency of chromosomal translocations in mES cells. Although Metnase evolved late in primate evolution [8], it is functional in mice, probably because it interacts with highly conserved proteins involved in DNA metabolism, including DNA Ligase IV, Topo IIα, and Pso4 [12–16]. This is an important demonstration of the ability of Metnase to insert seamlessly into a lower organism’s NHEJ DNA repair pathway, which lends credence to our hypothesis: that the fusion event that generated Metnase improved the stability of complex primate genomes. Thus, another benefit of this study is that it introduces a novel model to study the impact of primate-specific factors on the stability that is essential for the complexity of the primate genome. Of more clinical significance, our findings provide insight into the mechanism of translocation-dependent oncogenesis, considering that Metnase expression in cells could alter their propensity for aberrant NHEJ and the resulting translocations. They also suggest that Metnase may be useful as a biomarker to predict leukemia arising from chemotherapy-induced translocations [14, 19, 22], or as a genetic marker of general cancer predisposition.
Acknowledgments
S.-H.L. was supported by NIH grant CA92111 and the U.S. Army (DAMD17-00-1-0295), B.D.B. was supported by an NIH pre-doctoral training grant (T32 DK0075-20), the Walther Cancer Institute, and the IU Simon Cancer Center, J.A.N. was supported by NIH grant CA100862, and R.H. was supported by NIH grants HL093606 and CA140442.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annual review of genomics and human genetics. 2002;3:179–98. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 2.Nickoloff JA, De Haro LP, Wray J, Hromas R. Mechanisms of leukemia translocations. Current opinion in hematology. 2008;15:338–45. doi: 10.1097/MOH.0b013e328302f711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annual review of genomics and human genetics. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 4.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–80. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roman Y, Oshige M, Lee YJ, Goodwin K, Georgiadis MM, Hromas RA, Lee SH. Biochemical characterization of a SET and transposase fusion protein, Metnase: its DNA binding and DNA cleavage activity. Biochemistry. 2007;46:11369–76. doi: 10.1021/bi7005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miskey C, Papp B, Mates L, Sinzelle L, Keller H, Izsvak Z, Ivics Z. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol Cell Biol. 2007;27:4589–600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D, Bischerour J, Siddique A, Buisine N, Bigot Y, Chalmers R. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol Cell Biol. 2007;27:1125–32. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci U S A. 2006;103:8101–6. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miskey C, Izsvak Z, Kawakami K, Ivics Z. DNA transposons in vertebrate functional genomics. Cell Mol Life Sci. 2005;62:629–41. doi: 10.1007/s00018-004-4232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci U S A. 2005;102:18075–80. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williamson EA, Farrington J, Martinez L, Ness S, O’Rourke J, Lee SH, Nickoloff J, Hromas R. Expression levels of the human DNA repair protein metnase influence lentiviral genomic integration. Biochimie. 2008 doi: 10.1016/j.biochi.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hromas R, Wray J, Lee SH, Martinez L, Farrington J, Corwin LK, Ramsey H, Nickoloff JA, Williamson EA. The human set and transposase domain protein Metnase interacts with DNA Ligase IV and enhances the efficiency and accuracy of non-homologous end-joining. DNA Repair (Amst) 2008 doi: 10.1016/j.dnarep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck BD, Park SJ, Lee YJ, Roman Y, Hromas RA, Lee SH. Human Pso4 is a Metnase (SETMAR)-binding partner that regulates Metnase function in DNA repair. J Biol Chem. 2008;283:9023–30. doi: 10.1074/jbc.M800150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wray J, Williamson EA, Sheema S, Lee SH, Libby E, Willman CL, Nickoloff JA, Hromas R. Metnase mediates chromosome decatenation in acute leukemia cells. Blood. 2009;114:1852–8. doi: 10.1182/blood-2008-08-175760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wray J, Williamson EA, Royce M, Shaheen M, Beck BD, Lee SH, Nickoloff JA, Hromas R. Metnase mediates resistance to topoisomerase II inhibitors in breast cancer cells. PloS one. 2009;4:e5323. doi: 10.1371/journal.pone.0005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson EA, Rasila KK, Corwin LK, Wray J, Beck BD, Severns V, Mobarak C, Lee SH, Nickoloff JA, Hromas R. The SET and transposase domain protein Metnase enhances chromosome decatenation: regulation by automethylation. Nucleic Acids Res. 2008;36:5822–31. doi: 10.1093/nar/gkn560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–81. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–82. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lovett BD, Lo Nigro L, Rappaport EF, Blair IA, Osheroff N, Zheng N, Megonigal MD, Williams WR, Nowell PC, Felix CA. Near-precise interchromosomal recombination and functional DNA topoisomerase II cleavage sites at MLL and AF-4 genomic breakpoints in treatment-related acute lymphoblastic leukemia with t(4;11) translocation. Proc Natl Acad Sci U S A. 2001;98:9802–7. doi: 10.1073/pnas.171309898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aplan PD. Causes of oncogenic chromosomal translocation. Trends Genet. 2006;22:46–55. doi: 10.1016/j.tig.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins JJ, Pucilowska J, Lombardi RQ, Rooney JP. Candidate genes for recessive non-syndromic mental retardation on chromosome 3p (MRT2A) Clinical genetics. 2004;65:496–500. doi: 10.1111/j.0009-9163.2004.00267.x. [DOI] [PubMed] [Google Scholar]
- 22.Aplan PD. Chromosomal translocations involving the MLL gene: molecular mechanisms. DNA Repair (Amst) 2006;5:1265–72. doi: 10.1016/j.dnarep.2006.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]