Abstract
microRNAs (miRNAs) are a class of highly conserved small non-coding RNAs that negatively regulate gene expression post-transcriptionally. miRNAs are known to mediate myriad cell processes, including proliferation, differentiation, and apoptosis. With more than 600 miRNAs identified in humans, it is generally believed that many miRNAs function through simultaneously inhibiting multiple regulatory mRNA targets, suggesting that miRNAs participate in regulating the expression of many, if not all, genes. While many miRNAs are expressed ubiquitously, some are expressed in a tissue specific manner. The muscle specific miR-1, miR-133 and miR-206 are perhaps the most studied and best characterized miRNAs to date. Many studies demonstrate that these miRNAs are necessary for proper skeletal and cardiac muscle development and function, and have a profound influence on multiple myopathies, such as hypertrophy, dystrophy, and conduction defects.
Keywords: microRNA, muscle development, skeletal muscle, cardiac muscle
1. Introduction
miRNAs are short (~22 nucleotide), non-coding RNA molecules that influence gene expression, primarily post-transcriptionally. miRNAs negatively regulate gene expression through complementary base-pair binding of the miRNA “seed sequence” (nucleotides 2–7) to the 3′ untranslated region (UTR) of target mRNA, degrading or destabilizing the RNA message, or inhibiting protein translation, depending on the quantity of complimentary base-pair matches (Valencia-Sanchez, Liu, Hannon, & Parker, 2006) or the number of miRNA targeting sites within the 3′ UTR (Sandberg, Neilson, Sarma, Sharp, & Burge, 2008).
The muscle specific miR-1 and -206 are closely related in terms of expression and function, but differ based on chromosomal location, specific targets, and individual transcriptional activation. In general, both miR-1, found in both skeletal and cardiac muscle, and miR-206, specific for skeletal muscle, are shown to promote myoblast-to-myotube differentiation. By contrast, miR-133 promotes the proliferation of myoblasts, and inhibits their differentiation. miR-1-1 and miR-1–2 have identical mature nucleotide sequences, and miR-206 differs from this conserved miR-1 sequence by four nucleotides (Chen et al., 2006; Kim, Lee, Sivaprasad, Malhotra, & Dutta, 2006; Lagos-Quintana et al., 2002; Rosenberg, Georges, Asawachaicharn, Analau, & Tapscott, 2006). miR-133a-1 and miR-133a-2 share identical mature sequences, with miR-133b differing from this by a single nucleotide at the 3′ end (Chen et al., 2006; Niu, Li, Zhang, & Schwartz, 2007). The miR-1-1/miR-133a-2 and the miR-206/miR-133b clusters are transcribed from non-coding regions on mouse chromosomes 2 and 1, respectively, while the miR-1-2/miR-133a-1 cluster is transcribed from an intronic region of the ubiquitously expressed E3 ubiquitin ligase gene Mib1 (Chen et al., 2006). Therefore, while these three related clusters have similar sequences and expression patterns, each not only has distinct targets and functions, but also must rely on their own promoters for muscle specific expression.
2. Development and Function of Cardiac and Skeletal Muscle
There are three primary muscle types; cardiac, skeletal, and smooth. All are derived from the embryonic mesoderm in vertebrates. Muscle development is a coordinated process which involves cellular proliferation, differentiation, migration, and cell death. Regulation of gene expression at both transcriptional and translational levels is crucial for morphologic development of muscle tissues. Among the three muscle types, cardiac and skeletal muscle are striated and terminally differentiated. They are highly specific cells with the primary function of contraction. The heart is the first functioning organ in the embryo and is developed from lateral plate mesoderm, whereas skeletal muscle is derived from the paraxial mesoderm, forming embryonic somites which in turn become compartmentalized into the myotome (Buckingham, 2006; Garry & Olson, 2006).
Gene expression regulation is perhaps the most important factor governing proper development. Studies examining gene expression during muscle development have primarily focused on transcription factor-mediated regulation. Specifically, the family of myogenic regulatory factors (MRFs) including MyoD, myogenin, MRF4, and Myf5, as well as the myocyte enhancer factor 2 (MEF2) family of transcription factors are key regulators of skeletal muscle development (Tapscott, 2005), while the GATA4, Nkx2.5, myocardin, and serum response factor (SRF) are critical for myocardium development (Olson, 2006). Most recently, miRNAs have emerged as another class of key regulators for muscle development (Callis & Wang, 2008). Interestingly, many transcription factors are also affected by miRNA mediated repression, through positive and negative feedback loops.
3. Regulation and Expression of miRNAs During Muscle Development
Whereas many miRNAs are ubiquitously expressed, some miRNAs are expressed in a tissue-specific manner. Investigation into transcriptional regulation of miRNAs in muscle development comes from, in part, recent studies of the miR-1 and miR-133 families. These studies indicate that expression of miRNA genes is under transcriptional regulation, similar to that of protein-coding genes. The bicistronic pairs of cardiac and skeletal muscle specific miR-1-1 and miR-133a-2 (clustered on mouse chromosome 2) and miR-1-2 and miR-133a-1 (clustered on mouse chromosome 18) are regulated by SRF, MEF2, and other muscle associated transcription factors such as GATA4 and Nkx2-5 (Chen et al., 2006; Liu et al., 2007; Niu, Li, Zhang, & Schwartz, 2007; Zhao, Samal, & Srivastava, 2005). The bicistronic pair of skeletal muscle specific miR-206 and miR-133b are also regulated by MEF2, MyoD, and other factors (Kim, Lee, Sivaprasad, Malhotra, & Dutta, 2006; Rao, Kumar, Farkhondeh, Baskerville, & Lodish, 2006; Rosenberg, Georges, Asawachaicharn, Analau, & Tapscott, 2006). MyoD and myogenin provide a common temporal regulation through the induction of miR-1, -133, and -206 expression during myogenesis in C2C12 and human myoblast cells (Rao, Kumar, Farkhondeh, Baskerville, & Lodish, 2006). Other studies using gain and loss of function approaches indicate that Myf5 is both sufficient and necessary for the expression of miR-1 and miR-206, and that growth factors can negatively influence miRNA transcription through MRF repression (Sweetman et al., 2008). Interestingly, these miRNAs can act to repress there own regulators, such as SRF and histone deacetylase 4 (HDAC4), providing a regulatory mechanism for negative feedback (Chen et al., 2006; Lu, McKinsey, Zhang, & Olson, 2000).
4. Biological Roles of miRNA in Skeletal and Cardiac Muscle
To determine the global role of miRNAs in development, Bernstein and colleagues knocked out Dicer in mice, preventing the processing of miRNA precursor into functional mature molecules, and found that lethality occurs at embryonic day 7.5, with development likely halting in gastrulation (Bernstein et al., 2003). Further examination using tissue-specific Dicer deletion revealed that miRNAs are required for proper morphogenesis in skeletal muscle (O’Rourke et al., 2007) and cardiac muscle (Chen et al., 2008; Zhao et al., 2007). These studies provide convincing genetic evidences for the essential role of miRNAs in muscle development and function. However, though the deletion of Dicer demonstrated the requirement of miRNAs for proper development in many tissues and organs, these studies did not provide insights into the biological functions of individual miRNAs.
Skeletal Muscle
Skeletal muscle cells develop from the embryonic mesoderm during development, where they exist as proliferating myoblasts or terminally differentiated myotubes that have exited the cell cycle. Overexpression and knock-down experiments using the C2C12 cell line revealed that the roles of miR-1 and miR-206 contrast with those of miR-133 (Chen et al., 2006; Kim, Lee, Sivaprasad, Malhotra, & Dutta, 2006). Overexpression of miR-1 or miR-206 promoted myogenic differentiation, while miR-133 overexpression enhanced myoblast proliferation, but represses differentiation. Knock-down assays show the converse effects on proliferation and differentiation. miR-206 is thought to promote muscle differentiation by, in part, inhibiting a component of DNA polymerase (Kim, Lee, Sivaprasad, Malhotra, & Dutta, 2006) as well as follistatin-1 (Fstl1) and utrophin (Utrn), genes that are suppressed in fibroblasts converted to skeletal muscle cells (Rosenberg, Georges, Asawachaicharn, Analau, & Tapscott, 2006). miR-133 is also shown in C2C12 cells to repress the neuronal homologue of the polypyrimidine tract-binding protein (nPTB), an important alternative splicing factor, during myoblast differentiation (Boutz, Chawla, Stoilov, & Black, 2007). Misexpression of miR-206 and miR-133a has been observed in a mouse model of muscular dystrophy (McCarthy, Esser, & Andrade, 2007), while miR-1 and -133 have been implicated in skeletal muscle hypertrophy (McCarthy & Esser, 2007).
Interestingly, miR-1 and miR-133, though co-expressed from the same gene, are shown to have opposite effects on apoptosis (Xu et al., 2007) and embryonic stem cell differentiation into cardiomyocytes (Ivey et al., 2008). Targeting predictions, as well as experimental evidences, suggest a negative regulatory feedback mechanism that may help explain the contrasting affects of miR-1 and miR-206 with those of miR-133. Our lab observed that miR-1 repressed HDAC4, which in turn represses MEF2, contributing to the increase of differentiated myotubes during miR-1 overexpression. The same study showed that miR-133 repressed SRF, therefore leading to increased myoblast proliferation (Chen et al., 2006), and this has been reinforced by other studies (Niu, Li, Zhang, & Schwartz, 2007). This regulatory feedback mechanism may contribute to the specificity and regulation of muscle cell proliferation and differentiation during development.
Cardiac Muscle
Cardiac development depends on an intricate coordination of different cell types. As the heart is the first organ to develop in the embryo, strict regulation is essential at critical temporal points. Cardiac specific deletion of Dicer, using a Cre-recombinase driven by the alpha myosin heavy chain (αMHC) promoter, which is expressed in the atria at embryonic day (ED) 8 and in both the ventricles and atria at birth, leads to early postnatal lethality in mice. Mutant hearts displayed dilated cardiomyopathy with cardiomyocyte sarcomeres affected (Chen et al., 2008). Conversely, early cardiac deletion of Dicer using an Nkx2–5 driven Cre-recombinase, which is expressed as early as ED 7.5, resulted in embryonic lethality and developmental heart defects (Zhao et al., 2007). Together, these studies demonstrate an essential role of miRNAs in proper heart development and function.
Studies in both Drosophila (Kwon, Han, Olson, & Srivastava, 2005; Sokol & Ambros, 2005) and mice (Zhao, Samal, & Srivastava, 2005) demonstrated the importance of miR-1 (dmiR-1 in Drosophila) during cardiogenesis. Specifically, flies lacking dmiR-1 displayed a loss of differentiation in some (but not all) muscle, and this dmiR-1 deletion resulted in pools of undifferentiated progenitor cells. Conversely, dmiR-1 overexpression disrupted cardioblast patterning in fractions of embryos. The delta/Notch signaling pathway is proposed to mediate the morphologic consequences of dmiR-1 in Drosophila. Similar to these results, it was demonstrated that overexpression of miR-1 in mouse hearts causes a decrease of ventricular cardiomyocyte pools through, in part, the targeting of Hand2, ultimately leading to heart failure (Zhao, Samal, & Srivastava, 2005). In the absence of miR-1-2, mouse hearts became hyperplastic, indicating a role for miR-1-2 in negatively regulating proliferation in cardiogenesis. Mutant cardiomyocytes also underwent karyokinesis, indicative of cell cycle dysregulation. These mice also displayed ventricular septal defects (VSDs), possibly due to the subsequent upregulation of the Notch signaling mediator Hrt2/Hey2, and the bHLH transcription factors Hand 1 and 2 (Zhao et al., 2007).
In support of studies using mice and flies, injections of miR-1 into Xenopus laevis embryos at the one-cell stage resulted in abnormal cardiac development and decreased cell proliferation. Similar injections of miR-133 resulted in a highly disorganized heart, absent of any cardiac looping or chamber formation, as well as an increase in cell proliferation (Chen et al., 2006). Recent genetic studies in mice further demonstrate the essential role of miR-133a in cardiomyocyte proliferation and heart development (Liu et al., 2008).
In addition, cardiac conduction defects have been observed in recent studies of miR-1 and miR-133. When overexpressed in rat hearts, miR-1 appears to diminish conduction, leading to arrhythmias, apparently through repression of the potassium channel subunit Kcnj2 and the gap junction protein connexin 43 (Yang et al., 2007). miR-1–2 has also been shown to directly target Irx5, a repressor of the potassium channel Kcnd2, leading to increased arrhythmias (Zhao et al., 2007). In a rabbit model of diabetes (Xiao et al., 2007), it was reported that the cardiac potassium ion channel protein ether-a-go-go (ERG) was repressed in mice overexpressing miR-133, leading to slower repolarization and QT prolongation, resulting in cardiac arrhythmia.
5. Medical Significance
As the characterization of specific miRNAs has increased exponentially in recent years, miRNAs have been implicated in a vast array of diseases, ranging from cancer to cardiovascular disorders. Many recent studies have documented dysregulation and abnormal expression of miRNAs in human patients and/or animal models for human disease. Given that miRNAs are also evolutionary conserved, a picture has emerged which points to a fundamental requirement of miRNAs in cardiac and skeletal muscle development and function, implying their potential involvement in muscle-related disease.
In 2008, our lab reported a correlation between Dicer protein levels and cardiac disease in human patients. In hearts failing from dilated cardiomyopathy, Dicer protein levels were very low when compared to non-failing hearts. Interestingly, when the same patients were fitted with a left ventricular assist device (LVAD), a device used to provide mechanical support for end-stage failing hearts, Dicer levels rebounded to levels comparable with non-failing hearts (Chen et al., 2008).
miR-1 and miR-133 have been implicated in cardiac muscle remodeling and their expression is dysregulated during cardiac hypertrophy and heart failure (Care et al., 2007; Sayed, Hong, Chen, Lypowy, & Abdellatif, 2007; Tatsuguchi et al., 2007). In mice, miR-1 is downregulated immediately following surgically induced cardiac hypertrophy, suggesting a role in cardiac remodeling. (Sayed, Hong, Chen, Lypowy, & Abdellatif, 2007). Interestingly, miR-133 suppression was reported to induce hypertrophy in mice by directly targeting RhoA, Cdc42, and Nelf-A/WHSC2 (Care et al., 2007). This paradox of both miR-1 and -133 perhaps sharing a similar function in cardiac hypertrophy underscores the complexity of miRNA targeting events that ultimately lead to biological function.
Taken with the previously mentioned studies examining miRNAs in models of muscular dystrophy (McCarthy, Esser, & Andrade, 2007), diabetes (Xiao et al., 2007), and cardiac arrhythmias (Xiao et al., 2007; Yang et al., 2007; Zhao et al., 2007), it is clear that miRNAs can have profound influence by specifically regulating multiple targets that coordinate within a common biological function, maintaining the homeostasis required for normal processes (Leung & Sharp, 2007). We predict that miRNAs will have therapeutic potential either by being used to target specific genes, or by becoming therapeutic targets themselves.
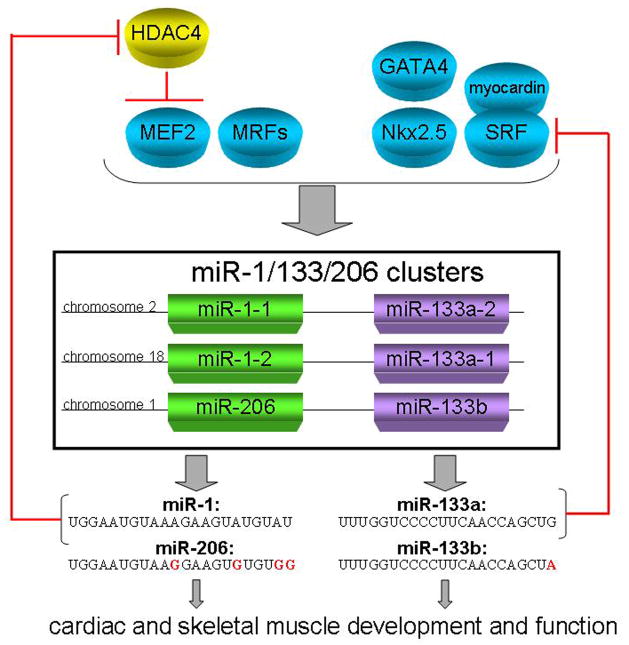
Figure 1. Regulation and function of muscle specific miRNAs.
Cardiac and skeletal transcription factors GATA4, Nkx2.5, myocardin, SRF, MEF2, and other myogenic regulatory factors (MRFs) regulate the expression of miR-1/133/206 polycistronic loci. The subsequent transcription of miR-1 promotes myocyte differentiation through the repression of HDAC4, which in turn represses MEF2. miR-133 promotes myocyte proliferation by repressing SRF. Thus, a feedback loop is created in which miR-1 and miR-133 and their respective transcription factors regulate each others expression.
TABLE 1.
miRNAs 1, 133, and 206 in cardiac and skeletal muscle development and disease.
| microRNA | Expression Pattern | Biological Roles | Validated Targets | References |
|---|---|---|---|---|
| miR-1 | Heart, Skeletal Muscle | Apoptosis, Cardiogenesis, Conduction, Myogenesis, Hypertrophy | Delta, Fibronectin, Hand2, HDAC4, Hsp60, Hsp70, RasGAP | (Chen et al., 2006; Kwon, Han, Olson, & Srivastava, 2005; Sokol & Ambros, 2005; Tatsuguchi et al., 2007) |
| miR-133 | Heart, Skeletal Muscle | Apoptosis, Conduction, Myogenesis, Hypertrophy | RhoA, SRF, Delta, Cdc42, Whsc2 | (Care et al., 2007; Chen et al., 2006; Kwon, Han, Olson, & Srivastava, 2005; Sokol & Ambros, 2005; Tatsuguchi et al., 2007) |
| miR-206 | Skeletal Muscle | Myogenesis | Fstl1, Pola1, Utrn | (Kim, Lee, Sivaprasad, Malhotra, & Dutta, 2006; Rosenberg, Georges, Asawachaicharn, Analau, & Tapscott, 2006) |
Abbreviations: Hand2, Heart and neural crest derivatives expressed 2; HDAC4, Histone deacetylase 4; Hsp60, heat-shock protein 60; Hsp70, heat-shock protein 70; RasGAP, Ras GTPase-activating protein; RhoA, Ras homolog A; SRF, Serum response factor; Fstl1, follistatin 1; Utrn, utrophin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21(1):71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev. 2006;16(5):525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Callis TE, Wang DZ. Taking microRNAs to heart. Trends Mol Med. 2008;14(6):254–260. doi: 10.1016/j.molmed.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105(6):2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. 2006;127(6):1101–1104. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2(3):219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174(5):677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005;102(52):18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130(4):581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22(23):3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A. 2007;104(52):20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000;6(2):233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102(1):306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am J Physiol Cell Physiol. 2007;293(1):C451–457. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol. 2007;19(6):618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, et al. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311(2):359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313(5795):1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103(23):8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175(1):77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100(3):416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19(19):2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, et al. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol. 2008;321(2):491–499. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132(12):2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, et al. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42(6):1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Xiao J, Luo X, Lin H, Zhang Y, Lu Y, Wang N, et al. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem. 2007;282(17):12363–12367. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120(Pt 17):3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13(4):486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]