Summary
microRNAs (miRNAs) are ~22 nucleotide regulatory RNAs derived from hairpins generated either by Drosha cleavage (canonical substrates) or by splicing and debranching of short introns (mirtrons). The 5′ end of the highly conserved Drosophila mirtron-like locus mir-1017 is coincident with the splice donor, but a substantial “tail” separates its hairpin from the 3′ splice acceptor. Genetic and biochemical studies define a biogenesis pathway involving splicing, lariat debranching, and RNA exosome-mediated “trimming”, followed by conventional dicing and loading into AGO1 to yield a miRNA that can repress seed-matched targets. Analysis of cloned small RNAs yielded six additional candidate 3′ tailed mirtrons in D. melanogaster. Altogether, these data reveal an unexpected role for the exosome in the biogenesis of miRNAs from hybrid mirtron substrates.
Introduction
Canonical miRNAs derive from primary miRNA (pri-miRNA) transcripts bearing one or more imperfect hairpins typically ~55–70 nt in length. In animals, pri-miRNAs are cleaved by the nuclear Drosha RNase III enzyme to release pre-miRNA hairpins, which are cleaved by the cytoplasmic Dicer RNase III enzyme to generate miRNA/miRNA* duplexes (Kim et al., 2009). One strand is preferentially selected for incorporation into an Argonaute complex, which uses the miRNA as a guide to identify mRNA targets for degradation and/or translational inhibition (Filipowicz et al., 2008). Animal miRNAs usually target partially complementary mRNAs, often involving 7 nt of Watson-Crick basepairing to positions 2–8 of the miRNA (the “seed”) (Bartel, 2009; Brennecke et al., 2005; Lai, 2002).
The repertoire of miRNA-class regulatory RNAs was expanded by the discovery of short hairpin introns known as mirtrons (Okamura et al., 2007; Ruby et al., 2007a). Mirtrons bypass Drosha cleavage by exploiting the spliceosome to generate their precursor ends. Following lariat debranching, linearized mirtrons adopt hairpin structures that are diced and loaded into Argonaute proteins as functional miRNAs. While best characterized in Drosophila, mirtrons exist in species as diverse as nematodes (Ruby et al., 2007a), mammals (Babiarz et al., 2008; Berezikov et al., 2007), avians (Glazov et al., 2008), and potentially plants (Zhu et al., 2008).
In the atypical Drosophila mirtron-like locus (mir-1017), only the 5′ hairpin terminus coincides with a splice junction; a substantial 3′ tail follows to its 3′ splice junction (Ruby et al., 2007a). We provide genetic and biochemical evidence that mir-1017 generates a miRNA-class regulatory RNA via a multistep process involving intron splicing and debranching, exosome-mediated trimming of the 3′ tail, and dicing. Analysis of Drosophila small RNA data revealed additional intronic hairpins bearing 3′ tails that are processed into miRNA/miRNA* duplexes, revealing a subfamily of miRNAs that transit an exosome-mediated biogenesis pathway.
Results
The tailed mirtron mir-1017 encodes a functional miRNA-class regulatory RNA
mir-1017 is located in an intron of the nicotinic acetylcholine receptor alpha subunit gene at 96AB (nAchRalpha-96Ab), previously known as the Dα2 subunit (Bossy et al., 1988). This locus was annotated, on the basis of ~150 small RNA reads, as a mirtron-like precursor for which only the 5′ hairpin end coincides with a splice junction; a ~100 nt extension ensues before the 3′ splice site (Ruby et al., 2007a). Small RNA data from male heads (Chung et al., 2008) containing ~6000 reads for mature miR-1017 now revealed 14 miR-1017* reads, which initiated precisely with the 5′ splice donor sequence. Together with mature miR-1017, these adopted a typical miRNA duplex exhibiting 3′ overhangs (Figure 1A). These features were consistent with its biogenesis via a splicing- and dicing-dependent mechanism.
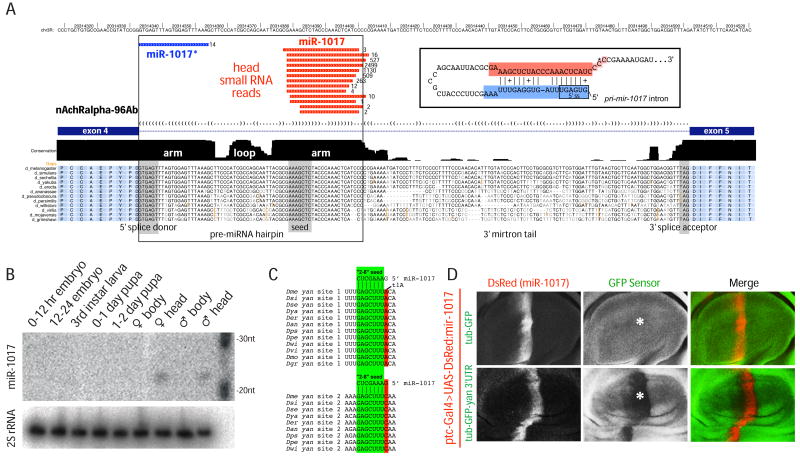
Figure 1.
Drosophila mir-1017 , a tailed mirtron locus. (A) Alignment of the exon 4–5 region of nAchalpha-96Ab across 12 Drosophilids (http://ucsc.genome.org). The pre-miRNA hairpin embedded at the 5′ end of the intron follows the characteristic pattern of miRNA evolution in that the hairpin arms are much more conserved than the terminal loop; the miRNA* arm is also slightly more diverged than the miRNA-encoding arm. A ~100 nt tail lacking sequence constraint separates the 3′ end of the hairpin from the 3′ splice acceptor site. Small RNA data from male heads revealed that miR-1017* reads (blue) initiate precisely with the 5′ splice donor, and together with its partner miR-1017 (red) form a typical miRNA/miRNA* duplex with 3′ overhangs (inset; the asterisk denotes most frequent 3p read terminus). (B) Northern analysis indicates the specific accumulation of mature miR-1017 in adult heads. (C) Conservation of two miR-1017 seed matches in the yan 3′ UTR. (D) Sensor assay for miRNA-type target repression by miR-1017. Shown are wing pouches of third instar wing imaginal discs bearing ptc-Gal4, UAS-DsRed-mir-1017, and either tub-GFP (above) or tub-GFP-yan 3′ UTR (below). DsRed fluorescence marks the domain of miR-1017 activity while the center panels show GFP staining; merged channels are shown to the right. Ectopic miR-1017 did not affect the control sensor but robustly and specifically repressed the GFP-yan 3′ UTR sensor (asterisk).
mir-1017 has been highly conserved across the 12 sequenced Drosophila species (Figure 1A), and its evolutionary profile follows the characteristic evolutionary pattern for conserved canonical miRNA and mirtron genes (Lai et al., 2003; Okamura et al., 2007). Specifically, its terminal loop is much more divergent than the stem, and the miRNA* is slightly less conserved than the mature miRNA. Pre-miRNA hairpins typically exhibit conservation in flanking 5′ and 3′ sequence, since these mediate accurate Drosha processing (in the case of canonical miRNAs) (Han et al., 2006) or have protein-coding potential (in the case of mirtrons). In contrast, the intronic sequence immediately downstream of the pre-mir-1017 hairpin has diverged substantially (Figure 1A), and no potential “AG” splice acceptor exists until the conserved, constitutive, intron-exon junction ~100 nt downstream. Developmental Northern analysis detected miR-1017 only in adult heads (Figure 1B), consistent with head-specific expression of its host gene (http://www.flyatlas.org). This implied that expression of miR-1017 is coupled to transcription of Dα2.
TargetScan (http://www.targetscan.org/) predicted the Ets-domain transcription factor encoded by anterior open/yan as a likely target of miR-1017. The yan 3′ UTR contains a miR-1017 8mer site (2–8 seed pairing+t1A) perfectly conserved across all 12 sequenced Drosophilids, and a “2–8” 7mer site conserved across 9 species (Figure 1C). We used an in vivo sensor assay (Stark et al., 2003) to ask whether miR-1017 could repress via the yan 3′ UTR (Li and Carthew, 2005). While activation of UAS-DsRed-mir-1017 using ptc-Gal4 did not affect a tub-GFP control transgene, it specifically and robustly repressed a tub-GFP-yan 3′ UTR transgene (Figure 1D). We confirmed the ability of mir-1017 to inhibit a renilla luciferase-yan 3′ UTR sensor in transfected S2R+ cells, which do not express miR-1017 endogenously (Supplementary Figure 1). This was due directly to the miR-1017 seed matches, since mutation of these sites in yan abrogated repression (Supplementary Figure 1). Therefore, miR-1017 exhibits typical miRNA activity in both S2 cells and in the animal.
Sequence requirements for the biogenesis of 3′ tailed mirtrons
We used structure-function studies to probe mir-1017 biogenesis (Figure 2A). The pri-mir-1017 intron tail has incurred abundant nucleotide changes, insertions, and deletions during Drosophilid radiation (Figure 1A), suggesting that substantial flexibility in “tail” sequence is compatible with access to the miRNA pathway. To verify this, we compared UAS-DsRed constructs carrying ~500 bp minigenes centered on the mir-1017 intron from Drosophila melanogaster (Dme), Drosphila simulans (Dsi) and Drosophila mojavensis (Dmo) in S2R+ cells. All three yielded mature miR-1017 (Figure 2B), verifying that different tailing sequences are compatible with trimming of pri-mir-1017. On the other hand, pri-mir-1017 definitively requires splicing, since point mutation of the 5′ splice site (Figure 2A, “mir-1017 5′ssMut”) abolished the appearance of all intermediate and mature forms of miR-1017 (Figure 2B). The splice-site mutant generated similar levels of DsRed as wild-type constructs, indicating normal expression (Supplementary Figure 2A).
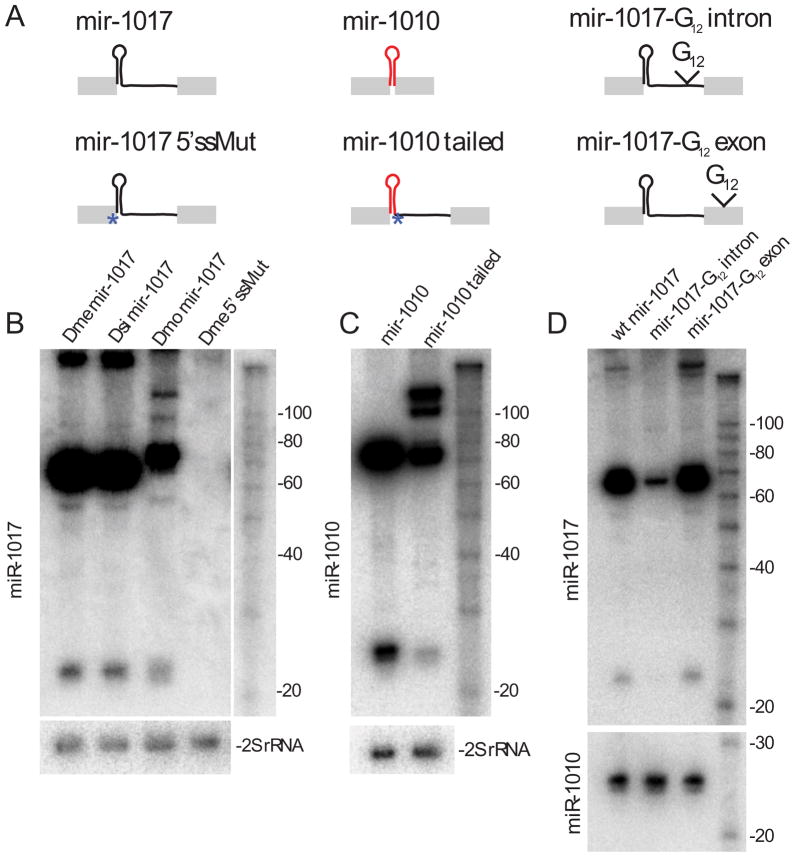
Figure 2.
Structure-function analysis of tailed mirtron substrates. These panels depict Northern analysis of S2R+ cells transfected with ub-Gal4 and UAS-DsRed-mir-1017 or mir-1010 constructs. (A) Constructs used in these tests. Wild-type mir-1017 plasmids contained ~500 bp including endogenous flanking exons from D. melanogaster (Dme), D. simulans (Dsi) and D. mojavensis (Dmo); a Dme 5′ splice site mutant was generated in this context. The “mir-1010” construct expresses this mirtron from a 500 bp context, while the “mir-1010 tailed” construct contains a point mutation in its endogenous 3′ splice acceptor, fused to the mir-1017 intron tail. The last constructs contain 12 guanine residues inserted into the intron or exon of pri-mir-1017. (B) Constructs derived from all three species yielded mature miR-1017. Mutation of the GU 5′ splice donor to “CC” abolished the production of all intermediate and mature Dme miR-1017 products. (C) The conventional mir-1010 mirtron can be converted into a functional tailed mirtron. (D) Insertion of G12 into the mir-1017 intron interferes with generation of the pre-miRNA hairpin and mature product, but biogenesis is unaffected when G12 was inserted into exon sequence. In this experiment, mir-1017 and mir-1010 plasmids were cotransfected and production of mature miR-1010 was assayed as a blotting control; other experiments assayed 30 nt 2S rRNA hybridization.
To assess a potential intrinsic requirement for the mir-1017 hairpin itself, we exchanged it for the mir-1010 mirtron hairpin (“mir-1010-tailed”) in the context of the mir-1017 intron; the endogenous 3′ splice site of mir-1010 was mutated to force usage of the Dα2/mir-1017 splice site. As with the normal mir-1010 construct (Okamura et al., 2007), the tailed variant supported the production of mature miR-1010 (Figure 2C). Therefore, access to the tailed mirtron biogenesis pathway is portable to synthetic substrates.
We inferred that after splicing, the tailed region following the mir-1017 hairpin must be removed by endonucleolytic or exonucleolytic cleavage. Putative 3′ end processing was consistent with the relatively imprecisely defined 3′ ends of cloned miR-1017 reads (Figure 1A). To examine this further, we inserted a dozen guanine residues into the dme-mir-1017 construct (Figure 2A); such a poly-G tract was previously used to block exonuclease processing (Anderson and Parker, 1998). We inserted the poly-G tract 20nt upstream of the 3′ splice site (G12 intron), and made a control insertion 30 nt downstream of the 3′ splice site (G12 exon). Placement of G12 into the intron, 80 nt downstream of the mir-1017 hairpin, severely disturbed the accumulation of hairpin and mature forms of mir-1017 (Figure 2D). In contrast, insertion of G12 into the 3′ exon did not interfere with mir-1017 biogenesis. rt-PCR confirmed effective splicing of both G12 intron and G12 exon variants, although both accumulated slightly more unspliced product relative to wildtype (Supplementary Figure 2B). The observation that the spliced pri-mir-1017-G12 intron was barely able to generate pre-miRNA hairpin, and produced almost no mature miRNA, implied that removal of the mir-1017 “tail” depends on exonuclease activity.
Biogenesis of tailed mirtrons requires components of the mirtron pathway
To determine specific factors involved in miR-1017 biogenesis, we depleted members of the miRNA/mirtron pathways from S2R+ cells and then co-transfected them with ub-Gal4 and UAS-DsRed-mir-1017 (Figure 3A). Harvested RNAs were subjected to Northern blotting with antisense probes to miR-1017, miR-276a (a canonical miRNA) and 2S rRNA as a control; knockdown efficiency was determined by qRT-PCR (Supplementary Figure 3A).
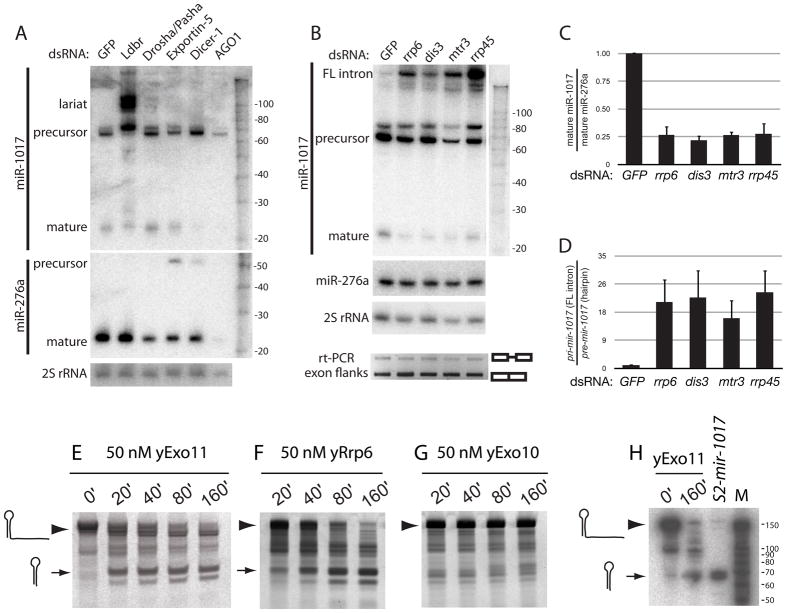
Figure 3.
Unique biogenesis of tailed mirtrons via an exosome-mediated pathway. (A) Depletion of Drosha and pasha in S2 cells reduced mature miR-276a but not miR-1017. Conversely, miR-1017 was highly sensitive to loss of lariat debranching enzyme (Ldbr); these cells accumulate branched intermediates (see also Supplementary Figure 4). Knockdown of Dicer-1 reduced the level of mature miR-1017 and miR-276a, and knockdown of AGO1 nearly eliminated these species. (B) Mature miR-1017 was reduced relative to miR-276a upon depletion of the rrp6, dis3, mtr3, and rrp45, while its full-length intron accumulated. rt-PCR across flanking exons indicated that exosome depletion did not alter transcription or splicing of pri-mir-1017. (C) miR-1017 was selectively depleted relative to miR-276a in exosome knockdown cells. Ratio of mature miR-1017 relative to miR-276a were normalized to their ratio in GFP knockdown cells. Standard error is shown; all differences in exosome knockdown cells were significant to <0.001 by pairwise T-test. (D) Accumulation of full-length pri-mir-1017 intron in exosome knockdown cells; all differences were significant to <0.025 by pairwise T-test. These graphs summarize data from 5 independent knockdown samples analyzed by Northern blot; error bars indicate standard error. (E–H) Full length in vitro transcribed mir-1017 intron was incubated with reconstituted 11-subunit yeast nuclear exosome holoenzyme (yExo11), yRrp6 alone, or 10-subunit yeast cytoplasmic exosome lackng yRrp6 (yExo10). Timecourse assays reveal the conversion of the full length substrate (arrowhead) into a relatively stable hairpin product (arrow), here detected with SYBR Gold staining of yExo11 (E) and yRrp6 (F) reactions. yExo10 failed to process the mir-1017 intron (G), but was active on other substrates (Supplementary Figure 5). (H) The yExo11-trimmed product co-migrates with in vivo processed mir-1017 pre-miRNA hairpin, as assayed by Northern blot; “M”, RNA size markers. The full-length 169 nt intron migrated slightly differently in this experiment than in panel 3B; see also Supplementary Figure 4.
The biogenesis of miR-1017 exhibited hallmarks of a conventional mirtron, in that it was clearly dependent on Ldbr but was little affected by Drosha/Pasha knockdown (Figure 3A). In addition to a 2-fold decrease in mature miR-1017, Ldbr knockdown cells accumulated several putatively branched intermediates (Figure 3A, top blot). Analysis of 16%, 12% and 8% PAGE revealed three species with variable mobility upon Ldbr depletion (Supplementary Figure 4), indicating their non-linear structure. Along with the splice site mutant test (Figure 2), these data confirmed the biogenesis of miR-1017 via a genuine mirtron pathway. However, mir-1017 biogenesis merged with the canonical miRNA pathway, since it required Dicer-1 and AGO1 (Figure 3A). These characteristics confirmed the classification of 3′ tailed mirtron products as genuine miRNAs.
Trimming of tailed-mirtrons is mediated by the RNA exosome
An attractive candidate to remove the mir-1017 tail is the RNA exosome, a multisubunit complex that serves as the major 3′–5′ exonuclease for RNA turnover in eukaryotic cells (Houseley et al., 2006). This multisubunit complex degrades or processes substrates via Dis3 (Rrp44), a processive 3′ to 5′ exoribonuclease, and Rrp6, a distributive 3′ to 5′ exoribonuclease. While Dis3 is a core constituent of both nuclear and cytoplasmic forms of the exosome, Rrp6 is believed to be specific to the nuclear exosome.
To test the requirement for the exosome in maturation of miR-1017, we depleted a panel of exosome subunits using dsRNAs to rrp6, dis3, mtr3, and rrp45 (Supplementary Figure 3A). We also observed accumulation of precursor rRNA with all exosome subunits excepting Rrp45 (Supplementary Figure 3B), providing additional evidence for functional exosome loss. Mature miR-1017 was reproducibly reduced in cells depleted of any of the four exosome subunits (Figure 3B), under conditions in which the accumulation of endogenous miR-276a (Figure 3B) and bantam miRNA (data not shown) were barely affected. The data from five independent knockdown experiments were quantified in Figure 3C. We also excluded that the knockdown conditions interfered with transcription and splicing across the mir-1017 host intron, since rt-PCR using primers in the flanking exons revealed similar amounts of unspliced and spliced products in cells treated with GFP or exosome dsRNAs (Figure 3B, bottom).
Treatment with exosome dsRNAs resulted in accumulation of full-length pri-mir-1017 intron (Figure 3B, top blot). The absolute amount of untrimmed intron was somewhat variable, and this may be due to variable amounts of residual exosome proteins in these knockdowns, which were not tolerated as well as other dsRNA incubations. However, quantification of the ratio of pre-mir-1017 hairpin to full-length pri-mir-1017 intron across multiple independent knockdowns demonstrated highly reproducible defects in the conversion of the linearized full-length intron into the pre-miRNA hairpin, upon depletion of any of the four exosome subunits (Figure 3D).
In vitro reconstitution of the 3′ tail trimming reaction
The extent of exosome- and Rrp6-mediated decay can be modulated by substrate structure (Liu et al., 2006), consistent with their characterized roles in maturing the 3′ ends of structured RNAs such as rRNAs, snRNAs and snoRNAs (Allmang et al., 1999). We hypothesized that the structure of the pre-miRNA hairpin embedded in the tailed mirtron precursor might permit its release from an engaged exosome, and sought to test this in vitro. To date, active 11-subunit RNA exosome holoenzyme (Exo11) has only been successfully reconstituted using purified components from S. cerevisiae (Liu et al., 2006). Nevertheless, given the highly conserved nature of the RNA exosome, we considered this to be a relevant experimental system.
We incubated purified, reconstituted yExo11 (Greimann and Lima, 2008) with full-length pri-mir-1017 intron produced by in vitro transcription, and analyzed products using SYBR Gold staining. These reactions yielded relatively stable accumulation of a ~70 nt intermediate corresponding to the pre-mir-1017 hairpin (Figure 3E). We obtained similar results in reactions with purified yRrp6 (Figure 3F). In contrast, purified yExo10, the cytoplasmic exosome complex containing Dis3 ribonuclease but lacking Rrp6, was unable to process the pri-mir-1017 intron (Figure 3G). Using aliquots from the same exosome preparations used for mir-1017 tests, we demonstrated all three purified complexes to have exoribonuclease activity using a control RNA substrate (Liu et al., 2006) (Supplementary Figure 5).
Northern analysis of the 0′ and 160′ yExo11 reactions with full length pri-mir-1017 intron showed that its product comigrated with the hairpin produced by in vivo processed UAS-DsRed-mir-1017 (Figure 3H). The liberation of a pre-miRNA-like hairpin, following digestion of the debranched tailed mirtron, illustrates how pre-mir-1017 avoids degradation during exosome-mediated biogenesis. As Rrp6 was necessary (as judged by comparing yExo10 and yExo11 reactions) and sufficient for removal of the pri-mir-1017 intron tail, we infer that trimming in vivo normally occurs in the nucleus and likely involves an Rrp6-containing exosome. However, we do not exclude the possibility of a handoff between different exosome complexes in vivo, or the possibility that Rrp6 operates alone during some aspect of miR-1017 maturation.
Genome survey for other Drosophila tailed mirtrons
We recently annotated the second intron of CG7927/mir-2501 as containing a mirtron (Berezikov et al., 2010); however, reconsideration of this locus indicated a 4–5 nt tail from the end of the 3p cloned read to the splice acceptor (Supplementary Figure 6). Since this tail should be removed for the hairpin to serve as an Exportin-5 substrate (Okada et al., 2009), we believe that this is also a tailed mirtron locus. We searched for other loci using a collection of ~15 million Drosophila short RNA sequences (Chung et al., 2008; Ruby et al., 2007b). We mapped reads to introns <200 nt in length, and generated candidate RNA folds. We then asked if any loci satisfied the criteria of having a hairpin at one end of the intron, for which >20 cloned short RNAs mapped to both arms of the hairpin as a duplex with 3′ overhangs. The most highly expressed locus was mir-1017, but six other loci generated 25–150 reads and warranted classification as tailed mirtrons (Figure 4 and Supplementary Figure 6). These loci are modestly expressed compared to miR-1017, and it is possible that they are less effective substrates of the tailed mirtron pathway. Nevertheless, the recovery of miRNA and miRNA* reads indicates their relatively specific processing by a Dicer enzyme (Chiang et al., 2010).
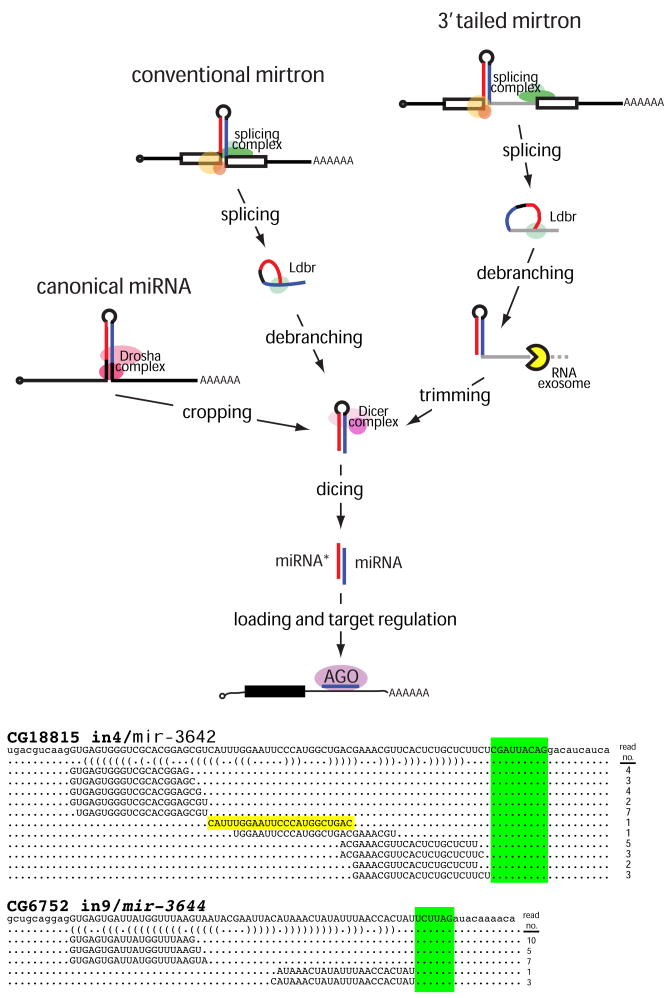
Figure 4.
Pathways that generate miRNA-class regulatory RNAs from short hairpins in Drosophila. In the canonical pathway, the RNase III enzyme Drosha “crops” a primary miRNA transcript to release the pre-miRNA. In the conventional mirtron pathway, the splicing complex liberates short hairpin introns from protein-coding genes. Following their linearization by lariat debranching enzyme (Ldbr), these can fold into pre-miRNA mimics. In the 3′ tailed mirtron pathway, a 3′ tail separates the hairpin from the 3′ splice acceptor site. These require Ldbr and the RNA exosome to remove the 3′ tail up to the hairpin, yielding the pre-miRNA. In all cases, pre-miRNA hairpins are cleaved by the cytoplasmic RNase III Dicer-1 to yield miRNA/miRNA* duplexes. One strand is predominantly selected for incorporation into an AGO1 complex to repress seed-matched targets. At the bottom are additional examples of D. melanogaster tailed mirtrons whose inferred trimmed tails are highlighted green (see also Supplementary Figure 6). Note that the precise terminal loop of the CG18815_in4/mir-3642 tailed mirtron was also cloned (yellow), providing additional evidence for Dicer cleavage.
Curiously, while our survey was open to the potential derivation of hairpins from either 5′ or 3′ intron ends, all of the loci that we recovered exhibited 3′ tails. mir-1017 proved to have the longest 3′ tail (~100 nt), followed by mlc-c/mir-3645 (45 nt); the remainder had ~10 nt tails (CG18815/mir-3642 and CG6370/mir-3643) or 4–6 nt tails (CG3630/mir-3641, CG7927/mir-2501, and CG6752/mir-3644). Even with the shorter tails, the miRNA/miRNA* duplexes terminate upstream of the 3′ splice site, indicative of a trimming reaction preceding their dicing. These data provide evidence for a family of exosome-processed mirtrons in Drosophila.
Discussion
Diverse pathways generate Dicer substrates and miRNA-class regulatory RNAs
We showed that a subset of Drosophila mirtrons encode a terminal extension 3′ of the pre-miRNA hairpin, which is “trimmed” by the RNA exosome (Figure 4). Otherwise, tailed mirtrons are similar to conventional mirtrons in that they bypass the Microprocessor by accessing the splicing and debranching pathway (Figure 4). Most of our studies focused on the 3′ tailed mirtron mir-1017, which is strictly conserved across Drosophilid evolution and regulates conserved target genes including yan. We also identified more recently-evolved substrates that appear to access the 3′ tailed mirtron pathway. Currently, we and others annotated 151 canonical miRNAs, 18 conventional mirtrons and 7 tailed mirtrons in Drosophila melanogaster (Berezikov et al., 2010; Ruby et al., 2007a; Ruby et al., 2007b; Sandmann and Cohen, 2007; Stark et al., 2007). We envision the conventional mirtron pathway as an “add-on” to the canonical miRNA pathway, in which splicing has evolved to generate substrates that exploit a pre-existing canonical pathway. Similarly, we hypothesize that the tailed mirtron pathway represents an “add-on” to the conventional mirtron pathway, whereby the RNA exosome has been recruited to permit access of an asymmetric mirtron into the canonical miRNA pathway.
The RNA exosome is well known for its role in the turnover of normal mRNAs and abnormal transcripts. However, this study provides additional evidence for positive roles of the exosome in the biogenesis of non-coding RNAs. In previous studies, the exosome was shown to be required for maturation of rRNA, snRNAs and snoRNAs through 3′–5′ trimming of terminal nucleotides. Thus, the consequence of blocking exosome processing of these substrates is the retention of undesired 3′ nucleotides (Allmang et al., 1999). The role of the exosome in biogenesis of 3′ tailed mirtrons is distinct in that substrate trimming is prerequisite for subsequent steps in substrate metabolism. In this sense it is reminiscent of processing of yeast 5.8S rRNA, which involves consecutive exonucleolytic processing reactions by the exosome followed by the Rex proteins (van Hoof et al., 2000). Our evidence that 3′ trimming is mediated by the nuclear Exo11 complex via Rrp6 (Figure 3E, F) suggests that the trimming and dicing reactions are compartmentalized in the cell. Removal of the 3′ tail may be requisite for efficient export by Exportin-5, which is selective for hairpins with a short 3′ overhang (Okada et al., 2009).
Other non-canonical substrates generate miRNA-class regulatory RNAs, including certain snoRNAs (Ender et al., 2008) and tRNA (Babiarz et al., 2008). More recently, a viral tRNA/miRNA fusion was found to use tRNaseZ to liberate a pre-miRNA hairpin (Bogerd et al., 2010). In addition, siRNA-class regulatory RNAs derive from other classes of inverted repeat transcripts, such as hairpin RNAs and endo-shRNAs (Babiarz et al., 2008; Czech et al., 2008; Kawamura et al., 2008; Okamura et al., 2008b). Finally, a variety of trans-encoded substrates, generating either perfect or imperfect dsRNA, access Dicer pathways to generate endo-siRNAs in Drosophila (Chung et al., 2008; Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008a; Okamura and Lai, 2008) and mammals (Babiarz et al., 2008; Tam et al., 2008; Watanabe et al., 2008). Altogether, a multitude of biogenesis pathways have emanated from the simple building blocks of cis- or trans-encoded dsRNA and a Dicer-class enzyme to generate diverse regulatory RNAs (Okamura et al., 2008c).
Experimental Procedures
We used previously published methods for knockdowns, transfections and Northern analysis (Okamura et al., 2007), exosome analysis (Greimann and Lima, 2008), and Drosophila immunohistochemistry (Lai and Rubin, 2001). A detailed description of the experimental methods, along with all of the primer sequences used for cloning or hybridization, are found in the Supplementary Text.
Supplementary Material
Acknowledgments
We thank Katsutomo Okamura for preliminary Northern analysis of miR-1017 and critical reading of the manuscript, and David Tyler for constructing UAS-DsRed-mir-1017 flies. We are grateful to Drs. Richard Carthew and Stephen Cohen for gifts of Drosophila stocks and plasmids that were critical for this study. J.C.G. and C.D.L. were supported in part by a grant from the National Institutes of Health (GM079196). E.C.L. was supported by the Sidney Kimmel Cancer Foundation, the Burroughs Wellcome Foundation, the Alfred Bressler Scholars Fund and the US National Institutes of Health (GM083300 and HG004261).
Footnotes
Supplemental Information includes 6 figures, Supplemental Experimental Procedures, and Supplemental References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. Embo J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. Embo J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung W-J, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Liu N, Flynt AS, Hodges E, Rooks M, Hannon GJ, Lai EC. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat Genet. 2010;42:6–9. doi: 10.1038/ng0110-6. author reply 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossy B, Ballivet M, Spierer P. Conservation of neural nicotinic acetylcholine receptors from Drosophila to vertebrate central nervous systems. Embo J. 1988;7:611–618. doi: 10.1002/j.1460-2075.1988.tb02854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of MicroRNA-Target Recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010 doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Current Biology. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, et al. An endogenous siRNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greimann JC, Lima CD. Reconstitution of RNA exosomes from human and Saccharomyces cerevisiae cloning, expression, purification, and activity assays. Methods Enzymol. 2008;448:185–210. doi: 10.1016/S0076-6879(08)02610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada T, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Lai EC. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai EC, Rubin GM. neuralized functions cell-autonomously to regulate a subset of Notch-dependent processes during adult Drosophila development. Dev Biol. 2001;231:217–233. doi: 10.1006/dbio.2000.0124. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42.41–R42.20. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Carthew RW. A microRNA Mediates EGF Receptor Signaling and Promotes Photoreceptor Differentiation in the Drosophila Eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila. Nat Struct Mol Biol. 2008a;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung W-J, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008b;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Lai EC. The long and short of inverted repeat genes in animals: microRNAs, mirtrons and hairpin RNAs. Cell Cycle. 2008c;7:2840–2845. doi: 10.4161/cc.7.18.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007a;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007b;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Cohen SM. Identification of novel Drosophila melanogaster microRNAs. PLoS ONE. 2007;2:e1265. doi: 10.1371/journal.pone.0001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA Targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. Embo J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.