Abstract
Background
Transcription cofactors related to Drosophila Schnurri facilitate the transcriptional programs regulated by BMP signaling in C. elegans, Drosophila, Xenopus, and mouse. In different systems, Schnurri homologs have been shown to act as either agonists or antagonists of Smad function, and as either positive or negative regulators of transcription. How Schnurri proteins achieve this diversity of activities is not clear. The C. elegans sma-9/schnurri locus undergoes alternative splicing, including an unusual trans-splicing event that could generate two non-overlapping shorter transcripts.
Results
We demonstrate here that the shorter transcripts are expressed in vivo. Furthermore, we find that one of the short transcripts plays a tissue-specific role in sma-9 function, contributing to the patterning of male-specific sensory rays, but not to the regulation of body size. Based on previous results, we suggest that this transcript encodes a C-terminal SMA-9 isoform that may provide transcriptional activation activity, while full length isoforms may mediate transcriptional repression and/or activation in a context-dependent manner.
Conclusion
The alternative trans-splicing of sma-9 may contribute to the diversity of functions necessary to mediate tissue-specific outputs of BMP signaling.
Background
The transforming growth factor β (TGFβ) superfamily comprises a large number of secreted peptide growth factors that have major regulatory effects on cell growth and differentiation [1,2]. Members of this superfamily include the TGFβs, the prototypes of the superfamily; the bone morphogenetic proteins (BMPs); and other members such as activin, inhibin, and Nodal. TGFβ superfamily ligands bind to a heteromeric receptor complex at the cell surface. This complex contains two related transmembrane serine/threonine kinases, the type I and type II receptors [3,4]. Signaling downstream of the receptors is mediated by the Smad proteins, which shuttle between the cytoplasm and nucleus to regulate target gene expression [5]. Type I receptors directly activate receptor-regulated Smads (R-Smads) by phosphorylation at C-terminal SXS sequences [6,7]. Phosphorylation of R-Smads promotes heterotrimeric complex formation with Co-Smads and accumulation in the nucleus to regulate gene transcription [7-11]. In mammals, five R-Smads are present: two (Smad2,3) that transduce TGFβ/activin/Nodal signals and three (Smad1,5,8) that transduce BMP signals. Strikingly, the TGFβ family ligands far outnumber the Smads available for signal transduction. Furthermore, many of these ligands are capable of eliciting diverse context-dependent responses. Thus, Smad complexes must be capable of mediating multiple diverse outcomes. It is thought that Smad complexes rely in part on transcription cofactors for appropriate regulation of target genes.
In the nematode Caenorhabditis elegans, the BMP-related factor DBL-1 regulates body size and male tail morphogenesis via a conserved receptor/Smad signaling pathway [12]. Using a genetic approach to uncover components of this pathway, we previously identified sma-9, a gene that is required for the body size and male tail patterning functions of DBL-1 [13]. sma-9 is also required for patterning of the mesodermal lineage, in which it acts antagonistically to the DBL-1 pathway [14]. sma-9 is predicted to encode multiple protein products homologous to Drosophila Schnurri, a large zinc finger transcription cofactor that functions in Dpp/BMP signaling [15,16]. Analysis of sma-9 therefore provides the opportunity to elucidate the requirements for transcription cofactor function in an in vivo model system during the course of development. In addition to Drosophila Schnurri and C. elegans SMA-9/Schnurri, three vertebrate Schnurri homologs have been identified. These bind the κB-binding site and function in T cell development [17-19]. Notably, vertebrate Schnurri homologs have more recently been demonstrated to mediate transcriptional regulation downstream of BMP and TGFβ ligands, indicating a conserved role for these family members in TGFβ signal transduction [20-22].
The sma-9 open reading frame (ORF) predicted from genomic sequence encodes a protein of 2170 aa consisting of an N-terminal Gln-rich domain (encoded by predicted exons 1-3) and a C-terminal domain containing seven Zn fingers (in predicted exons 12-22). The sequencing of cDNA clones, however, revealed at least eight different mRNA species with alternative protein coding regions [13]. Interestingly, like sma-9, human Shn-1 and Shn-3 genes undergo alternative splicing [23,24], but the functional consequences of this processing have not been addressed. The sma-9 cDNA clones were classified based on their potential to code for the seven C-terminal Zn fingers: class I encodes all seven Zn fingers (ZF1-7), class II encodes the first pair and the triplet of Zn fingers (ZF1-5), and class III encodes only the first pair (ZF1-2). To determine whether these domains have different functions in vivo, Foehr et al. created genomic/cDNA hybrid constructs and tested their ability to rescue the body size and mesodermal patterning defects of sma-9 mutants [14]. They found that constructs encoding class I and class II C-termini were capable of rescuing both phenotypes, while the class III construct was not, suggesting that the presence of the Zn finger triplet is critical for function in body size and mesodermal lineage regulation.
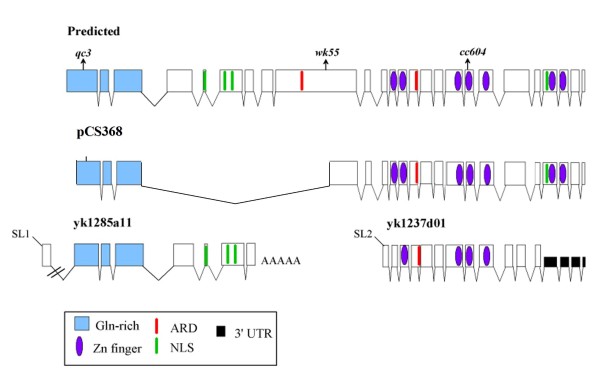
Since sma-9 is a large gene, most of the existing cDNA clones contain incomplete coding sequences that are missing the 5' end of the gene. Two cDNA clones, yk1285a11 and yk1237d01, however, are notably different (Figure 1). Although these two cDNA clones are less than half the length of the predicted transcript, both of them have all of the hallmarks of a full-length cDNA: trans-spliced leader sequence, poly(A) tail, and a complete ORF. In C. elegans, many transcripts are processed at the 5' end by trans-splicing, which results in the addition of a 22-nucleotide splice leader sequence, SL1 or SL2 [25]. The mechanism of trans-splicing is similar to that of cis-splicing (intron removal), except that the splice donor sequences are provided by the SL1 and SL2 genes, rather than being contained within the context of the individual gene [25]. About half of all C. elegans genes are subjected to SL1 trans-splicing at the 5' end. A smaller subset of C. elegans genes are transcribed in polycistronic operons. In an operon, the 5'-most gene generally receives the SL1 splice leader. The individual downstream genes in an operon are then separated via SL2 trans-splicing to the 5' ends of each of the downstream genes. SL2 trans-splicing is accompanied by polyadenylation to create the 3' end of the neighboring gene upstream. yk1285a11 contains the SL1 splice leader and sequences from predicted exons 1-7. yk1237d01 contains the SL2 splice leader and sequences from predicted exons 11-25. As previously noted [13], the putative intercistronic region contains features reminiscent of the identified signals for trans-splicing, including a U-rich sequence, but is atypically long [26,27]. Thus, the structures of the sma-9 cDNA clones yk1285a11 and yk1237d01 suggest that they may represent two transcripts processed from a single longer transcript by SL2 trans-splicing and polyadenylation as normally occurs in a C. elegans polycistronic operon.
Figure 1.
Structure of sma-9 transcript variants. The predicted sma-9 intron-exon structure is shown above. Arrows mark positions of nonsense codons in three sma-9 alleles. pCS386 is the most complete cDNA variant isolated to date. The two short variants represented by cDNA clones yk1285a11 (A11) and yk1237d01 (D01) are shown below. ARD: acidic residue-rich domain; NLS: nuclear localization signal.
Since each of these trans-spliced short sma-9 transcripts was only represented by a single cDNA clone, we could not be certain that the clones were not due to cloning artifacts or a rare spurious event. Furthermore, our previous analysis did not address whether these short transcripts provide any function required for DBL-1 signal transduction. We therefore address here several remaining questions about these predicted transcripts. First, can we verify that the mRNA variants represented by yk1285a11 and yk1237d01 are expressed in vivo, rather than being artifacts produced during cDNA library construction? If so, then how is the expression of these variants regulated? Finally, do these variants show functional differences between each other and/or relative to full-length transcripts in vivo? In this work, we will refer to the splice variants represented by clones yk1285a11 (5') and yk1237d01 (3') as A11 and D01, respectively.
Results
Isolation of cDNAs spanning upstream and downstream regions
The sma-9 ORF predicted from genomic sequence is encoded by 25 exons ([13]; Figure 1). Previously characterized cDNA clones, however, contained only a subset of these 25 exons and, in particular, none of the existing cDNA clones spanned a region including both the N-terminal Gln-rich domain and the C-terminal Zn finger domains. We therefore used primers in exons 1 and 25 with the potential to generate nearly full-length cDNA inserts by RT-PCR. After RT-PCR, the clones with the longest inserts were selected for sequencing. The most complete cDNA clone isolated by this approach was pCS368, in which exons 4-8 and part of exon 9 are spliced out (Figure 1). This transcript form is reminiscent of a form identified by the ORFeome project, in which exon 3 becomes spliced to exon 20 [28]. These results thus support the existence of sma-9 transcripts capable of encoding both the Gln-rich and the Zn finger domains. Additional support derives from RT-PCR data using exon 9 primers (see below).
Expression of sma-9 variant transcripts
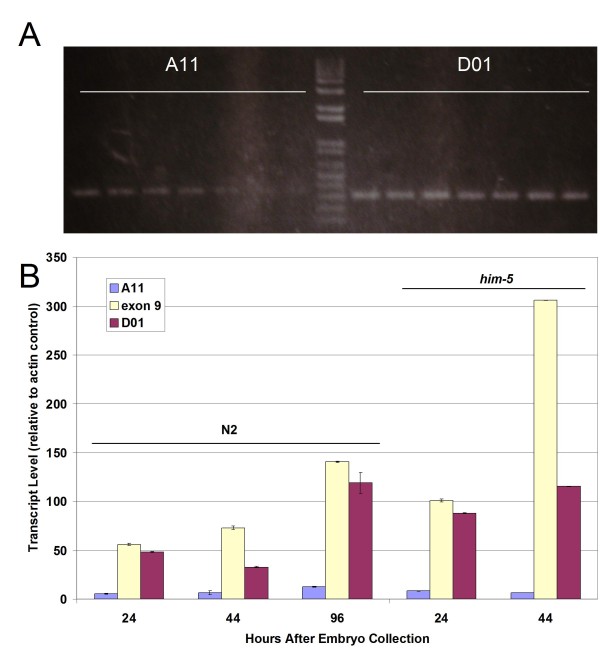
We next sought to obtain evidence that the short transcripts represented by cDNA clones yk1285a11 and yk1237d01 (A11 and D01) are expressed in vivo, rather than cDNA artifacts. To address this question, we needed an approach to distinguish these short transcripts from full-length transcripts containing the same internal sequences. Unfortunately, we were unable to detect sma-9 mRNAs by Northern blot (data not shown). Instead, we used the unique termini of the short transcripts to design variant-specific primer pairs for RT-PCR. For A11 we employed an oligo-dT-containing reverse primer anchored by two nucleotides from sma-9 exon 7 with a gene-specific internal forward primer; for D01 we used an SL2-containing primer anchored by two nucleotides from sma-9 exon 11 with a gene-specific internal reverse primer. To determine empirically whether these primer pairs are specific for A11 and D01, we performed RT-PCR using a gradient of annealing temperatures (50°C - 55°C) and analyzed the products (Figure 2A). In both cases, the primer pairs amplified a single band of the correct size (330 bp for A11 and 300 bp for D01), indicating that the primers specifically detect the desired transcripts. Since the higher annealing temperatures resulted in reduced yield, we used 50°C for all subsequent analyses. Based on these results, we conclude that transcripts polyadenylated downstream of exon 7 and transcripts containing the SL2 splice leader sequence upstream of exon 11 are represented in the pool of sma-9 mRNAs in vivo.
Figure 2.
Expression of sma-9 transcript variants. A. Detection of A11 and D01 transcripts using variant-specific RT-PCR over a gradient of annealing temperatures (left to right: 50°C - 55°C). The expected size of the PCR product is 330 bp for A11 and 300 bp for D01. B. Developmental profile of sma-9 transcript expression levels, as determined by qRT-PCR. Data are shown as transcript abundance relative to act-1 actin gene control, and error bars show standard deviation. See Table 1 for quantitation of transcript abundance relative to exon-9-containing control.
The initial RT-PCR experiments were performed non-quantitatively on mixed-stage RNA preparations. We subsequently used real-time quantitative RT-PCR (qRT-PCR) to determine the relative expression levels of sma-9 variant transcripts and whether they are developmentally or sex-specifically regulated. Since we expect full-length transcripts, but not the short transcripts, to contain sequences from exon 9, we used exon 9 internal primers to determine the expression levels of potentially full-length transcripts and calculated the abundance of the short transcripts relative to exon-9-containing transcripts. Three developmental time points were assayed: 24 hr (first larval stage - L1), 44 hr (approximately L3), and 96 hr (adults). We find that exon-9-containing transcript levels increase during larval development and are highest in adulthood (Figure 2B). D01 transcripts accumulate in wild-type strain N2 at 45% - 85% of the level of exon-9-containing transcripts at all developmental time points examined, while A11 transcripts accumulate at approximately 10% of the level of exon-9-containing transcripts (Figure 2B; Table 1). If A11 and D01 are produced concurrently by trans-splicing of full-length transcripts, then the reduced accumulation of A11 may be due to differential stability of the transcripts. One possible mechanism for reduced stability of the A11 transcript is the unusual location of the termination codon TAA, in which the terminal adenines are introduced during polyadenylation and are contiguous with the poly(A) tail. To test for sex-specific expression of these transcripts, him-5 (high incidence of males) mutant populations that contain both hermaphrodites and males were compared to wild-type N2 hermaphrodite populations. In him-5 populations, the full-length and the D01 transcripts, but not A11, accumulate to a 2-4-fold higher level than in hermaphrodite populations, suggesting that these transcripts are expressed at a higher level in males. The short transcripts are not male-specific, however, since they are also expressed in hermaphrodites. Thus, the A11 and D01 cDNA clones represent detectable mRNAs that are expressed during larval and adult stages when DBL-1 is active in regulating body size and male tail patterning.
Table 1.
Relative expression levels of sma-9 transcript variants
| Strain | Developmental Timepoint |
A11 Transcript Level |
D01 Transcript Level |
|---|---|---|---|
| N2 | L1 | 10.12 ± 1.06 | 86.35 ± 1.17 |
| N2 | L3 | 8.88 ± 2.58 | 45.00 ± 0.87 |
| N2 | adult | 8.97 ± 0.43 | 84.58 ± 7.82 |
| him-5 | L1 | 8.23 ± 0.28 | 87.01 ± 0.37 |
| him-5 | L3 | 2.18 ± 0.00 | 37.73 ± 0.11 |
| sma-9(qc3);him-5 | L3 | 6.20 ± 0.01 | 49.55 ± 1.29 |
| sma-9(wk55);him-5 | L3 | 1.81 ± 0.09 | 88.50 ± 1.32 |
| sma-9(cc604);him-5 | L3 | 9.40 ± 0.05 | 62.70 ± 1.26 |
Mean expression levels are give as percent of the expression level of exon-9-containing control, plus or minus standard deviation.
Loss of function of sma-9 variant transcripts
Previous work has shown that sma-9 constructs lacking the Zn finger triplet (ZF3-5) are unable to rescue the body size and mesodermal patterning defects of a sma-9 mutant [14]. These experiments did not test, however, for differential requirements of short vs. full-length transcripts. To test whether sma-9 variant transcripts have specific functional roles, we performed RNAi targeting three different regions of the sma-9 gene. In previous experiments, dsRNA was introduced by microinjection [13,14], but we have found that RNAi by feeding results in a higher penetrance of male tail defects. We have therefore repeated these experiments using the feeding technique [29], and extended them by targeting the internal exon 9 in addition to the 5' and 3' ends of the gene. The effectiveness of each RNAi treatment was monitored by the appearance of the characteristic small body size phenotype (data not shown).
As a component in the DBL-1 pathway, sma-9 mutant phenotypes include small body size and male abnormal phenotypes [13]. One aspect of the male abnormal phenotype in sma-9 is the fusion of male tail sensory rays 8 and 9, two of the nine bilaterally symmetrical pairs of male-specific sensory organs. In wild-type animals, ray 8-9 fusions are also observed, but at a lower frequency. Treatment of wild-type animals with RNAi targeting exons 1-7 caused no increase in the frequency of ray 8-9 fusions (Table 2), although the small body size phenotype was produced. This treatment is predicted to knockdown expression of full-length and A11 variants but not the D01 variant. Conversely, RNAi targeting exons 21-25, predicted to knockdown the D01 and full-length variants, resulted in a two-fold increase in the frequency of ray 8-9 fusions (Table 2). These results are consistent with our previous findings that RNAi of the 3' end of the gene results in more severe male abnormal phenotypes than inactivation of the 5' end [13]. We next compared the male tail phenotype produced by RNAi targeting the central exon 9, predicted to inhibit full-length transcripts but not the short transcripts. This treatment resulted in a male tail defect of lower penetrance than that produced by RNAi targeting the 3' end (Table 2). Since the most severe male tail defect occurred when D01 was targeted, these experiments suggest that D01 has a role in regulating sensory ray patterning. Full-length sma-9 products are also predicted to function in sensory ray patterning, since the inhibition of a central exon resulted in a partial male abnormal phenotype.
Table 2.
sma-9 loss of function phenotypes in male tail patterning and body size
| RNAi (Exon) or Mutant | Forms Predicted to be Blocked | Frequency of Ray 8-9 Fusions1 (n) | Body Length2 (n) |
|---|---|---|---|
| none | none | 18% (50) | ND |
| 1-7 | full-length, A11 | 20% (50) | ND |
| 9 | full-length only | 29% (100) | ND |
| 21-25 | full-length, D01 | 36% (50) | ND |
| sma-9(qc3) | full-length, A11 | 50% (50) | 692 ± 85 (43) |
| sma-9(wk55) | full-length only | 66% (50) | 566 ± 34 (31) |
| sma-9(cc604) | full-length, D01 | 72% (50) | 716 ± 61 (30) |
1All male tail phenotypes are scored with him-5(e1490) in the background to increase the frequency of males.
2Body length is given in μm ± standard deviation at 96 hours after embryo collection.
ND: not determined.
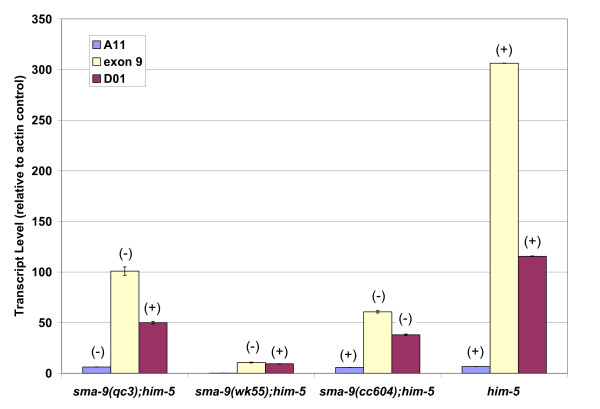
We sought to repeat this analysis using genetic mutants, since the RNAi treatments might cause some nonspecific effects due to targeting of unspliced pre-mRNA or to RNAi spreading [30,31]. We took advantage of three sma-9 alleles containing premature termination codons at different locations in the gene (Figure 1; [13,14]): qc3 in predicted exon 1 (within A11), wk55 in predicted exon 9 (between the short variants), and cc604 in predicted exon 18 (within D01). Therefore, in qc3, only the D01 transcript is capable of encoding a functional gene product; in wk55, both A11 and D01, but not the full-length transcript, are capable of encoding functional gene products; and in cc604, only the A11 transcript is capable of encoding a functional gene product. Since additional disruptions may occur due to nonsense-mediated decay [32], we determined the transcript expression profile for these strains at the L3 stage (Figure 3; Table 1). In all three mutant backgrounds, the transcript levels are somewhat reduced compared to the him-5 control. The reduction in transcript levels is most striking in the sma-9(wk55) background (Figure 3). This severe reduction in transcript levels is consistent with our observations that this allele causes the most severe body size phenotype (Table 2; [13]). Most importantly for the interpretation of our experiments, sma-9(qc3) and sma-9(cc604) animals express the short transcripts at detectable levels near the normal range (Figure 3).
Figure 3.
Expression levels of sma-9 transcripts in mutant backgrounds. Expression levels of sma-9 transcripts as determined by qRT-PCR. Animals were collected at the L3 stage. Data are shown as transcript abundance relative to act-1 actin gene control, and error bars show standard deviation. See Table 1 for quantitation of transcript abundance relative to exon-9-containing transcripts. (-): transcript harbors a premature termination codon. (+): transcript sequence is normal.
The usual expectation for a series of nonsense mutations is that the earliest premature termination codon will have the most severe mutant phenotype while later termination codons may cause less severe defects if those alleles encode partially functional gene products. For the sma-9 alleles, the opposite result is seen in the male tail. The allele encoding the latest termination codon, cc604, causes the most severe male tail defect, with a frequency of ray 8-9 fusions greater than any of the previously characterized alleles (Table 2, [13]). Since different investigators typically vary in their quantitation of ray fusion frequencies, we also repeated measurement of the male tail defects of qc3 and wk55 for direct comparison. Consistent with previous results, qc3, the allele containing the earliest termination codon, causes a mild male tail defect, while wk55, containing a termination codon in exon 9, results in an intermediate frequency of ray fusions (Table 2). Thus, the severity of the sensory ray defect is inversely correlated with predicted D01 activity. The most severe defect is manifested in cc604 mutants, in which the D01 isoform is disrupted by mutation. An intermediate frequency of ray fusions occurs in wk55 mutants, in which the D01 transcript is expressed at reduced levels but is not disrupted by mutation. A mild male tail defect is seen in qc3 mutants, in which levels of D01 transcripts are lower than in wild type but higher than in wk55 (Figure 3). Overall, with regard to male tail patterning, these mutations form an allelic series in which their levels of activity are the opposite of that expected.
In body size, a different result was obtained with these three mutants (Table 2). The sma-9(wk55) mutant shows the smallest body size. In contrast, both qc3 and cc604 produce less severe body length phenotypes that are mutually indistinguishable (Table 2). Since qc3 and cc604 produce indistinguishable body size defects, this analysis does not support a specific role for either the A11 or the D01 transcript in the regulation of body size. Furthermore, the more severe body size phenotype of wk55 mutants could be due to the lower overall level of sma-9 expression in these mutants (Figure 3). The results of body size analysis thus suggest that full-length isoforms, rather than A11 or D01, are critical for promoting growth. Based on our RNAi data and on the differential effects of the three nonsense alleles on body size and male tail development, we hypothesize that the D01 short transcript has a tissue-specific function in sensory ray development.
Rescue of sma-9 mutant phenotypes by overexpression of D01 cDNA
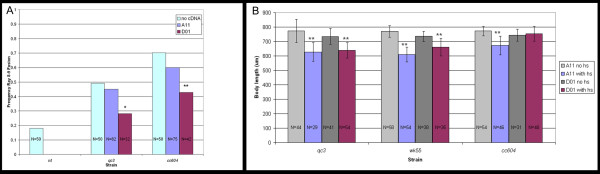
Consistent with our hypothesis, we have previously demonstrated that overexpression of the D01 cDNA, but not of A11, from a heat shock promoter can partially rescue sma-9(wk55) mutant male tail defects [33]. In comparison, expression of sma-9 from a cosmid genomic clone containing the entire coding region resulted in nearly complete rescue of the ray fusion defect [13]. We next asked whether expression of sma-9 short transcripts is sufficient to provide normal gene function in qc3 and in cc604 mutants, in which the A11 and the D01 transcript, respectively, are disrupted by premature termination codons. As before, transgenics carrying hs-sma-9 constructs were subjected to heat shock during the L3 stage when sensory ray identities are being established [34] and males were scored in adulthood. In cc604 mutants, which contain a premature termination codon within the D01 transcript, expression of D01 resulted in partial rescue of the male tail defect (Figure 4A). In contrast, overexpression of A11 in cc604 caused a slight reduction in frequency of sensory ray fusions, but this effect was not statistically significant (p = 0.16). In qc3 mutants, in which the A11 transcript contains a premature termination codon, we were surprised to find that overexpression of A11 produced no change in male tail phenotype, whereas overexpression of D01 resulted in significant rescue of the male tail defect (Figure 4A). Based on the rescue of male tail defects by D01 in both qc3 and cc604 mutants, we conclude that increased expression of D01 may be able to compensate partially for defective full-length sma-9 transcripts.
Figure 4.
Phenotypic rescue by overexpression of sma-9 short transcripts. A. Male tail phenotypes are given as frequency of fusion of rays 8 and 9 per male tail side scored. p values were calculated using a Student's t-test. *: significant difference from no cDNA control (p < 0.05). **: highly significant difference from no cDNA control (p < 0.01). The p values for experiments with A11 cDNA overexpression showed no significant difference from controls (p = 0.59 for qc3; p = 0.16 for cc604). All strains contain him-5(e1490) to increase the proportion of males. B. Body size phenotypes. Mean body length at 96 hours after embryo collection (adult stage) is shown. Error bars show standard deviation. Note that these strains are transgenics expressing the rol-6 marker that influences body length, so all comparisons are made between heat-shocked animals and no heat shock control rather than using a nontransgenic control. p values were calculated using a Student's t-test. **: highly significant difference from no heat shock control (p < 0.01).
Finally, we looked for evidence of A11 or D01 function in body size regulation. The body size phenotypes of qc3 and cc604 did not support a specific role for either A11 or D01 in body size regulation, since these two mutants have indistinguishable sizes (Table 2). To determine whether increased expression of A11 and/or D01 can promote growth, we used our heat shock constructs to drive their expression. Heat shock of transgenics in the L3 stage produced no evident changes in body size (data not shown), so we performed heat shock earlier in development, in L1 animals, and measured body length in adulthood. In contrast to the rescue of male abnormal phenotypes by D01, heat shock of A11 and D01 transgenics led either to a reduction in body size or to no change in body size (Figure 4B). These results suggest that A11 and D01 do not contribute to the growth promoting activity of sma-9. If anything, increased expression of A11 and D01 may interfere with the ability of gene products from full-length transcripts to promote increases in body size.
Discussion
We have shown here that the C. elegans sma-9/schnurri locus undergoes a novel trans-splicing process to generate two shorter transcripts, one of which has a tissue-specific function in DBL-1/BMP signaling. First, we have demonstrated that an SL2 trans-spliced, truncated cDNA form, yk1237d01 (D01), represents an actual mRNA that is detectable by RT-PCR. Second, we have verified that longer transcripts containing both the 5' and 3' ends of the gene are also detectable by RT-PCR. Third, loss-of-function experiments (RNAi and premature termination mutants) demonstrate that the short transcript D01 is necessary but not sufficient for male tail patterning, since depletion of both D01 and full-length transcripts causes a more severe male tail patterning defect than depletion of full-length transcripts alone. Fourth, gain-of-function experiments (heat shock-induced overexpression) indicate that the short transcript D01 can partially rescue male tail but not body size defects of sma-9 mutants. In contrast to the results on D01, we find for the yk1285a11 (A11) transcript little or no evidence of an in vivo function, as well as low abundance throughout development. In addition, class I and II full-length transcripts and the D01 short transcript, but not the A11 transcript, encode the Zn finger triplet domain shown to be critical in the regulation of body size and mesodermal patterning [14]. In Drosophila, this domain is also critical in mediating Dpp-responsive target gene regulation [35]. The gene product encoded by the A11 transcript lacks this domain and may therefore not be capable of being recruited to target genes. Why then is the A11 transcript detected at all? One possibility is that it is generated as a byproduct of the trans-splicing mechanism that generates D01.
Since we have shown that full-length transcripts and D01 transcripts are each necessary but not sufficient for male tail patterning, we must consider what differences in molecular function are found in the respective gene products. In other organisms, Schnurri homologs have been shown to act either as transcriptional activators [20,21,36] or as transcriptional repressors [16,37] mediating BMP-dependent transcription. Strikingly, Yao et al. [21] have shown by swapping Drosophila and Xenopus Schnurri homologs that transcriptional activation or repression activities may be context-dependent rather than an intrinsic property of the protein. In C. elegans, we have previously analyzed intrinsic transcriptional activities of SMA-9 protein domains using a heterologous transcription assay [33]. In these experiments, the acidic residue-rich domain (ARD) in exon 14 exhibited transcriptional activator activity, while the N-terminal region of SMA-9 displayed intrinsic transcriptional repressor activity [33]. Full-length sma-9 transcripts should encode protein products containing both of these transcriptional domains, so that their transcriptional activities may be context-dependent as was described for Drosophila and Xenopus Schnurri [21]. On the other hand, D01 transcripts encode products containing solely the transcriptional activation domain (Figure 1). We therefore hypothesize that full-length SMA-9 isoforms function as transcriptional repressors or activators depending on context, while D01-encoded isoforms may be obligate transcriptional activators. This transcriptional activation activity may be necessary for the robust activation of sma-9 target genes in sensory ray development, but may be dispensable in body size regulation. Consistent with this hypothesis, a SMA-9 Zn finger domain fusion with a known transcriptional activator, VP16, can partially rescue the male tail defects but not the body size defects of sma-9 mutants. In contrast to the SMA-9::VP16 fusion, a SMA-9::enR repression domain fusion rescues body size, suggesting that transcriptional repression is more important than activation for SMA-9 activity in body size regulation [33]. Overall, the in vivo activity of the SMA-9::VP16 fusion is strikingly similar to that of the D01 isoform.
Alternative splicing is regulated by trans-acting factors that influence the choice of splice sites. It will be of interest to determine the trans-acting factors that regulate alternative trans-splicing of sma-9. In preliminary experiments, we tested whether sma-9 trans-splicing depends on mec-8, which encodes an RNA recognition motif-containing protein that regulates alternative splicing in the hypodermis [38,39]. We found that A11, D01, and exon 9-containing transcript levels were all reduced 2- to 5-fold in mec-8 mutants relative to N2 (data not shown). If mec-8 were directly involved in processing the short transcripts from a longer precursor, we expected to find reciprocal changes in accumulation of the full-length transcripts relative to short transcripts. Our results are therefore not sufficiently clear to draw a conclusion as to whether MEC-8 is directly involved in sma-9 trans-splicing, and additional investigations will be necessary to identify the trans-acting factors that mediate this reaction.
A variety of genetic mechanisms exist for generating diverse functions from a single genetic locus, including those that affect transcript structure such as alternative splicing, alternative transcriptional start sites, and alternative polyadenylation. One prominent example of alternative splicing is the regulation of sex determination in Drosophila by Sex-lethal [40]. This system also uses developmentally regulated alternative promoters. In C. elegans, extensive use of alternative promoters leading to different protein isoforms has been documented [41]. Alternative polyadenylation can also lead to the production of truncated protein variants, such as in the daf-4 BMP receptor gene in C. elegans, in which alternative polyadenylation leads to the production of a secreted negative inhibitor of DAF-4 signaling [42]. To our knowledge, alternative trans-splicing as we have characterized it for sma-9 has not previously been reported for other loci in C. elegans. The unique feature of this trans-splicing event is the choice between processing the transcript via cis- or trans-splicing at the splice acceptor site of predicted exon 11. In an organism without trans-splicing, a similar truncation could be achieved via use of a downstream transcriptional start site or via proteolytic cleavage. For example, in the Hedgehog signaling pathway, full-length transcription factors of the Ci/Gli family are transcriptional activators that, upon proteolytic cleavage, are converted to transcriptional repressors [43]. We speculate that since the trans-splicing machinery exists in C. elegans, it was available to be recruited for this unusual role in the sma-9 locus.
Conclusions
Both C. elegans sma-9/schnurri [13] and the human homologs Shn-1 and Shn-3 [23,24] display alternative splicing that is predicted to result in multiple protein isoforms. For most of these isoforms, however, no specific in vivo function has been described. We have now demonstrated a functional role for an alternatively trans-spliced sma-9 short transcript represented by cDNA clone yk1237d01 (D01). This transcript mediates a tissue-specific role, since it is required for sensory ray development but not for body size regulation. Our results provide insight into how a single genetic locus can contribute to diverse protein functions.
Methods
C. elegans strains and culture
Nematodes were cultured using standard methods and grown at 20°C unless otherwise noted [44]. In addition to strains generated in this work, the following strains were used: N2 (wild type); LG V, him-5(e1490); LG X, sma-9(qc3, wk55, cc604).
RT-PCR
RT-PCR was performed in two steps using SuperScript III First-Strand Synthesis System (Invitrogen) and RNA extracted from mixed-stage N2 wild-type animals. Temperature gradient PCR was performed on an Eppendorf gradient thermal cycler (MasterCycler) following the manufacturer's instructions.
Real time RT-PCR was performed on a LightCycler 2.0 (Roche). Data analysis software used was LightCycler software version 4.0. Animals were collected at desired time points and total RNA extracted by Trizol as described [13]. SuperScriptTM III platinum two-step qRT-PCR kit with SYBR Green (Invitrogen) was used to perform RT-PCR. Actin gene act-1 was used as standard control. A primer in an intron of the act-1 gene was used to confirm the absence of genomic DNA in RNA preps. Primer sequences for all RT-PCR experiments are available upon request.
RNAi
RNAi feeding method was performed as described [29] after cloning sma-9 fragments into RNAi feeding vector pPD129.36 (a gift from Dr. A. Fire). To score sensory ray patterning, only adult males displaying a small phenotype were collected and scored to ensure that the animals analyzed had undergone a knockdown of sma-9 activity.
Heat-shock clone constructs and generation of transgenic animals
Two sma-9 splice variants yk1285a11 and yk1237d01 (GenBank Accession Numbers: AY390537 and AY390550) were identified previously [13], which were SL1- and SL2- trans-spliced cDNA clones that appeared to represent shorter but complete mRNAs. Heat-shock vector pPD49.83 was a kind gift from Dr. J. Liu. yk1285a11 and yk1237d01 were cloned into pPD49.83 at Nhe I and Kpn I sites. Transgenic nematodes were generated by microinjection of constructs (10 ng/ul) into the gonadal syncytia of him-5 hermaphrodites, with rol-6 (100 ng/ul) as a marker [45]. Mutant strains carrying transgenic constructs were generated by appropriate crosses between the mutant strains and transgenic lines.
Heat-shock experiments
Gravid hermaphrodites were washed off plates with M9 buffer and collected in an Eppendorf tube. Hermaphrodites were destroyed with sodium hypochlorite solution, leaving eggs that were collected and inoculated onto OP50-seeded plates. For body size measurement, 24 hours after egg collection, worms were collected and transferred to a siliconized Eppendorf tube containing 100 ul of M9 buffer [44], which was placed in a circulating water bath under the heat-shock conditions specified. After heat-shock, worms were recovered, placed at 25°C on OP50-seeded plates and allowed to develop to adulthood. The length of individual worms was measured using Image-Pro Express software (Sigma) with Nomarski optics. For male tail ray examination, worms were collected 72 hours after egg collection and transferred to a siliconized Eppendorf tube containing 100 ul of M9 buffer [44], which was placed in a circulating water bath under the heat-shock conditions specified. After heat-shock, worms were recovered, placed at 25°C on OP50-seeded plates and allowed to develop to adulthood and then scored for ray patterning.
Authors' contributions
JY performed the heat shock and RT-PCR experiments and participated in interpretation of data. LY performed the RNAi experiments. CSD conceived of the study, participated in its design and coordination, analyzed mutant phenotypes, and drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jianghua Yin, Email: vortexyin@yahoo.com.cn.
Ling Yu, Email: lingyu_us@hotmail.com.
Cathy Savage-Dunn, Email: cathy.savagedunn@qc.cuny.edu.
Acknowledgements
We thank Jun Kelly Liu for sharing reagents and for helpful discussions. Some of the experiments were done with equipment from the Core Facilities for Imaging, Cellular and Molecular Biology at Queens College. Some C. elegans mutant strains were obtained from the Caenorhabditis Genetics Center, which is supported by the NIH National Center for Research Resources (NCRR). This work was carried out in partial fulfillment of the requirements for the Ph.D. degree from the Graduate Center of City University of New York (J.Y.). This research was supported by RSG-98-230-04-DDC from the American Cancer Society to C.S.-D.
References
- Roberts AB, Sporn MB. Physiological actions and clinical applications of transforming growth factor-beta (TGF-beta) Growth Factors. 1993;8(1):1–9. doi: 10.3109/08977199309029129. [DOI] [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14(6):627–644. [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massague J. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71(6):1003–1014. doi: 10.1016/0092-8674(92)90395-S. [DOI] [PubMed] [Google Scholar]
- Liu F, Ventura F, Doody J, Massague J. Human type II receptor for bone morphogenic proteins (BMPs): extension of the two-kinase receptor model to the BMPs. Mol Cell Biol. 1995;15(7):3479–3486. doi: 10.1128/mcb.15.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell. 2002;10(2):283–294. doi: 10.1016/S1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87(7):1215–1224. doi: 10.1016/S0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Liu F, Hata A, Doody J, Massague J. The TGF-beta family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 1997;11(8):984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- Wu RY, Zhang Y, Feng XH, Derynck R. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol Cell Biol. 1997;17(5):2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature. 1996;383(6596):168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Hill CS. Stoichiometry of active smad-transcription factor complexes on DNA. J Biol Chem. 2002;277(52):51008–51016. doi: 10.1074/jbc.M208532200. [DOI] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature. 1996;383(6603):832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- Savage-Dunn C. In: WormBook. The C. elegans Research Community, editor. Vol. 9. 2005. TGF-β signaling; pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Lints R, Foehr ML, Tokarz R, Yu L, Emmons SW, Liu J, Savage-Dunn C. The Caenorhabditis elegans schnurri homolog sma-9 mediates stage- and cell type-specific responses to DBL-1 BMP-related signaling. Development. 2003;130(26):6453–6464. doi: 10.1242/dev.00863. [DOI] [PubMed] [Google Scholar]
- Foehr ML, Lindy AS, Fairbank RC, Amin NM, Xu M, Yanowitz J, Fire AZ, Liu J. An antagonistic role for the C. elegans Schnurri homolog SMA-9 in modulating TGFbeta signaling during mesodermal patterning. Development. 2006;133(15):2887–2896. doi: 10.1242/dev.02476. [DOI] [PubMed] [Google Scholar]
- Dai H, Hogan C, Gopalakrishnan B, Torres-Vazquez J, Nguyen M, Park S, Raftery LA, Warrior R, Arora K. The zinc finger protein schnurri acts as a Smad partner in mediating the transcriptional response to decapentaplegic. Dev Biol. 2000;227(2):373–387. doi: 10.1006/dbio.2000.9901. [DOI] [PubMed] [Google Scholar]
- Marty T, Muller B, Basler K, Affolter M. Schnurri mediates Dpp-dependent repression of brinker transcription. Nat Cell Biol. 2000;2(10):745–749. doi: 10.1038/35036383. [DOI] [PubMed] [Google Scholar]
- Takagi T, Harada J, Ishii S. Murine Schnurri-2 is required for positive selection of thymocytes. Nat Immunol. 2001;2(11):1048–1053. doi: 10.1038/ni728. [DOI] [PubMed] [Google Scholar]
- Oukka M, Kim ST, Lugo G, Sun J, Wu LC, Glimcher LH. A mammalian homolog of Drosophila schnurri, KRC, regulates TNF receptor-driven responses and interacts with TRAF2. Mol Cell. 2002;9(1):121–131. doi: 10.1016/S1097-2765(01)00434-8. [DOI] [PubMed] [Google Scholar]
- Wu LC. ZAS: C2H2 zinc finger proteins involved in growth and development. Gene Expr. 2002;10(4):137–152. doi: 10.3727/000000002783992479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Takagi T, Kanesashi SN, Kurahashi T, Nomura T, Harada J, Ishii S. Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev Cell. 2006;10(4):461–471. doi: 10.1016/j.devcel.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Yao LC, Blitz IL, Peiffer DA, Phin S, Wang Y, Ogata S, Cho KW, Arora K, Warrior R. Schnurri transcription factors from Drosophila and vertebrates can mediate Bmp signaling through a phylogenetically conserved mechanism. Development. 2006;133(20):4025–4034. doi: 10.1242/dev.02561. [DOI] [PubMed] [Google Scholar]
- Shukla A, Malik M, Cataisson C, Ho Y, Friesen T, Suh KS, Yuspa SH. TGF-beta signalling is regulated by Schnurri-2-dependent nuclear translocation of CLIC4 and consequent stabilization of phospho-Smad2 and 3. Nat Cell Biol. 2009;11(6):777–784. doi: 10.1038/ncb1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Seeler JS, Nirula A, Shurland DL, Gaynor RB. Regulation of human immunodeficiency virus enhancer function by PRDII-BF1 and c-rel gene products. J Virol. 1992;66(1):244–250. doi: 10.1128/jvi.66.1.244-250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicar MD, Liu Y, Allen CE, Wu LC. Structure of the human zinc finger protein HIVEP3: molecular cloning, expression, exon-intron structure, and comparison with paralogous genes HIVEP1 and HIVEP2. Genomics. 2001;71(1):89–100. doi: 10.1006/geno.2000.6425. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. In: WormBook. The C. elegans Research Community, editor. Vol. 25. 2005. Trans-splicing and operons; pp. 1–9. [DOI] [PubMed] [Google Scholar]
- Huang T, Kuersten S, Deshpande AM, Spieth J, MacMorris M, Blumenthal T. Intercistronic region required for polycistronic pre-mRNA processing in Caenorhabditis elegans. Mol Cell Biol. 2001;21(4):1111–1120. doi: 10.1128/MCB.21.4.1111-1120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, Kiraly M. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417(6891):851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Milstein S, Hao T, Rosenberg J, Li N, Sequerra R, Bosak S, Doucette-Stamm L, Vandenhaute J, Hill DE. C. elegans ORFeome version 3.1: increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. 2004;14(10B):2064–2069. doi: 10.1101/gr.2496804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–321. doi: 10.1016/S1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107(4):465–476. doi: 10.1016/S0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Bosher JM, Dufourcq P, Sookhareea S, Labouesse M. RNA interference can target pre-mRNA: consequences for gene expression in a Caenorhabditis elegans operon. Genetics. 1999;153(3):1245–1256. doi: 10.1093/genetics/153.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango SE. Stop making nonSense: the C. elegans smg genes. Trends Genet. 2001;17(11):646–653. doi: 10.1016/S0168-9525(01)02479-9. [DOI] [PubMed] [Google Scholar]
- Liang J, Yu L, Yin J, Savage-Dunn C. Transcriptional repressor and activator activities of SMA-9 contribute differentially to BMP-related signaling outputs. Dev Biol. 2007;305(2):714–725. doi: 10.1016/j.ydbio.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Lints R, Emmons SW. Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFbeta family signaling pathway and a Hox gene. Development. 1999;126(24):5819–5831. doi: 10.1242/dev.126.24.5819. [DOI] [PubMed] [Google Scholar]
- Muller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003;113(2):221–233. doi: 10.1016/S0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Torres-Vazquez J, Park S, Warrior R, Arora K. The transcription factor Schnurri plays a dual role in mediating Dpp signaling during embryogenesis. Development. 2001;128(9):1657–1670. doi: 10.1242/dev.128.9.1657. [DOI] [PubMed] [Google Scholar]
- Pyrowolakis G, Hartmann B, Muller B, Basler K, Affolter M. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev Cell. 2004;7(2):229–240. doi: 10.1016/j.devcel.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Spike CA, Davies AG, Shaw JE, Herman RK. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development. 2002;129(21):4999–5008. doi: 10.1242/dev.129.21.4999. [DOI] [PubMed] [Google Scholar]
- Lundquist EA, Herman RK, Rogalski TM, Mullen GP, Moerman DG, Shaw JE. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development. 1996;122(5):1601–1610. doi: 10.1242/dev.122.5.1601. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Choi J, Newman AP. A two-promoter system of gene expression in C. elegans. Dev Biol. 2006;296(2):537–544. doi: 10.1016/j.ydbio.2006.04.470. [DOI] [PubMed] [Google Scholar]
- Gunther CV, Riddle DL. Alternative polyadenylation results in a truncated daf-4 BMP receptor that antagonizes DAF-7-mediated development in Caenorhabditis elegans. J Biol Chem. 2004;279(38):39555–39564. doi: 10.1074/jbc.M407602200. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133(1):3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]