Abstract
Background
The genetic etiology of eosinophilic esophagitis (EE) has been largely unexplored until a recent genome-wide association study identified a disease susceptibility locus on 5q22, a region that harbors the thymic stromal lymphopoietin (TSLP) gene. However, it is unclear whether the observed genetic associations with EE are disease-specific or confounded by the high rate of allergy in EE patients. In addition, the genetic contributions of other allergy associated genes to EE risk have not been explored.
Objective
We aimed to delineate single nucleotide polymorphisms (SNP)s that associated with EE apart from allergy.
Methods
We utilized a custom array containing 738 SNPs in 53 genes implicated in allergic and/or immune responses to genotype 220 allergic or 246 non-allergic controls and a discovery cohort of 170 EE patients. We replicated a statistically significant SNP association in an independent case-control cohort and examined the induction of the candidate gene in primary esophageal epithelial cells.
Results
A single SNP residing in the TSLP gene reached Bonferroni LD adjusted significance and only when EE cases were compared with allergic controls (rs10062929, P = 4.11 × 10−5, odds ratio = 0.35). A non-synonymous polymorphism in the TSLP receptor on Xp22.3 and Yp11.3 was significantly associated with disease only in male EE patients. Primary esophageal epithelial cells expressed TSLP mRNA following Toll-like receptor 3 (TLR3) stimulation.
Conclusion
These data collectively identify TSLP as a candidate gene critically involved in EE susceptibility beyond its role in promoting Th2 responses.
Keywords: Eosinophilic esophagitis, thymic stromal lymphopoietin, single nucleotide polymorphism, allergy, cytokine receptor-like factor 2, Toll-like receptor 3
Introduction
Eosinophilic esophagitis (EE) is a chronic Th2-associated inflammatory disease of the esophagus that affects at least 4 in 10,000 individuals1. Although symptomatically resembling gastroesophageal reflux disease (GERD), EE is clinically defined as esophageal eosinophilia (≥15 intraepithelial eosinophils per high-powered field) in the absence of abnormal acid reflux disease (assessed by normal pH monitoring of the distal esophagus or persistent esophagitis while on high-dose acid suppression therapy) 2. Another distinguishing feature is the high rate of atopic diseases including asthma, eczema, and allergic rhinitis and sensitivities to environmental and food allergens within both pediatric and adult EE populations3, 4. Consistent with an immune based mechanism of disease induction, the most effective therapies used to manage EE are food antigen avoidance and swallowed glucocorticoid treatment. However, some EE patients are refractory to glucocorticoid treatment5, suggesting an inherent resistance with a potential genetic basis.
Reports of EE and esophageal dilatation in relatives of EE patients suggest that the incidence of EE and associated esophageal dysfunction is common among related individuals1, 6. Molecular analysis of the genetics behind EE was initiated in a study by Blanchard et al., which identified a characteristic gene expression profile within the esophagus of EE patients, termed the EE transcriptome that was distinct from that observed in other forms of chronic esophagitis7. Interestingly, the EE transcriptome was consistent across gender, age, familial or non-familial inheritance patterns and was independent of atopic status, suggesting a common disease mechanism despite phenotypic variations6, 7. The most highly expressed gene in the EE transcriptome is eotaxin-3 (53-fold increase), an eosinophil and mast cell chemoattractant; indeed, eotaxin-3 levels correlate with both esophageal eosinophil and mast cell levels7. Furthermore, evidence is mounting that disease pathogenesis is mediated by the interaction of Th2 cells and esophageal epithelial cells as the Th2 cytokine IL-13 induces a large fraction of the EE transcriptome (including eotaxin-3) in esophageal epithelial cells8, 9. A recent genome wide association study (GWAS) identified 5q22 as a susceptibility locus for pediatric EE10. Notably, although two genes (TSLP and WDR36) were present in one haplotype block that associated with EE, evidence was provided that TSLP was the stronger candidate gene because of its overexpression in the esophagus of EE patients and its known biological activity as a key regulator of allergic sensitization10. Yet TSLP has previously been implicated in various atopic responses11–13 and thus it is not clear whether the EE association with TSLP is reflective of a general association with atopic processes or if it is specific to EE.
In the present study, we sought to identify genetic variants that associated with EE using a broad spectrum candidate gene approach involving a panel of 738 SNPs in allergy associated molecules, especially epithelial gene products including TSLP. Furthermore, we aimed to identify genetic variants that associated with EE independent of allergy. To address this question we genetically compared EE patients to a set of clinically-defined allergic and non-allergic control groups. Compared with all genetic variants tested, we demonstrate that genetic variants within TSLP are associated with EE independent of allergy. Additionally, we present further evidence for the importance of TSLP in EE by demonstrating that a genetic variant in the TSLP receptor (TSLPR) also contributes to EE susceptibility. Furthermore, we show that primary esophageal epithelial cells express TSLP mRNA in response to TLR3 activation.
Methods
Study Participants
The discovery EE case cohort consisted of 172 patients who were recruited by the Cincinnati Center for Eosinophilic Disorders (CCED). DNA samples were collected at time of endoscopy or at follow-up from blood or saliva specimens. Patients were identified with clinically diagnosed EE (≥24 eosinophils per high power field). All EE patients self reported race as white. The male to female ratio was 2.18 and the mean age at diagnosis was 9.39 (SD 8.60) years. Approximately 85% of the discovery cohort was composed of patients characterized in a recent EE genome-wide association study10. The percentages of patients with diagnosed asthma, allergic rhinitis, or eczema were 31%, 53%, and 39%, respectively. Approximately 50% of these EE patients were on acid-suppressive therapy and 18% were on swallowed glucocorticoid treatment at the time of endoscopy. The discovery control cohorts were recruited through the Greater Cincinnati Pediatric Clinic Repository (GCPCR) and Genomic Control Cohort (GCC) at Cincinnati Children’s Hospital Medical Center (CCHMC). All consenting patients visiting various clinics at CCHMC including the allergy, immunology and pulmonary clinics provided buccal swabs or saliva samples for DNA isolation. Information regarding patient history was collected through questionnaires pertaining to allergy symptoms. All controls had self reported race as white. The allergic control cohort (n=227) was defined as non-asthmatics having clinically-diagnosed atopic rhinitis or atopic dermatitis or a self-reported history of environmental allergies, hay fever, or eczema. The non-allergic control group (n=246) had no personal or family history of asthma, or a personal history of environmental allergies, allergic rhinitis or atopic dermatitis.
The replication EE case cohort consisted of 122 independent patients with similar characteristics as the EE discovery cohort. This cohort was also collected at the CCED using the procedures described above. All EE patients in the replication cohort self reported race as white and 72% of these patients were included in the EE genome-wide association study. The replication control cohort consisted of 119 individuals. This cohort was comprised of consenting CCHMC employees who reported race as white with no self-reported personal history of GERD or other gastrointestinal symptoms. The percent of individuals within this group with self reported allergy was approximately 44%. These studies were approved by the CCHMC Institutional Review Board.
Candidate Gene and SNP selection
For the discovery phase, we used a custom Illumina SNP chip (nSNP = 768) designed to interrogate genes with a role in allergy-related signaling pathways and/or epithelial cell function based on current literature searches as part of NIH grant U19 AI070235. The TSLP gene was submitted for inclusion in the initial chip design by Dr. Yong-Jun Liu (University of Texas, M.D. Anderson Cancer Center, Houston, Texas, USA); a total of nine TSLP SNPs were included on the chip. In addition to candidate gene SNPs, 30 ancestry informative markers (AIMs) were also included. Genotyping using the Illumina GoldenGate Assay (http://www.illumina.com) system was performed at the CCHMC Genetic Variation and Gene Discovery Core. Genotypes were assigned using Illumina’s BeadStudio 2 Software (San Diego, CA). SNPs that exhibited differences in minor allele frequencies (MAF > 0.1) between plates were manually examined to ensure consistent clustering patterns. For the replication set, TSLP SNPs were genotyped using the ABI TaqMan™ allelic discrimination assays (Applied Biosystems). The initial TSLPR sequencing was performed using the ABI 3700 DNA Analyzer™. TSLPR SNPs were genotyped using the ABI TaqMan™ allelic discrimination assays (Applied Biosystems) in the replication case-control cohorts as well as additional EE patient and normal control DNA samples collected through the CCED.
Statistical Analyses
All analyses were performed separately among the allergic and non-allergic control cohorts. No TSLP SNPs failed Hardy Weinberg equilibrium in the control dataset (P < 10−3), were removed for poor genotype calling (missing rate greater than 10%) or were excluded with minor allele frequencies below 10%. A single TSLP SNP was removed prior to analysis due to poor clustering. To adjust for multiple testing, we applied a Bonferroni adjustment after correcting for LD correlation among the SNPs which passed quality control. This approach is less conservative than the standard Bonferroni adjustment as it incorporates an LD block-based correction to reduce the total number of independent tests, thus preventing type I error inflation14. Filtering of individuals with greater than 20% missing genotypic information over the chip resulted in the removal of 2 EE patients and 7 allergic controls prior to analysis, yielding a total of 170 cases and 220 allergic and 246 non-allergic controls that entered the analyses. To account for potential population stratification/confounding or admixture in these samples, principal component analyses (PCA) was performed using 30 AIMs and the EIGENSTRAT software. The principal component score for each individual was included as a covariate in all models along with age and gender in logistic regression models. As a general association screen, we tested for the additive models of single SNP analysis, which assume that each copy of the risk allele will increase disease prevalence. Unconditional logistic regression was used to calculate P-values and odds ratios for each SNP using the software PLINK (V1.05)15. Statistical analyses of the TSLPR SNPs (dominant model) and the TSLP SNPs (Cochran-Armitage trend test) in the replication cohorts were also performed in PLINK. Fisher’s method16, which calculates the combined probability from independent tests addressing the same hypothesis, was used in the meta-analysis of the discovery and replication cohorts.
TSLP Expression in Stimulated Esophageal Epithelial Cells
Primary esophageal epithelial cells were cultured from patient esophageal biopsies as previously described9. Cells were grown in hydrocortisone-free media 24 to 48 hours prior to stimulation. Cells were then treated with 10 or 100 µg/ml poly I:C (InvivoGen) for 3 or 8 hours. RNA was isolated using TRIzol (Invitrogen) and cDNA was synthesized using iScript (Bio-Rad). RT-PCR analysis for TSLP expression was performed using the following primers: forward (5’-cccaggctattcggaaactca-3’), reverse (5’-acgccacaatccttgtaattgtg-3’). Graphed data were normalized to GAPDH expression from 3 to 4 independent experiments performed in duplicate.
Results
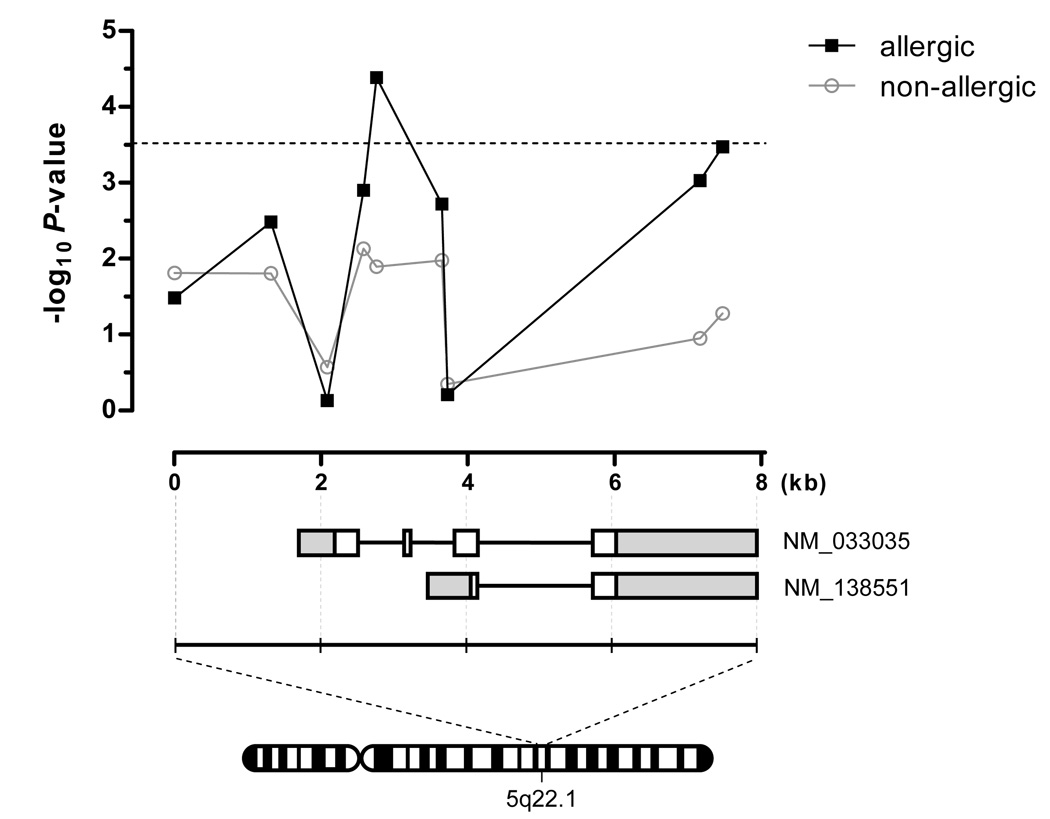
In our effort to identify EE-specific genetic susceptibility loci, we utilized a custom Illumina SNP genotyping chip to screen for variants within candidate genes involved in allergy and/or epithelial cell function. In order to account for multiple testing, the Bonferroni LD adjusted P-value required for statistical significance in this study was determined to be P = 3 × 10−4. As the coincidence of allergy is high (approximately 70%) in EE, we first chose to investigate the specificity of any potential associations of candidate gene SNPs by stratifying the discovery control cohort into two phenotypically defined groups based on a history of allergy (see Methods). When comparing EE to the allergic controls, we found that only TSLP harbored variants which reached statistical significance (Table I and Fig. 1, and Supplementary Table I). Indeed, seven SNPs in TSLP exhibited association with EE (4.11 × 10−5 ≤ P ≤ 3.29 × 10−3). However, when comparing EE to the non-allergic controls, no genes exhibited variants which reached statistical significance; the strongest observed TSLP SNP association only reached P = 7.35 × 10−3 in this analysis. When the two control groups were combined (allergic and non-allergic), the association strengthened, most likely owing to the increase in sample size of the control group. Interestingly, polymorphisms in IL4, a known allergy susceptibility gene, were associated with EE (best P = 1.33 × 10−3, OR = 1.91) using the non-allergic controls but not with the allergic controls (best P = 0.27) (see Supplementary Table I), emphasizing the importance of using phenotypic matched controls for identifying risk variants associated with primary disease. Moreover, these data highlight the specificity of the TSLP genetic association with EE.
Table I.
TSLP SNPs are associated with EE as compared to allergic and/or non-allergic controls
| EE Cases (n=170) |
Allergic Controls (n=220) |
Non-allergic Controls (n=246) |
Allergic and Non-allergic Controls (n=466) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP1 | BP2 | MAF3 Cases |
MAF Controls |
OR4 | P-value | MAF Controls |
OR | P-value | MAF Controls |
OR | P-value |
| rs3806932 | 110433574 | 0.33 | 0.40 | 0.72 | 3.27E-02 | 0.41 | 0.70 | 1.55E-02 | 0.41 | 0.70 | 9.02E-03 |
| rs3806933 | 110434641 | 0.31 | 0.41 | 0.63 | 3.29E-03 | 0.40 | 0.70 | 1.56E-02 | 0.40 | 0.66 | 2.38E-03 |
| rs2289276 | 110435406 | 0.24 | 0.25 | 0.94 | 7.40E-01 | 0.27 | 0.83 | 2.69E-01 | 0.26 | 0.86 | 3.43E-01 |
| rs1898671 | 110435901 | 0.45 | 0.33 | 1.61 | 1.26E-03 | 0.35 | 1.48 | 7.35E-03 | 0.34 | 1.57 | 5.52E-04 |
| rs10062929 | 110436078 | 0.08 | 0.17 | 0.35 | 4.11E-05 | 0.13 | 0.54 | 1.27E-02 | 0.15 | 0.45 | 5.79E-04 |
| rs2289277 | 110436966 | 0.31 | 0.42 | 0.61 | 1.90E-03 | 0.40 | 0.69 | 1.05E-02 | 0.41 | 0.64 | 1.35E-03 |
| rs11466749 | 110440484 | 0.11 | 0.19 | 0.48 | 9.29E-04 | 0.15 | 0.71 | 1.12E-01 | 0.17 | 0.59 | 8.47E-03 |
| rs11466750 | 110440793 | 0.09 | 0.18 | 0.43 | 3.37E-04 | 0.13 | 0.63 | 5.25E-02 | 0.15 | 0.52 | 3.08E-03 |
Presented are the associated P-values for TSLP SNPs between EE cases vs. three control groups (allergic, non-allergic, or combined)
dbSNP Build 130 rs number
BP, base pair; chromosomal location NBCI Build 36
MAF, minor allele frequency in EE cases or the specified control group
OR, odds ratio for the specified control group compared to EE cases
Figure 1. Associations of TSLP SNPs with EE are independent of allergy phenotypes.
Upper panel, the associated −log10 P-values for the genotyped TSLP SNPs from each analysis (EE vs. allergic or non-allergic controls) are plotted by relative base pair location. The dashed line represents the Bonferroni threshold for significance (P = 3 × 10−4). Lower panel, the TSLP SNPs reside within an approximate 8 kilobase interval on 5q22.1 encoding the TSLP gene isoforms (NM_ 033035 and NM_138551). Represented are exons (white boxes), introns (bold lines), and untranslated regions (gray boxes).
We next aimed to replicate the observed genetic association with TSLP variants using a completely independent replication cohort of EE cases and controls (Table II). Although the replication control cohort was composed of both allergic and non-allergic individuals, three TSLP SNPs were significantly associated with EE (Table II) in this independent analysis. In a meta-analysis of the associated TSLP variants from both the discovery and replication cohorts, the association of rs10062929 strengthened (combined P = 3.16 × 10−6) approximately 2-logs above the threshold of the Bonferroni adjustment. Moreover, the combined P-values for rs11466750 and rs11466749 were nominally significant (combined P = 8.48 × 10−4 and 1.86 × 10−3, respectively).
Table II.
Replication of TSLP SNP association with EE in two independent case-control cohorts
| Discovery Cohort1 | Replication Cohort2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | MAF Cases |
MAF Controls |
OR | P-value | MAF Cases |
MAF Controls |
OR | P-value | Combined P-value3 |
| rs3806933 | 0.31 | 0.40 | 0.66 | 2.38E-03 | 0.31 | 0.41 | 0.66 | 6.10E-02 | 1.43E-03 |
| rs2289276 | 0.24 | 0.26 | 0.86 | 3.43E-01 | 0.26 | 0.23 | 1.17 | 5.01E-01 | 4.74E-01 |
| rs10062929 | 0.08 | 0.15 | 0.45 | 5.79E-04 | 0.09 | 0.19 | 0.36 | 3.31E-04 | 3.16E-06 |
| rs2289277 | 0.31 | 0.41 | 0.64 | 1.35E-03 | 0.36 | 0.43 | 0.75 | 1.69E-01 | 2.14E-03 |
| rs11466749 | 0.11 | 0.17 | 0.52 | 3.08E-03 | 0.11 | 0.21 | 0.46 | 5.00E-03 | 1.86E-04 |
| rs11466750 | 0.09 | 0.15 | 0.70 | 9.02E-03 | 0.10 | 0.19 | 0.46 | 9.00E-03 | 8.48E-04 |
The discovery cohort was composed of 170 EE cases and 466 allergic and non-allergic controls as described in Table I
The replication cohort was composed of either 87 EE cases and 114 allergic and non-allergic controls or 87 EE cases and 122 allergic and non-allergic controls for SNP rs10062929
Combined P-values from the discovery and replication cohorts using Fisher’s method
The prevalence of EE in males is approximately 2.5-fold higher than in females. To determine whether TSLP variants contribute to this gender bias, we performed a gender-stratified analysis of EE cases and allergic controls from the discovery cohort and determined the SNPs with the greatest change in P-value in either the male or female cohorts (See Supplementary Table II). Of the nine TSLP SNPs, only rs10062929 and rs11466749 also associated in the female EE cohort (P = 0.016 and P = 0.048 in EE vs. female atopic controls, respectively), while the P-values of most variants in TSLP remained unchanged in the male-only analyses.
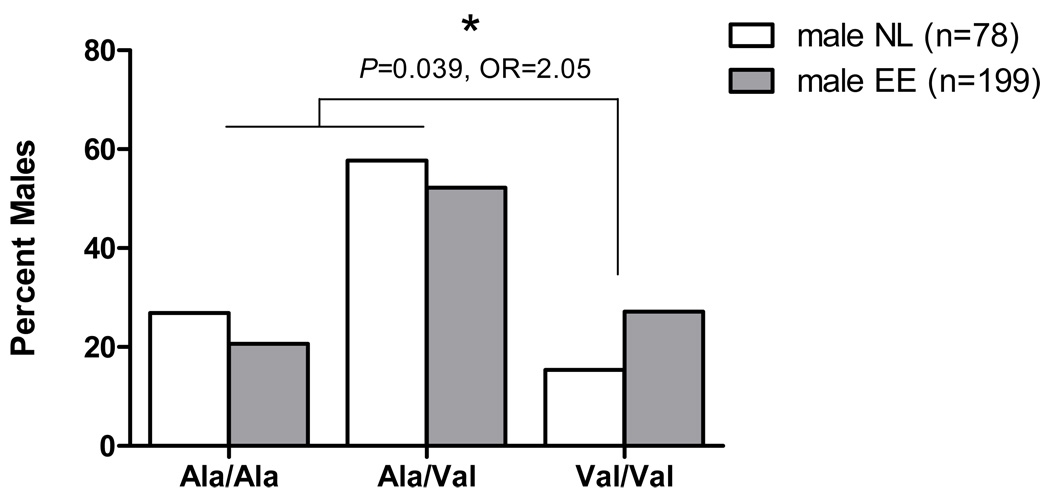
Interestingly, the TSLP receptor (TSLPR) is encoded on a pseudoautosomal region on Xp22.3 and Yp11.3, which further underscores the potential significance of finding SNPs in the TSLP/TSLPR pathway observed among males with EE17. We were interested in testing the hypothesis that variants in the TSLPR may also associate with EE in a gender-specific manner. Therefore, we genotyped three coding SNPs recently validated from direct sequencing of the TSLPR gene (P.G., unpublished data): rs36139698, rs36177645, and rs36133495. Of these, the SNP rs36133495 (Ala to Val) was significantly associated with disease risk in a cohort of male EE patients and normal male controls (% Val/Val, 27% in male EE cases vs. 15% in male controls) with a P = 0.039, OR = 2.05 (Fig. 2 and Table III). Conversely, rs36133495 was not significantly associated in a female case-control analysis (P = 0.929, OR = 1.22). Taken together, these data present evidence that the TSLP signaling pathway may contribute to the male bias in EE.
Figure 2. A non-synonymous SNP in the TSLPR associates with EE in a gender-specific manner.
A non-synonymous SNP (rs36133495) in the TSLPR, which results in an Ala to Val coding change shows a gender-based association with increased disease risk in male EE patients, with Val/Val and Ala/Val individuals at higher risk (OR = 2.05) compared to Ala/Ala individuals. Genotyped were 199 male EE patients and 78 male normal (NL) controls.
Table III.
A gender-specific association of a TSLPR SNP in male EE patients
| Male EE Cases (n=199) Male Controls (n=78) |
Female EE Cases (n=105) Female Controls (n=78) |
|||||||
|---|---|---|---|---|---|---|---|---|
| SNP1 | MAF Cases |
MAF Controls |
OR | P-value2 | MAF Cases |
MAF Controls |
OR | P-value2 |
| rs36133495 | 0.53 | 0.44 | 2.05 | 0.039 | 0.47 | 0.42 | 1.22 | 0.929 |
dbSNP Build 130 rs number
P-value using a dominant model of association
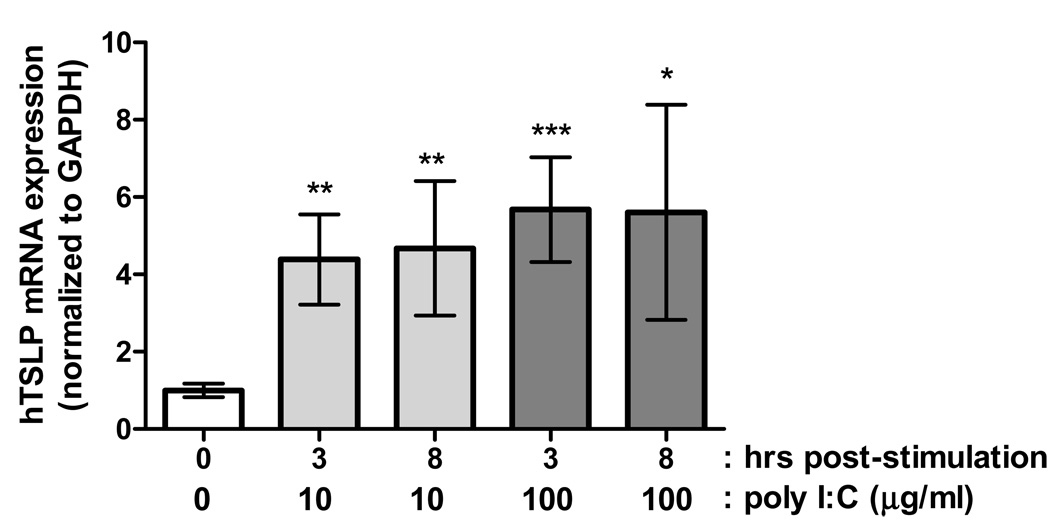
In the lung and skin, TSLP is produced primarily by epithelial cells in response to Th2 cytokines or TLR3 agonists, subsequently targeting dendritic cells to develop Th2-polarizing activity including cytokine and chemokine production13, 18; however, it has been observed that activated mast cells can also express TSLP to similarly drive Th2 responses12, 19, 20. Indeed, we have recently observed increased expression of TSLP mRNA in the esophagus of EE patients compared with control individuals10. Accordingly, we aimed to determine if primary esophageal epithelial cells could produce TSLP. Notably, exposure to the TLR3 ligand poly I:C (a dsRNA mimetic) robustly increased TSLP mRNA expression in as little as 3 hours post-treatment with either 10 or 100 µg/ml poly I:C (Fig. 3). These data suggest that the esophageal epithelium is a source of TSLP expression in EE.
Figure 3. TLR signaling stimulates TSLP expression in primary esophageal epithelial cells.
Treatment of primary esophageal epithelial cells with the TLR3 agonist poly I:C induces a robust increase in TSLP mRNA levels. The data shown are the mean ± SEM from 4–5 independent experiments performed in duplicate (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Discussion
Herein, we report that TSLP is the most dominant genetic variant associated with EE risk using a large SNP panel within relevant allergy and epithelial gene products. We demonstrate that the genetic association of EE with TSLP occurs largely independent of allergy, providing compelling evidence in support of our recent GWAS analysis that identified the 5q22 locus as a risk locus for EE susceptibility10. It is notable that the direction of the disease risk (OR < 1) was similar in the discovery and replication cohorts in our present study and recent EE GWAS, supporting the genetic association to this region. However, given that the more common allele is associated with disease susceptibility, the causative allele in EE is unlikely to have been identified. That the same genetic region (5q22) has been linked to blood eosinophilia further implicates a potential relationship between TSLP and eosinophilic disease21.
In exploring potential mechanisms for the male predilection observed in EE, we performed a gender stratified analysis for TSLP and TSLPR polymorphisms. Indeed, a sex-specific interaction was supported for TSLP variants as well as a non-synonymous SNP within the TSLPR on Xp22.3/Yp11.3 in male EE patients. Bioinformatic analysis of the variant suggests that the mutant TSLPR has increased stability compared to wild type protein, which could conceivably prolong TSLP-induced signal transduction22. In a recent study by Mullighan et al., comparative genomic hybridization analysis in B-progenitor acute lymphoblastic leukemia patients identified a fusion event between exon 1 of the P2RY8 receptor and TSLPR, resulting in a constitutive activation of the Jak-STAT pathway23, indicating the profound ability of the TSLP pathway to induce disease.
We also demonstrate that TSLP mRNA expression is induced in primary esophageal epithelial cells following activation of the TLR3 pathway; it is notable that viral gastroenteritis-like symptoms often precede the onset of EE. TSLP was originally identified as a pro-survival factor that could induce pre-B cell differentiation and proliferation24. Constitutive phosphorylation of STAT5 has been linked with lymphoproliferative diseases and malignancies25. Perhaps TSLP overexpression could contribute to both the inflammatory and hyperproliferative states observed in the esophageal epithelium of EE patients.
Beyond elucidating a role for TSLP in EE, these results have broad implications for the discovery of rare disease risk variants. Specifically, these results demonstrate the importance of appropriate control cohort selection. Consideration of co-morbid diseases may provide insight into the role of genetic risk variants, particularly when a rare disease phenotype (EE) tracks with a more common phenotype (allergy). In the present study, we show that use of well characterized control populations in genetic association studies can overcome relatively small sample sizes to identify true risk variants.
In conclusion, we have identified a specific genetic contribution of TSLP to EE susceptibility, a finding which provides key insight into disease pathogenesis, and likely explains, at least in part, the male gender bias in this disease. We propose that activation of the innate immune system in the esophageal epithelium involving TSLP is likely to have a key role in the subsequent adaptive allergic response, ultimately triggered by food antigens. These findings present TSLP as a potential molecular target for therapeutic intervention and thus highlight the key role of innate immunity in the development of specific allergic disease.
Key Messages
Polymorphisms in TSLP are risk factors for EE independent of underlying allergy phenotypes.
A gender-specific association between SNPs in TSLP as well as a non-synonymous SNP in the TSLPR suggests a mechanism for the male predilection of the EE.
Primary esophageal epithelial cells express TSLP mRNA in response to TLR3 signaling.
Acknowledgements
This work was supported in part by NIH U19 AI070235, NIH R01 DK076893, the PHS Grant P30 DK0789392, Department of Defense, Food Allergy Project, the Buckeye Foundation and the Campaign Urging Research for Eosinophilic Disorders (CURED) Foundation. J.D.S. is supported by a T32 NIH training grant (HL091805). P.G., B.S.B. and M.E.R. were supported by a Dana Foundation Human Immunology Consortium Grant. M.E.R. is a consultant for Merck and Ception Therapeutics.
We would like to thank all of the participating families, patients, physicians, and nurses as well as B. Buckmeier Butz, A. Ahrens, S. Jameson, M. Palazzolo, H. Foote, A. Ernstberger, N. Wang and M. Mingler for assistance with patient enrollment, DNA preparation and/or database management at the Cincinnati Center for Eosinophilic Disorders. In addition, we also thank the physicians, nurses and staff of Cincinnati Children’s Hospital Medical Center Allergy and Immunology, Pulmonary, Dermatology, Headache Center, Dental and Orthopedic clinics and Emergency Department for their contributions to the Greater Cincinnati Pediatric Clinic Repository as well as the investigators and staff of the Genomic Control Cohort. We thank Tesfaye Mersha Baye, Jayanta Gupta, Mark Lindsey and Tia Patterson for key assistance with the control DNA cohorts. Lastly, we thank Drs. Jonathan Spergel, Hakon Hakonarson and Kathleen Barnes for insightful discussions and review of this manuscript, as well as Dr. Marsha Wills-Karp for contributing to the custom SNP chip.
Abbreviations
- EE
eosinophilic esophagitis
- GERD
gastroesophageal reflux disease
- TSLP
thymic stromal lymphopoietin
- SNP
single nucleotide polymorphism
- OR
odds ratio
- MAF
minor allele frequency
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Capsule Summary
This work defines specific genetic associations between EE and polymorphisms in the TSLP and TSLPR genes. Notably, these associations are not driven by patient allergic status and occur mainly within male patients, respectively.
References
- 1.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;351:940–941. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 2.Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–1363. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 3.Assa'ad AH, Putnam PE, Collins MH, Akers RM, Jameson SC, Kirby CL, et al. Pediatric patients with eosinophilic esophagitis: an 8-year follow-up. J Allergy Clin Immunol. 2007;119:731–738. doi: 10.1016/j.jaci.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 4.Roy-Ghanta S, Larosa DF, Katzka DA. Atopic characteristics of adult patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2008;6:531–535. doi: 10.1016/j.cgh.2007.12.045. [DOI] [PubMed] [Google Scholar]
- 5.Konikoff MR, Noel RJ, Blanchard C, Kirby C, Jameson SC, Buckmeier BK, et al. A randomized, double-blind, placebo-controlled trial of fluticasone propionate for pediatric eosinophilic esophagitis. Gastroenterology. 2006;131:1381–1391. doi: 10.1053/j.gastro.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 6.Collins MH, Blanchard C, Abonia JP, Kirby C, Akers R, Wang N, et al. Clinical, pathologic, and molecular characterization of familial eosinophilic esophagitis compared with sporadic cases. Clin Gastroenterol Hepatol. 2008;6:621–629. doi: 10.1016/j.cgh.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid-Grendelmeier P, Altznauer F, Fischer B, Bizer C, Straumann A, Menz G, et al. Eosinophils express functional IL-13 in eosinophilic inflammatory diseases. J Immunol. 2002;169:1021–1027. doi: 10.4049/jimmunol.169.2.1021. [DOI] [PubMed] [Google Scholar]
- 9.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 10.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. doi: 10.1038/ng.547. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harada M, Hirota T, Jodo AI, Doi S, Kameda M, Fujita K, et al. Functional analysis of the thymic stromal lymphopoietin variants in human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2009;40:368–374. doi: 10.1165/rcmb.2008-0041OC. [DOI] [PubMed] [Google Scholar]
- 12.Soumelis V, Reche PA, Kanzler H, Yuan W, Edward G, Homey B, et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol. 2002;3:673–680. doi: 10.1038/ni805. [DOI] [PubMed] [Google Scholar]
- 13.Bogiatzi SI, Fernandez I, Bichet JC, Marloie-Provost MA, Volpe E, Sastre X, et al. Cutting Edge: Proinflammatory and Th2 cytokines synergize to induce thymic stromal lymphopoietin production by human skin keratinocytes. J Immunol. 2007;178:3373–3377. doi: 10.4049/jimmunol.178.6.3373. [DOI] [PubMed] [Google Scholar]
- 14.Nicodemus KK, Liu W, Chase GA, Tsai YY, Fallin MD. Comparison of type I error for multiple test corrections in large single-nucleotide polymorphism studies using principal components versus haplotype blocking algorithms. BMC Genet. 2005;6 Suppl 1:S78. doi: 10.1186/1471-2156-6-S1-S78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher RA. Statistical methods for research workers. 14th ed. Edinburgh: Oliver and Boyd; 1970. [Google Scholar]
- 17.Tonozuka Y, Fujio K, Sugiyama T, Nosaka T, Hirai M, Kitamura T. Molecular cloning of a human novel type I cytokine receptor related to delta1/TSLPR. Cytogenet Cell Genet. 2001;93:23–25. doi: 10.1159/000056941. [DOI] [PubMed] [Google Scholar]
- 18.Kato A, Favoreto S, Jr, Avila PC, Schleimer RP. TLR3- and Th2 cytokine-dependent production of thymic stromal lymphopoietin in human airway epithelial cells. J Immunol. 2007;179:1080–1087. doi: 10.4049/jimmunol.179.2.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knisz J, Banks A, McKeag L, Metcalfe DD, Rothman PB, Brown JM. Loss of SOCS7 in mice results in severe cutaneous disease and increased mast cell activation. Clin Immunol. 2009;132:277–284. doi: 10.1016/j.clim.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Okayama Y, Okumura S, Sagara H, Yuki K, Sasaki T, Watanabe N, et al. FcepsilonRI-mediated thymic stromal lymphopoietin production by interleukin-4-primed human mast cells. Eur Respir J. 2009;34:425–435. doi: 10.1183/09031936.00121008. [DOI] [PubMed] [Google Scholar]
- 21.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 22.Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 23.Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009 doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asakawa M, Kinoshita Y, Kimura S, Hato F, Wada S, Nishijima T, et al. Restorative effects of thymosin fraction 5 on reduction in responsiveness of rat thymocytes to allogenic lymphocytes during bladder tumor induction. Cell Mol Biol. 1990;36:265–274. [PubMed] [Google Scholar]
- 25.Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]