SUMMARY
Within dendritic spines, actin is presumed to anchor receptors in the postsynaptic density and play numerous roles regulating synaptic transmission. However, the submicron dimensions of spines have limited examination of actin dynamics within spines, and prevented live-cell discrimination of perisynaptic actin filaments. Using photoactivated localization microscopy, we measured movement of individual actin molecules within living spines. Velocity of single actin molecules along filaments, an index of filament polymerization rate, was highly heterogeneous within individual spines. Most strikingly, molecular velocity was elevated in discrete, well-separated foci occurring not principally at the spine tip, but in subdomains throughout the spine, including the neck. Whereas actin velocity on filaments at the synapse was substantially elevated, those at the endocytic zone showed no enhanced polymerization activity. We conclude that actin subserves spatially diverse, independently regulated processes throughout spines. Perisynaptic actin forms a uniquely dynamic structure well suited for direct, active regulation of the synapse.
INTRODUCTION
Synapses are inherently plastic, and undergo persistent changes in both strength and postsynaptic composition which are thought to underlie many forms of experience-driven learning. Remarkably, plasticity at individual synapses is not coupled to changes at nearby synapses (Malenka and Bear, 2004). Such autonomy apparently is possible because dendritic spines, the micron-sized dendritic protrusions that are the site of most synaptic contacts in the brain, contain molecular machinery to permit modification independent of neighboring spines (Newpher and Ehlers, 2009).
A critical component of this machinery is actin, the major cytoskeletal element of dendritic spines known to play a variety of roles regulating the synapse. Most prominently, actin has long been recognized as the principle determinant of spine morphology (Caceres et al., 1983; Penzes et al., 2008), and it is clear that regulation of actin drives spine structural plasticity over time scales of minutes to hours (Matus, 2000). Rapid morphological change is possible because actin within spines undergoes continuous turnover, driven by the broadly conserved mechanism of new monomer addition at the barbed end of existing filaments, and removal from the pointed end (Honkura et al., 2008; Hotulainen and Hoogenraad, 2010; Star et al., 2002).
Except in the cases of synaptogenesis or synapse disassembly, the physiological role of actin-driven changes in spine morphology is surprisingly unclear. In fact, it is probable that control of spine morphology per se is not the most important function of ongoing actin polymerization within spines. This is clear from observations that spine morphology is in many cases dissociable from plasticity of synaptic strength (Bagal et al., 2005; Sdrulla and Linden, 2007; Wang et al., 2007; Zhou et al., 2004), whereas ongoing actin polymerization is critical for the induction and maintenance of long-term potentiation (Fukazawa et al., 2003; Krucker et al., 2000; Rex et al., 2009). On the other hand, accumulating evidence suggests that actin plays a variety of other, specific roles in spines, most notably at the postsynaptic density (PSD). A number of actin binding proteins interact with neurotransmitter receptors or PSD scaffold molecules (Cingolani and Goda, 2008; Penzes et al., 2008), supporting the notion that actin serves to “anchor” receptors in the synapse. However, disruption of actin polymerization with latrunculin results in a reduction of AMPA receptors in synapses (Zhou et al., 2001), suggesting that this role of actin may involve dynamic filaments instead of static anchorage. Consistent with this, ongoing polymerization exerts direct control over the structure and composition of the PSD itself, both by driving ongoing structural rearrangement (Blanpied et al., 2008) and maintaining a defined subset of scaffolding proteins within the PSD (Allison et al., 2000; Kuriu et al., 2006). Actin filaments have in addition been suggested to regulate both AMPA receptor exocytic mobilization and internalization (Correia et al., 2008; Osterweil et al., 2005; Wang et al., 2008), actions which are likely distinct from effects at the PSD. Thus, actin polymerization serves multiple roles in spines to regulate synaptic function, in particular by regulating components of the synapse directly.
Despite the importance of actin polymerization in controlling synaptic function, the nature of perisynaptic actin remains undetermined. Electron microscopy has provided a static picture of branching actin filaments, which reach throughout the spine and appear to interact with the PSD (Landis and Reese, 1983), but has not revealed any potential dynamic nature of these structures. Conversely, numerous studies have investigated the regulation of actin in spines of living neurons (Honkura et al., 2008; Hotulainen et al., 2009; Okamoto et al., 2004; Star et al., 2002), but generally lacked the resolution for measurement of actin dynamics at spine subregions. Further, though actin regulates AMPAR internalization, and endocytosis in many systems is actin-dependent (Kaksonen et al., 2006; Merrifield et al., 2002), whether actin is directly associated with the endocytic zone is not clear. Thus, it is not known whether actin at the PSD or endocytic zone is static or dynamic, and more generally whether actin polymerization in single spines is regulated globally throughout the spine or more precisely within functional spine subdomains.
A major obstacle to discerning the spatial organization of actin polymerization within spines is their small size. Here, we reasoned that tracking single molecules of intracellular proteins within the small confines of living neurons could resolve spine organizational substructure by surpassing the limits imposed by diffraction. Using Photoactivated Localization Microscopy (PALM) (Betzig et al., 2006; Hess et al., 2006; Manley et al., 2008) to track single actin molecules in the spines of living neurons, we find that spines contain highly localized but spatially diverse foci of polymerization, frequently but not exclusively at the PSD.
RESULTS
Distributed Sites of Actin Polymerization Within Single Spines
To identify regions of actin polymerization within dendritic spines, we transfected cultured hippocampal neurons with actin tagged with photoactivatable-GFP (PA-actin). Actin molecules in single spines were labeled selectively by two-photon photoactivation. Photoactivation within the targeted region was complete with a single activation (Fig. S1A–C). Photoactivated PA-actin exited the spine rapidly, following a time course described well by a single exponential decay (τ = 17.94 ± 0.70 s; r2 = 0.991 ± 0.001, n = 32) with 4.1 ± 0.6 % remaining after 96 seconds. We, like others (Honkura et al., 2008; Star et al., 2002) find that this rate is far slower than the diffusion of monomeric PA-actin (Fig. S1D–F) or PA-GFP (Fig. S1G), and instead reflects treadmilling cycles of actin polymerization (elongating filaments through addition of actin monomers to their free, barbed ends) and depolymerization (Fig. S1H, I; Movie S1).
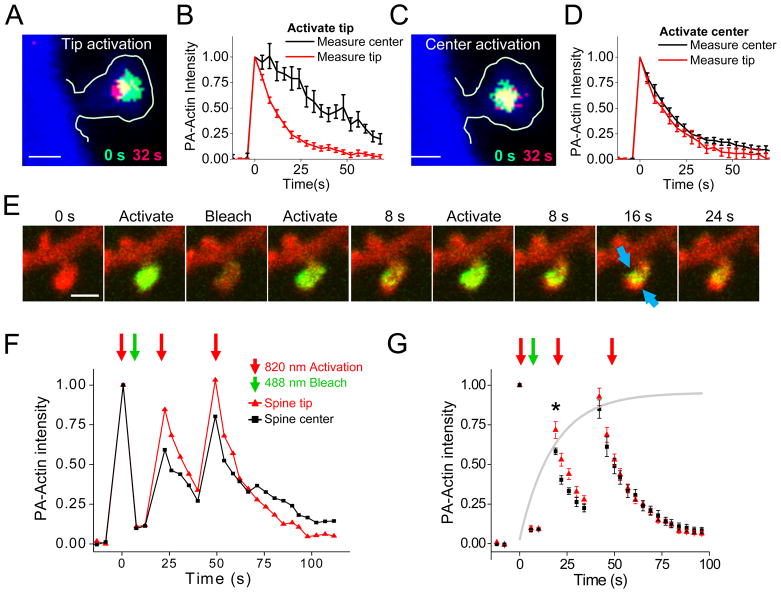
The rate of decay was faster at the spine tip than in the center (Figs. S1J, K; τtip = 17.55 ± 1.42 s, τcenter = 20.50 ± 1.09 s; two sample t test p≪0.01), and nearly all spines (26 of 32) exhibited faster fractional loss of PA-actin at the tip (Fig. S1L). To determine whether this difference in kinetics was due to constitutively slower treadmilling of filaments in the spine center, we sequentially targeted the tip and center of individual spines. Following tip activation, we observed a delayed peak of intensity at the center which occurred 9.3 ± 1.6 s following the peak at the tip (Fig. 1A, B). Overall, fluorescence at the center diminished dramatically more slowly than at the tip (time to 50% intensity: 37.20 ± 5.70 s vs. 10.55 ± 0.91 s; N=9, p≪0.01). In contrast, following targeting of the center (Fig. 1C, D) of the same spines, we observed neither a delayed peak at the tip, nor a tendency for tip kinetics to be slowed (τcenter = 19.55 ± 2.73 s; τtip = 18.66 ± 2.51 s). Following spine tip activation, we measured PA-actin fluorescence intensity move toward the center at speeds between 0.5 ± 0.1 μm/min and 1.1 ± .04 μm/min (Fig. S1M-O), consistent with the speed of polymerization-driven actin flow (Danuser and Waterman-Storer, 2006; Schaefer et al., 2002), and suggesting (Honkura et al., 2008) that actin dynamics at the spine tip drive flow toward the center.
Figure 1. Heterogeneous actin polymerization rates within single spines.
A. PA-actin fluorescence immediately after (green) or 32 s after (red) photoactivation at the spine tip. Intensity was thresholded to reveal the peak 25% of the fluorescence at each time point. PA-actin distribution is shown superimposed on cell morphology obtained from tdTomato image (blue). Scale bar, 0.5 μm in A and C.
B. PA-actin intensity measured within tip (red) or center (black) regions, following photoactivation in spine tip. Intensities are normalized within-region. n = 8 spines, separate neurons.
C. As in A, except photoactivation was targeted at the spine center.
D. As in B, except photoactivation was targeted at the spine center.
E. Optical monomer incorporation assay to identify sites of ongoing polymerization. Actin in single dendritic spines was entirely photoactivated and then photobleached. After a pause to allow additional, unactivated monomers to enter the spine, spines were photoactivated again. Newly incorporated actin moved inward from the spine membrane, with restricted regions of high and low flow along the membrane (arrows).
F. Analysis of example shown in E. Fractional PA-Actin intensity was normalized to the intensity after the first activation step within ROIs at the spine tip (red) and center (black).
G. Group data from monomer incorporation assay (n = 9 spines, separate neurons), showing higher fractional intensity in the spine tip following subsequent photoactivation steps. The grey line represents the expected time course of new monomer incorporation within whole spines, estimated from the time course (measured in Fig. 1b) of actin loss from whole spines following photoactivation. *, p=0.014.
Sites of actin polymerization are important points of regulatory control over the cytoskeleton, so measuring these sites is necessary for clarifying spine organization. To test whether overall flow is driven by addition of monomers preferentially at the tip, we designed an experiment to measure relative rates of monomer incorporation throughout the spine without the spatial bias of targeted photoactivation (Fig. 1E–G). Whole spines were first photoactivated but then immediately photobleached. A delay of roughly the time constant of actin turnover (19 s) then permitted the spine to refill with new monomers which can become polymerized, and the spine was again photoactivated. Because non-polymerized monomers diffuse rapidly out of the spine (Fig. S1FD–G), the pattern of fluorescence intensity following the second activation reveals sites at which new monomers were incorporated into filaments during the pause. We measured intensity at the tip and the center in each spine through this assay, and plotted these values normalized to the region intensity following initial photoactivation (Fig. 1F, G). In each spine, relative incorporation was greater at the spine tip than at the center. Overall, incorporation was 71.7 ±6.4% complete within 19 sec at the tip but only 58.2 ± 2.5% at the center (n=9, p=0.014, paired sample t test). However, reincorporation was not restricted to the spine tip (Fig. 1E, Movie S3) but broadly distributed within the spine head, apparent in some cases along its circumference. Reincorporation was blocked by addition of jasplakinolide (Fig. S1P). Thus, this assay directly confirms that the spine tip is a site of ongoing actin polymerization, but clearly suggests that actin dynamics within spines are more spatially heterogeneous than can be resolved using confocal microscopy.
Tracking the Behavior of Single Actin Molecules within Living Neurons
To obtain higher resolution than achievable through traditional light microscopy, we turned to photoactivated localization microscopy (PALM), which uses the sequential localization of activatable probes at very low spatial density to provide nanometer resolution of macromolecular structures (Betzig et al., 2006; Hess et al., 2006). We reasoned that tracking polymerized actin molecules would allow measurement of orientation and dynamics of filaments within the small confines of dendritic spines. Importantly, whereas the image of any individual molecule is diffracted to a broad peak as expected by the PSF of the microscope objective, the error in our estimate of the location of the molecule is expected to be much smaller (~10x smaller) than the width of this blurred image (Thompson et al., 2002). Tracking individual molecules over time in a living cell provides not only nm-scale localization but also direct observation of molecular motion within the cell. We therefore acquired single-molecule images of actin in the spines of cultured neurons by adapting a previous approach (Manley et al., 2008).
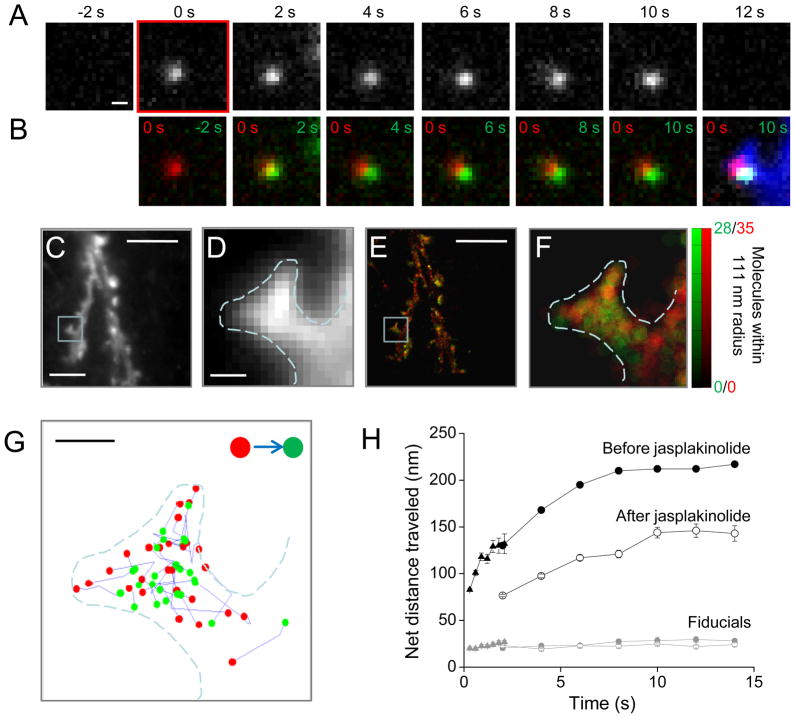
Cells were transfected with actin fused to mEos2 (McKinney et al., 2009), which fluoresces green until exposed to UV light that irreversibly converts its emission to red. As expected, transfection with actin-mEos2 did not affect spine number or head size (Fig S2A–D). Molecules were activated with very low intensity 405 nm excitation, providing roughly 0.010 to 0.025 molecules per μm2 in a given frame. Using oblique excitation illumination, molecules could frequently be followed by eye over consecutive frames before their stepwise disappearance (Fig. 2A, B). We adopted several strategies for preferentially measuring the dynamics of polymerized actin (see Supplementary Methods), most notably a long frame exposure time (Watanabe and Mitchison, 2002) with gaps between frames. By acquiring time-lapse datasets that included 100s or 1000s of images, we were able to compile trajectories for tens of thousands of localized molecules within transfected cells (Fig. 2C–G), many of which were tracked over multiple frames. Consistent with the distribution of phalloidin staining of F-actin in neurons (Allison et al., 1998), tracked molecules were primarily found in spines and spine-like protrusions from the dendritic shaft. To quantify the distribution of localized molecules, we mapped their density within subresolution spatial bins (111 nm bins, 1 camera pixel). The molecular density of tracked molecules provided a map of the polymerized actin distribution within unperturbed, living neurons (Fig. 2E, F, green). Importantly, the distribution of untracked molecules (those appearing in only one frame) in addition revealed the extent of the cell cytosol, providing a map of cell morphology derived from super-resolved molecular localization information (Fig. 2E, F, red). Within spines identified from these maps, the trajectories of tracked molecules showed a propensity for inward polarization often readily discernible without further analysis (Fig. 2G, S2E).
Figure 2. Single actin molecules tracked with PALM within dendritic spines.
A. Time series from sequential images, showing a molecule which appears at time = 0 s and persists for 6 frames before disappearing. Scale bar, 200 nm.
B. Overlays showing the molecule when it first appeared (red) and in each of the ensuing frames (green) before its disappearance. The final image shows the position of the molecule at time = 0 s (red) and 10 s (green) superimposed over the mean intensity image of all 1531 frames (blue).
C. Mean intensity projection of all 1531 frames provides a diffraction-limited image of the neuronal dendrite. Scale bar, C&E, 5 μm.
D. High magnification of boxed region in D. Dashed line represents the border of the image in E. Scale bar, D&F, 250 nm.
E. The local density of polymerized actin calculated as the total number of molecules which appeared within a radius of 111 nm (1 camera pixel) and that were tracked over two or more frames (green) or localized in only a single frame (red). Output plotted in 27.8 nm bins (1/4 the search radius).
F. High magnification view of the highlighted region in E. The dashed line in D, F, and G represents the outline of this image.
G. The first localized position (red) and last localized position (green) of all molecules tracked for ≥3 frames and over a distance greater than 100 nm are displayed. Tracks are concentrated in the spine head, and point predominantly away from the spine membrane. Scale bar, 250 nm.
H. Net distance traveled by molecules in a representative cell plotted as a function of the time since their appearance. 150 ms exposures were collected at 0.5 Hz (triangles) or 3.3 Hz (circles). 1978 frames were collected at 3.3 Hz, in which 7608 molecules were localized in only a single frame, and 3278 were tracked over 2 frames or more. 1025 frames were collected at 0.5 Hz, in which 12708 molecules were localized for a single frame and 3662 were tracked for at least 2 frames. Of these, 54.9% were tracked for 2 frames, and only 3.1% persisted >5 frames. Following jasplakinolide (open circles), 704 frames were collected at 0.5 Hz, in which 2599 molecules were localized in a single frame and 1921 were tracked for at least 2 frames. Of these tracked molecules, 44.0% persisted at least 2 frames, and 12.8% persisted >5 frames. Fiducial beads showed little motion (gray symbols).
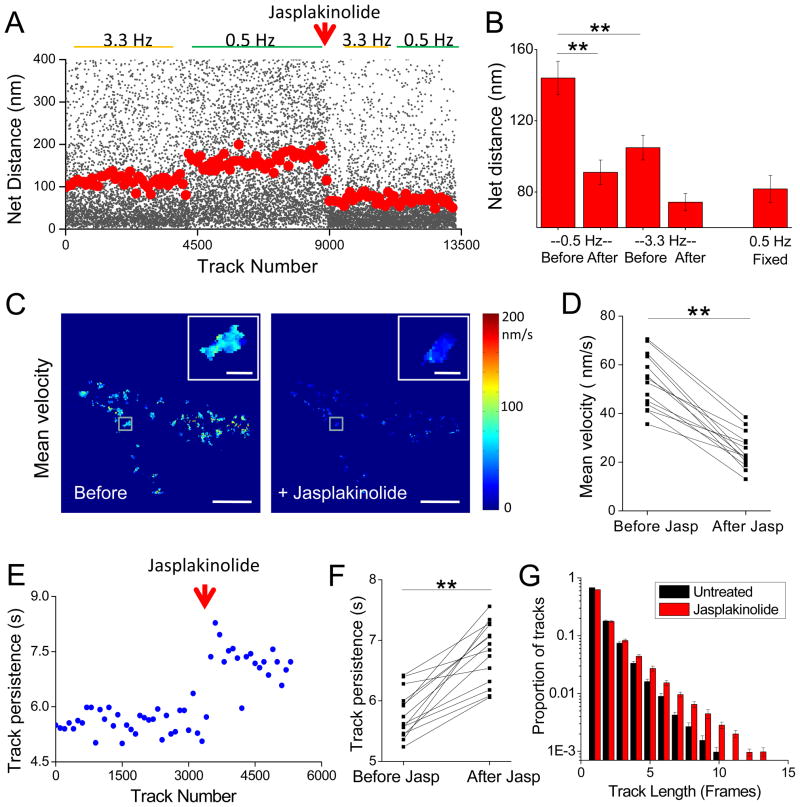
To quantify molecular dynamics, we plotted the mean distance traveled by each molecule as a function of its time since appearance (Fig. 2H). Because photobleaching limits the number of frames for which molecules can be imaged, we tracked populations of molecules over short and longer periods by acquiring images with first a rapid (3.3 Hz) then a slow (0.5 Hz) frame rate. The distance traveled by molecules tracked at 3.3 Hz over 2.1 s was essentially the same as molecules tracked at 0.5 Hz over 2 s (3 Hz at 2.1 s: 137.5 ± 12.6 nm; 0.5 Hz at 2.0 s: 137.0 ± 6.9 nm, n = 11), verifying our tracking criteria (see Supplementary Information). Molecules tracked over longer periods of time moved greater distances, as expected. The initial slope of this relationship is expected to reflect the average velocity of molecular motion along filaments. Tracks in images acquired at 3.3 Hz revealed that molecules moved on average 38.8 nm/s (2.3 μm/min). This value is considerably higher than the measured bulk flow velocity (Fig. S1M-O), suggesting that bulk flow obscures faster flow rates along individual fibers of different orientations. However, the slope decreased in longer tracks (13.5 nm/s between 2 and 8 s), and approached 0 for the longest tracks between 8 and 14 sec long (0.9 nm/s). The asymptote of 205.4 ± 11.7 (mean length of tracks 5 to 8 frames in length at 0.5 Hz, n = 14 neurons) is potentially explained if filaments within spines rarely extend beyond this length, similar to a confinement radius describing a limited area over which an otherwise freely diffusing molecule can roam, as derived from single-molecule mean squared displacement over time (Saxton and Jacobson, 1997). Alternatively, these data may reflect that flow velocity on spine filaments is heterogeneous, such that slower moving molecules remain on filaments for longer periods of time.
We tested whether our analysis selectively tracked the motion of polymerized actin molecules by examining the effects of jasplakinolide, an actin filament stabilizer. Jasplakinolide greatly restricted the motion of tracked molecules (Fig. 2H), reducing the distance traveled during the initial 2 s (130 nm to 76.6 nm) as well as the slope of the relationship between 2 and 8 s (13.5 nm/s before; 8.7 nm/s after jasplakinolide). Jasplakinolide reduced the mean distance traveled by tracked molecules to a level indistinguishable from that seen in fixed cells (i.e. from the value arising from instrumentation noise) at both rapid and slow frame rates (Fig. 3A, B; Fig. S3A–C). The fraction of molecules that traveled beyond 100 nm was also decreased following jasplakinolide to a level similar to that seen in fixed cells (Fig. S3D), and the mean net distance for tracks 5 to 8 frames in length was reduced to 110.1 ± 8.2 nm (from 205.4 ± 11.7 prior to treatment, p ≪ 0.01; paired t test, n = 14). Maps of mean local molecular velocity were created by averaging the velocity (net distance/track duration in sec) of all molecules which originated within 111 nm (1 camera pixel) of each 55.5 nm output pixel (Fig. 3C). Mean local molecular velocity was reduced by jasplakinolide throughout each of 14 neurons (Fig. 3C, D; Fig. S3E–F; 53.2 ± 11.2 nm/s before, 24.8 ± 7.3 nm/s after, n=14, p ≪.01). The value after jasplakinolide was essentially indistinguishable from that in fixed cells (24.6 ± 2.3 nm/s, n=7; Fig. S3G), indicating that any remaining motion was within the limit of our detection (Thompson et al., 2002).
Figure 3. Single molecule motion describes the behavior of polymerized actin monomers.
A. A neuron expressing actin-mEos2 was imaged sequentially at 3.3 Hz and 0.5 Hz. Following the addition of jasplakinolide, the neuron was again imaged at 3.3 Hz and 0.5 Hz. Each black point represents the distance traveled by a tracked molecule. Red circles represent the mean distance traveled of 100 consecutive tracked molecules. Note that the time-dependent increase in molecule motion is abrogated by the addition of jasplakinolide.
B. Local mean net distance traveled by localized molecules within cells which were consecutively imaged at 3.3 Hz and 0.5 Hz before and after jasplakinolide (n = 7) as in A, and separate cells which were fixed in 4% paraformaldehyde before imaging at 0.5 Hz (n = 7 cells from 4 cultures). Two-way ANOVA; ** p < 0.001.
C. Molecule velocity map of the same neuron before (left) and after (right) jasplakinolide. Molecule velocity decreased in all spines following treatment. Inset: High magnification of a representative spine from indicated region. Scale bar: 5 μm, inset 1 μm.
D. Local mean velocity was calculated by averaging the velocity of each track originating within a 111 nm radius of each 55.5 nm output pixel (as in A). Local mean velocity was decreased in 14/14 cells (8 cultures) following jasplakinolide (averaged per pixel). **, p < 0.001.
E. Representative example showing average time over which molecules were tracked (in seconds; tracked at 0.5 Hz; same cell as S3A, C) before and after addition of jasplakinolide. Each point represents the average track length of 100 tracked molecules.
F. The mean time over which molecules were tracked increased after the addition of jasplakinolide in 14/14 cells (8 cultures). Paired t-test; ** p < .0001.
G. Analysis of all tracked molecules before and after the addition of jasplakinolide (14 neurons, 8 cultures). Total molecules tracked before jasplakinolide was 172,879. Total molecules tracked after the addition of jasplakinolide was 96,240. The fraction of tracks persisting over the designated number of frames is plotted.
If continuous polymerization is responsible for driving most actin flow along spine filaments, depolymerization and subsequent free diffusion of molecules would be expected to terminate tracks. Indeed, the mean persistence of tracked molecules (in seconds) increased after jasplakinolide treatment in 14/14 cells (Fig. 3E–G, Fig. S3H). Together with the rapid time constant for turnover of photoactivated PA-actin (Fig. 1), these data indicate that many tracked molecules reach the end of filaments and are depolymerized before photobleaching, consistent with a population of short filaments within spines. The strong effects of jasplakinolide on molecular velocity suggest that polymerization drives the large majority of actin flow in spines. It is likely that the remaining motion (above the error-derived values in fixed cells) reflects the role of myosin-based contractility.
Resolution of Perisynaptic Polymerized Actin
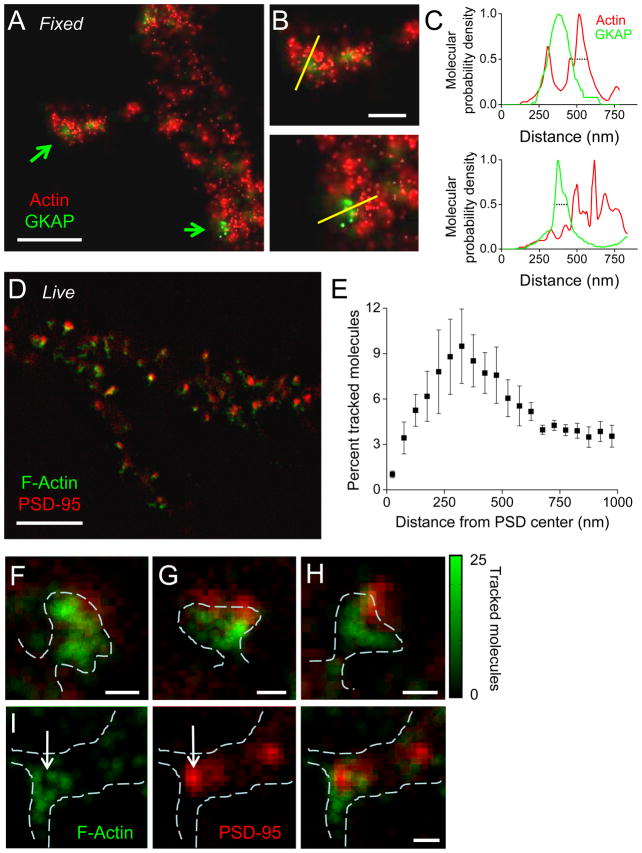
To measure the distribution of actin near synapses, we used two-color PALM (Shroff et al., 2007) of fixed neurons co-transfected with actin-tdEos and the postsynaptic density protein GKAP (Welch et al., 2004) tagged with the photoswitchable green protein dronpa (Fig. 4A–C). Molecules of each were photoactivated, and the locations and localization uncertainty were plotted to encode the probability of finding a molecule at any position (Betzig et al., 2006). The resulting images showed areas of high actin density abutting the PSD of synapses both in spines and on the dendritic shaft (Fig. 4B). Line profiles through areas of interest confirmed the low probability of finding actin molecules within the PSD itself (Fig. 4C).
Figure 4. Spatially restricted distribution of polymerized actin visualized with PALM near synapses.
A. Two-color PALM image of actin-tdEos and GKAP-dronpa in a fixed neuron. Molecules localized with error less than 30 nm (Actin-tdEos, red) or 100 nm (GKAP-Dronpa, green) are shown mapped with a pixel size of 5 nm. Scale bar, 1 μm.
B. Closer view of synapses on a spine (top) and shaft (bottom), showing lines along which measurements were made in C. Scale bar, 500 nm.
C. Relative molecular density probability of actin-tdEos (green) and GKAP-Dronpa (red) along 75-nm wide lines (30 pixels) shown in B. Dotted lines highlight the width of the profile at a probability of 0.5 (top, 121 nm; bottom, 87 nm).
D. The local density of polymerized actin calculated as the number of tracked actin-mEos2 molecules within a 111 nm radius. Local density (green) was plotted at ¼ this scale (27.8 nm bins), and overlaid on the diffraction-limited image of PSD-95-cerulean (red). Scale bar, 5 μm.
E. The distribution of tracked actin-mEos2 molecules as a function of distance from the center of PSDs identified by localization of PSD-95-cerulean (n = 4 neurons, 148 PSDs).
F–H. Three synapses from D, revealing close association of polymerizing actin with the PSD. Scale bars, 500 nm.
I. Actin distribution at a shaft synapse showing actin (left) encircling the PSD (middle). Scale bar, 500 nm. Green color bar represents local density of polymerized actin in A–E.
To examine polymerized actin density in relation to the PSD in living cells, we acquired single-molecule time lapses of actin-mEos2 followed by diffraction-limited images of PSD95-cerulean (Fig. 4D), and compared the molecular density maps created from tracked actin molecules to the position of PSD95-cerulean. This analysis revealed a striking peak density of polymerized actin near the PSD (Fig. 4E). Examining individual images confirmed that polymerized actin density was notably higher near the edge of the PSD (Fig 4F–H), even in the case of morphologically complex PSDs. Some actin molecules were localized overlaying the PSD, though the finite thickness of the optical section precludes determining whether these molecules were above or truly within the PSD. These observations using PALM thus confirm EM of fixed cells showing filaments near or touching the PSD (Capani et al., 2001; Fifkova and Delay, 1982). Further analysis (see below) extends these results significantly by demonstrating directly that such perisynaptic actin is highly dynamic rather than merely a stable or structural scaffold.
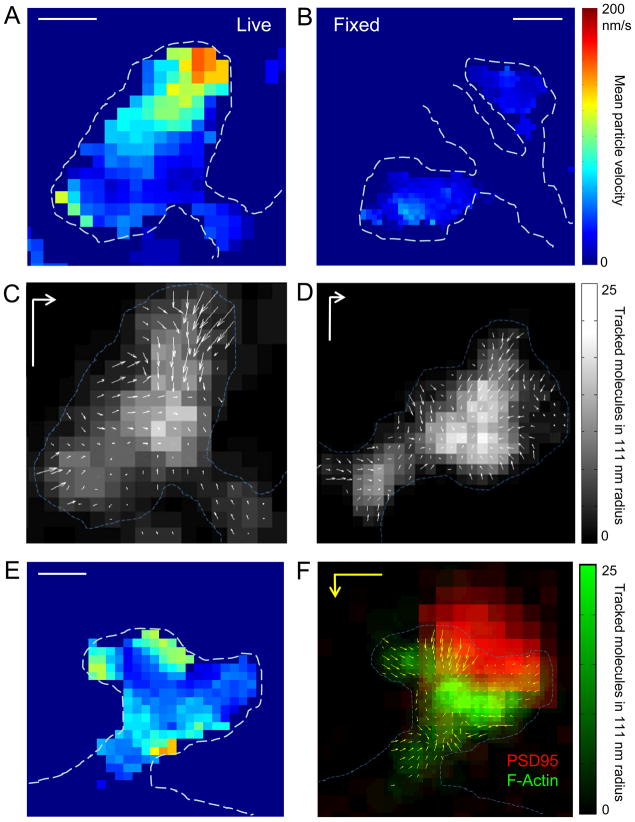
Heterogeneous Actin Dynamics Within Spine Subregions
Tracking single molecules permitted us to measure the spatial heterogeneity of actin dynamics across the inner extent of spines, by mapping characteristics of molecules whose tracks began within defined spatial bins. Molecular velocity was strikingly heterogeneous within spines (Fig. 5A). Most notable were distinct regions of high velocity motion. Such regions were typically found along restricted portions of the spine membrane. The narrow lateral extent and the depth into the cell interior argue against the possibility that they represent an edge effect, since such an effect should be uniform along the membrane and not affect molecules farther interior than the error in molecule localization. In fixed cells and neurons treated with jasplakinolide, mean local velocities decreased to the level of localization-error-derived background noise (Fig. 5B; Fig. S3E, F). Vector plots representing local average molecular direction and velocity were constructed to map flow at high spatial resolution. This local flow was overwhelmingly directed inward from the spine membrane (Fig. 5C, D; also Fig. S4). The interior of all spines comprised dense actin of somewhat slower velocity and notably reduced local flow, most easily explained by heterogeneous filament orientation. Perhaps most strikingly, at irregular points along the membrane, regions of inwardly directed, high net velocity were observed, which we take to be sites of dramatically enhanced polymerization. These sites presumably underlie the distributed monomer incorporation observed following unbiased confocal photoactivation (Fig. 1E–G).
Figure 5. Heterogeneous actin dynamics within individual spines.
A. Map of actin molecule velocity across the inner extent of a dendritic spine. The mean velocity of molecules within 111 nm was plotted at each 55.5 nm pixel for which there were ≥3 molecules within this radius. Restricted areas of high velocity are clearly visible. Scale bar, 250 nm
B. Actin velocity map of fixed spines, showing that error stemming from finite molecular localization precision is minimal and spatially unmodulated. Scale bar, 500 nm.
C–D. Representative spines showing inward orientation of actin flow. Arrow length represents relative velocity. Gray scale represents tracked molecular density. Scale bar, 200 nm; vector, 100 nm/s.
E. Actin velocity map and (F) locally averaged molecular movement vector plotted superimposed on local tracked molecule density (green) and the deconvolved widefield image of PSD-95-cerulean (red). Some but not all foci of high velocity motion are closely associated with the synapse. Scale bar, 250 nm; vector, 200 nm/s. Color bar represents tracked molecular density for F.
Surprisingly, the inward orientation of flow was observed even at the opening of the spine neck. Actin within the spine neck has been difficult to resolve in live cells, but we found that the high sensitivity and spatial resolution of single-molecule tracking enabled measurement of molecular motion along filaments in the neck (Fig. 5C, D, F; Fig. S4). This provides direct evidence supporting the existence of dynamic actin filaments in the spine neck. Further, all spines with a clearly identified neck contained molecules with a net inward flow, though overall, roughly one-half as many actin molecules flowed outward through the neck as flowed inward (67±5% of filaments inward; n = 13 spines from 13 neurons in 4 cultures).
Maps of actin molecular velocity in many cases revealed multiple foci in broadly spaced regions of the spine (Fig. 5E). To examine filament orientation within these regions, we constructed vector plots of local net flow direction and magnitude (Fig. 5C, D, F; Fig. S4). This analysis indicated that foci of high velocity motion were associated with more strongly congruent directionality, i.e. a fast local flow vector. Though not all regions of relatively highly correlated motion direction were within regions of high velocity, such association was seen in nearly all of >100 foci examined. Importantly, by overlaying vector plots of net flow with widefield images of PSD-95-cerulean, it was clear that foci of high velocity motion often occurred near or overlapping with the diffraction-limited image of the PSD (Fig. 5F; Fig. S4; and see below).
Distribution of focal sites of polymerization within spines
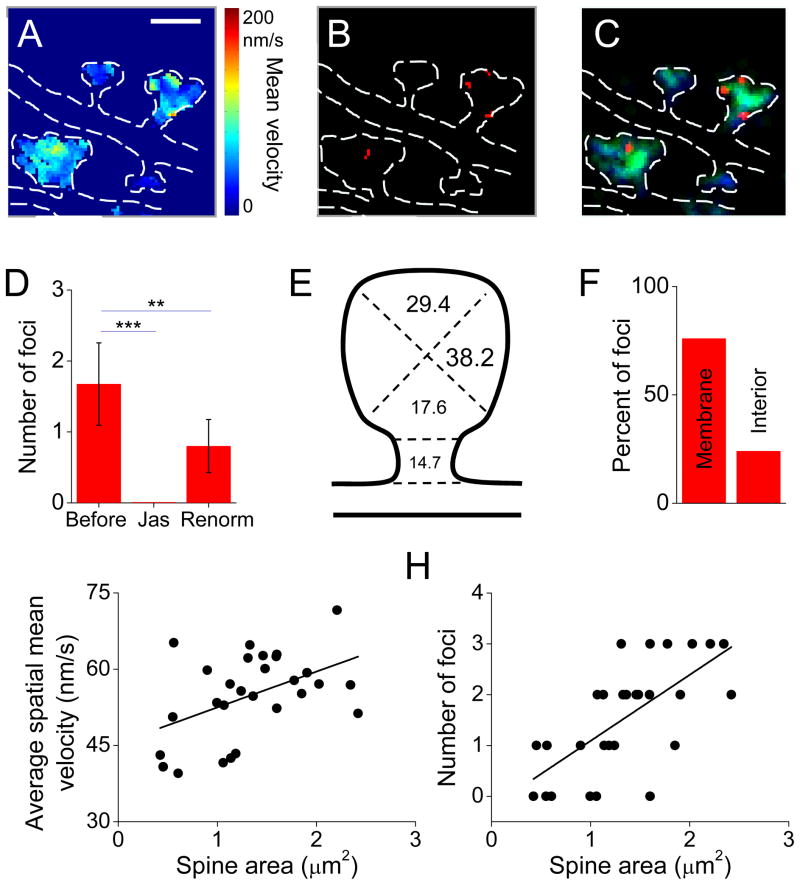
To quantitatively assess the distribution within spines of focal sites of enhanced polymerization, we identified these regions first by thresholding each cell’s velocity map (Fig. 6A–C; see Experimental Procedures). Spines contained on average 1.7 ± 0.6 foci (n = 68 foci in 30 spines in 5 neurons, Fig. 6C, D). All foci were eliminated by application of jasplakinolide, and rescaling the threshold based on the extremely low velocity following jasplakinolide did not rescue the number of foci, confirming their dissipation in the absence of normal polymerization (Fig. 6D). Foci were found distributed widely through the spine, with a preponderance in the lateral or tip regions of the spines (Fig. 6E). The large majority were within 150 nm of the spine membrane (Fig. 6F), (the location of which was deduced from the distribution of all localized actin molecules). Like others (Honkura et al., 2008), we found that the average velocity of actin flow was weakly correlated with the size of the spine (Fig 6G; correlation coefficient=0.43, p<0.05, n = 28). More notably, however, larger spines had more foci (Fig. 6H; correlation coefficient=0.67, p<0.01, n = 28). To examine this further, we compared the number of foci in spines from neurons overexpressing clathrin-cerulean, which has no effects on spine size (Blanpied et al., 2002; Petrini et al., 2009), with those overexpressing PSD-95, which is known to increase synapse strength and spine size (Bredt and Nicoll, 2003). PSD-95 overexpression increased the number of foci by 1.6-fold (p<0.05, n = 24 and 27 spines from 4 cells each for PSD-95 and clathrin, respectively). These data indicate that the subspine actin velocity landscape is more intricate in large spines, and suggests that as synapses strengthen, they acquire unique organizational or functional complexity.
Figure 6. Widespread distribution of focal points of high velocity flow within spines.
A–C. Definition of polymerization foci. (A) Spatially averaged velocity maps were thresholded to their mean + 1.5 times their standard deviation, to produce binary maps of pixels with high average velocity (B). These were band pass filtered to exclude and combine single pixels. (C) Foci (red) superimposed on an image map showing density of both tracked (green) and untracked molecules (blue).
F. Average number of foci per spine before and after 5 min treatment with jasplakinolide, and then after recalculation of the velocity map threshold following jasplakinolide (** p < .01, *** p < .0001, repeated measures ANOVA; n = 12 spines from 5 neurons, 5 cultures).
G. Distribution of foci positions within spine subregions delineated as sketched, (n = 68 foci, 30 spines from 5 neurons, 5 cultures). The majority were at the tip and lateral regions.
H. The distribution of focal regions from E, with respect to the plasma membrane.
I. The average velocity of all pixels within individual spines (n = 32 spines, 6 neurons, 6 cultures). Solid line shows the linear regression.
J. Number of foci per spine as a function of spine area (n = 32 spines, 6 neurons, 6 cultures). Solid line shows the linear regression.
Differing actin polymerization dynamics at the synapse and endocytic zone
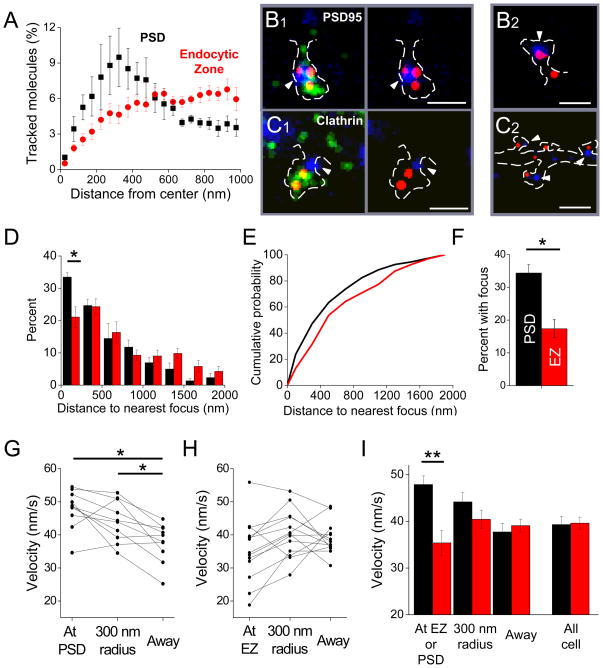
Sites of enhanced polymerization could be randomly distributed within spines and thus presumably mediate only morphological change, or could be tied to functional domains such as the synapse or the endocytic zone (EZ). To distinguish between these possibilities, we first examined spines from neurons triply transfected with actin-mCherry, PSD-95-GFP to mark synapses, and clathrin-cerulean to mark the EZ (Blanpied et al., 2002). Intriguingly, confocal images of these spines suggested that the density of actin near PSDs was greater than near the EZ (Fig. S7A), but the limited resolution prevented detailed analysis of actin dynamics in relation to each structure. We therefore measured the distribution of polymerized actin via smtPALM in relation to widefield images of clathrin-cerulean acquired after tracking. Strikingly, the distributions of 1000s of tracked (polymerized) molecules surrounding the center of the EZ was quite different than that surrounding the PSD (Fig. 7A). Whereas the peak density occurred only 300 nm from the imaged center of the PSD, tracked actin molecules showed no prominent peak density even within 1 μm. We therefore examined directly whether polymerization foci occurred preferentially near synapses or the EZ, by overlaying maps of foci derived from single-molecule tracking with widefield images of the PSD or EZ (Fig. 7B, C). PSDs typically were found with one or more actin foci on or almost directed apposed to them. On the other hand, actin foci appeared less frequently to abut the EZ marked with clathrin (Fig. 7C). We calculated a nearest neighbor distribution by measuring the distance from the center of each PSD or clathrin punctum to the nearest actin polymerization focus. A histogram of these nearest neighbors (Fig 7D) revealed that actin foci lay significantly further from the EZ than from the PSD (median distance 565±43 nm for clathrin vs. 420±14 for PSD-95, p<0.05). Furthermore, the most striking difference in the distributions was in the closest bin: a significantly greater fraction of PSDs were associated with an actin polymerization focus within 250 nm (p<0.05, t-test). This closer relationship was apparent also in the left-shifted cumulative nearest neighbor distance (Fig. 7E; p<0.005, Man-Whitney U test). To confirm this automated analysis, we manually counted foci to score whether they overlapped 1 or more image pixels with thresholded images of PSDs or EZs. The results were very similar (Fig. 7F), indicating that actin foci occurred at PSDs with roughly twice the likelihood as at EZs (p<0.01).
Figure 7. The PSD and the endocytic zone show distinct patterns of polymerization dynamics.
A. The distribution of tracked actin-mEos2 molecules as a function of distance from the center of PSDs (black) or EZs (red). (n = 148 PSDs, 4 neurons, 4 cultures; 354 EZ, 4 neurons, 2 cultures)
B. Frequent colocalization in spines of PSD-95-cerulean (blue) with focal areas of high velocity actin motion (red). The density of tracked actin-mEos2 is shown (green). Arrowheads indicate PSDs. B1 and B2 show spines from two different neurons. Scale bars, 500 nm.
C. Infrequent and less direct colocalization of clathrin-cerulean3 (blue) with actin polymerization foci (red). The density of tracked actin-mEos2 is shown (green). Arrowheads indicate EZs. C1 and C2 show spines from two different neurons. Scale bars, 500 nm in C1, 1 μm in C2.
D. Histogram of the distribution of distances from localized PSDs (black) or EZs (red) to the nearest focal point of high-velocity actin motion. A significantly higher fraction of PSDs had associated foci within the closest 250 nm (p < .05, t-test). The average PSD was closer to a focus of high velocity particle motion than the average EZ (median 420.0 ± 14.4 vs 565.5 ± 42.3 nm, p < .05 t-test). Data from 4 cells from 4 cultures.
E. Cumulative distribution plot of individual PSDs (n=148) or EZs (n=354) from the cells in D. These curves were different (p<0.01, Mann-Whitney U test).
F. Percent of PSDs or EZs that colocalized (overlapped pixels with) actin polymerization foci (*, p < .05, t test; n = 100 PSDs, 4 neurons, 4 cultures; 148 EZs, 4 neurons, 2 cultures).
G. Average velocity of tracked particles originating within thresholded PSDs, within 300 nm of the PSD, or elsewhere in each of 10 cells. Tracked molecules originating at or near the PSD moved significantly faster than molecules originating elsewhere in the cell (p < .05; paired t-test with Bonferroni correction).
H. Average velocity of tracked particles originating within thresholded EZs, within 300 nm of the EZ, or elsewhere in each of 13 cells. Tracked molecules originating at or near the EZ did not move significantly differently than molecules originating elsewhere in the cell (p > .05, paired t-test with Bonferroni correction).
I. Average velocity of tracked particles at the PSD or EZ, within a 300 nm radius, or elsewhere in the cell. The velocity of tracked actin molecules originating at the PSD was significantly higher than that of tracked actin molecules originating at the EZ (p < .01, unpaired t-test with Bonferroni correction).
To test in an independent way whether the behavior of polymerized actin molecules in the vicinity of the PSD or EZ differed, but without relying on definition of discrete foci, we analyzed the mean molecular velocity of actin in spatial bins surrounding the functional markers. Molecules whose tracks began closest to the synapse (i.e. their tracks initiated within the borders of the PSD as determined from deconvolved, widefield images of PSD-95-cerulean) or those within a 300 nm annulus around the PSD had significantly higher mean velocities than molecules whose tracks initiated further than 300 nm away from the synapse (Fig. 7G). In contrast, molecules whose tracks initiated within the boundaries of the EZ were no faster than those lying immediately outside the EZ or more than 300 nm from it (Fig. 7H). Accordingly, even though the average molecular velocity across the entire cells did not differ for neurons expressing PSD-95-cerulean or clathrin-cerulean, the velocity of molecules at the synapse was significantly greater than those at the EZ (Fig. 7I; p<0.01, t-test followed by Bonferroni correction for multiple comparisons; Fig S5B–D).
DISCUSSION
We have used a variety of confocal and single-molecule tracking measurements to analyze actin dynamics within the submicron dimensions of single living dendritic spines. Our results make clear that spines contain a dense and highly dynamic perisynaptic actin network, tightly localized foci of polymerization both at and away from the synapse, a dense central core of heterogeneous filament orientation, and actin filaments in the spine neck frequently oriented with barbed ends toward the dendritic shaft.
Dynamics of perisynaptic actin
We find continuously polymerizing actin immediately apposed to the synapse, and the density of actin filaments reaches a peak within 300 nm of the PSD center. Recent indications that filament structure observed by EM is highly sensitive to fixation conditions (Urban et al., 2010) emphasize the need for further live-cell measurement of cytoskeleton behavior and function. Nevertheless, the distribution we observed is consistent with EM localization of filaments near or abutting the PSD (Caceres et al., 1983; Capani et al., 2001; Rostaing et al., 2006). Indeed, Fifkova et al. (1982) found filaments with barbed ends oriented toward the PSD, as well as filaments of differing orientation near the PSD, even running parallel to it, which is consistent with the highly localized and heterogeneous organization we have observed. This complex assembly pattern likely arises from the concentration of the Arp2/3 branching nucleation factor only ~200 nm from the PSD edge (Racz and Weinberg, 2008), and further suggests that the perisynaptic actin network is a uniquely nimble collection of highly dynamic filaments.
The tight synaptic association of the cytoskeleton presumably underlies its role in maintaining synaptic glutamate receptors (Allison et al., 1998; Kim and Lisman, 1999; Zhou et al., 2001). We find that polymerization-driven flow along filaments at the synapse is faster than elsewhere in the spine, suggesting that receptor “anchoring” by the cytoskeleton may in fact involve an active process mediated by polymerizing filaments, not stable ones. Dynamic perisynaptic filaments also provide a straightforward explanation for the pronounced, actin-driven morphological distortion that most PSDs undergo on a continual basis (Blanpied et al., 2008). Direct cytoskeletal control of the morphology of the synapse proper, rather than the spine in general, may contribute to subsynaptic receptor positioning with respect to sites of neurotransmitter release important for synaptic strength (Raghavachari and Lisman, 2004). Indeed, the latrunculin sensitivity of PSD scaffold protein content (Allison et al., 2000; Kuriu et al., 2006) suggests that actin-dependent receptor anchoring may stem from cytoskeletal interaction with the proteins of the PSD rather than receptors directly. As actin polymerization also regulates the lateral diffusion of membrane proteins into and out of the dendritic spine (Richards et al., 2004), it is tempting to speculate that a dynamically regulated cytoskeleton surrounding the PSD may restrict the territory explored by freely diffusing receptors, potentially gating receptor exit or entry to synapses (Yang et al., 2008).
Roles of polymerized actin in spine subdomains
Using targeted photoactivation we observed as reported previously (Honkura et al., 2008) that spines possess a general tip-to-base orientation of actin flow. However, both an optical monomer-incorporation assay and single-molecule tracking revealed that this does not result strictly from preferential polymerization activity at the spine tip or at the synapse. Rather, sites of high polymerization activity are broadly distributed and found at spine tips, in lateral domains, and even in or near the neck. These distributed sites likely represent points of regulated control over filament density, length, and turnover. Thus, the tip-to-base flow structure that can be resolved via relatively low resolution confocal microscopy appears to be an emergent phenomenon that masks a more intricate and functionally revealing underlying organization. Synapses but not endocytic zones were prominently associated with these foci, we take this non-random distribution as clear evidence in favor of the notion that actin dynamics regulate spine function through specific mechanisms aside from influencing spine morphology. Indeed, larger spines were not merely expanded versions of small spines: they were more intricate, containing more foci. Thus, we conclude that for spines, strength is associated not merely with size but with organizational complexity that may support computational sophistication.
The high resolution of PALM allowed us to identify unexpected characteristics of actin dynamics within two clearly identified subdomains away from the synapse. The endocytic zone (EZ), a region of clathrin assembly reliably positioned 100s of nm away from the synapse (Blanpied et al., 2002; Lu et al., 2007; Racz et al., 2004), is an important potential site of actin polymerization within the spine. However, by a number of tests, foci of polymerization were not closely associated with clathrin puncta within the spine, and the velocity of actin monomers on filaments near the EZ was not different than the surrounding spine milieu. This is particularly surprising given that the endocytic zone contains numerous actin-binding molecules which likely regulate endocytosis (Engqvist-Goldstein and Drubin, 2003; Rocca et al., 2008; Yarar et al., 2005). However, consistent with a limited tonic role of actin polymerization at the spine EZ, clathrin puncta are not disassembled or disrupted during latrunculin application (Blanpied et al., 2002), whereas AMPA receptors and PSD scaffold proteins are quickly lost (Kuriu et al., 2006; Zhou et al., 2001). Our results are thus consistent with the idea that actin polymerization at the EZ is either highly transient during endocytosis (Merrifield et al., 2002) or even negatively regulated in spine subregions (Rocca et al., 2008). PALM offers the possibility to resolve whether actin de/polymerization during induction of long-term depression occurs at the synapse, at the EZ, or at intervening points of the membrane traversed by receptors destined for endocytosis.
Platinum replica electron microscopy recently identified the presence of the Arp2/3 branching nucleator and a network rather than a bundle of filaments at the point of emergence of the spine from the shaft (Korobova and Svitkina, 2009), suggesting that filament organization within spine necks is not a simple bundle. Our results confirm and extend these observations by providing live-cell measurements indicating that mature spine necks contain a complex filament organization of mixed directionality. The molecular basis for this remains to be determined, but functionally, the mixed orientation of neck filaments suggests that myosins which translocate to either the barbed end or pointed end of actin filaments could mediate vesicular traffic both to and from the spine head (Correia et al., 2008; Osterweil et al., 2005; Wang et al., 2008).
Regulation of filament length through depolymerization is mediated by cytosolic proteins that are unlikely to discriminate between filaments emanating from different, distant locations. On the other hand, there are a number of mechanisms by which active spatial regulation of polymerization would allow filaments of unique length or structure to be organized at discrete locations. At the synapse, for instance, cortactin (Hering and Sheng, 2003; Uruno et al., 2001) and Abp1 (Haeckel et al., 2008) interact with the PSD protein Shank through SH3 domains and activate the Arp2/3 complex. Thus, it will be essential to measure within the spine the distribution of proteins that control polymerization, the mechanisms of their positioning over both short and long time scales, and potential means of compartmentalizing regulatory signaling cascades.
Dynamics of spine filament organization
On average, the distance traveled by tracked actin molecules increased with time for only approximately 8 seconds, reaching a plateau near 200 nm, suggesting that the large majority of filaments are shorter than 200 nm. Consistent with this interpretation, both classical and more recent EM methods show a filamentous actin network which stretches throughout the spine head, but with few filaments extending unambiguously longer than 250 nm (Fifkova and Delay, 1982; Korobova and Svitkina, 2009; Rostaing et al., 2006). Furthermore, immuno-EM localization of actin-binding proteins within dendritic spines shows that the branch regulator Arp2/3 concentrates within 100 nm of the spine plasma membrane (Racz and Weinberg, 2008) and the filament-severing protein cofilin concentrates within 200 nm of the plasma membrane (Racz and Weinberg, 2006), suggesting tight regulation of filament dynamics near the membrane. Thus, one significant difference between the actin networks of dendritic spines and lamellipodia is the considerably shorter filament length that exists in the spine head. Because neural activity is well known to regulate spine actin dynamics (Cingolani and Goda, 2008; Honkura et al., 2008; Okamoto et al., 2004; Penzes et al., 2008; Star et al., 2002), one intriguing interpretation of this difference is that a network of short filaments optimizes both the temporal and spatial response characteristics of actin within the spine following receptor activation.
Filament dynamics in the spine interior were complex, with frequently high single-molecule velocities yet little correlation of the local direction of motion. Interestingly, both photoactivation and particle tracking indicated that few filaments penetrate from the spine head into the spine neck. This suggests that the deep interior of the spine near the spine neck represents a zone of depolymerization where filaments converge from broadly distributed regions of the spine head and neck. One potential implication of such an organization is that depolymerization of filaments at this point would offer coordinated regulatory control over both spine morphology and diverse actin functions in the spine. The actin severing protein cofilin is enriched in spines (Racz and Weinberg, 2006), and with its regulators LIM kinase and slingshot is known to exert powerful control over spine morphology (Meng et al., 2002) during long-term depression (Wang et al., 2007; Zhou et al., 2004) and potentiation (Rex et al., 2009). Such control may be facilitated by attacking filaments at this key organizational point.
Single-molecule tracking of intracellular proteins
The high contrast ratio of activated to non-activated fluorescence of mEos2 (Shroff et al., 2007) has allowed us to track single molecules with very high precision in the x–y plane. Spine geometry thus permitted determination of molecular motion with highest confidence along the spine’s most peripheral membrane, where internally directed flow is parallel to the plane of focus. In addition, spine necks are thin enough to be contained within the illuminated volume, and the predominant flow towards the head is unlikely to be contaminated by 3d ambiguity. Within the spine central core, filament orientation is clearly more complex and irregular, but is also more difficult to determine unambiguously due to the existence of axially oriented filaments in which molecular velocity will be underestimated. Future advances in tracking tools may allow detailed analysis of molecules in three dimensions. Accurate 3d tracking of molecule motion within living spines or other cell regions will complement other emerging 3d super-resolution techniques such as STED (Nagerl et al., 2008), though creating volumetric instead of planar maps through optical and analytical strategies for 3d localization will naturally require a dramatic increase in the total number of molecules tracked, at the expense of temporal resolution.
Our results demonstrate that single-molecule tracking offers the potential to dramatically facilitate analysis of signaling molecules, scaffolds, and other critical proteins whose dynamic behavior has remained obscure due to intricate cell morphology and highly compartmentalized function. Antibody-based methods to track single proteins in the plasma membrane have been remarkably successful (Borgdorff and Choquet, 2002; Ehlers et al., 2007; Sako et al., 2000). However, tracking intracellular proteins has presented a very steep challenge, because antibodies are generally not suitable to label single proteins within cells whereas bulk-expressed GFP-tagged proteins cannot easily be discriminated as single molecules. Massively parallel, dual-probe (Subach et al., 2009), single-molecule tracking with PALM appears to provide a general tool for mapping the dynamics of key constituents in synapses and other small domains of many cell types.
EXPERIMENTAL PROCEDURES
Hippocampal neuronal cultures were prepared from E18 rats (Blanpied et al., 2002). Transfections were performed using Lipofectamine 2000 after 13–21 days in culture and imaging was performed 48 hours later. Multiphoton photoactivation was conducted on an LSM510 Meta (Zeiss), using 820 nm excitation from a MaiTai Ti:Sapphire laser (SpectraPhysics) and a 60x, 1.4NA objective. PALM imaging of fixed neurons expressing actin-tdEos and GKAP-Dronpa was conducted essentially as described (Shroff et al., 2007). sptPALM was carried out by modifying the approach of (Manley et al., 2008). For tracking single actin molecules (Tatavarty et al., 2009), images were acquired using 150 ms exposures every 300 ms or 2 s as noted. Following acquisition of sptPALM images, widefield images of PSD95- or clathrin-cerulean were acquired using a Xenon lamp and appropriate filters (Shroff et al., 2007) at multiple Z positions surrounding the sptPALM focal position, deconvolved in ImageJ, and summed. Molecules were localized by fitting a 2D elliptical Gaussian function to a 9×9 pixel array surrounding the peak, and locations were assembled into tracks using freely available algorithms (Manley et al., 2008). All subsequent analysis was written in Matlab. Molecules were segregated into those which were tracked over multiple frames, and those which appeared in a single frame only. The distribution of tracked molecules representing polymerized actin was calculated based on the density of molecules within a search radius of 1 camera pixel (111 nm). Local mean velocities and local flow rates were calculated using the first and last points of tracks originating within this radius. Velocities and vectors were mapped only in pixels with 3 or more tracks originating within the search radius.
Supplementary Material
Acknowledgments
We thank the following for supplying plasmids: S. McKinney (mEos2-actin), B. Imhof (GFP-actin), G. Patterson (PA-GFP), M. Rizzo (cerulean), J. Lippincott-Schwartz (tdEos-paxillin), D. Bredt (PSD-95-GFP), R. Y. Tsien (tdTomato, mCherry), G. Feng (GKAP), and M. Davidson (Dronpa). We thank also J. Kerr and H. Lu for helpful discussions, and M. Ehlers, J. Bear, and S. Thompson for comments on the manuscript. Support was provided by a Summer Training in Aging Research Topics - Mental Health (Start-MH) fellowship, Training Grant NIGMS T32GM008181, and NRSA F30MH086185 (NAF), and NIMH R01MH080046, The Dana Foundation, and the Katherine D. and Theodore J. Carski Foundation (TAB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison DW, Chervin AS, Gelfand VI, Craig AM. Postsynaptic scaffolds of excitatory and inhibitory synapses in hippocampal neurons: maintenance of core components independent of actin filaments and microtubules. J Neurosci. 2000;20:4545–4554. doi: 10.1523/JNEUROSCI.20-12-04545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DW, Gelfand VI, Spector I, Craig AM. Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: differential attachment of NMDA versus AMPA receptors. J Neurosci. 1998;18:2423–2436. doi: 10.1523/JNEUROSCI.18-07-02423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagal AA, Kao JP, Tang CM, Thompson SM. Long-term potentiation of exogenous glutamate responses at single dendritic spines. Proc Natl Acad Sci U S A. 2005;102:14434–14439. doi: 10.1073/pnas.0501956102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Blanpied TA, Kerr JM, Ehlers MD. Structural plasticity with preserved topology in the postsynaptic protein network. Proc Natl Acad Sci U S A. 2008;105:12587–12592. doi: 10.1073/pnas.0711669105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Scott DB, Ehlers MD. Regulation and dynamics of clathrin at specialized endocytic zones in dendrites and spines. Neuron. 2002;36:435–449. doi: 10.1016/s0896-6273(02)00979-0. [DOI] [PubMed] [Google Scholar]
- Borgdorff AJ, Choquet D. Regulation of AMPA receptor lateral movements. Nature. 2002;417:649–653. doi: 10.1038/nature00780. [DOI] [PubMed] [Google Scholar]
- Bredt DS, Nicoll RA. AMPA receptor trafficking at excitatory synapses. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Caceres A, Payne MR, Binder LI, Steward O. Immunocytochemical Localization of Actin and Microtubule-Associated Protein MAP2 in Dendritic Spines. Proceedings of the National Academy of Sciences. 1983;80:1738–1742. doi: 10.1073/pnas.80.6.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capani F, Martone ME, Deerinck TJ, Ellisman MH. Selective localization of high concentrations of F-actin in subpopulations of dendritic spines in rat central nervous system: a three-dimensional electron microscopic study. J Comp Neurol. 2001;435:156–170. doi: 10.1002/cne.1199. [DOI] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- Correia SS, Bassani S, Brown TC, Lise MF, Backos DS, El-Husseini A, Passafaro M, Esteban JA. Motor protein-dependent transport of AMPA receptors into spines during long-term potentiation. Nat Neurosci. 2008;11:457–466. doi: 10.1038/nn2063. [DOI] [PubMed] [Google Scholar]
- Danuser G, Waterman-Storer CM. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu Rev Biophys Biomol Struct. 2006;35:361–387. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- Ehlers MD, Heine M, Groc L, Lee MC, Choquet D. Diffusional trapping of GluR1 AMPA receptors by input-specific synaptic activity. Neuron. 2007;54:447–460. doi: 10.1016/j.neuron.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Drubin DG. Actin Assembly and Endocytosis: From Yeast to Mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- Fifkova E, Delay RJ. Cytoplasmic actin in neuronal processes as a possible mediator of synaptic plasticity. J Cell Biol. 1982;95:345–350. doi: 10.1083/jcb.95.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y, Saitoh Y, Ozawa F, Ohta Y, Mizuno K, Inokuchi K. Hippocampal LTP Is Accompanied by Enhanced F-Actin Content within the Dendritic Spine that Is Essential for Late LTP Maintenance In Vivo. Neuron. 2003;38:447–460. doi: 10.1016/s0896-6273(03)00206-x. [DOI] [PubMed] [Google Scholar]
- Haeckel A, Ahuja R, Gundelfinger ED, Qualmann B, Kessels MM. The actin-binding protein Abp1 controls dendritic spine morphology and is important for spine head and synapse formation. J Neurosci. 2008;28:10031–10044. doi: 10.1523/JNEUROSCI.0336-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering H, Sheng M. Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci. 2003;23:11759–11769. doi: 10.1523/JNEUROSCI.23-37-11759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. J Cell Biol. 2010;xx:xxx–yyy. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Llano O, Smirnov S, Tanhuanpaa K, Faix J, Rivera C, Lappalainen P. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- Kim CH, Lisman JE. A role of actin filament in synaptic transmission and long-term potentiation. J Neurosci. 1999;19:4314–4324. doi: 10.1523/JNEUROSCI.19-11-04314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Molecular Architecture of Synaptic Actin Cytoskeleton in Hippocampal Neurons Reveals a Mechanism of Dendritic Spine Morphogenesis. Mol Biol Cell. 2009 doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krucker T, Siggins GR, Halpain S. Dynamic actin filaments are required for stable long-term potentiation (LTP) in area CA1 of the hippocampus. Proc Natl Acad Sci U S A. 2000;97:6856–6861. doi: 10.1073/pnas.100139797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriu T, Inoue A, Bito H, Sobue K, Okabe S. Differential Control of Postsynaptic Density Scaffolds via Actin-Dependent and -Independent Mechanisms. J Neurosci. 2006;26:7693–7706. doi: 10.1523/JNEUROSCI.0522-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis DM, Reese TS. Cytoplasmic organization in cerebellar dendritic spines. J Cell Biol. 1983;97:1169–1178. doi: 10.1083/jcb.97.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Helton TD, Blanpied TA, Racz B, Newpher TM, Weinberg RJ, Ehlers MD. Postsynaptic positioning of endocytic zones and AMPA receptor cycling by physical coupling of dynamin-3 to homer. Neuron. 2007;55:874–889. doi: 10.1016/j.neuron.2007.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, MacDonald JF, Wang JY, Falls DL, Jia Z. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- Nagerl UV, Willig KI, Hein B, Hell SW, Bonhoeffer T. Live-cell imaging of dendritic spines by STED microscopy. Proc Natl Acad Sci U S A. 2008;105:18982–18987. doi: 10.1073/pnas.0810028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher TM, Ehlers MD. Spine microdomains for postsynaptic signaling and plasticity. Trends Cell Biol. 2009 doi: 10.1016/j.tcb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Nagai T, Miyawaki A, Hayashi Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat Neurosci. 2004;7:1104–1112. doi: 10.1038/nn1311. [DOI] [PubMed] [Google Scholar]
- Osterweil E, Wells DG, Mooseker MS. A role for myosin VI in postsynaptic structure and glutamate receptor endocytosis. J Cell Biol. 2005;168:329–338. doi: 10.1083/jcb.200410091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, Srivastava DP. Convergent CaMK and RacGEF signals control dendritic structure and function. Trends Cell Biol. 2008;18:405–413. doi: 10.1016/j.tcb.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Petrini EM, Lu J, Cognet L, Lounis B, Ehlers MD, Choquet D. Endocytic trafficking and recycling maintain a pool of mobile surface AMPA receptors required for synaptic potentiation. Neuron. 2009;63:92–105. doi: 10.1016/j.neuron.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racz B, Blanpied TA, Ehlers MD, Weinberg RJ. Lateral organization of endocytic machinery in dendritic spines. Nat Neurosci. 2004;7:917–918. doi: 10.1038/nn1303. [DOI] [PubMed] [Google Scholar]
- Racz B, Weinberg RJ. Spatial organization of cofilin in dendritic spines. Neuroscience. 2006;138:447. doi: 10.1016/j.neuroscience.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Racz B, Weinberg RJ. Organization of the Arp2/3 complex in hippocampal spines. J Neurosci. 2008;28:5654–5659. doi: 10.1523/JNEUROSCI.0756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavachari S, Lisman JE. Properties of quantal transmission at CA1 synapses. J Neurophysiol. 2004;92:2456–2467. doi: 10.1152/jn.00258.2004. [DOI] [PubMed] [Google Scholar]
- Rex CS, Chen LY, Sharma A, Liu J, Babayan AH, Gall CM, Lynch G. Different Rho GTPase-dependent signaling pathways initiate sequential steps in the consolidation of long-term potentiation. J Cell Biol. 2009;186:85–97. doi: 10.1083/jcb.200901084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DA, De Paola V, Caroni P, Gahwiler BH, McKinney RA. AMPA-receptor activation regulates the diffusion of a membrane marker in parallel with dendritic spine motility in the mouse hippocampus. J Physiol. 2004;558:503–512. doi: 10.1113/jphysiol.2004.062091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostaing P, Real E, Siksou L, Lechaire JP, Boudier T, Boeckers TM, Gertler F, Gundelfinger ED, Triller A, Marty S. Analysis of synaptic ultrastructure without fixative using high-pressure freezing and tomography. Eur J Neurosci. 2006;24:3463–3474. doi: 10.1111/j.1460-9568.2006.05234.x. [DOI] [PubMed] [Google Scholar]
- Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K. Single-particle tracking: Applications to membrane dynamics. Annual Review of Biophysics and Biomolecular Structure. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Schaefer AW, Kabir N, Forscher P. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J Cell Biol. 2002;158:139–152. doi: 10.1083/jcb.200203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdrulla AD, Linden DJ. Double dissociation between long-term depression and dendritic spine morphology in cerebellar Purkinje cells. Nat Neurosci. 2007;10:546–548. doi: 10.1038/nn1889. [DOI] [PubMed] [Google Scholar]
- Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J, Olenych S, Davidson MW, Betzig E. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci U S A. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat Neurosci. 2002;5:239–246. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatavarty V, Kim EJ, Rodionov V, Yu J. Investigating sub-spine actin dynamics in rat hippocampal neurons with super-resolution optical imaging. PLoS One. 2009;4:e7724. doi: 10.1371/journal.pone.0007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Electron tomography reveals unbranched networks of actin filaments in lamellipodia. Nat Cell Biol. 2010;12:429–435. doi: 10.1038/ncb2044. [DOI] [PubMed] [Google Scholar]
- Uruno T, Liu J, Zhang P, Fan Y, Egile C, Li R, Mueller SC, Zhan X. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat Cell Biol. 2001;3:259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- Wang XB, Yang Y, Zhou Q. Independent expression of synaptic and morphological plasticity associated with long-term depression. J Neurosci. 2007;27:12419–12429. doi: 10.1523/JNEUROSCI.2015-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Edwards JG, Riley N, Provance DW, Jr, Karcher R, Li XD, Davison IG, Ikebe M, Mercer JA, Kauer JA, Ehlers MD. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Mitchison TJ. Single-Molecule Speckle Analysis of Actin Filament Turnover in Lamellipodia. Science. 2002;295:1083–1087. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- Welch JM, Wang D, Feng G. Differential mRNA expression and protein localization of the SAP90/PSD-95-associated proteins (SAPAPs) in the nervous system of the mouse. J Comp Neurol. 2004;472:24–39. doi: 10.1002/cne.20060. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang XB, Frerking M, Zhou Q. Delivery of AMPA receptors to perisynaptic sites precedes the full expression of long-term potentiation. Proc Natl Acad Sci U S A. 2008;105:11388–11393. doi: 10.1073/pnas.0802978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL. A Dynamic Actin Cytoskeleton Functions at Multiple Stages of Clathrin-mediated Endocytosis. Mol Biol Cell. 2005;16:964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Xiao M, Nicoll RA. Contribution of cytoskeleton to the internalization of AMPA receptors. Proc Natl Acad Sci U S A. 2001;98:1261–1266. doi: 10.1073/pnas.031573798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.