Abstract
A number of arylamides have been synthesized and found to exhibit potent antimicrobial activities against a broad spectrum of Gram-positive and Gram-negative bacteria while low toxicity towards eukaryotic cells. These facially amphiphilic foldamers have a relatively rigid intramolecular hydrogen-bonded arylamide as a framework, which places trifluormethyl versus positively charged amino and guanidino groups along opposite faces of the elongated molecule, facilitating interactions with lipid membranes. To better understand the mechanism of action of these antimicrobial foldamers, we have investigated the lipid interaction, depth of insertion, orientation and dynamics of an arylamide, PMX30016, using 31P and 19F solid-state NMR spectroscopy. Static 31P NMR lineshapes of lipid membranes with a range of compositions indicate that PMX30016 does not disrupt the lamellar order of the lipid bilayer, but perturbs the lipid headgroup conformation. This headgroup perturbation, manifested as systematic 31P chemical shift anisotropy increases, is consistent with the well documented “electrometer” effect of lipid membranes in response to the addition of positive charges to membrane surfaces. Paramagnetic relaxation enhancement experiments indicate that the arylamide inserts to the membrane-water interface, just below the headgroup region. Measurement of 19F-19F dipolar couplings within each CF3 moiety revealed that PMX30016 is oriented with the molecular plane 20° and 30° from the membrane normal of neutral and anionic bilayers, respectively, and the long molecular axis lies parallel to the membrane plane. Thus, this arylamide inserts into the bilayer in a knife-like fashion, consistent with previous vibrational spectroscopy results. Moreover, 19F NMR lineshapes indicate that this molecular knife undergoes fast uniaxial rotation around the bilayer normal. These results suggest that antimicrobial arylamides destabilize anionic lipid membranes primarily by altering the membrane electric potential profile, and the spinning molecular knife may additionally create transient defects in the lipid membrane. Compared to typical antimicrobial peptides, this arylamide has more subtle effects on and is less disruptive of the structure of lipid bilayers.
Keywords: antimicrobial molecules, arylamides, 19F solid-state NMR, orientation, bicelle
Introduction
Antimicrobial peptides (AMPs) have attracted much attention in the last two decades as potentially alternative antibiotics against drug-resistant bacteria 1. Many naturally occurring AMPs have been discovered that possess broad-spectrum and potent antimicrobial activities against many bacteria. However, despite the large number of AMPs studied, therapeutic applications have been limited by their relatively large size, toxicity and difficulties associated with large-scale synthesis of these molecules 1. Thus, it is important to design smaller and more structurally robust synthetic antimicrobial molecules with the essential structural requirements for potent antimicrobial activity and low toxicity. It is by now well known that AMP activities often correlate with their amphiphilic structures and high cationic charge densities, which enable them to selectively target and disrupt the negatively charged lipid membranes of microbial cells 2. Based on these considerations, short sequences of synthetic antimicrobial foldamers have been designed that contain arylamide 3, phenylene ethynylene 4,5, polynorbornene 6,7 and polymethacrylate 8 backbones. The arylamide series has been particularly well studied. The arylamide backbone is decorated with basic guanidinium sidechains and hydrophobic moieties, and the molecules are conformationally rigid by virtue of thioether-based intramolecular hydrogen bonds and amide groups 9,10. Structure-activity relationship studies revealed that the amphiphilicity and conformational rigidity of these arylamide foldamers are essential for their high antimicrobial potency and low toxicity, and members of this class of foldamers have been found to have low minimum inhibitory concentrations (MICs) but high EC50 or HC50 values towards eukaryotic cells 9,11.
While the antimicrobial activities of arylamide foldamers have been extensively studied, the physical mechanism of their interaction with the lipid membrane is still not well understood. Determining the three-dimensional structure and topology of these antimicrobial molecules in the lipid bilayer is essential for elucidating their mechanisms of action. So far, sum frequency generation (SFG) vibrational spectroscopy 12,13 and molecular dynamics (MD) simulations 14,15 have been used to deduce the topological structure and dynamics of the arylamides in the membrane. These studies suggest that the arylamide oligomers insert into the membrane perpendicular to the bilayer surface, and do not cause the formation of water pores in the membrane. However, high-resolution orientational constraints, depth of insertion, and the mobility of the arylamides have not been reported.
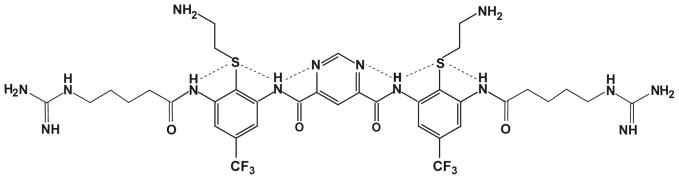
The chemical structure of one of the arylamide foldamers, PMX30016, is shown in Fig. 1. The molecule consists of a central pyrimidine ring flanked by two phenylene diamine units. A thioether moiety allows the attachment of basic groups and forms intramolecular hydrogen bonds with neighboring amides. The phenylene diamine rings are also decorated with a trifluoromethyl group and a terminal guanidine-pentanoyl sidechain that increases the positive charge density of the molecule. PMX30016 was found to have excellent therapeutic indices: its MICs are 0.1 μM against Gram-negative E. coli, 0.2 μM against Gram-positive tetracyclin- and streptomycin-resistant S. aureus, and its HC50 against red blood cells is 440 μM 9,10.
Figure 1.
Molecular structure of PMX30016. Dash lines indicate the intra-molecular hydrogen bonds.
In this work, we use 31P and 19F solid-state NMR spectroscopy to determine the lipid interaction and membrane topology of this arylamide foldamer in lipid bilayers of varying compositions. 31P NMR is a sensitive probe of the membrane morphology and disorder induced by antimicrobial molecules 16–18. 19F NMR is a highly sensitive and background-free indicator of membrane-active molecules, and has been used extensively for determining the orientation 19,20 and quaternary structure 21–23 of oligomeric membrane peptides and proteins. While the three 19F spins of a trifluoromethyl group are chemically equivalent, dipolar couplings among the three spins are observable and depend on the orientation of the C-CF3 bond with respect to the membrane normal 24. We used a simple 2D dipolar-chemical-shift correlation technique to resolve the orientation-dependent 19F-19F dipolar coupling from the 19F chemical shift pattern. Combined with paramagnetic relaxation enhancement experiments, we have determined both the orientation and the depth of insertion of PMX30016 in lipid bilayers, and propose its membrane disruptive mechanism.
Materials and Methods
Lipids and arylamide compound
All lipids, including 1-palmitoyl -2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylserine (POPS), 1,2-dimyristoyl-sn-glycero-3-phosphochloline (DMPC), 1,2-dimyristoyl-sn-glycero-3-phosphatidylglycerol (DMPG), dihexylphosphatidylcholine (6-O-PC) and cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL) and used without further purification. PMX30016 was synthesized by PolyMedix (Radnor, PA).
Membrane sample preparation
Glass-plate oriented lipid membranes containing varying concentrations of PMX30016 were prepared using a naphthalene-incorporated method 25. After co-dissolving PMX30016, lipids and naphthalene at desired molar ratios in MeOH/CHCl3 (1:2, v/v), the solution was deposited drop-wise onto 6 × 12 mm glass plates (Marienfeld, Germany). After drying overnight in a lyophilizer, the lipid films were hydrated first by directly applying 5 μl water on each plate, followed by incubation in a 97% humidity chamber containing saturated K2SO4 solution at 20°C for 4–5 days. Finally, the glass plates were stacked and wrapped with parafilm for static 31P NMR experiments. We prepared four membrane series with different lipid compositions: POPE/POPG (3: 1), POPC/POPG (3: 1), POPC/POPS (4.5: 1), and POPC/POPS/cholesterol (4.5: 1: 2.4).
The bicelle samples for orientation determination were prepared using published protocols 26. Briefly, the zwitterionic lipid DMPC was mixed with the ether lipid 6-O-PC at a DMPC/6-O-PC molar ratio of q = 3.2 to a total lipid concentration of 35% (w/v) in pH 7.0 phosphate buffer. The mixture was vortexed, heated to 42°C and cooled to 5°C until a clear, homogeneous and viscous solution was obtained. Static 31P NMR spectra confirmed that the lipid mixture was well aligned in the membrane with the bilayer normal perpendicular to the magnetic field at 306 K. PMX30016 was added to the bicelle solution to a drug: lipid molar ratio of 1: 15, then subjected to another round of vortexing, heating and cooling cycles, and the alignment of the resulting bicelle was again checked by 31P NMR.
Unoriented PMX30016-containing lipid membranes were prepared by mixing PMX30016 with DMPC/DMPG (3:1) vesicles at a molar ratio of 1:15. The solution was incubated overnight and then spun down to obtain a hydrated membrane pellet. UV-VIS spectroscopy showed that >96% of PMX30016 was bound to the lipids.
For paramagnet relaxation enhancement (PRE) experiments, 5 mol% Mn2+ relative to the total molar amount of lipids were added to hydrated DMPC/DMPG vesicles containing PMX30016. In general, Mn2+ ions added to a lipid vesicle solution cannot cross a lipid bilayer that is free of any defects or pores. Thus, all Mn2+ ions should be distributed on the outer surface of the bilayer, as demonstrated by NMR 27. However, freeze-thawing a one-sided Mn2+-bound membrane sample redistributes the ions to both surfaces of the membrane due to the fragmentation of the bilayer by ice. We prepared both one-sided and two-sided Mn2+-bound membrane samples containing PMX30016 to assess the depth and polarity of insertion of the arylamide compound.
Solid-state NMR experiments
All solid-state NMR experiments were carried out on a Bruker DSX-400 spectrometer (Karlsruhe, Germany) operating at a resonance frequency 376.8 MHz for 19F and 162.1 MHz for 31P. Typical radiofrequency (rf) pulse lengths were 5 μs for 1H and 31P and 6 μs for 19F. 31P chemical shifts were referenced to the liquid phosphoric acid peak at 0 ppm for the oriented membrane experiments, and to the isotropic signal of hydroxyapatite at +2.73 ppm on the phosphoric acid scale for unoriented membrane experiments. The 19F chemical shifts were referenced to the Teflon isotropic 19F signal at −122 ppm.
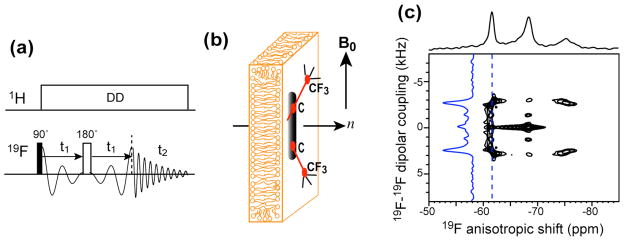
A double-resonance probe equipped with a custom-designed 6 × 12 × 5 mm rectangular coil was used for the 31P NMR experiments on glass-plate oriented samples. The samples were positioned in the magnet with the alignment axis parallel to the magnetic field. The 31P spectra were measured at 296 K by single pulse excitation. A 1H/19F/X magic-angle-spinning (MAS) probe was used for all 19F experiments. Static 1D 31P and 19F spectra of bicelle samples were measured at 306 K, at which bicelle maintains good magnetic alignment. A 2D 1H-decoupled correlation experiment (Fig. 5a) was implemented to correlate the 19F-19F dipolar coupling with the 19F chemical shift anisotropy. A dwell time of 20 μs and a total t1 evolution time of 6.4 ms were used.
Figure 5.
Orientation determination of PMX30016 in neutral lipid bilayers. (a) Static 2D 19F dipolar chemical-shift correlation pulse sequence for measuring 19F-19F dipolar couplings. (b) Geometry of magnetically oriented bicelles containing PMX30016. (c) Static 2D 19F homonuclear DIPSHIFT spectrum of PMX30016 in aligned DMPC/6-O-PC bicelles. The dipolar cross section at −62 ppm is shown as a blue trace.
Orientation calculation and 19F NMR lineshape analysis
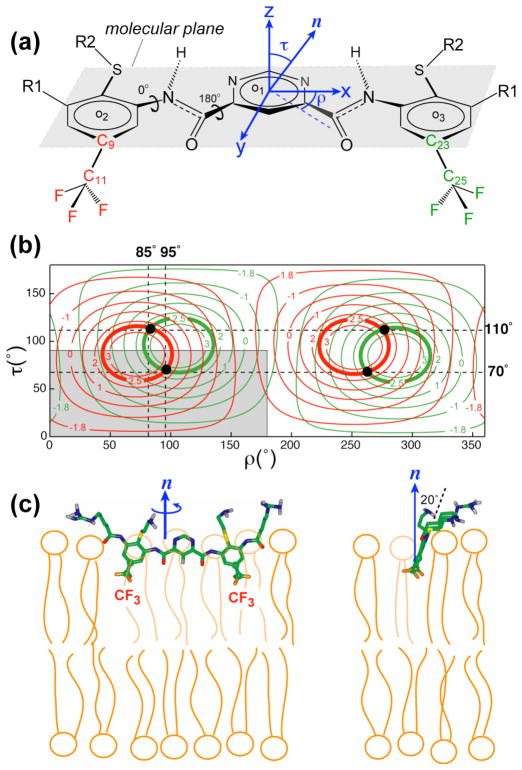
The two C-CF3 bonds lie mostly in the molecular plane formed by the centers of the three aromatic rings (Fig. 6a). The angle between the C-CF3 bond and the long molecular axis, which is defined as the vector connecting the two carbons ortho to the nitrogen atoms of the pyrimidine ring, is 72°.
Figure 6.
Extraction of the orientation of PMX30016 in neutral DMPC/6-O-PC bicelles. (a) Definitions of the molecular coordinate system and (τ, ρ) angles. (b) Calculated 19F-19F dipolar couplings as a function of (τ, ρ) for the CF3 groups associated with the C9–C11 bond (red) and the C23–C25 bond (green). Contour lines for the measured value of 2.6 kHz are bolded. The best-fit (τ, ρ) = (70°, 95°) and its degenerate solutions are indicated as black dots. The non-degenerate region of the orientation plot is shown in grey. (c) Front (left) and side (right) views of the orientation of PMX30016 in DMPC bilayers. The plane of PMX30016 is tilted by 20° from the bilayer normal, which corresponds to τ = 70°.
For orientation calculations, we defined a molecule-fixed frame where the x-axis is the long molecular axis described above, the y-axis lies in the plane of the central aromatic ring, perpendicular to the ortho C-C vector, and the z-axis is perpendicular to the ring (Fig. 6a). The direction of the bilayer normal relative to the molecule is defined by a tilt angle τ from the z-axis and a rotation angle ρ between the x-axis and the projection of the bilayer normal onto the x-y plane. By rotating the bilayer normal through all combinations of (τ, ρ) from 0° to 360°, we can obtain the corresponding C-CF3 bond orientations to the bilayer normal and thus calculate the 19F-19F dipolar couplings as averaged by the CF3 rotation to be along the C-CF3 bond.
The chemical shift anisotropy (CSA) tensor is defined by three principal values, δxx, δyy, δzz, whose average is the isotropic shift δiso = (δxx + δyy + δzz)/3. The anisotropy parameter δ is defined as δ = δzz − δiso, and the asymmetry parameter η is defined as η = (δyy − δxx)/(δzz − δiso). The δyy principal value is the closest to the isotropic chemical shift while δzz is the furthest. Powder patterns with η = 0 due to identical δyy and δxx frequencies are called uniaxial powder patterns. Another indicator of the size of the CSA is the span Δσ = δzz − δxx. For η = 0 CSA patterns, which are observed for all uniaxially diffusive molecules in lipid bilayers, the frequency position δxx = δyy is called the 90° edge since they result from molecules whose bilayer normal is perpendicular to the magnetic field, while the δzz frequency is called the 0° edge.
Results
The PMX30016 structure has a two-fold symmetry with respect to the central ring (Fig. 1). The pyrimidine-4,6-dicarboxylic center is connected to an m-phenylenediamine group on each side. A CF3 group and a thioether group are attached to each m-phenylenediamine to increase the amphiphilicity of the molecule. The molecule is terminated at the two ends by a hydrophilic guanidine-pentanoyl side chain. The aromatic rings, amide groups and hydrogen bonds create a highly rigid molecule, which was found to be important for antimicrobial potency and selectivity 10. The compound has a molecular weight of 1063 Da, which is smaller than that of most antimicrobial peptides, which have molecular weights of 2–5 kDa.
Membrane perturbation by PMX30016
Static 31P NMR lineshapes of aligned lipid membranes are sensitive reporters of the types of lipid morphology caused by antimicrobial molecules. Both the anisotropic frequency and the intensity distribution reflect the membrane morphology and the lipid headgroup packing. Lamellar bilayers aligned with the bilayer normal parallel to the magnetic field exhibit a single narrow peak at ~30 ppm. Unoriented bilayers show a broad powder pattern with maximum intensity at about −12 ppm. Isotropic vesicles and micelles give a sharp isotropic signal near 0 ppm (Fig. 2a). Membranes whose orientational order is perturbed by antimicrobial peptides often exhibit residual powder patterns and intensities at the 90° edge. Thus the area fraction of the disordered region reflects the extent of the membrane disruption 16,18.
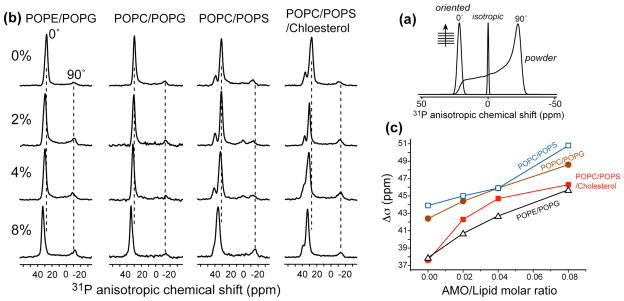
Figure 2.
(a) Static 31P lineshapes of representative membrane morphologies. (b) Static 31P spectra of oriented lipid membranes of different compositions in the presence of PMX30016 at 296 K. The PMX30016 molar concentrations are 0%, 2%, 4% and 8% from top to bottom. The membrane compositions are POPE/POPG, POPC/POPG (brown), POPC/POPS (blue), and POPC/POPS/cholesterol (red). (c) 31P chemical shift anisotropy (CSA) change of different lipid membranes as a function of PMX30016 concentration.
Fig. 2b shows the static 31P spectra of four series of oriented membranes in the presence of varying concentrations of PMX30016. Three series combine a zwitterionic lipid (POPE or POPC) with a negatively charged lipid (POPG or POPS) to mimic the bacterial cell membrane composition, while the fourth series contains cholesterol along with POPC and POPS to mimic the eukaryotic membrane composition. Interestingly, we did not observe any significant residual powder patterns nor isotropic intensities in these spectra up to a peptide molar concentration of 16%. The fractional disorder is 20–35% at 8% PMX30016, which is small compared to most antimicrobial peptides 16,18. Thus, PMX30016 does not disrupt the overall lamellar morphology of the bilayer. However, in all four series the 0° 31P peak shifted downfield (to larger chemical shifts) with increasing concentrations of PMX30016, indicating that the electronic environment of the 31P was altered by the drug. Using the difference between the 0° and 90° edges of the powder pattern as an indicator of the magnitude of the 31P CSA, we found that the span increased by 6.2 – 8.7 ppm, or 15–23%, for the four membrane series between 0 and 8% arylamide (Fig. 2c). Upon addition of 16% PMX30016, the most bacteria-mimetic membrane, POPE/POPG, showed a span increase of 9.3 ppm (26%), while the most eukaryote-mimetic POPC/POPS/cholesterol membrane exhibited a span increase of 13.2 ppm (32%).
Binding of PMX30016 to the membrane-water interface
To determine the depth of insertion of PMX30016 in lipid membranes, we carried out a paramagnetic resonance enhancement (PRE) experiment using Mn2+ ions. Mn2+ ions bind to the surfaces of lipid bilayers and enhance the T2 relaxation rates of nuclear spins in a distance-dependent fashion. The closer the nuclear spins to the paramagnetic center, the broader and lower the signal intensities 28. The ratio of the peak height in the presence of Mn2+ to the full intensity in the absence of Mn2+ reflects the distance of the nuclear spins to the paramagnetic ions on the membrane surface. By comparing the PRE effect of the peptide signals with that of lipid functional groups, whose depths are known 29, we can thus determine the insertion depth of PMX30016.
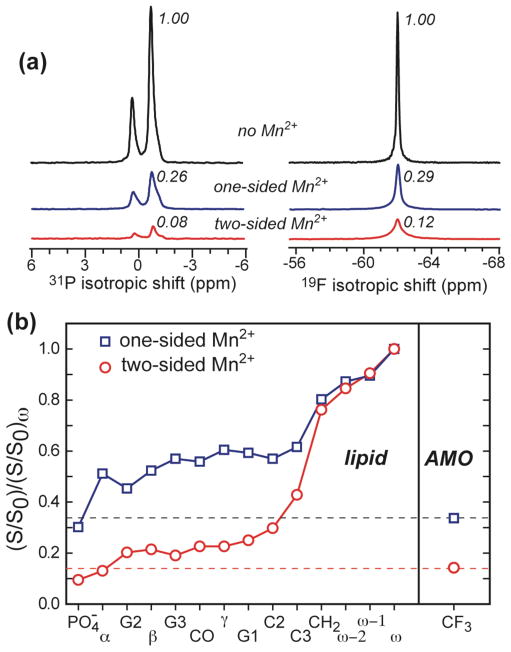
Fig. 3a shows the single-pulse 31P spectra of the DMPC/DMPG bilayer and 19F spectra of PMX30016. Three spectra, without Mn2+, with Mn2+ on the outer surface of the bilayer, and with Mn2+ on both surfaces of the bilayer, are compared. The one-side Mn2+ sample reduced the 31P intensity to 26% and the 19F signal of the arylamide to a similar level of 29%. The addition of Mn2+ to both surfaces of the bilayer further decreased the 31P and 19F signals. Again, the residual 19F signal height (12%) is larger than the residual 31P signal (8%), indicating that the CF3 groups are buried more deeply than the phosphate group. The lower NMR signals of the one-side Mn2+ sample compared to the two-side Mn2+ sample indicates that the arylamide, just like the lipid, is bound to both leaflets of the bilayer rather than to only the outer leaflet, since the distance from the outer leaflet to the inner surface of the membrane is sufficiently large that purely outer-leaflet bound molecules would not experience sufficient PRE effect from Mn2+ ions on the inner membrane surface 27. The difference between the two spectra also clearly demonstrates that Mn2+ is not freely permeable to the inside of the vesicle, thus ruling out the possibility that the arylamide forms large stable pores.
Figure 3.
Depth of insertion of PMX30016 in lipid membranes from Mn2+ paramagnetic relaxation enhancement experiments. (a) 31P and 19F MAS spectra of PMX30016-containing DMPC/DMPG bilayers without Mn2+ (black), with Mn2+ on the outer membrane surface (blue), and with Mn2+ on both membrane surfaces. The intensities normalized to the Mn2+-free samples are indicated. (b) PRE effects of PMX30016 in DMPC/DMPG (3:1) bilayers containing 5% Mn2+. The ratio of the lipid 13C intensities (not shown) between the Mn2+-containing sample (S) and a Mn2+-free control sample (S0) is S/S0. This dephasing value was further normalized to the S/S0 of the acyl-chain terminal methyl group ω. The 19F intensity of PMX30016 is comparable to the glycerol and headgroup carbons, indicating that PMX30016 is associated to the membrane-water interface, close to the surface water.
To obtain more quantitative depth information, we compared the PRE effects of all lipid 13C signals in the one-sided and two- sided Mn2+ samples with the 19F PRE of PMX30016 (Fig. 3b). The lipid signals show the expected trend of increasing residual intensity (weaker PRE) with increasing distance from the membrane surface, and the two-side Mn2+ samples have lower residual intensities than the one-side Mn2+ sample. The 19F intensities are comparable to that of the headgroup carbons for each sample, indicating that PMX30016 is shallowly inserted into the membrane-water interface, between the phosphate groups and the glycerol backbone region.
Orientation and uniaxial rotation of PMX30016 in lipid bilayers
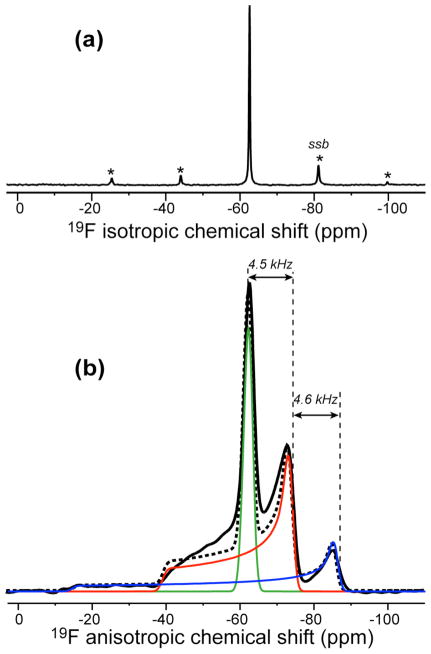
The 19F MAS spectrum of PMX30016 in DMPC/DMPG bilayers (Fig. 4a) shows only a single 19F peak (−62.6 ppm), indicating that the two trifluoromethyl groups have the same chemical environment in lipid membranes. Under the static condition, the 19F spectrum shows a triplet powder pattern with three distinct 90° edges at −62.6 ppm, −73.4 ppm and −85.4 ppm (Fig. 4b). This triplet pattern is well known for CF3 groups as resulting from 19F-19F dipolar splitting of each 19F by the two other fluorines in the trifluoromethyl group and has been explored extensively for orientation measurements of membrane peptides 19,20. The three components of the triplet correspond to the unperturbed chemical shift spectrum and the sum and difference of the anisotropic dipolar and chemical shift frequencies: ωCSA − ωD, ωCSA, and ωCSA + ωD. Analogous to the C-H J-splitting of a CH2 group, the integrated intensities of the three sub-spectra have the ratio of 1: 2: 1. Fig. 4b shows that the central component of the PMX30016 triplet has a uniaxial lineshape (η = 0), indicating that the molecule undergoes fast uniaxial rotation around the bilayer normal. This motion averages both the 19F-19F dipolar coupling and the 19F CSA to be uniaxial and collinear with the bilayer normal. As a result, the sum and difference spectra are particularly simple: they have η = 0 lineshapes and anisotropy parameters of δ̄CSA + δ̄D and δ̄CSA − δ̄D. Since the central 90° edge of the triplet (−73.4 ppm) is 4.4 kHz from the two other 90° edges, the motionally averaged 19F-19F dipolar coupling constant δ̄D is 4.4 kHz. Fig. 4b shows simulated 19F spectrum based on a 1: 2: 1 superposition of three η = 0 powder patterns with motionally averaged anisotropy parameters of 0.5, 23.6, and 46.7 ppm, respectively. The simulation has excellent agreement with the experimental spectrum, confirming the nature of the triplet.
Figure 4.
19F spectrum of PMX30016 in unoriented DMPC/DMPG (3:1) bilayers. (a) MAS spectrum. (b) Static spectrum (black line). Three simulated powder patterns corresponding to ωCSA − ωD (green), ωCSA (red), and ωCSA + ωD (blue), at 1: 2: 1 area ratios are shown. The sum of the three simulated patterns is shown as dashed line. A line broadening of 10 ppm was used for each sub-spectrum. The 90° edges of the triplet are spaced by a 19F-19F dipolar coupling of 4.5 kHz.
To better resolve the 19F-19F dipolar coupling from the 19F CSA, we implemented a 2D dipolar chemical-shift (DIPSHIFT) correlation experiment where a 180° pulse in the middle of the t1 evolution period encodes pure 19F-19F dipolar coupling in the indirect dimension and correlates it with the mixed CSA-dipolar spectrum in the direct dimension (Fig. 5a). Although the directly detected dimension is not a pure chemical shift spectrum, for simplicity we use the terminology of homonuclear DIPSHIFT to refer to this experiment. Pure 19F dipolar spectra have been previously extracted mainly using the 1D Carr-Purcell-Meiboom-Gill (CPMG) experiment 30,31, where the 19F signals were detected in the windows of long multiple-pulse trains 19,24,32. Compared to the multiple-pulse experiment, this 2D Hahn-echo based homonuclear DIPSHIFT experiment has the benefits of a lower radio-frequency duty cycle and a simpler signal acquisition method, since windowed detection entails scaling of the spectral width and is sensitive to the cumulative effects of pulse imperfections.
We first applied the static 2D homonuclear DIPSHIFT experiment to PMX30016 in magnetically aligned DMPC/6-O-PC bicelles. The alignment axis of the bicelle is perpendicular to the magnetic field, thus scaling all measured couplings by −0.5 (Fig. 5b). The 2D spectrum shows the expected correlation of the triplet pattern in the ω2 dimension with 19F-19F dipolar coupling in the ω1 dimension. Both the most downfield (−61.5 ppm) and the most upfield (−75.3 ppm) 19F peaks exhibit a doublet splitting of 5.2 kHz, as expected because these two 19F peaks correlate with dipolar couplings with the same magnitude, δ̄CSA − δ̄D and δ̄CSA + δ̄D, and the dipolar spectrum is sign-insensitive. Also as expected, the central 19F peak (−68.5 ppm) does not exhibit a splitting because it corresponds to the unperturbed CSA, δ̄CSA. The dipolar coupling constant of interest is half the splitting, which is thus 2.6 kHz.
We now consider the orientation dependence of the motionally averaged 19F-19F dipolar coupling of the CF3 group. For two 19F spins separated by 2.09 Å as in a CF3 group, the homonuclear 19F-19F dipolar coupling in the absence of motion is 17.6 kHz. The three-site jumps of the CF3 scales the coupling by a factor of (3cos290 − 1)/2 = −0.5 due to the 90° angle between each F-F vector and the C-CF3 axis. Thus, the 19F-19F coupling of a rotating CF3 group without any other motion is 8.8 kHz. If the C-CF3 axis undergoes rotation around the bilayer normal, then the dipolar couplings will be scaled by an orientation-dependent factor SCC,n = (3cos2θCC,n −1)/2.
All lipids and peptides in aligned bicelles rotate around the bicelle normal, which is perpendicular to the magnetic field in our case. The bicelle normal usually exhibits a small degree of wobbling, as described by an order parameter Sbicelle. These two effects further reduce the dipolar coupling, so that the total measured 19F-19F coupling is:
| (1) |
In the above equation we have used a Sbicelle value of 0.89, which was directly measured from the 31P spectrum according to
| (2) |
where δiso = −0.9 ppm, δ90° = −14.9 ppm, δobs = −13.3 ppm. This Sbicelle value is consistent with the literature range of 0.75 – 0.94, which depends on the q ratio, the hydration level and temperature of the bicelle 26,33.
In unoriented membranes where the molecule of interest undergoes uniaxial diffusion, the observed 19F-19F dipolar coupling constant is larger due to the lack of two scaling factors:
| (3) |
Equations (1) and (3) show that the 19F-19F dipolar coupling of the CF3 group ultimately depends only on the angle between the C-CF3 bond and the bilayer normal. Since the C-CF3 bond is rigidly held to the rest of the arylamide ring plane, this vector orientation reveals the orientation of the rigid molecular plane with respect to the membrane normal. Thus, simulation of the 19F-19F dipolar coupling can yield the orientation of PMX30016 in the lipid bilayer.
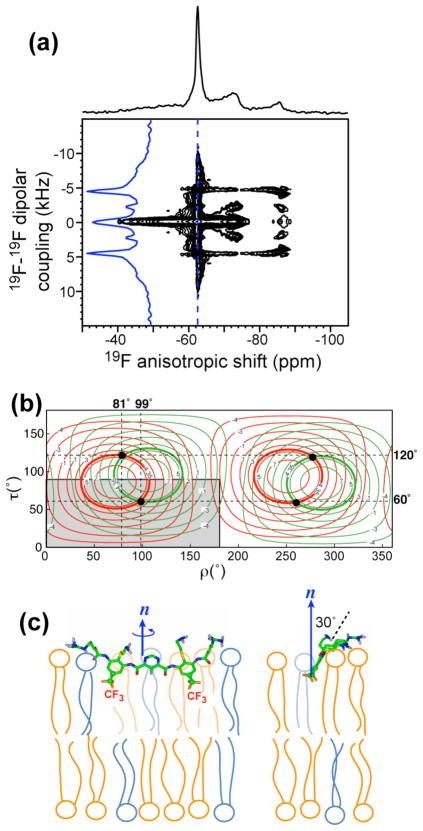
Fig. 6b shows the calculated 19F-19F dipolar couplings as a function of the tilt angle and rotation angle of the arylamide with respect to the bilayer normal. A tilt angle of 0° and 90° means, respectively, that the normal of the molecular plane is parallel and perpendicular to the bilayer normal. A rotation angle of 0° and 90° corresponds to the long molecular axis being parallel and perpendicular to the bilayer normal, respectively. The observation of only one 19F-19F dipolar coupling in the static spectrum indicates that the two C-CF3 bonds have the same orientation with respect to the bilayer normal. The calculated dipolar couplings of the two CF3 groups are shown in red and green contour lines, and the contour lines corresponding to the observed 2.6 kHz coupling are bolded. Since both CF3 groups exhibit the same coupling, only a single solution of (τ, ρ) = (70°, 95°) was identified within the non-degenerate quadrant of 0–180° for ρ and 0°–90° for τ. All other solutions such as (τ, ρ) = (110°, 85°) are degenerate. Fig. 6c shows the (τ =70°, ρ =95°) orientation of PMX30016 in the DMPC bilayer. The molecular plane is almost perpendicular to the bilayer plane, reflecting the tilt angle, while the long axis of the molecule is nearly parallel to the bilayer surface, reflecting the rotation angle. This orientation corresponds to a knife-like insertion of the arylamide into the membrane, with the aromatic plane only 20° from the vertical bilayer normal.
To determine whether the orientation is affected by the presence of negatively charged lipids, we measured the 2D 19F DIPSHIFT spectrum of PMX30016 in unoriented DMPC/DMPG bilayers. Fig. 7a shows a similar triplet pattern in the direct dimension, but it is now correlated with a larger dipolar splitting of 8.6 kHz due to the absence of two bicelle scaling factors (Equation 3). The dipolar coupling constant of 4.35 kHz was calculated to yield a unique orientation solution of (τ = 60°, ρ =99°) (Fig. 7b), indicating that now the molecular plane is moderately more tilted from the bilayer normal (30°) than the case in the neutral DMPC bilayer, but the long molecular axis remains largely parallel to the membrane plane. Overall, the knife-like insertion motif is maintained, as shown in Fig. 7c.
Figure 7.
Orientation of PMX30016 in unoriented DMPC/DMPG membranes. (a) 2D 19F homonuclear DIPSHIFT spectrum of PMX30016 in DMPC/DMPG (3:1) membranes. (b) Calculated 19F-19F dipolar couplings as a function of (τ, ρ) angles. The intercepts of the two CF3 groups’ couplings at 4.35 kHz give the best-fit orientation, which is (τ, ρ) = (60°, 99°). The non-degenerate region of the orientation plot is shown in grey. (c) Best-fit orientation of PMX30016 in anionic lipid membranes.
Discussion
The above solid-state NMR data provides the most comprehensive information to date about the lipid interaction, depth of insertion, orientation, and dynamics of an arylamide oligomer. We first discuss the membrane topology of this arylamide foldamer. In both neutral DMPC bilayers and anionic DMPC/DMPG bilayers, the long molecular backbone lies parallel to the bilayer surface, as reflected by ρ ≈ 90°. Our 19F NMR method allows precise measurement of the orientation of the phenyl rings of the arylamide molecule. We did not measure the orientation of the central pyrimidyl ring, but previous studies showed it to be constrained to lie coplanar with the phenyl rings through the formation of an extended hydrogen-bonded network (Fig. 1) 9. The two phenyl groups are oriented with their rigid aryl planes nearly perpendicular to the membrane surface (τ = 60°,70°). The phenyl ring’s deviation from perpendicularity is 20° for the neutral bilayer and 30° for the anionic bilayer. This orientation resembles a knife cutting into the bilayer, which makes perfect sense because it matches the amphiphilic structure of the arylamide with the amphiphilic structure of the lipid bilayer, by pointing the basic guanidinium and the amino-ethyl thioether substituents to the aqueous surface of the membrane while the hydrophobic groups towards the lipid chains. The parallel orientation of the long molecular axis with the membrane plane satisfies the two-fold symmetry of the molecule and maintains the same intermolecular potentials for the two sides of the pyrimidine ring, which are chemically identical. The moderate difference in the tilt angle between the neutral and anionic membranes can be understood by the fact that the less vertical molecular plane enables the positively charged guanidinium groups to interact more strongly with the negatively charged DMPG headgroups on the membrane surface (Fig. 7c). In other words, competition between water and the negative charge density of the membrane surface may cause small modulation of the tilt angle of the arylamide plane.
The knife-like orientation of PMX30016 obtained here is in excellent agreement with the results of SFG vibrational spectra of an analogous arylamide 12. There, analysis of the symmetric and asymmetric stretching modes of tert-butyl sidechains adsorbed onto DPPG bilayers concluded an angle of 0° – 35° for the three-fold axis of the tert-butyl. This orientation, while of the limiting nature, is consistent with the 19F NMR result for the DMPC/DMPG-bound PMX30016, where the molecular plane is 30° from the bilayer normal (Fig. 7c). Since the two arylamide molecules differ in their basic sidechains and the hydrophobic functional groups, the similarity of the results indicates that the facial amphiphilicity and restrained backbone structure of the molecules are sufficient to determine their orientations. The 19F NMR constraints obtained here have higher angular resolution and more complete information than the SFG results, since as long as the arylamide plane is not exactly perpendicular to the bilayer plane, a second angle defining the long axis direction is necessary to fully describe the molecular orientation.
Paramagnetic relaxation enhancement data indicate that the knife-like arylamide inserts only shallowly into the membrane, just below the phosphate headgroups and not reaching into the acyl chain region (Fig. 3). The shallow depth was obtained at a drug: lipid molar ratio of 1: 15, at which the arylamide was already distributed into both leaflets of the bilayer. Direct comparisons of the drug: lipid molar ratios in the hydrated pastes of the solid-state NMR samples versus the aqueous solutions of antimicrobial assays are fraught with uncertainties. Nevertheless, we believe at higher arylamide concentrations the equilibrium structure is unlikely to be a deeper transmembrane insertion, and the mechanism of action of the arylamide should be considered in the context of the observed shallow depth and knife-like orientation. This topology should be combined with the fact that the drug undergoes fast uniaxial diffusion at rates greater than 105 s−1. Thus, the rigid arylamide knife is by no means static, but spins around the membrane normal with its full extended length, which has the potential to perturb and destabilize a large area of the membrane.
The topology and dynamics of PMX30016 in the lipid bilayer are inconsistent with the three main classical antimicrobial structural models, namely, the barrel-stave 34,35, the toroidal pore 36,37, and the carpet models 38. The arylamide foldamer is not transmembrane, and does not cause lipid orientational disorders of the toroidal type. The knife-like topology is also distinct from the carpet model, since the arylamide does not aggregate into extended immobile assemblies.
One of the most intriguing aspects of the arylamide interaction with the lipid membrane is the increased 31P CSA and the nearly complete absence of orientational disorder as the concentration of the foldamer increases. These features indicate that the doubly charged molecule influences the headgroup conformation of the phospholipids without causing large-scale orientational disorder of the bilayer. It is well known that the polar lipid headgroups act as molecular electrometers to membrane surface charges 39. The addition of positively charged metal ions, hydrophobic ions, amphiphiles, and peptides increases the magnitude of the 31P CSA and the 2H quadrupolar coupling of the headgroup β-CD2, but decreases the 2H coupling of the α-CD2 39–42. The addition of negatively charged species to the membrane surface creates the opposite effects. Based on analysis of the 31P chemical shift tensor and 2H quadrupolar couplings, it was proposed that cationic species change the lipid headgroup conformation such that the N+ end of the −P–N+ dipole moves towards the water phase. The PMX30016-induced 31P CSA increase is consistent with this electrometer effect both qualitatively and quantitatively. A cationic amphiphile, sodium dialkyl phosphate, was found to increase the 31P CSA of POPC lipids by 8 ppm at 20 mol% amphiphile 40. Similarly, at 8 mol% arylamide, which corresponds to 16 mol% positive charge density, the 31P CSA span of the POPE/POPG membrane increased by 7.8 ppm (Fig. 2).
The 19F NMR results of the arylamide and the 31P NMR spectra of the lipids, taken together, suggest that the antimicrobial arylamide disrupts the membrane barrier function of microbial cells primarily by altering the membrane electric potential profile. While the spinning molecular knife would certainly perturb lipid packing on the molecular level, it appears to do so only transiently and leaves no long-lasting physical damages behind. The lack of permanent membrane disorder by PMX30016, manifested by the 31P spectra, is in sharp contrast to many other antimicrobial peptides such as PG-1 18,43 and magainin 44. The absence of permanent disruption is more akin to tachyplesin, which, interestingly, also exhibits fast uniaxial rotation with an in-plane orientation 17,45. Instead, the spinning arylamide knife, with its positively charged guanidinium groups, significantly alters the lipid headgroup conformation, an effect that is absent for tachyplesins. We propose that this perturbation of the lipid headgroup conformation changes the membrane surface potential and eventually leads to membrane permeabilization, possibly by interacting with protein components of bacterial membranes.
At higher drug concentrations than used in this study, it is possible that the spinning arylamide may increasingly cause more pronounced physical disruption of the membrane, so that a balance between the two effects –perturbation of the membrane potential versus the physical disruption – may be responsible for bacterial killing. Such a mixed mechanistic scenario has been suggested before for analogous antimicrobial arylamides based on the different concentrations and kinetics of membrane permeabilization and bacteria killing 10. The possibility of a mixture of mechanistic modes has also been gleaned for some antimicrobial peptides. For example, magainin showed clear signs of physical disruption of lipid membranes based on 31P NMR spectra 44, but it also dissipates the electrical potential across lipid membranes and has been proposed to kill bacteria by decreasing the membrane potential and interfering with free-energy transduction in microbial cells 46.
The present study illustrates the robustness and utility of 19F solid-state NMR for elucidating the topology and dynamics of pharmaceutical compounds. The high sensitivity and lack of background of 19F spins, combined with the large chemical shift anisotropy and 19F-19F dipolar couplings, make 19F NMR exquisitely sensitive to molecular orientation and dynamics. The simple 2D 19F homonuclear DIPSHIFT experiment, while analogous to several other NMR techniques 47–49, has not been employed before for determining orientation-dependent dipolar couplings, and promises to facilitate high-resolution structural analysis of CF3-containing pharmaceutical compounds.
Conclusion
The orientation, depth of insertion, mobility and lipid interaction of an antimicrobial arylamide has been determined using 19F and 31P solid-state NMR. The arylamide inserts into the lipid membrane just below the headgroups with the molecular plane nearly perpendicular to the bilayer surface and the long axis parallel to the bilayer. In this knife-like manner, the molecule undergoes fast uniaxial rotation around the bilayer normal. Interestingly, this spinning molecular knife does not directly cause permanent damage to the lamellar integrity of the lipid bilayer, but rather changes the lipid headgroup conformation, specifically, the P-N dipole orientation, through interaction of the positively charged guanidinium ions with the lipid phosphate groups. This conformational change, manifested as 31P chemical shift increases, was detected in all negatively charged lipid membranes studied here. We propose that the antimicrobial arylamide destroys the barrier function of microbial cell membrane mainly by altering its electrical potential, and the spinning molecular knife may further create transient defects in the membrane.
Acknowledgments
This work is funded by NIH grants GM66976 to M.H., AI75866 to W.F.D. and UL1RR024134 from the National Center For Research Resources.
References
- 1.Zasloff M. Nature. 2002:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Epand RM, Vogel HJ. Biochim Biophys Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 3.Tew GN, Liu D, Chen B, Doerksen RJ, Kaplan J, Carroll PJ, Klein ML, DeGrado WF. Proc Natl Acad Sci U S A. 2002;99:5110–5114. doi: 10.1073/pnas.082046199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tew GN, Clements D, Tang H, Arnt L, Scott RW. Biochim Biophys Acta. 2006;1758:1387–1392. doi: 10.1016/j.bbamem.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Gordon VD, Trinkle DR, Schmidt NW, Davis MA, DeVries C, Som A, Cronan JE, Tew GN, Wong GC. Proc Natl Acad Sci U S A. 2008;105:20595–20600. doi: 10.1073/pnas.0806456105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabriel GJ, Pool JG, Som A, Dabkowski JM, Coughlin EB, Muthukumar M, Tew GN. Langmuir. 2008;24:12489–12495. doi: 10.1021/la802232p. [DOI] [PubMed] [Google Scholar]
- 7.Som A, Vemparala S, Ivanov I, Tew GN. Biopolymers. 2008;90:83–93. doi: 10.1002/bip.20970. [DOI] [PubMed] [Google Scholar]
- 8.Kuroda K, DeGrado WF. J Am Chem Soc. 2005;127:4128–4129. doi: 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]
- 9.Tew GN, Scott RW, Klein ML, DeGrado WF. Acc Chem Res. 2010;43:30–39. doi: 10.1021/ar900036b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi S, Isaacs A, Clements D, Liu D, Kim H, Scott RW, Winkler JD, DeGrado WF. Proc Natl Acad Sci U S A. 2009;106:6968–6973. doi: 10.1073/pnas.0811818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott RW, DeGrado WF, Tew GN. Curr Opin Biotechnol. 2008;19:620–627. doi: 10.1016/j.copbio.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Tang H, Even MA, Wang J, Tew GN, Chen Z. J Am Chem Soc. 2006;128:2711–2714. doi: 10.1021/ja057029t. [DOI] [PubMed] [Google Scholar]
- 13.Avery CW, Som A, Xu Y, Tew GN, Chen Z. Anal Chem. 2009;81:8365–8372. doi: 10.1021/ac901271f. [DOI] [PubMed] [Google Scholar]
- 14.Lopez CF, Nielsen SO, Srinivas G, DeGrado WF, Klein ML. J Chem Theory Comput. 2006;2:649–655. doi: 10.1021/ct050298p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, Choi S, Chen B, Doerksen RJ, Clements DJ, Winkler JD, Klein ML, DeGrado WF. Angew Chem Int Ed Engl. 2004;43:1033. [Google Scholar]
- 16.Buffy JJ, McCormick MJ, Wi S, Waring AJ, Lehrer RI, Hong M. Biochemistry. 2004;43:9800–9812. doi: 10.1021/bi036243w. [DOI] [PubMed] [Google Scholar]
- 17.Doherty T, Waring AJ, Hong M. Biochim Biophys Acta. 2006:1285–1291. doi: 10.1016/j.bbamem.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 18.Mani R, Buffy JJ, Waring AJ, Lehrer RI, Hong M. Biochemistry. 2004;43:13839–13848. doi: 10.1021/bi048650t. [DOI] [PubMed] [Google Scholar]
- 19.Ulrich AS. Prog Nucl Magn Reson Spectrosc. 2005:1–21. [Google Scholar]
- 20.Glaser RW, Sachse C, Dürr UH, Wadhwani P, Afonin S, Strandberg E, Ulrich AS. Biophys J. 2005;88:3392–3397. doi: 10.1529/biophysj.104.056424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo W, Hong M. J Am Chem Soc. 2006:7242–7251. doi: 10.1021/ja0603406. [DOI] [PubMed] [Google Scholar]
- 22.Luo W, Mani R, Hong M. J Phys Chem. 2007;111:10825–10832. doi: 10.1021/jp073823k. [DOI] [PubMed] [Google Scholar]
- 23.Hong M. J Phys Chem B. 2007:10340–10351. doi: 10.1021/jp073652j. [DOI] [PubMed] [Google Scholar]
- 24.Grage SL, Ulrich AS. J Magn Reson. 2000:81–88. doi: 10.1006/jmre.2000.2127. [DOI] [PubMed] [Google Scholar]
- 25.Hallock KJ, Henzler-Wildman KA, Lee DK, Ramamoorthy A. Biophys J. 2002:2499–2503. doi: 10.1016/S0006-3495(02)75592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Angelis A, Opella SJ. Nat Protoc. 2007;2:2332–2338. doi: 10.1038/nprot.2007.329. [DOI] [PubMed] [Google Scholar]
- 27.Su Y, Mani R, Hong M. J Am Chem Soc. 2008;130:8856–8864. doi: 10.1021/ja802383t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buffy JJ, Hong T, Yamaguchi S, Waring AJ, Lehrer RI, Hong M. Biophys J. 2003;85:2363–2373. doi: 10.1016/s0006-3495(03)74660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiener MC, White SH. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carr HY, Purcell EM. Phys Rev. 1954;94:630–638. [Google Scholar]
- 31.Meiboom S, Gill D. Rev Sci Instrum. 1958;29:688–691. [Google Scholar]
- 32.Grage SL, Ulrich AS. J Magn Reson. 1999:98–106. doi: 10.1006/jmre.1999.1726. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto K, Soong R, Ramamoorthy A. Langmuir. 2009;25:7012–7018. doi: 10.1021/la900200s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baumann G, Mueller P. J Supramol Struct. 1974;2:538–557. doi: 10.1002/jss.400020504. [DOI] [PubMed] [Google Scholar]
- 35.He K, Ludtke SJ, Worcester DL, Huang HW. Biophys J. 1996;70:2659–2666. doi: 10.1016/S0006-3495(96)79835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 37.Matsuzaki K, Murase O, Fujii N, Miyajima K. Biochemistry. 1996;35:11361–11368. doi: 10.1021/bi960016v. [DOI] [PubMed] [Google Scholar]
- 38.Pouny Y, Rapaport D, Mor A, Nicolas P, Shai Y. Biochemistry. 1992;31:12416–12423. doi: 10.1021/bi00164a017. [DOI] [PubMed] [Google Scholar]
- 39.Seelig J, MacDonald PM, Scherer PG. Biochemistry. 1987;26:7535–7541. doi: 10.1021/bi00398a001. [DOI] [PubMed] [Google Scholar]
- 40.Scherer PG, Seelig J. Biochemistry. 1989;28:7720–7728. doi: 10.1021/bi00445a030. [DOI] [PubMed] [Google Scholar]
- 41.MacDonald PM, Leisen J, Marassi FM. Biochemistry. 1991;30:3558–3566. doi: 10.1021/bi00228a029. [DOI] [PubMed] [Google Scholar]
- 42.Ziegler A, Blatter XL, Seelig AJS. Biochemistry. 2003;42:9185–9194. doi: 10.1021/bi0346805. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi S, Waring A, Hong T, Lehrer R, Hong M. Biochemistry. 2002;41:9852–9862. doi: 10.1021/bi0257991. [DOI] [PubMed] [Google Scholar]
- 44.Bechinger B. Biochim Biophys Acta. 2005;1712:101–108. doi: 10.1016/j.bbamem.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Doherty T, Waring AJ, Hong M. Biochemistry. 2008;47:1105–1116. doi: 10.1021/bi701390t. [DOI] [PubMed] [Google Scholar]
- 46.Westerhoff HV, Juretić D, Hendler RW, Zasloff M. Proc Natl Acad Sci U S A. 1989;86:6597–6601. doi: 10.1073/pnas.86.17.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennett AE, Becerra LR, Griffin RG. J Chem Phys. 1994;100:812–814. [Google Scholar]
- 48.Wu CH, Ramamoorthy A, Opella SJ. J Magn Reson. 1994;109:270–272. [Google Scholar]
- 49.Munowitz MG, Griffin RG, Bodenhausen G, Huang TH. J Am Chem Soc. 1981;103:2529–2533. [Google Scholar]