Abstract
Atrophic changes in early Alzheimer’s disease (AD) and amnestic mild cognitive impairment (MCI) have been proposed as biomarkers for detection and monitoring. We analyzed MRI atrophy rate from baseline to 1-year in 4 groups of participants from the Alzheimer’s Disease Neuroimaging Initiative (ADNI): AD (n=152), converters from MCI to probable AD (MCI-C, n=60), stable MCI (MCI-S, n=261), and healthy controls (HC, n=200). Scans were analyzed using multiple methods, including voxel-based morphometry (VBM), regions of interest (ROIs), and automated parcellation, permitting comparison of annual percent change (APC) in neurodegeneration markers. Effect sizes and the sample required to detect 25% reduction in atrophy rates were calculated. The influence of APOE genotype on APC was also evaluated. AD and MCI-C patients demonstrated high atrophy APCs across regions compared to minimal change in HC. MCI-S showed intermediate atrophy. APOE genotype was associated with APC in key regions. In sum, APC rates are influenced by APOE genotype, imminent MCI to AD conversion, and AD-related neurodegeneration.
Keywords: Alzheimer’s Disease Neuroimaging Initiative (ADNI), magnetic resonance imaging (MRI), voxel-based morphometry (VBM), mild cognitive impairment (MCI), hippocampus, longitudinal change, genetic factors, apolipoprotein E (APOE) epsilon 4 allele
Introduction
Alzheimer’s disease (AD) is the most common age-related neurodegenerative disease affecting nearly 25 million people worldwide, a number expected to triple in the next 50 years (Ferri, et al., 2005, Wimo, et al., 2003). Patients with AD show significant impairment in multiple cognitive domains, including deficits in memory and executive functioning. Progress in the early clinical diagnosis of AD has led to the characterization of a prodromal syndrome featuring relatively isolated memory deficits termed “amnestic mild cognitive impairment” (MCI) (Petersen, et al., 2001, Petersen and Negash, 2008). Amnestic MCI is conceptualized as a preliminary stage of AD-associated neurodegeneration with the majority of patients eventually progressing to AD at a rate of 10–15% per year (Petersen, 2000, Petersen, et al., 1999).
The increasing recognition that early diagnosis and therapeutic intervention will be necessary to prevent the development of AD underscores the need to develop sensitive and specific biomarkers for detecting and monitoring MCI and AD. Structural magnetic resonance imaging (MRI) has shown significant promise as a biomarker to detect early MCI and AD associated changes, as well as to predict the rate of disease progression (de Leon, et al., 2007, Jack, et al., 1999, Risacher and Saykin, in press). Cross-sectional studies evaluating the utility of structural MRI in detecting neurodegeneration have identified significant brain atrophy in patients with MCI and AD, particularly in regions of the medial temporal lobe (MTL) using regional volumetric extraction tools such as manual tracing of regions of interest (ROIs) (de Leon, et al., 2007, De Toledo-Morrell, et al., 2000, Dickerson, et al., 2001, Du, et al., 2001, Jack, et al., 1992, Killiany, et al., 2002, Pennanen, et al., 2004, Saykin, et al., 2006, Xu, et al., 2000), and more recently, automated segmentation and parcellation of target regions (Becker, et al., 2006, Colliot, et al., 2008, Du, et al., 2007, Fischl and Dale, 2000a, Risacher, et al., 2009). Other semi-automated tools which provide three-dimensional mapping of brain morphology, including voxel-based morphometry (VBM), tensor-based morphometry (TBM) and related techniques have also identified significant global and local tissue changes in patients with MCI and AD, including decreased whole brain, hippocampal, and temporal lobar grey matter (GM) density (Busatto, et al., 2003, Chetelat, et al., 2002, Frisoni, et al., 2002, Jack, et al., 2008b, Karas, et al., 2003, Pennanen, et al., 2005, Saykin, et al., 2006, Trivedi, et al., 2006). Structural MRI techniques have also been shown to provide sensitive prediction of disease progression. Hippocampal volume and GM density, as well as measures of MTL volume and cortical thickness, have been identified as sensitive biomarkers for predicting conversion from MCI to probable AD (Apostolova, et al., 2006, Bozzali, et al., 2006, Chetelat, et al., 2005, Devanand, et al., 2007, Jack, et al., 1999, Kinkingnehun, et al., 2008, Risacher, et al., 2009, Visser, et al., 2002, Whitwell, et al., 2008b).
Longitudinal monitoring of rate of decline on MRI measures has also proven sensitive to AD-related changes. Increased rates of whole brain and MTL atrophy in patients with MCI and AD relative to healthy elderly controls (HC) are routinely reported in studies of brain aging and dementia (for recent review, see Frisoni, et al., 2010). Manual tracing or automated ROI techniques and analysis of deformation fields reflecting brain shrinkage are the most commonly employed methods for evaluating longitudinal changes in global and regional volume, particularly in the MTL. Previous studies have reported rates of hippocampal annual decline of −4.5% in patients with AD and −3% in patients with MCI in contrast to −1% in controls (for meta-analysis see Barnes, et al., 2009). Furthermore, increased atrophy rates can also predict future decline, including MCI to probable AD conversion (Erten-Lyons, et al., 2006, Jack, et al., 2000, Jack, et al., 2004, Jack, et al., 2005, Sluimer, et al., 2008, Stoub, et al., 2008), with patients who convert from MCI to probable AD showing higher rates of hippocampal atrophy compared to patients with a stable diagnosis of MCI, reported as −3.5% and −2.2%, respectively (Jack, et al., 2000, Jack, et al., 2004).
Genetic factors play a significant role in the development and progression of MCI and AD. Genetic variation in the apolipoprotein E gene (APOE) is the most commonly reported genetic risk factor associated with AD, with the presence of a single ε4 allele conferring a 2-fold or 3-fold increased risk of developing AD and two ε4 alleles associated with nearly an 11-fold increased risk (Bertram and Tanzi, 2008, Farrer, et al., 1997, Gatz, et al., 2006). In addition to an increased risk of AD, the presence of an ε4 allele has been associated with imaging markers, including significantly greater hippocampal atrophy and an increased rate of hippocampal and whole brain atrophy in ε4 carriers has been reported in non-demented individuals, as well as in MCI and AD patients, in some studies (Cohen, et al., 2001, Fjell, et al., 2010a, Fleisher, et al., 2005, Hamalainen, et al., 2008, Jack, et al., 2008c, Jack, et al., 2008d, Mori, et al., 2002, Morra, et al., 2009, Schuff, et al., 2009, Wang, et al., 2006) but not in others (Du, et al., 2006, Wang, et al., 2009).
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) is a 5-year consortium study designed to assess the utility of various biomarkers for detecting early changes associated with MCI and AD and predicting disease course over time, including cross-sectional and longitudinal neuroimaging biomarkers from structural MRI and positron emission tomography (PET), genetic factors, psychometric scores, CSF markers, and other variables. A number of studies utilizing MRI data from this cohort have been published within the last year. Using both ROI and three-dimensional mapping techniques the expected differences in structural MRI markers have been found between diagnostic (AD, MCI, HC) groups at baseline assessment, including atrophy in hippocampal and other MTL regions and enlarged ventricles in patients with AD and MCI (Chou, et al., 2009, Chupin, et al., 2009, Fan, et al., 2008, Fennema-Notestine, et al., 2009, Nestor, et al., 2008, Querbes, et al., 2009, Risacher, et al., 2009, Vemuri, et al., 2009, Walhovd, et al., 2008). Hippocampal volume has also been found to be sensitive and specific for predicting 1-year conversion from MCI to probable AD (Calvini, et al., 2009, Chupin, et al., 2009, McEvoy, et al., 2009, Misra, et al., 2009, Nestor, et al., 2008, Querbes, et al., 2009, Risacher, et al., 2009). MRI studies of the ADNI cohort have also examined longitudinal change in brain volumes using ROI and whole-brain structural change techniques (e.g., Jacobian determinants, boundary shift integral), and have detected differences in annual change in whole brain volume, hippocampal volume, and ventricular volume as a function of baseline diagnostic group (AD, MCI, HC) (Evans, et al., 2009, Fjell, et al., 2010b, Ho, et al., 2009, Holland, et al., 2009, Hua, et al., 2009, Jack, et al., 2009, Leow, et al., 2009, McDonald, et al., 2009, McEvoy, et al., 2009, Misra, et al., 2009, Morra, et al., 2009, Nestor, et al., 2008, Schuff, et al., 2009) and of APOE ε4 genotype (Fjell, et al., 2010a, Morra, et al., 2009, Nestor, et al., 2008, Schuff, et al., 2009). Several studies have reported larger declines in whole brain and regional volumes, as well as larger ventricular volume increases in MCI to AD converters than MCI non-converters (Evans, et al., 2009, Leow, et al., 2009, Misra, et al., 2009, Nestor, et al., 2008).
In order to better evaluate the effectiveness of future disease modifying therapeutics, biomarkers of disease state and progression are likely to be more sensitive and reliable than clinical measures, which may be highly variable within and between participants. When designing clinical trials, an important consideration is the sample size needed to detect a therapeutic effect that is both statistically significant and clinically meaningful in a target biomarker with 80% or 90% power. Several previous studies in the ADNI cohort have calculated the relative sample size needed to detect a hypothetical treatment-induced 25% reduction in brain atrophy for various regional MRI markers and have suggested that to achieve 80% power approximately 35–100 AD and 100–200 MCI participants are required (Ho, et al., 2009, Holland, et al., 2009, Hua, et al., 2009, Nestor, et al., 2008).
Despite the extensive MRI analyses in AD and MCI, prior studies have not directly compared the relative sensitivity of longitudinal changes in GM density and volume, cortical thickness and ROI volumes in relation to changes in clinical status. In ADNI, longitudinal studies have primarily focused on baseline diagnostic groups rather than one year clinical conversion status. The present study was designed to compare the APC of different types of structural MRI markers in groups defined by baseline diagnosis and 1-year MCI to AD conversion status using the final 1-year sample. We hypothesized that patients with more advanced clinical indicators of disease progression, particularly AD and MCI-C participants, would show significantly greater APC in major structural MRI markers. We also evaluated the relative sensitivity of these markers to progression of atrophy over time. Because of the important implications for design of future therapeutic trials of disease modifying agents, we also calculated the sample size needed to detect a 25% reduction in atrophy rate for selected markers. We hypothesized that the MTL changes would constitute the most sensitive regional markers of progression and therefore require the smallest potential sample sizes. Prior ADNI reports have not evaluated the sample size needed for trials in rapidly progressing MCI participants (MCI-Converters) compared to stable MCI participants, an important distinction for trial design. Additionally, previous reports have focused primarily on sample sizes needed for MRI markers that were extracted using a single technique. In the present study, we compared GM density and volume, cortical thickness and ROI volumetric markers. Finally, we assessed the impact of APOE genotype on the APC in several key target regions, which to-date has not been examined in patients who converted from MCI to probable AD the ADNI cohort to our knowledge. We hypothesized that the presence of an ε4 allele would increase the annual rate of decline in selected MRI markers of MTL integrity.
Methods
ADNI
ADNI is a consortium study initiated in 2004 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations. More than 800 participants age 55–90 have been recruited from 59 sites across the U.S. and Canada to be followed for 2–3 years, with repeated structural MRI and PET scans and functional, psychological, and psychometric test data collected every 6 or 12 months. For additional information about ADNI, see www.adni-info.org and (S. G. Mueller, et al., 2005a, S. G. Mueller, et al., 2005b).
MRI Scans
Raw baseline 1.5T MRI scans from 820 participants were downloaded from the ADNI public website (http://www.loni.ucla.edu/ADNI/) onto local servers at Indiana University School of Medicine between January and April 2008 and processed using Freesurfer and VBM as implemented in SPM5 as previously described (Risacher, et al., 2009). All available 1.5T MP-RAGE scans collected at the 1-year follow-up visit (“12mo scans”) were also downloaded for all participants (n=693) as of 06/2009. A minimum of two MP-RAGE images were acquired at each time point for each participant, using a standard MP-RAGE protocol that was selected and tested by ADNI (Jack, et al., 2008a).
Participants were only included in the present analysis if their baseline and 12mo MRI scans were successfully preprocessed. 4 participants failed Freesurfer processing and were not included in any analyses. 30 additional participants were excluded from only the VBM analyses due to failed processing of scans from either the baseline or 12mo visit. Participants who did not have either baseline (n=2) or 12mo (n=124) scans were also excluded. Included participants (n=673 for Freesurfer analyses, n=643 for VBM analyses) were divided into groups by baseline and 1-year clinical diagnosis and 12 month MCI to probable AD conversion status, resulting in 4 groups: (1) participants with a stable AD diagnosis (AD; n=152 for Freesurfer analyses, n=143 for VBM analyses); (2) participants with an MCI diagnosis at baseline who converted to a diagnosis of probable AD at either the 6 month or 12 month (MCI-Converters (MCI-C); n=60 for Freesurfer analyses, n=57 for VBM analyses); (3) participants with a stable diagnosis of MCI (MCI-Stable (MCI-S); n=261 for Freesurfer analyses, n=253 for VBM analyses); (4) participants with a stable designation of healthy elderly control (HC; n=200 for Freesurfer analyses, n=190 for VBM analyses). Participants who showed other forms of conversion, reversion, or otherwise unstable diagnostic designation were excluded (e.g. conversion from HC to MCI at the 6 month visit, followed by a reversion from MCI to HC at the 12 month visit, etc.; n=16). Details of the ADNI design, participant recruitment, clinical testing, and additional methods have been published previously (Fleisher, et al., 2008, S. G. Mueller, et al., 2005a, S. G. Mueller, et al., 2005b, Petersen, et al., 2010b) and at www.adni-info.org.
Demographic and Clinical Data
Demographic information, APOE genotype, neuropsychological test scores, and diagnosis information for all analyzed visits were downloaded from the ADNI clinical data repository (http://www.loni.ucla.edu/ADNI/Data/ADCS_Download.jsp). The “8-09-09” version of the ADNI clinical database was used for all analyses. By this time all 1-year clinical and scan data was complete. Participants were classified into groups based on baseline and 12mo diagnoses as reported in the conversion/reversion database.
In order to evaluate the impact of APOE genotype on annual rate of atrophy, we also classified participants by the presence or absence of an APOE ε4 allele. Given the unknown impact of having an ε2ε4 genotype (i.e. possessing a potential protective allele (ε2) and a risk allele (ε4) for AD), we chose to run analyses both including and excluding the ε2ε4 participants (n=13; 3 AD, 7 MCI-S, 3 HC). We found similar results from the two comparisons (data not shown), and thus, chose to use the largest available sample in the results presented in this report. For the evaluation of hippocampal volume and EC thickness, 673 participants were included: 99 AD, 35 MCI-C, 143 MCI-S, and 56 HC who were APOE ε4 positive (ε2ε4, ε3ε4 or ε4ε4 genotypes) and 53 AD, 25 MCI-C, 118 MCI-S, and 144 HC who were APOE ε4 negative (ε2ε2, ε2ε3, or ε3ε3 genotypes). 30 participants were excluded due to failed VBM processing, as previously described. Thus, the analysis of the effect of APOE ε4 genotype on bilateral mean hippocampal GM density and volume included the following participants (n=643): 95 AD, 34 MCI-C, 142 MCI-S, and 53 HC who were APOE ε4 positive and 48 AD, 23 MCI-C, 111 MCI-S, and 137 HC who were APOE ε4 negative.
Image Processing
VBM
Scans were processed with VBM in SPM5 (http://www.fil.ion.ucl.ac.uk/spm/), using previously described methods (Ashburner and Friston, 2000, Good, et al., 2001, Mechelli, et al., 2005). Briefly, after conversion from DICOM to NIfTI, both baseline MP-RAGE scans were aligned to the T1 template and both 12mo scans were co-registered to the T1-aligned baseline scans. After alignment, all scans were bias corrected and segmented into GM, WM, and CSF compartments using standard SPM5 templates. GM maps were normalized to MNI atlas space as 1×1×1 mm voxels and smoothed using a 10 mm FWHM Gaussian kernel. Both modulated and unmodulated GM maps were generated. In order to maximize signal and minimize variability in the imaging markers, we chose to create a mean GM image of the two independent MP-RAGE-derived GM maps using SPM5. These mean GM volumes were then employed in all subsequent VBM analyses. This process was completed for both unmodulated and modulated normalized GM maps from each individual, yielding a mean GM density image and a mean GM volume image, respectively.
Regions of Interest (ROIs)
A hippocampal ROI template was created by manual tracing of the left and right hippocampi in an independent sample of 40 HC participants enrolled in a study of brain aging and MCI (McHugh, et al., 2007, Saykin, et al., 2006, Shen, et al., 2010). Hippocampal GM density and GM volume values were extracted from baseline and 12mo mean GM maps from VBM as previously described (Risacher, et al., 2009). Additionally, mean GM density and mean GM volume were extracted from 90 cortical and 26 cerebellar regions using MarsBaR ROI templates (Brett, et al., 2002). Mean lobar measures from MarsBaR regions were calculated from target ROIs as follows: mean frontal lobe is the mean of GM density values from inferior frontal operculum and triangularis, inferior, medial, middle and superior orbital frontal, middle and superior frontal, and medial superior frontal regions; mean parietal lobe is the mean of inferior and superior parietal, angular gyrus, supramarginal gyrus, and precuneus GM density values; and mean temporal lobe value is the mean of GM density values from the amygdala and hippocampus, middle and superior temporal pole, inferior, middle and superior temporal gyri, and fusiform, Heschl’s, lingual, olfactory, and parahippocampal gyri.
Automated Parcellation
Bilateral volumetric and cortical thickness estimates from the baseline and 12mo scans were extracted using Freesurfer V4 (Dale, et al., 1999, Fischl and Dale, 2000b, Fischl, et al., 2002, Fischl, et al., 1999, Shen, et al., 2010) as previously described (Risacher, et al., 2009). Each scan from each time point was processed independently. The final extracted values were then used to calculate a mean volume or cortical thickness for each region for both the baseline and 12mo time points. Mean lobar cortical thickness measures were calculated from selected ROI mean cortical thicknesses from Freesurfer as follows: mean frontal lobe was the mean of caudal midfrontal, rostral midfrontal, lateral orbitofrontal, medial orbitofrontal, and superior frontal gyri, pars opercularis, oribitalis, and triangularis, and frontal pole thicknesses; mean parietal lobe was the mean of inferior parietal, superior parietal and supramarginal gyri, and precuneus thicknesses; and mean temporal lobe was the mean of the fusiform, lingual, parahippocampal, inferior temporal, middle temporal, and lateral temporal gyri, as well as temporal and transverse temporal pole thicknesses.
VBM Statistical Analysis
A two-way ANOVA assessing time and group membership (AD, MCI-C, MCI-S, HC) was performed to compare the change over 1-year between groups using the smoothed, unmodulated normalized mean GM maps. Statistical analyses were performed on a voxel-by-voxel basis using a general linear model (GLM) approach implemented in SPM5. A threshold of p<0.0001 (uncorrected for multiple comparisons) and minimum cluster size (k) of 27 voxels was considered significant. We chose to show the VBM comparison images at this threshold (p<0.0001 unc.) for display purposes although all comparisons, except for AD vs. MCI-C, survive p<0.05 with a false discovery rate (FDR) correction for multiple comparisons and all 6 comparisons have at least 1 cluster which survives p<0.01 with a family-wise error (FWE) multiple comparison correction. Baseline age, gender, years of education, handedness, and baseline mean intracranial volume (ICV) were included as covariates, and an explicit GM mask was used to restrict analyses to GM regions.
Other Statistical Analyses
Annual percent change (APC) estimates were calculated using mean values from left and right ROIs from baseline and 12 month scans for each participant using the following equation:
A one-way multivariate ANOVA was used to assess differences in mean MRI change measures between groups. Baseline age, gender, education, handedness, and baseline mean ICV were included as covariates. Pairwise comparisons with a Bonferroni adjustment for multiple comparisons were also used to assess differences between individual group pairs. One-way ANOVA and chi-square tests were used to determine group differences in demographic variables, as well as baseline values and annual change of psychometric test scores. SPSS (version 17.0.2) was used for statistical analysis.
The sample size needed to detect a 25% reduction in mean APC (two-sided t-test; α=0.05) with 80% or 90% power was also calculated using Microsoft Excel 2007 for the absolute change over 1 year of all target variables for all four diagnostic groups to determine the relative sensitivity of MRI change measures for monitoring atrophy progression. Only participants with values for all analyzed regions were included in these calculations (n=643; 143 AD, 57 MCI-C, 253 MCI-S, 190 HC). Sample size was calculated using the following equation:
where n is the target sample size, α = 0.05, β is the adjusted mean absolute change, σ is the standard deviation of the measure, and za is the value from the standard distribution for 80% or 90% power (Ho, et al., 2009, Hua, et al., 2009, Rosner, 1990).
Effect sizes for the comparisons between pairs of diagnostic groups were also calculated for bilateral mean APC and baseline values of selected imaging markers. Left and right adjusted means, covaried for baseline age, gender, education, handedness, and baseline mean ICV, were averaged to yield a bilateral estimate. These bilateral mean values were then used to calculate the effect size (Cohen’s d) between group pairs for all imaging measures in Microsoft Excel 2007 as follows:
where, for a target marker, M1 = mean value for group 1, M2 = mean value for group 2, σ1 = standard deviation for group 1, and σ2 = standard deviation for group 2 (Cohen, 1988). In order to accurately compare the resulting effect sizes, only participants with values for all analyzed regions were included in this comparison (n=643; 143 AD, 57 MCI-C, 253 MCI-S, 190 HC).
Finally, a two-way ANOVA was used to assess the impact of diagnostic group and APOE ε4 genotype on the most sensitive imaging phenotypes as determined by effect size in the comparison of MCI-C and MCI-S participants, namely the APC in bilateral mean hippocampal GM density and volume, hippocampal volume, and EC thickness. Additionally, two-sample t-tests were used to evaluate the influence of APOE ε4 genotype within each of the 4 diagnostic groups on MTL change measures. Age, gender, education, handedness, and baseline ICV were included as covariates in all analyses. All graphs were created using SigmaPlot (version 10.0).
Results
Group Characteristics and Change in Psychometric Scores
Demographic information and the baseline values and change in selected psychometric scores over the first year are found in Table 1. Significant differences were demonstrated in education level (F=6.53, p<0.001) and APOE genotype (percentage positive for at least 1 ε4 allele; χ2=56.64, p<0.001), specifically between HCs and patient groups. Expected differences between groups in psychometric test scores were found to be significant for both baseline scores and annual change in scores on the clinical dementia rating sum of boxes (CDR-SoB; baseline, F=532.91, p<0.001; annual change, F=21.42, p<0.001), mini-mental state exam (MMSE; baseline, F=342.97, p<0.001; annual change, F=23.14, p<0.001), and Rey Auditory Verbal Learning Test (RAVLT; baseline, F=193.85, p<0.001; annual change, F=8.02, p<0.001). Paired comparisons between groups also indicated significant differences in both baseline values and annual change as shown in Table 1. No significant difference between groups was detected in baseline or 12 month age, gender distribution, handedness distribution, or baseline mean intracranial volume (ICV).
Table 1.
Demographic Information and Neuropsychological Test Scores (Mean (SE))
| AD (n=152) |
MCI-C (n=60) |
MCI-S (n=261) |
HC (n=200) |
p-value | Significant Pair Comparisons (p<0.05) |
|
|---|---|---|---|---|---|---|
| Baseline Age | 75.33 (0.5) | 74.04 (0.9) | 75.07 (0.4) | 75.95 (0.5) | NS | none |
| 12mo Age | 76.41 (0.5) | 75.11 (0.9) | 76.15 (0.4) | 77.04 (0.5) | NS | none |
| Education | 14.82 (0.2) | 15.15 (0.4) | 15.89 (0.2) | 16.08 (0.2) | p<0.001 | HC, MCI-S>AD |
| Gender (M, F) | 80, 72 | 35, 25 | 166, 95 | 105, 95 | p=0.06 | MCI-S vs. AD, HC |
| Handedness (R, L) | 144, 8 | 55, 5 | 239, 22 | 185, 15 | NS | none |
| ApoE Genotype (% ApoE4+) | 65.13% | 58.33% | 54.79% | 28.00% | p<0.001 | AD, MCI-C, MCI-S>HC |
| Baseline CDR-SoB | 4.18 (0.1) | 2.00 (0.1) | 1.52 (0.1) | 0.02 (0.1) | p<0.001 | AD>MCI-C>MCI-S>HC |
| 12mo Change in CDR-SoBa | +2.13 (0.1) | +2.64 (0.2) | +1.26 (0.1) | +1.1 (0.1) | p<0.001 | AD, MCI-C>MCI-S, HC |
| Baseline MMSE | 23.53 (0.1) | 26.58 (0.2) | 27.11 (0.1) | 29.11 (0.1) | p<0.001 | HC>MCI-S, MCI-C >AD |
| 12mo Change in MMSEb | −1.88 (0.7) | −2.56 (0.4) | −0.35 (0.2) | 0.00 (0.2) | p<0.001 | HC, MCI-S>MCI-C, AD |
| Baseline RAVLTc | 23.25 (0.7) | 26.20 (1.1) | 32.00 (0.5) | 43.65 (0.6) | p<0.001 | HC>MCI-S>MCI-C, AD |
| 12mo Change in RAVLTd | −2.91 (0.5) | −2.68 (0.8) | −0.81 (0.4) | +0.3 (0.5) | p<0.001 | HC>MCI-C, AD; MCI-S>AD |
| Baseline ICV | 1552677.32 (13807.5) |
1566297.83 (21976.7) |
1570616.06 (10537.0) |
1534944.90 (12037.1) |
NS | none |
7 participants missing data (2 AD, 1 MCI-S, 4 HC)
2 participants missing data (1 MCI-S, 1 HC)
3 participants missing data (1 AD, 2 HC)
14 participants missing data (8 AD, 3 MCI-S, 3 HC)
VBM Comparisons
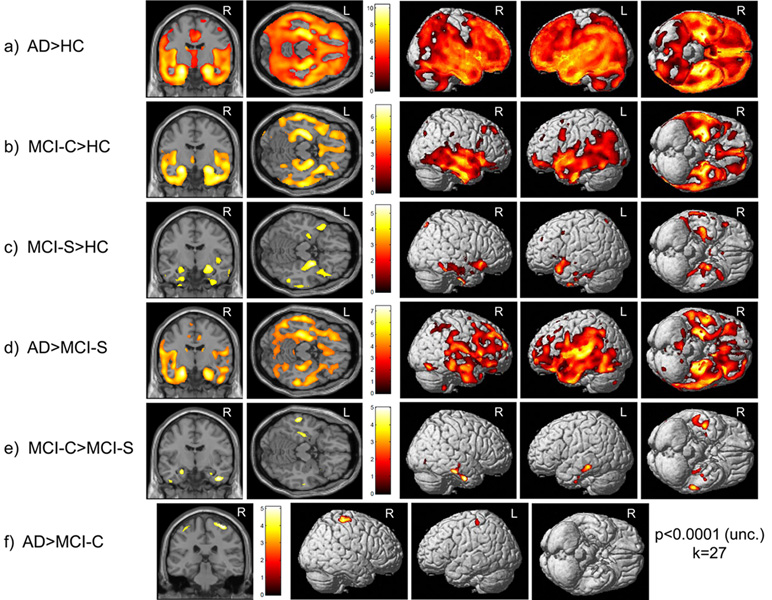
AD participants showed greater decline in global GM density than HCs (Figure 1a, p<0.0001(unc.), k=27) and MCI-S participants (Figure 1d, p<0.0001(unc.), k=27) in widespread regions including bilateral medial and lateral temporal lobe, frontal lobe, and parietal lobe, with maximal differences found in the left MTL. MCI-C participants also showed greater decline in global GM density relative to HCs (Figure 1b, p<0.0001 (unc.), k=27) in bilateral medial and lateral temporal lobes, and this was maximal in the left MTL (global peak). Differences in decline in GM density were also detected in bilateral hippocampal regions between HC and MCI-S participants and between MCI-C and MCI-S participants (Figure 1c & 1e, p<0.0001 (unc.), k=27). Finally, greater decline in global GM density was detected for AD participants relative to MCI-C in a small cluster of voxels in the anterior parietal/posterior frontal lobe region (Figure 1f, p<0.0001 (unc.), k=27).
Figure 1. Group Differences in Pattern of Reduction in Grey Matter (GM) Density over 12 Months in the ADNI Cohort.
Time x diagnosis group interactions demonstrate differences in atrophy progression reflected by reduction in GM density from baseline to 1-year in the ADNI cohort (n=643*; 143 AD, 57 MCI-C, 253 MCI-S, 190 HC). Interaction contrasts are displayed at a threshold of p<0.0001 (unc.) with a minimum cluster size (k) = 27 voxels. Cross-sections in (a–e) are (0, −9, 0, coronal) and (0, −23, −16, axial), left to right. Cross-section in (f) is (34, −29, 64, coronal). (*30 participants removed from comparisons due to failed image processing)
Target Region Comparisons
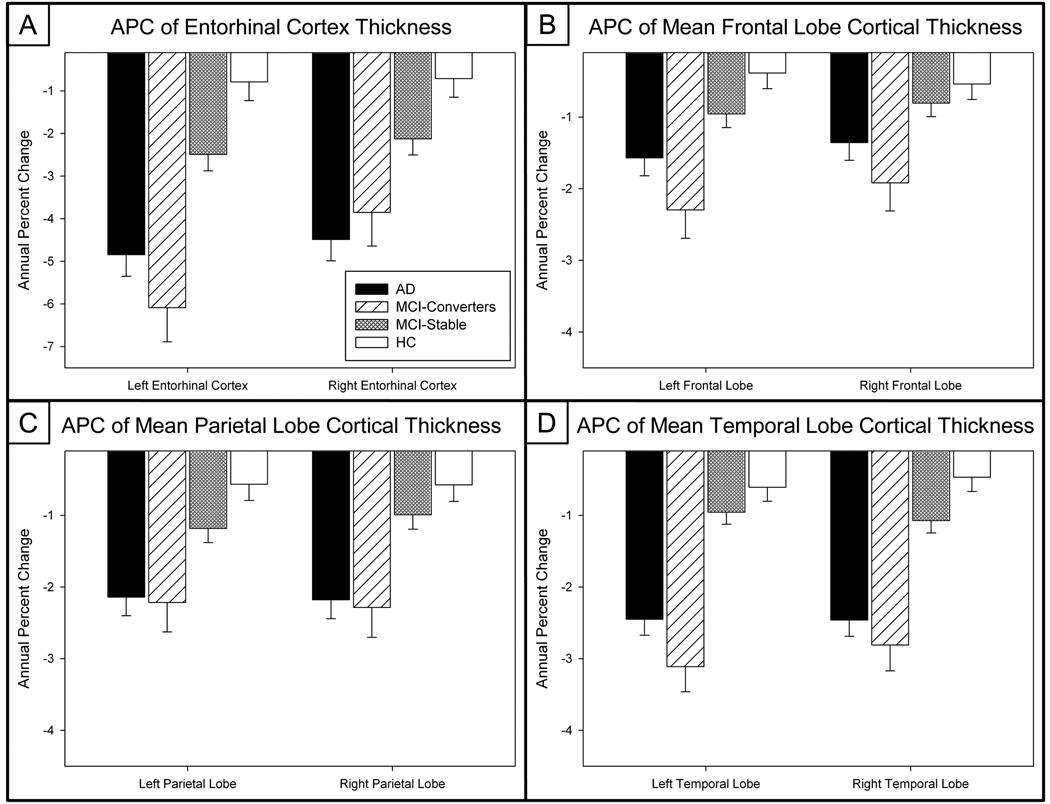
Results from regional assessments of GM density and volume, as well as cortical thickness and volumetric measures, show a similar magnitude and anatomical pattern of decline over 12 months by group as seen in the results from the VBM comparisons. The APC values for all selected ROIs, including hippocampal GM density and GM volume extracted using two ROI methods (Brett, et al., 2002, McHugh, et al., 2007, Saykin, et al., 2006, Shen, et al., 2010), hippocampal volume, entorhinal cortex (EC) thickness and mean lobar thickness values extracted using Freesurfer (Dale, et al., 1999, Fischl and Dale, 2000a, Fischl, et al., 2002, Fischl, et al., 1999), and mean lobar GM density and GM volume extracted using MarsBaR ROIs (Brett, et al., 2002) are found in Table 2 and Figures 2–4. All APC values were significantly different across groups (p<0.001). Significant post-hoc paired comparisons using a Bonferroni correction are indicated in Table 2.
Table 2.
APC of Selected Imaging Biomarkers (Mean (SE))
| AD (n=152) |
MCI-C (n=60) |
MCI-S (n=261) |
HC (n=200) |
p-value | Significant Pair Comparisons (p<0.05) |
||
|---|---|---|---|---|---|---|---|
| Hippocampal GM Density (Indep. ROI)a,b |
L | −4.51 (0.2) | −3.94 (0.4) | −2.05 (0.2) | −0.88 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD |
| R | −4.51 (0.2) | −3.82 (0.4) | −2.18 (0.2) | −0.83 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD | |
| Hippocampal GM Density (MarsBaR)a,b |
L | −4.57 (0.2) | −4.02 (0.4) | −2.16 (0.2) | −0.97 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD |
| R | −4.62 (0.2) | −4.05 (0.4) | −2.33 (0.2) | −0.85 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD | |
| Hippocampal GM Volume (Indep. ROI)a,b |
L | −4.69 (0.2) | −4.55 (0.4) | −2.07 (0.2) | −1.18 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD |
| R | −4.71 (0.2) | −4.30 (0.4) | −2.25 (0.2) | −1.07 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD | |
| Hippocampal GM Volume (MarsBaR)a,b |
L | −4.67 (0.2) | −4.55 (0.4) | −2.14 (0.2) | −2.24 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD |
| R | −4.85 (0.3) | −4.55 (0.4) | −2.39 (0.2) | −1.07 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD | |
| Hippocampal Volume (FreeSurfer)b |
L | −4.12 (0.3) | −4.20 (0.5) | −2.55 (0.2) | −0.94 (0.3) | p<0.001 | HC>MCI-S>MCI-C, AD |
| R | −3.76 (0.3) | −3.99 (0.5) | −2.71 (0.2) | −1.38 (0.3) | p<0.001 | HC>MCI-S, MCI-C, AD | |
| EC Thickness (FreeSurfer)b | L | −4.84 (0.5) | −6.09 (0.8) | −2.49 (0.4) | −0.79 (0.4) | p<0.001 | HC>MCI-S>MCI-C, AD |
| R | −4.49 (0.5) | −3.85 (0.8) | −2.16 (0.4) | −0.71 (0.4) | p<0.001 | HC>MCI-C, AD; MCI-S>AD | |
| Mean Frontal Cortical Thickness (FreeSurfer)b |
L | −1.57 (0.3) | −2.30 (0.4) | −0.96 (0.2) | −0.39 (0.2) | p<0.001 | HC, MCI-S>MCI-C; HC>AD |
| R | −1.36 (0.2) | −1.92 (0.4) | −0.81 (0.2) | −0.54 (0.2) | p=0.006 | HC>MCI-C | |
| Mean Parietal Cortical Thickness (FreeSurfer)b |
L | −2.14 (0.3) | −2.22 (0.4) | −1.18 (0.2) | −0.57 (0.2) | p<0.001 | HC>MCI-C, AD; MCI-S>AD |
| R | −2.18 (0.3) | −2.28 (0.4) | −0.99 (0.2) | −0.58 (0.2) | p<0.001 | HC, MCI-S>MCI-C, AD | |
| Mean Temporal Cortical Thickness (FreeSurfer)b |
L | −2.45 (0.2) | −3.11 (0.4) | −0.96 (0.2) | −0.61 (0.2) | p<0.001 | HC, MCI-S>MCI-C, AD |
| R | −2.46 (0.2) | −2.81 (0.4) | −1.07 (0.2) | −0.47 (0.2) | p<0.001 | HC, MCI-S>MCI-C, AD | |
| Mean Frontal GM Density (MarsBaR)a,b |
L | −3.02 (0.2) | −2.47 (0.4) | −1.48 (0.2) | −0.65 (0.2) | p<0.001 | HC>MCI-S>AD; HC>MCI-C |
| R | −2.91 (0.2) | −2.25 (0.4) | −1.37 (0.2) | −0.59 (0.2) | p<0.001 | HC>MCI-S>AD; HC>MCI-C | |
| Mean Frontal GM Volume (MarsBaR)a,b |
L | −2.92 (0.2) | −2.59 (0.4) | −1.46 (0.2) | −0.86 (0.2) | p<0.001 | HC, MCI-S>MCI-C, AD |
| R | −2.84 (0.2) | −2.37 (0.4) | −1.36 (0.2) | −0.86 (0.2) | p<0.001 | HC>MCI-S>AD; HC>MCI-C | |
| Mean Parietal GM Density (MarsBaR)a,b |
L | −3.21 (0.3) | −2.40 (0.5) | −1.53 (0.2) | −0.51 (0.3) | p<0.001 | HC>MCI-S>AD; HC>MCI-C |
| R | −3.02 (0.3) | −1.95 (0.5) | −1.41 (0.2) | −0.55 (0.3) | p<0.001 | HC, MCI-S>AD | |
| Mean Parietal GM Volume (MarsBaR)a,b |
L | −3.60 (0.3) | −2.71 (0.4) | −1.76 (0.2) | −0.90 (0.2) | p<0.001 | HC>MCI-S>AD; HC>MCI-C |
| R | −3.64 (0.3) | −2.45 (0.5) | −1.51 (0.2) | −0.76 (0.3) | p<0.001 | HC>MCI-S>AD; HC>MCI-C | |
| Mean Temporal GM Density (MarsBaR)a,b |
L | −3.33 (0.2) | −2.81 (0.3) | −1.63 (0.1) | −0.76 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD |
| R | −3.04 (0.2) | −2.66 (0.3) | −1.59 (0.1) | −0.64 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD | |
| Mean Temporal GM Volume (MarsBaR)a,b |
L | −3.32 (0.2) | −3.30 (0.3) | −1.52 (0.2) | −0.89 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD |
| R | −3.14 (0.2) | −3.00 (0.3) | −1.52 (0.2) | −0.73 (0.2) | p<0.001 | HC>MCI-S>MCI-C, AD | |
30 participants excluded because of failed processing (9 AD, 3 MCI-C, 9 MCI-S, 9 HC)
Covaried for baseline age, gender, education, handedness, and baseline ICV
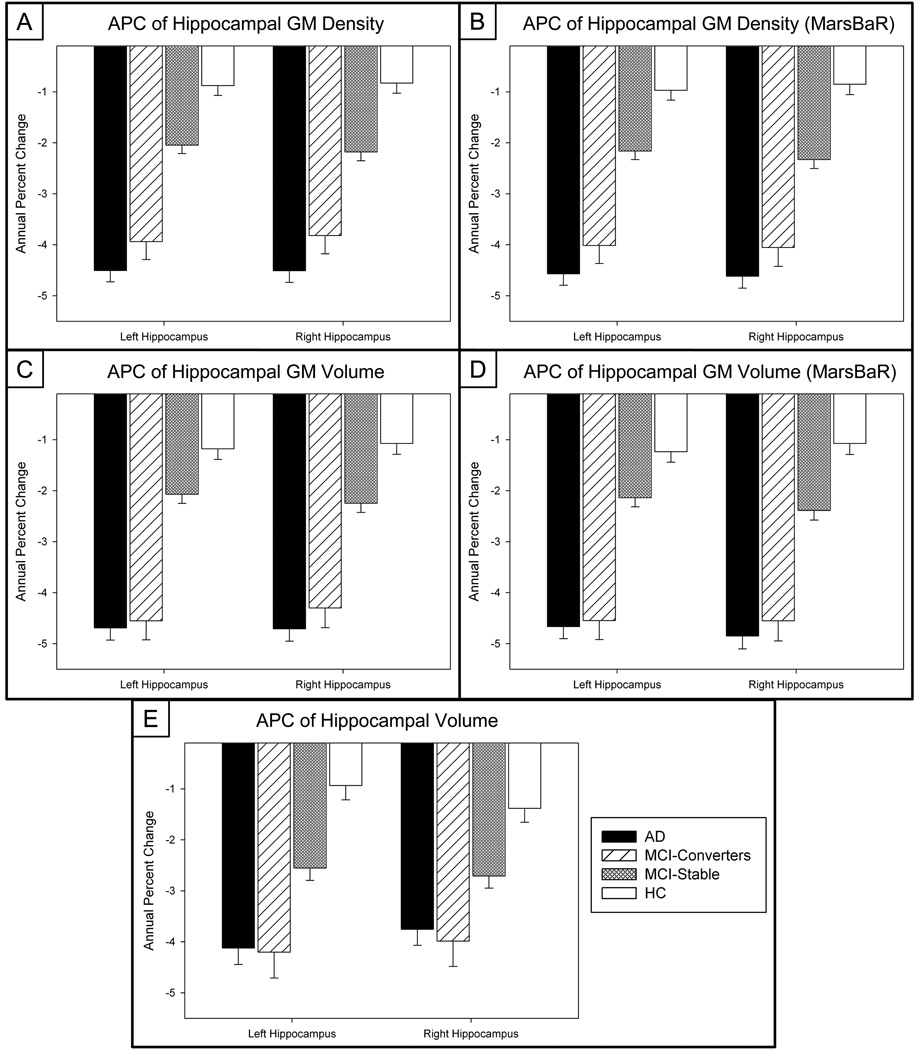
Figure 2. Annual Percent Change (APC) and Decline over 12 months of Selected MTL Imaging Biomarkers.
Plots of the mean APC in (a, b) hippocampal GM density and (c, d) GM volume extracted using a hippocampal ROI extracted using a template derived on an independent sample of 40 healthy elderly controls (McHugh, et al., 2007, Saykin, et al., 2006, Shen, et al., 2010) and from MarsBaR, respectively (n=643*; 143 AD, 57 MCI-C, 253 MCI-S, 190 HC). AD and MCI-C participants show significantly greater APC in hippocampal GM density and hippocampal GM volume relative to MCI-S and HC. The APC in (e) hippocampal volume (n=673; 152 AD, 60 MCI-C, 261 MCI-S, 200 HC) extracted using Freesurfer showed a similar trend. (*30 participants removed from comparisons due to failed image processing)
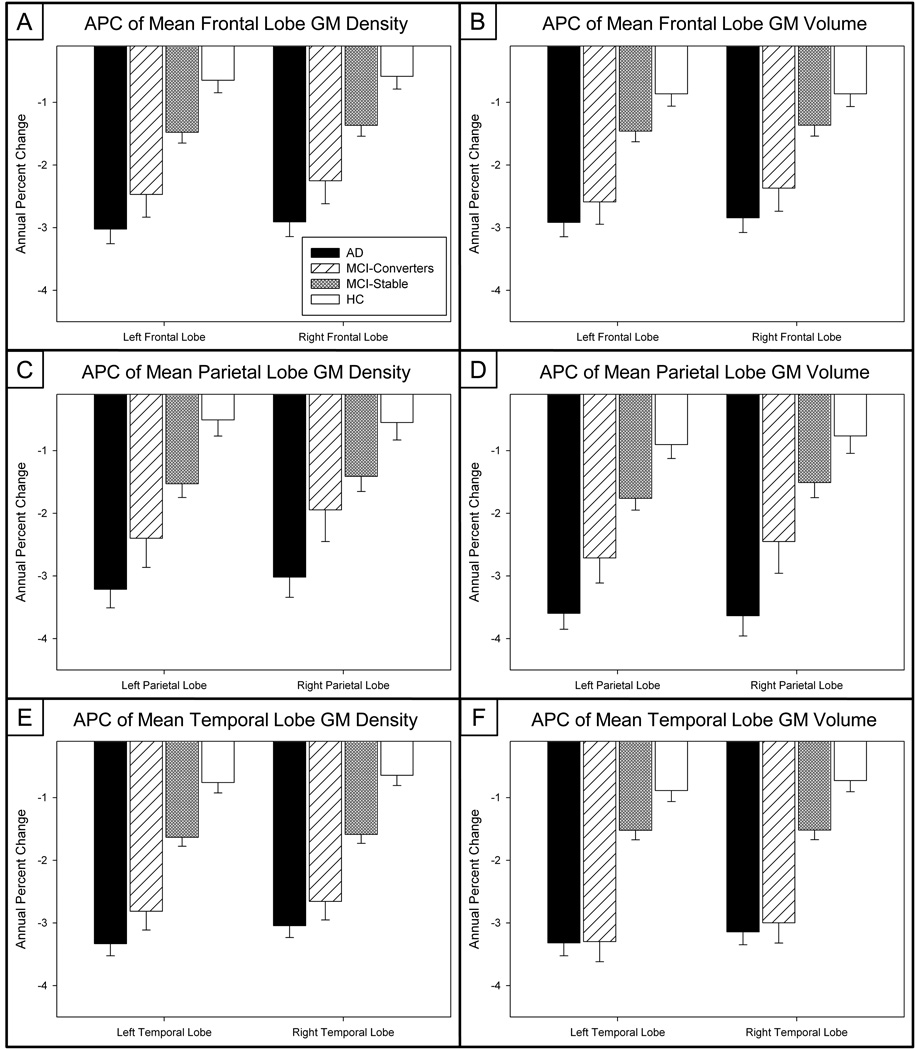
Figure 4. Annual Percent Change (APC) of Mean Frontal, Parietal, and Temporal Lobe GM Density and Volume Measures.
The APC in mean frontal lobe (a) GM density and (b) GM volume, mean parietal lobe (c) GM density and (d) GM volume, and mean temporal lobe (e) GM density and (f) GM volume are significantly different across groups (n=643*; 143 AD, 57 MCI-C, 253 MCI-S, 190 HC; *30 participants removed due to failed image processing).
Sample Sizes
The sample size needed to detect a 25% reduction in APC of MRI biomarkers was calculated for 80% or 90% power and a type I error (α) of p<0.05 for significant regions assessed in the present analysis (Table 3). Mean bilateral hippocampal GM density and GM volume estimates measured using either the independent or MarsBaR ROIs would require the smallest sample size to detect the desired reduction for all of the target groups. Other relatively sensitive ROIs for detecting a reduction in regional brain atrophy include hippocampal volume extracted using Freesurfer, mean temporal lobar GM density and GM volume, mean temporal lobe cortical thickness (MCI-C only), and mean frontal lobar GM density and GM volume. A full list of sample sizes needed to detect a 25% decline in brain atrophy at either 80% or 90% power for selected ROIs is found in Table 3.
Table 3.
Sample Sizes to Detect 25% Reduction in APC of Selected MRI Biomarkers
| AD | MCI-C | MCI-S | HC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Power: | 0.8 | 0.9 | 0.8 | 0.9 | 0.8 | 0.9 | 0.8 | 0.9 | |
| Hipp. GM Density (Indep. ROI) | 134 | 180 | 95 | 128 | 307 | 411 | 1243 | 1665 | |
| Hipp. GM Density (MarsBaR ROI) | 129 | 173 | 96 | 129 | 290 | 388 | 1204 | 1612 | |
| Hipp. GM Volume (Indep. ROI) | 133 | 178 | 77 | 103 | 400 | 535 | 745 | 998 | |
| Hipp. GM Volume (MarsBaR ROI) | 135 | 181 | 74 | 100 | 378 | 507 | 767 | 1028 | |
| Hipp. Volume (FreeSurfer) | 242 | 324 | 129 | 173 | 452 | 606 | 1136 | 1521 | |
| EC Thickness (FreeSurfer) | 328 | 439 | 214 | 286 | 1156 | 1548 | 7948 | 10648 | |
| Mean Frontal Lobe Cortical Thickness (FreeSurfer) | 1037 | 1389 | 369 | 494 | 2488 | 3333 | 4727 | 6333 | |
| Mean Parietal Lobe Cortical Thickness (FreeSurfer) | 629 | 842 | 515 | 689 | 1848 | 2475 | 3142 | 4209 | |
| Mean Temp. Lobe Cortical Thickness (FreeSurfer) | 403 | 539 | 121 | 162 | 1405 | 1882 | 3031 | 4061 | |
| Mean Frontal Lobe GM Density (MarsBaR ROIs) | 284 | 381 | 222 | 297 | 788 | 1056 | 3682 | 4932 | |
| Mean Frontal Lobe GM Volume (MarsBaR ROIs) | 280 | 376 | 263 | 353 | 856 | 1147 | 1475 | 1976 | |
| Mean Parietal Lobe GM Density (MarsBaR ROIs) | 384 | 514 | 373 | 499 | 1437 | 1925 | 7260 | 9726 | |
| Mean Parietal Lobe GM Volume (MarsBaR ROIs) | 238 | 319 | 315 | 422 | 976 | 1308 | 1977 | 2649 | |
| Mean Temp. Lobe GM Density (MarsBaR ROIs) | 157 | 210 | 129 | 173 | 456 | 610 | 1850 | 2479 | |
| Mean Temp. Lobe GM Volume (MarsBaR ROIs) | 158 | 212 | 100 | 134 | 646 | 866 | 1427 | 1911 | |
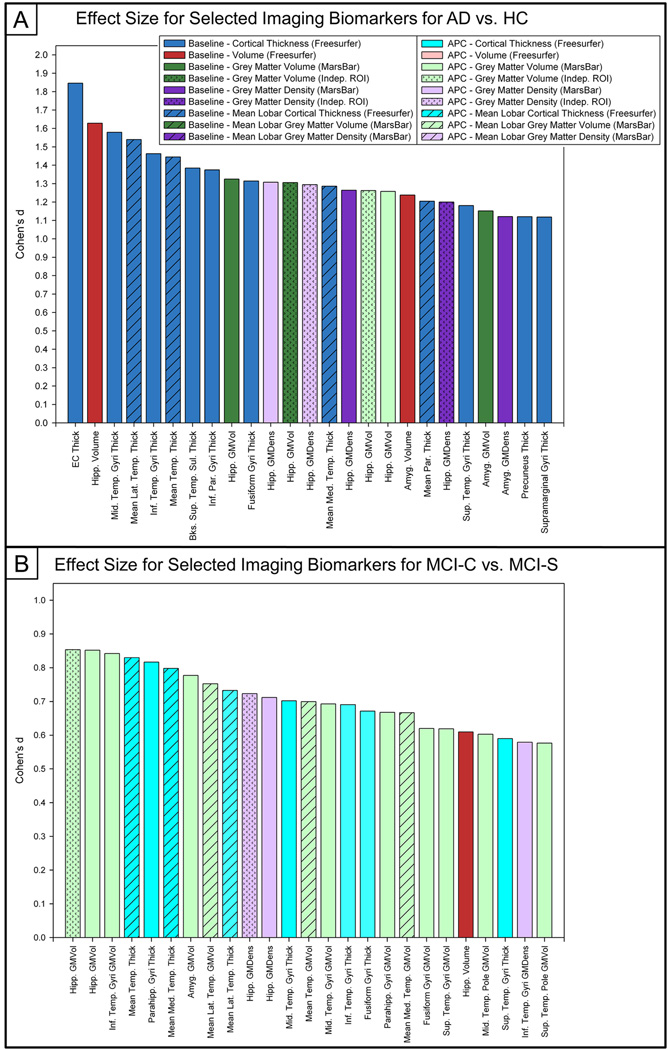
Effect Sizes
In order to effectively compare the relative sensitivity of MRI markers to distinguish between groups, we calculated the effect size for all available baseline and APC ROIs from VBM and Freesurfer for each group pair. Effect sizes for the comparison of AD and HC participants and MCI-C and MCI-S participants are found in Figure 5, while those for other pairs (MCI-C vs. HC; MCI-S vs. HC; AD vs. MCI-S; AD vs. MCI-C) are found in Supplementary Figure 1. Baseline medial temporal lobe biomarkers, including EC thickness, hippocampal volume, and middle temporal gyri cortical thickness measures, had the highest effect sizes for the comparison of AD vs. HC (Figure 5a), with Cohen’s d values of 1.846, 1.628, and 1.579, respectively. The APC in hippocampal GM density extracted using the MarsBaR ROIs had the highest effect size of the APC measures for AD vs. HC with a Cohen’s d of 1.308. Measures with maximal effect sizes for the comparison of MCI-C and MCI-S participants included APC in hippocampal GM volume (independent sample ROI, Cohen’s d=0.853; MarsBaR ROI, Cohen’s d=0.852), APC in inferior temporal gyri GM volume (Cohen’s d=0.842), and APC in mean temporal lobe GM volume (Cohen’s d=0.830). Medial temporal lobe ROIs also had high effect sizes for some of the other comparisons with baseline hippocampal volume showing the highest effect sizes for MCI-C vs. HC (Suppl. Figure 1a, Cohen’s d=1.652) and MCI-S vs. HC (Suppl. Figure 1b, Cohen’s d=0.958), and baseline middle temporal gyri thickness having the highest effect size for AD vs. MCI-S (Suppl. Figure 1c, Cohen’s d=0.890). APC in superior parietal gyri GM volume demonstrated the highest effect size for AD vs. MCI-C with a Cohen’s d of 0.456 (Suppl. Figure 1d).
Figure 5. Effect Sizes of Comparisons between AD & HC and MCI-C & MCI-S for Selected Imaging Biomarkers.
The effect sizes for selected baseline and APC values for the comparison of (a) AD and HC participants and (b) MCI-C and MCI-S participants are shown. Baseline MTL regions had the greatest effect sizes when comparing AD and HC, while APC in MTL regions demonstrated the greatest effect sizes in the MCI-C vs. MCI-S comparison. (n=643*; 143 AD, 57 MCI-C, 253 MCI-S, 190 HC; *30 participants removed due to failed image processing)
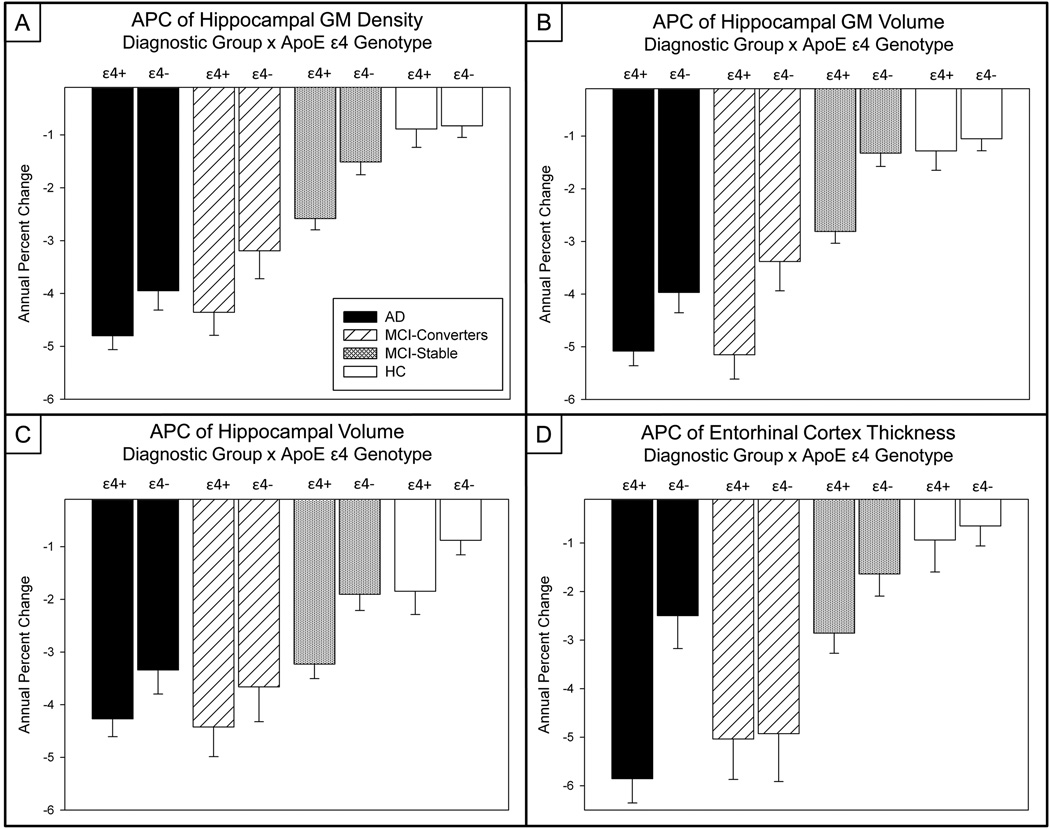
Influence of APOE ε4 Genotype
The presence of one or more APOE ε4 alleles increased APC atrophy markers for hippocampal GM density (p=0.001, Figure 6a), hippocampal GM volume (p<0.001; Figure 6b), hippocampal volume (p=0.001; Figure 6c), and EC thickness (p=0.003; Figure 6d). Additionally, a significant interaction between diagnosis group and APOE ε4 genotype was observed for the APC in EC thickness (p=0.029). Subsequent analyses within diagnostic group demonstrated that for AD patients APOE ε4 carriers showed greater decline in hippocampal GM volume (p=0.031) and EC thickness (p=0.002). For MCI-C, APOE ε4 positive participants also showed greater rate of atrophy in hippocampal GM density (p=0.031) and GM volume (p=0.001). For the MCI-S group, the atrophy rate in all evaluated regions was greater in APOE ε4 positive than negative participants, including APCs for hippocampal GM density (p=0.004), hippocampal GM volume (p<0.001), hippocampal volume (p=0.006), and EC thickness (p=0.004). Finally, APOE ε4 positive HC participants showed a significantly greater APC in hippocampal volume than those who were APOE ε4 negative (p=0.004).
Figure 6. Impact of APOE ε4 Genotype on Annual Percent Change (APC) of Selected MTL Measures.
The APC in bilateral mean hippocampal (a) GM density and (b) GM volume (n=643*; 143 AD, 57 MCI-C, 253 MCI-S, 190 HC), extracted using an independent sample of 40 healthy elderly controls (McHugh, et al., 2007, Saykin, et al., 2006, Shen, et al., 2010), as well as (c) hippocampal volume, and (d) EC thickness extracted using automated parcellation (n=673; 152 AD, 60 MCI-C, 261 MCI-S, 200 HC) show a significant effect of both diagnostic group and APOE ε4 genotype. (*30 participants removed due to failed image processing)
Discussion
Our main goal was to assess a detailed panel of longitudinal MRI atrophy markers in the ADNI cohort, including patients with probable AD, MCI to AD converters (within 12 months), stable MCI (over 12 months) and control participants. Our main findings were that AD and MCI-C groups had a significantly higher rate of annual decline in global and hippocampal GM density and GM volume, hippocampal total volume, EC thickness, and mean frontal, parietal and temporal lobar GM density, GM volume and cortical thickness measures than MCI-S and HC participants. Sample size calculations indicated that hippocampal GM density and GM volume required the smallest samples to detect a 25% reduction in rate of regional brain atrophy. Finally, effect size estimates indicated that dynamic measures, including APC in MTL volumes and cortical thickness, showed the most discrimination between MCI-C and MCI-S participants. However, at baseline, hippocampal volume and GM density, as well as temporal lobe cortical thickness measures, demonstrated the greatest effect size when comparing AD and HC participants. This pattern suggests that structural MRI markers may have differential utility as a function of stage of disease or role within a clinical trial. Where hippocampal volume and GM density are powerful tools for assessing baseline neurodegeneration, annual change rate in MTL volumes and cortical thickness may be most useful for comparing stable vs. rapidly progressing individuals, and may be the best choice for surrogate markers in trials of disease modifying agents.
Our estimates of APC in hippocampal volume, including −3.95% for AD patients, −4.10% for MCI-C participants, −2.65% for MCI-S participants, and −1.12% for HCs, were similar to estimates from previous reports in the ADNI cohort, as well as other samples (Table 1; (Barnes, et al., 2009)). These results demonstrate a significantly accelerated rate of brain atrophy in participants diagnosed with AD, as well as those who show rapid clinical decline from MCI to AD. Participants who show stable clinical diagnoses (both MCI and HC) also show relatively stable brain volume and cortical thickness measures, as well as minimal change in psychometric variables (Table 1).
We examined the influence of APOE ε4 genotype on annual atrophy rate in selected MTL MRI markers given the mixed prior findings, including significant effects of APOE on brain atrophy in some reports (Jack, et al., 2008c, Jack, et al., 2008d), whereas others found no effect (Du, et al., 2006, Wang, et al., 2009). In the present study, we observed a modest but significant effect of APOE ε4 genotype on annualized hippocampal and EC atrophy rates. This effect was maximal in MCI-S participants, with ε4 positive participants demonstrating significantly greater APC in all measures evaluated. However, the effect of APOE genotype in AD and MCI-C groups was only observed on some measures, suggesting a more moderate yet still detectable effect of genotype. Finally, APOE ε4 positive HC participants showed an enhanced rate of atrophy relative to ε4 negative participants only on hippocampal volume. Our results support the complicated nature of the relationship between APOE genotype and MRI markers of degeneration and suggest that the magnitude of the effect may differ by diagnostic stage, as has been previously reported in the ADNI cohort (Nestor, et al., 2008, Schuff, et al., 2009). Future studies will further characterize the impact of APOE, as well as that of variation in other candidate genes, on MRI and other ADNI biomarkers which may assist in elucidating the role of genetic factors in the neuropathology of AD (Saykin, et al., 2010).
This report adds to the body of research demonstrating the utility of MRI metrics in detecting and monitoring atrophy associated with AD and MCI, and extends prior research by focusing on identifying differences between rapidly declining MCI to AD converters and individuals with relatively stable MCI. Reports in other smaller samples have led to similar conclusions regarding the utility of MRI extracted measures of global and local brain volume, cortical thickness, and morphometry in detecting and monitoring brain atrophy associated with AD and MCI (Barnes, et al., 2007, Cardenas, et al., 2003, Erten-Lyons, et al., 2006, Fox and Freeborough, 1997, Jack, et al., 2000, Jack, et al., 2004, Jack, et al., 2005, Mungas, et al., 2005, Sluimer, et al., 2008, Stoub, et al., 2008, Thompson, et al., 2004, Whitwell, et al., 2008a). As previously reported in the ADNI sample, baseline values of hippocampal GM density and volume, amygdalar volume, EC thickness, and temporal and parietal lobe cortical thickness measures are significantly different between MCI-C and MCI-S participants (Risacher, et al., 2009). In fact, MCI-C and AD participants show nearly equivalent atrophy at baseline, up to 12 months prior to equivalent clinical diagnoses, indicating that MRI can serve as an antecedent biomarker. Measures of annual decline provide further evidence that MCI to AD converters have characteristic cross-sectional and longitudinal brain atrophy more similar to AD patients than to those with stable MCI. The different longitudinal phenotypes warrant investigation and may be useful in examining genetic variation associated with rate of decline (Jack, et al., 2008c, Saykin, et al., 2009).
Our results are generally consistent with previous reports using subsets of the ADNI cohort and alternative methods. Four studies employed Freesurfer based ROI techniques to estimate APC in selected cortical and sub-cortical regions and reported similar APC values and differences between diagnostic groups as those observed in the present analysis (Fjell, et al., 2010b, Holland, et al., 2009, McDonald, et al., 2009, McEvoy, et al., 2009). Furthermore, two of these studies divided the MCI group by baseline CDR-SoB (McDonald, et al., 2009) and by atrophy pattern (AD-like vs. HC-like, (McEvoy, et al., 2009)) and showed variability of APC values within the MCI group, similar to that seen in the present report between MCI-Converters and MCI-Stable participants. Two studies used various hippocampal ROIs and reported significantly greater APC in hippocampal volume in AD participants relative to MCI and HC participants (Morra, et al., 2009, Schuff, et al., 2009). Two studies examined changes in ventricular volume, demonstrating greater rates of ventricular enlargement in AD and MCI participants relative to HCs, as well as greater ventricular enlargement in participants who converted from MCI to AD within the first 6 months of the study relative to MCI-Stable participants (Jack, et al., 2009, Nestor, et al., 2008). Three additional studies employed TBM and Jacobian maps to investigate whole brain and temporal lobe atrophy rates and found a similar pattern of differences between participants as seen in the present study (Ho, et al., 2009, Hua, et al., 2009, Leow, et al., 2009). One of these studies also reported a higher rate of atrophy in MCI-C relative to MCI-S participants, albeit in a significantly smaller sample (7 MCI-C and 32 MCI-S) than used in the present analysis (Leow, et al., 2009). Misra et al (2009) also reported significant differences in atrophy rate between MCI-C and MCI-S participants using a VBM-like technique (RAVENS), although differences were limited to periventricular WM and the temporal horn (Misra, et al., 2009). Finally, another study used a boundary shift integral technique to evaluate annual rates of whole brain atrophy and ventricular enlargement (Evans, et al., 2009). This study reported greater annual rates of whole brain atrophy and ventricular enlargement in AD participants relative to MCI and HC participants, as well as in MCI-Converters relative to MCI-Stable participants. In fact, Evans et al. (2009) noted that MCI-Converters demonstrated nearly equivalent rates of atrophy as seen in the AD participants, similar to the pattern reported in the present study. Overall, the results of the present study extend this line of research by providing one of the first direct comparisons of annual atrophy rates for an ensemble of state of the art MRI morphometric, volumetric and cortical thickness variables in the ADNI cohort, particularly focusing on participants who converted from MCI to AD within the first 12 months of the study.
There are several limitations of the present study. First, we were unable to account for some other variables which may have impacted the results. Since the ADNI is an observational study, many participants were taking a number of medications prescribed for AD, MCI or other conditions that could have affected the results. Additionally, differences in disease severity beyond clinical diagnostic classification (i.e. AD, MCI, HC) was not considered in the present analyses. Although diagnostic classification and conversion status incorporates information from psychometric performance, the present report does not explicitly examine the relationship between changes in MRI variables and changes in psychometric performance. Secondly, the inclusion of only two timepoints separated by approximately 1 year in the present study limited the specificity and accuracy of the APC estimations. One of the major advantages of the ADNI project is the extensive longitudinal data collection. Therefore, as full datasets from the 2-year and 3-year timepoints become available, we plan to expand our analysis of annual atrophy rates in patients with AD, MCI-Converters, MCI-Stables, and healthy elderly. Furthermore, we will employ more advanced statistical modeling to compare the atrophy rates between MCI-Converters from several timepoints. With 3 or more timepoints non-linearities can be detected. Finally, this study was limited by the nature of the methods employed to measure atrophy. Specifically, some variability in segmentation and extraction of ROIs is likely, based on the interaction between scan quality or other properties and specific image processing algorithms which may have resulted in variation in the accuracy of the annual change estimates. However, the largely automated methodology employed in these analyses provides for little or no rater bias inherent in manually directed tools of volume extraction. Furthermore, other analysis techniques (e.g. TBM, BSI) may provide additional and complementary information to that extracted in the present study using VBM and automated parcellation. Although a comprehensive and direct comparison of the relative sensitivity and specificity of target MRI-based atrophy measures extracted using different methods has not been completed to our knowledge, ADNI provides an ideal cohort for investigating this issue. Finally, better methods for visualization and display of longitudinal changes on a voxel-wise basis would also be advantageous. Statistical parametric maps of time by group interactions, like those presented in the present study, do not illustrate the percent change at all significant locations. Development of improved tools for visualization of magnitude of changes as a function of diagnostic group would be useful.
In summary, these results used a combination of analysis methods to confirm that MRI based morphometric markers detect dynamic changes in rate of atrophy in patients with AD and MCI, compared to controls and are highly sensitive to the likelihood of clinical progression with one year. Measures of GM density change within medial and lateral temporal regions have been employed less than volumetric measures to date but appear particularly promising and complementary to more standard measures such as hippocampal volumetry. The sensitivity of automated and unbiased methods for detecting differences in rate of neurodegenerative changes encourages their use in clinical trials of disease modifying agents and in prevention trials.
Supplementary Material
The effect sizes for selected baseline and APC values for the comparison of (a) MCI-C and HC participants, (b) MCI-S and HC participants, (c) AD and MCI-S participants, and (d) AD and MCI-C participants are shown. (n=643*; 143 AD, 57 MCI-C, 253 MCI-S, 190 HC; *30 participants removed due to failed image processing)
Figure 3. Annual Percent Change (APC) of Entorhinal Cortex, Mean Frontal, Parietal, and Temporal Lobe Cortical Thickness Measures.
APC in (a) entorhinal cortex thickness, and mean (b) frontal, (c) parietal, and (d) temporal lobar cortical thickness are significantly different across groups (n=673; 152 AD, 60 MCI-C, 261 MCI-S, 200 HC).
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grants U01 AG024904 and RC2 AG036535, PI: Michael W. Weiner, MD). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., and Wyeth, as well as non-profit partners the Alzheimer's Association and Alzheimer's Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation. The National Cell Repository for Alzheimer's Disease (NIH grant U24 AG021886) provided support for DNA and cell line banking and processing for ADNI.
Data analysis was supported in part by the following grants from the National Institutes of Health: NIA R01 AG19771 to AJS and P30 AG10133-18S1 to B. Ghetti and AJS, and NIBIB R03 EB008674 to LS; and by the Indiana Economic Development Corporation (IEDC #87884 to AJS). The Freesurfer analyses were performed on a 112-node parallel computing environment called Quarry at Indiana University. We thank the University Information Technology Services at Indiana University and Randy Heiland, MA, MS for their support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. For a complete list of investigators involved in ADNI see: http://www.loni.ucla.edu/ADNI/Data/ADNI_Authorship_List.pdf.
Disclosures
SL Risacher reports no disclosures.
Dr. Shen reports no disclosures.
Dr. Kim reports no disclosures.
JD West reports no disclosures.
Dr. McDonald reports no disclosures.
Dr. Beckett reports no disclosures.
Dr. Harvey reports no disclosures.
Dr. Jack served on a scientific advisory board for Elan Corporation; receives research support from Pfizer Inc., the Mayo U of MN Biotechnology Partnership, and holds stock in GE Healthcare.
Dr. Weiner serves on scientific advisory boards for Bayer Schering Pharma, Eli Lilly and Company, CoMentis, Inc., Neurochem Inc, Eisai Inc., Avid Radiopharmaceuticals Inc., Aegis Therapies, Genentech, Inc., Allergan, Inc., Lippincott Williams & Wilkins, Bristol-Myers Squibb, Forest Laboratories, Inc., Pfizer Inc, McKinsey & Company, Mitsubishi Tanabe Pharma Corporation, and Novartis; has received funding for travel from Nestle´ and Kenes International and to attend conferences not funded by industry; has received honoraria from the Rotman Research Institute and BOLT International; serves as a consultant for Elan Corporation; receives research support from Merck & Co., Radiopharmaceuticals Inc., and holds stock in Synarc and Elan Corporation.
Dr. Saykin receives support from the NIH (R01 CA101318, R01 AG19771, RC2 AG036535, P30 AG10133-18S1, U01 AG032984), Indiana Economic Development Corporation (IEDC #87884), and from Siemens Medical Solutions and Welch Allyn, Inc.
Informed consent was obtained from all ADNI participants according to the Helsinki Declaration and necessary approval was received from Ethical Committees at each of the participating research institutions. Further information about ADNI can be found in (Jack, et al., 2008a, Susanne G. Mueller, et al., 2005, Petersen, et al., 2010a) and at www.adni-info.org.
Contributor Information
SL Risacher, Email: srisache@iupui.edu.
L Shen, Email: shenli@iupui.edu.
JD West, Email: jdwest@iupui.edu.
S Kim, Email: sk31@iupui.edu.
BC McDonald, Email: mcdonalb@iupui.edu.
LA Beckett, Email: labeckett@ucdavis.edu.
DJ Harvey, Email: djharvey@phs.ucdavis.edu.
CR Jack, Jr, Email: jack.clifford@mayo.edu.
MW Weiner, Email: Michael.Weiner@ucsf.edu.
References
- Apostolova L, Dutton R, Dinov I, Hayashi K, Toga A, Cummings J, Thompson P. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol. 2006;63(5):693–699. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11(6 Pt 1):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Barnes J, Bartlett JW, van de Pol LA, Loy CT, Scahill RI, Frost C, Thompson P, Fox NC. A meta-analysis of hippocampal atrophy rates in Alzheimer's disease. Neurobiol Aging. 2009;30(11):1711–1723. doi: 10.1016/j.neurobiolaging.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Boyes RG, Lewis EB, Schott JM, Frost C, Scahill RI, Fox NC. Automatic calculation of hippocampal atrophy rates using a hippocampal template and the boundary shift integral. Neurobiol Aging. 2007;28(11):1657–1663. doi: 10.1016/j.neurobiolaging.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Becker JT, Davis SW, Hayashi KM, Meltzer CC, Toga AW, Lopez OL, Thompson PM. Three-dimensional patterns of hippocampal atrophy in mild cognitive impairment. Arch Neurol. 2006;63(1):97–101. doi: 10.1001/archneur.63.1.97. [DOI] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9(10):768–778. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- Bozzali M, Filippi M, Magnani G, Cercignani M, Franceschi M, Schiatti E, Castiglioni S, Mossini R, Falautano M, Scotti G, Comi G, Falini A. The contribution of voxel-based morphometry in staging patients with mild cognitive impairment. Neurology. 2006;67(3):453–460. doi: 10.1212/01.wnl.0000228243.56665.c2. [DOI] [PubMed] [Google Scholar]
- Brett M, Anton J-L, Valabregue R, Poline J-B. Region of interest analysis using an SPM toolbox [abstract]. Presented at the 8th International Conference on Functional Mapping of the Human Brain; June 2–6, 2002; Sendai, Japan. 2002. [Google Scholar]
- Busatto GF, Garrido GE, Almeida OP, Castro CC, Camargo CH, Cid CG, Buchpiguel CA, Furuie S, Bottino CM. A voxel-based morphometry study of temporal lobe gray matter reductions in Alzheimer's disease. Neurobiol Aging. 2003;24(2):221–231. doi: 10.1016/s0197-4580(02)00084-2. [DOI] [PubMed] [Google Scholar]
- Calvini P, Chincarini A, Gemme G, Penco MA, Squarcia S, Nobili F, Rodriguez G, Bellotti R, Catanzariti E, Cerello P, De Mitri I, Fantacci ME. Automatic analysis of medial temporal lobe atrophy from structural MRIs for the early assessment of Alzheimer disease. Med Phys. 2009;36(8):3737–3747. doi: 10.1118/1.3171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas VA, Du AT, Hardin D, Ezekiel F, Weber P, Jagust WJ, Chui HC, Schuff N, Weiner MW. Comparison of methods for measuring longitudinal brain change in cognitive impairment and dementia. Neurobiol Aging. 2003;24(4):537–544. doi: 10.1016/s0197-4580(02)00130-6. [DOI] [PubMed] [Google Scholar]
- Chetelat G, Desgranges B, De La Sayette V, Viader F, Eustache F, Baron JC. Mapping gray matter loss with voxel-based morphometry in mild cognitive impairment. Neuroreport. 2002;13(15):1939–1943. doi: 10.1097/00001756-200210280-00022. [DOI] [PubMed] [Google Scholar]
- Chetelat G, Landeau B, Eustache F, Mezenge F, Viader F, de la Sayette V, Desgranges B, Baron JC. Using voxel-based morphometry to map the structural changes associated with rapid conversion in MCI: a longitudinal MRI study. Neuroimage. 2005;27(4):934–946. doi: 10.1016/j.neuroimage.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Chou YY, Lepore N, Avedissian C, Madsen SK, Parikshak N, Hua X, Shaw LM, Trojanowski JQ, Weiner MW, Toga AW, Thompson PM. Mapping correlations between ventricular expansion and CSF amyloid and tau biomarkers in 240 subjects with Alzheimer's disease, mild cognitive impairment and elderly controls. Neuroimage. 2009;46(2):394–410. doi: 10.1016/j.neuroimage.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupin M, Gerardin E, Cuingnet R, Boutet C, Lemieux L, Lehericy S, Benali H, Garnero L, Colliot O. Fully automatic hippocampus segmentation and classification in Alzheimer's disease and mild cognitive impairment applied on data from ADNI. Hippocampus. 2009;19(6):579–587. doi: 10.1002/hipo.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed. Hillsdale, NJ: Lawrence Erlbaum Associates, Inc.; 1988. [Google Scholar]
- Cohen RM, Small C, Lalonde F, Friz J, Sunderland T. Effect of apolipoprotein E genotype on hippocampal volume loss in aging healthy women. Neurology. 2001;57(12):2223–2228. doi: 10.1212/wnl.57.12.2223. [DOI] [PubMed] [Google Scholar]
- Colliot O, Chetelat G, Chupin M, Desgranges B, Magnin B, Benali H, Dubois B, Garnero L, Eustache F, Lehericy S. Discrimination between Alzheimer disease, mild cognitive impairment, and normal aging by using automated segmentation of the hippocampus. Radiology. 2008;248(1):194–201. doi: 10.1148/radiol.2481070876. [DOI] [PubMed] [Google Scholar]
- Dale A, Fischl B, Sereno M. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- de Leon MJ, Mosconi L, Blennow K, DeSanti S, Zinkowski R, Mehta PD, Pratico D, Tsui W, Saint Louis LA, Sobanska L, Brys M, Li Y, Rich K, Rinne J, Rusinek H. Imaging and CSF studies in the preclinical diagnosis of Alzheimer's disease. Annals of the New York Academy of Sciences. 2007;1097:114–145. doi: 10.1196/annals.1379.012. [DOI] [PubMed] [Google Scholar]
- De Toledo-Morrell L, Goncharova I, Dickerson B, Wilson RS, Bennett DA. From healthy aging to early Alzheimer's disease: in vivo detection of entorhinal cortex atrophy. Ann N Y Acad Sci. 2000;911:240–253. doi: 10.1111/j.1749-6632.2000.tb06730.x. [DOI] [PubMed] [Google Scholar]
- Devanand DP, Pradhaban G, Liu X, Khandji A, De Santi S, Segal S, Rusinek H, Pelton GH, Honig LS, Mayeux R, Stern Y, Tabert MH, de Leon MJ. Hippocampal and entorhinal atrophy in mild cognitive impairment: prediction of Alzheimer disease. Neurology. 2007;68(11):828–836. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Goncharova I, Sullivan MP, Forchetti C, Wilson RS, Bennett DA, Beckett LA, deToledo-Morrell L. MRI-derived entorhinal and hippocampal atrophy in incipient and very mild Alzheimer's disease. Neurobiology of Aging. 2001;22(5):747–754. doi: 10.1016/s0197-4580(01)00271-8. [DOI] [PubMed] [Google Scholar]
- Du AT, Schuff N, Amend D, Laakso MP, Hsu YY, Jagust WJ, Yaffe K, Kramer JH, Reed B, Norman D, Chui HC, Weiner MW. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2001;71(4):441–447. doi: 10.1136/jnnp.71.4.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du AT, Schuff N, Chao LL, Kornak J, Jagust WJ, Kramer JH, Reed BR, Miller BL, Norman D, Chui HC, Weiner MW. Age effects on atrophy rates of entorhinal cortex and hippocampus. Neurobiol Aging. 2006;27(5):733–740. doi: 10.1016/j.neurobiolaging.2005.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du AT, Schuff N, Kramer JH, Rosen HJ, Gorno-Tempini ML, Rankin K, Miller BL, Weiner MW. Different regional patterns of cortical thinning in Alzheimer's disease and frontotemporal dementia. Brain. 2007;130(Pt 4):1159–1166. doi: 10.1093/brain/awm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erten-Lyons D, Howieson D, Moore MM, Quinn J, Sexton G, Silbert L, Kaye J. Brain volume loss in MCI predicts dementia. Neurology. 2006;66(2):233–235. doi: 10.1212/01.wnl.0000194213.50222.1a. [DOI] [PubMed] [Google Scholar]
- Evans MC, Barnes J, Nielsen C, Kim LG, Clegg SL, Blair M, Leung KK, Douiri A, Boyes RG, Ourselin S, Fox NC. Volume changes in Alzheimer's disease and mild cognitive impairment: cognitive associations. Eur Radiol. 2009 doi: 10.1007/s00330-009-1581-5. [DOI] [PubMed] [Google Scholar]
- Fan Y, Batmanghelich N, Clark CM, Davatzikos C. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. Neuroimage. 2008;39(4):1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278(16):1349–1356. [PubMed] [Google Scholar]
- Fennema-Notestine C, Hagler DJ, Jr, McEvoy LK, Fleisher AS, Wu EH, Karow DS, Dale AM. Structural MRI biomarkers for preclinical and mild Alzheimer's disease. Hum Brain Mapp. 2009;30(10):3238–3253. doi: 10.1002/hbm.20744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, Hall K, Hasegawa K, Hendrie H, Huang Y, Jorm A, Mathers C, Menezes PR, Rimmer E, Scazufca M. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005;366(9503):2112–2117. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000a;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences of the United States of America. 2000b;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat D, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale A. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno M, Dale A. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Blennow K, Brewer JB, Dale AM. Brain Atrophy in Healthy Aging Is Related to CSF Levels of A{beta}1-42. Cereb Cortex. 2010a doi: 10.1093/cercor/bhp279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Brewer J, Dale A ADNI. CSF Biomarkers in Prediction of Cerebral and Clinical Change in Mild Cognitive Impairment and Alzheimer's Disease. Journal of Neuroscience. 2010b;30(6):2088–2101. doi: 10.1523/JNEUROSCI.3785-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisher A, Grundman M, Jack CR, Jr, Petersen RC, Taylor C, Kim HT, Schiller DH, Bagwell V, Sencakova D, Weiner MF, DeCarli C, DeKosky ST, van Dyck CH, Thal LJ. Sex, apolipoprotein E epsilon 4 status, and hippocampal volume in mild cognitive impairment. Arch Neurol. 2005;62(6):953–957. doi: 10.1001/archneur.62.6.953. [DOI] [PubMed] [Google Scholar]
- Fleisher AS, Sun S, Taylor C, Ward CP, Gamst AC, Petersen RC, Jack CR, Jr, Aisen PS, Thal LJ. Volumetric MRI vs clinical predictors of Alzheimer disease in mild cognitive impairment. Neurology. 2008;70(3):191–199. doi: 10.1212/01.wnl.0000287091.57376.65. [DOI] [PubMed] [Google Scholar]
- Fox NC, Freeborough PA. Brain atrophy progression measured from registered serial MRI: validation and application to Alzheimer's disease. J Magn Reson Imaging. 1997;7(6):1069–1075. doi: 10.1002/jmri.1880070620. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Fox NC, Jack CR, Jr, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010;6(2):67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni GB, Testa C, Zorzan A, Sabattoli F, Beltramello A, Soininen H, Laakso MP. Detection of grey matter loss in mild Alzheimer's disease with voxel based morphometry. J Neurol Neurosurg Psychiatry. 2002;73(6):657–664. doi: 10.1136/jnnp.73.6.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14(1 Pt 1):21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Hamalainen A, Grau-Olivares M, Tervo S, Niskanen E, Pennanen C, Huuskonen J, Kivipelto M, Hanninen T, Tapiola M, Vanhanen M, Hallikainen M, Helkala EL, Nissinen A, Vanninen RL, Soininen H. Apolipoprotein E epsilon4 allele is associated with increased atrophy in progressive mild cognitive impairment: a voxel-based morphometric study. Neurodegener. 2008;5(3–4):186–189. doi: 10.1159/000113698. [DOI] [PubMed] [Google Scholar]
- Ho AJ, Hua X, Lee S, Leow AD, Yanovsky I, Gutman B, Dinov ID, Lepore N, Stein JL, Toga AW, Jack CR, Jr, Bernstein MA, Reiman EM, Harvey DJ, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM. Comparing 3 T and 1.5 T MRI for tracking Alzheimer's disease progression with tensor-based morphometry. Hum Brain Mapp. 2009 doi: 10.1002/hbm.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland D, Brewer JB, Hagler DJ, Fenema-Notestine C, Dale AM, Weiner M, Thal L, Petersen R, Jack CR, Jr, Jagust W, Trojanowki J, Toga AW, Beckett L ADNI. Subregional neuroanatomical change as a biomarker for Alzheimer's disease. Proc Natl Acad Sci U S A. 2009;106(49):20954–20959. doi: 10.1073/pnas.0906053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Lee S, Yanovsky I, Leow AD, Chou YY, Ho AJ, Gutman B, Toga AW, Jack CR, Jr, Bernstein MA, Reiman EM, Harvey DJ, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM. Optimizing power to track brain degeneration in Alzheimer's disease and mild cognitive impairment with tensor-based morphometry: an ADNI study of 515 subjects. Neuroimage. 2009;48(4):668–681. doi: 10.1016/j.neuroimage.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, J LW, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008a;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Lowe VJ, Senjem ML, Weigand SD, Kemp BJ, Shiung MM, Knopman DS, Boeve BF, Klunk WE, Mathis CA, Petersen RC. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer's disease and amnestic mild cognitive impairment. Brain. 2008b;131(Pt 3):665–680. doi: 10.1093/brain/awm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Lowe VJ, Weigand SD, Wiste HJ, Senjem ML, Knopman DS, Shiung MM, Gunter JL, Boeve BF, Kemp BJ, Weiner M, Petersen RC. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer's disease: implications for sequence of pathological events in Alzheimer's disease. Brain. 2009;132(Pt 5):1355–1365. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Grundman M, Jin S, Gamst A, Ward CP, Sencakova D, Doody RS, Thal LJ. Longitudinal MRI findings from the vitamin E and donepezil treatment study for MCI. Neurobiol Aging. 2008c;29(9):1285–1295. doi: 10.1016/j.neurobiolaging.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, O'Brien PC, Tangalos EG. MR-based hippocampal volumetry in the diagnosis of Alzheimer's disease. Neurology. 1992;42(1):183–188. doi: 10.1212/wnl.42.1.183. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu Y, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Tangalos EG, Kokmen E. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000;55(4):484–489. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu YC, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52(7):1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Shiung MM, Gunter JL, O'Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, Tangalos EG, Petersen RC. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62(4):591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Shiung MM, Weigand SD, O'Brien PC, Gunter JL, Boeve BF, Knopman DS, Smith GE, Ivnik RJ, Tangalos EG, Petersen RC. Brain atrophy rates predict subsequent clinical conversion in normal elderly and amnestic MCI. Neurology. 2005;65(8):1227–1231. doi: 10.1212/01.wnl.0000180958.22678.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Weigand SD, Shiung MM, Przybelski SA, O'Brien PC, Gunter JL, Knopman DS, Boeve BF, Smith GE, Petersen RC. Atrophy rates accelerate in amnestic mild cognitive impairment. Neurology. 2008d;70(19 Pt 2):1740–1752. doi: 10.1212/01.wnl.0000281688.77598.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas GB, Burton EJ, Rombouts SA, van Schijndel RA, O'Brien JT, Scheltens P, McKeith IG, Williams D, Ballard C, Barkhof F. A comprehensive study of gray matter loss in patients with Alzheimer's disease using optimized voxel-based morphometry. Neuroimage. 2003;18(4):895–907. doi: 10.1016/s1053-8119(03)00041-7. [DOI] [PubMed] [Google Scholar]
- Killiany RJ, Hyman BT, Gomez-Isla T, Moss MB, Kikinis R, Jolesz F, Tanzi R, Jones K, Albert MS. MRI measures of entorhinal cortex vs hippocampus in preclinical AD. Neurology. 2002;58(8):1188–1196. doi: 10.1212/wnl.58.8.1188. [DOI] [PubMed] [Google Scholar]
- Kinkingnehun S, Sarazin M, Lehericy S, Guichart-Gomez E, Hergueta T, Dubois B. VBM anticipates the rate of progression of Alzheimer disease: a 3-year longitudinal study. Neurology. 2008;70(23):2201–2211. doi: 10.1212/01.wnl.0000303960.01039.43. [DOI] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, Parikshak N, Hua X, Lee S, Toga AW, Jack CR, Jr, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Borowski B, Shaw LM, Trojanowski JQ, Fleisher AS, Harvey D, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM. Alzheimer's Disease Neuroimaging Initiative: A one-year follow up study using Tensor-Based Morphometry correlating degenerative rates, biomarkers and cognition. Neuroimage. 2009 doi: 10.1016/j.neuroimage.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CR, McEvoy LK, Gharapetian L, Fennema-Notestine C, Hagler DJ, Jr, Holland D, Koyama A, Brewer JB, Dale AM. Regional rates of neocortical atrophy from normal aging to early Alzheimer disease. Neurology. 2009;73(6):457–465. doi: 10.1212/WNL.0b013e3181b16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy LK, Fennema-Notestine C, Roddey JC, Hagler DJ, Jr, Holland D, Karow DS, Pung CJ, Brewer JB, Dale AM. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251(1):195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh T, Saykin A, Wishart H, Flashman L, Cleavinger H, Rabin L, Mamourian A, Shen L. Hippocampal volume and shape analysis in an older adult population. Clin Neuropsychol. 2007;21(1):130–145. doi: 10.1080/13854040601064534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechelli A, Price CJ, Friston KJ, Ashburner J. Voxel-Based Morphometry of the Human Brain: Methods and Applications. Current Medical Imaging Reviews. 2005;I(1):1–9. [Google Scholar]
- Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neuroimage. 2009;44(4):1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori E, Lee K, Yasuda M, Hashimoto M, Kazui H, Hirono N, Matsui M. Accelerated hippocampal atrophy in Alzheimer's disease with apolipoprotein E epsilon4 allele. Ann Neurol. 2002;51(2):209–214. doi: 10.1002/ana.10093. [DOI] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR, Jr, Schuff N, Weiner MW, Thompson PM. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Neuroimage. 2009;45(1 Suppl):S3–S15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer's disease neuroimaging initiative. Neuroimaging Clin N Am. 2005a;15(4):869–877. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer's Disease Neuroimaging Initiative. Neuroimaging Clinics of North America. 2005;15(4):869–877. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer's disease: The Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005b;1(1):55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungas D, Harvey D, Reed BR, Jagust WJ, DeCarli C, Beckett L, Mack WJ, Kramer JH, Weiner MW, Schuff N, Chui HC. Longitudinal volumetric MRI change and rate of cognitive decline. Neurology. 2005;65(4):565–571. doi: 10.1212/01.wnl.0000172913.88973.0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor SM, Rupsingh R, Borrie M, Smith M, Accomazzi V, Wells JL, Fogarty J, Bartha R. Ventricular enlargement as a possible measure of Alzheimer's disease progression validated using the Alzheimer's disease neuroimaging initiative database. Brain. 2008;131(Pt 9):2443–2454. doi: 10.1093/brain/awn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennanen C, Kivipelto M, Tuomainen S, Hartikainen P, Hanninen T, Laakso MP, Hallikainen M, Vanhanen M, Nissinen A, Helkala EL, Vainio P, Vanninen R, Partanen K, Soininen H. Hippocampus and entorhinal cortex in mild cognitive impairment and early AD. Neurobiol Aging. 2004;25(3):303–310. doi: 10.1016/S0197-4580(03)00084-8. [DOI] [PubMed] [Google Scholar]
- Pennanen C, Testa C, Laakso MP, Hallikainen M, Helkala EL, Hanninen T, Kivipelto M, Kononen M, Nissinen A, Tervo S, Vanhanen M, Vanninen R, Frisoni GB, Soininen H. A voxel based morphometry study on mild cognitive impairment. Journal of Neurology, Neurosurgery & Psychiatry. 2005;76(1):11–14. doi: 10.1136/jnnp.2004.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC. Mild cognitive impairment: transition between aging and Alzheimer's disease. Neurologia. 2000;15(3):93–101. [PubMed] [Google Scholar]
- Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR, Jr, Jagust WJ, Shaw LM, Toga AW, Trojanowski JQ, Weiner MW. Alzheimer's Disease Neuroimaging Initiative (ADNI): Clinical characterization. Neurology. 2010a;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR, Jr, Jagust WJ, Shaw LM, Toga AW, Trojanowski JQ, Weiner MW. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010b;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch Neurol. 2001;58(12):1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Negash S. Mild cognitive impairment: an overview. CNS Spectr. 2008;13(1):45–53. doi: 10.1017/s1092852900016151. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56(3):303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- Querbes O, Aubry F, Pariente J, Lotterie JA, Demonet JF, Duret V, Puel M, Berry I, Fort JC, Celsis P. Early diagnosis of Alzheimer's disease using cortical thickness: impact of cognitive reserve. Brain. 2009;132(Pt 8):2036–2047. doi: 10.1093/brain/awp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher SL, Saykin AJ. Neuroimaging of Alzheimer's Disease, Mild Cognitive Impairment and Other Dementias. In: Sweet LH, Cohen RA, editors. Brain Imaging in Behavioral Medicine and Neuropsychology. New York: Springer; in press. [Google Scholar]
- Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res. 2009;6(4):347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner B. Fundamentals of Biostatistics. Boston: PWS-Kent Publishing Company; 1990. [Google Scholar]
- Saykin A, Wishart H, Rabin L, Santulli R, Flashman L, West J, McHugh T, Mamourian A. Older adults with cognitive complaints show brain atrophy similar to that of amnestic MCI. Neurology. 2006;67(5):834–842. doi: 10.1212/01.wnl.0000234032.77541.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Foroud T, Potkin SG, Swaminathan S, Kim S, Risacher SL, Nho K, Huentelman MJ, Craig DW, Thompson PM, Stein JL, Moore JH, Farrer LA, Green RC, Bertram L, Jack CR, Weiner MW ADNI. Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer's & Dementia. 2010:1–9. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Risacher SL, Kim S, Nho K, West JD, Foroud T ADNI. Genetic predictors of 12 month change in MRI hippocampal volume in the Alzheimer's Disease Neuroimaging Initiative cohort: Analysis of leading candidates from the AlzGene database. Alzheimer's & Dementia. 2009;5(4 Supplement):P3. [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Jr, Weiner MW. MRI of hippocampal volume loss in early Alzheimer's disease in relation to ApoE genotype and biomarkers. Brain. 2009;132(Pt 4):1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Saykin AJ, Kim S, Firpi H, West JD, Risacher SL, McDonald BC, McHugh TL, Wishart HA, Flashman LA. Comparison of manual and automated determination of hippocampal volumes in MCI and older adults with cognitive complaints. Brain Imaging and Behavior. 2010 doi: 10.1007/s11682-010-9088-x. DOI: 10.1007/s11682-010-9088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluimer JD, Vrenken H, Blankenstein MA, Fox NC, Scheltens P, Barkhof F, van der Flier WM. Whole-brain atrophy rate in Alzheimer disease: identifying fast progressors. Neurology. 2008;70(19 Pt 2):1836–1841. doi: 10.1212/01.wnl.0000311446.61861.e3. [DOI] [PubMed] [Google Scholar]
- Stoub TR, Rogalski EJ, Leurgans S, Bennett DA, Detoledo-Morrell L. Rate of entorhinal and hippocampal atrophy in incipient and mild AD: Relation to memory function. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, De Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 2004;22(4):1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Trivedi MA, Wichmann AK, Torgerson BM, Ward MA, Schmitz TW, Ries ML, Koscik RL, Asthana S, Johnson SC. Structural MRI discriminates individuals with Mild Cognitive Impairment from age-matched controls: A combined neuropsychological and voxel based morphometry study. Alzheimer's & Dementia. 2006;2:296–302. doi: 10.1016/j.jalz.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, Knopman DS, Petersen RC, Jack CR., Jr MRI and CSF biomarkers in normal, MCI, and AD subjects: diagnostic discrimination and cognitive correlations. Neurology. 2009;73(4):287–293. doi: 10.1212/WNL.0b013e3181af79e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser PJ, Verhey FR, Hofman PA, Scheltens P, Jolles J. Medial temporal lobe atrophy predicts Alzheimer's disease in patients with minor cognitive impairment. Journal of Neurology, Neurosurgery & Psychiatry. 2002;72(4):491–497. doi: 10.1136/jnnp.72.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Dale AM, McEvoy LK, Brewer J, Karow DS, Salmon DP, Fennema-Notestine C. Multi-modal imaging predicts memory performance in normal aging and cognitive decline. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PN, Lirng JF, Lin KN, Chang FC, Liu HC. Prediction of Alzheimer's disease in mild cognitive impairment: a prospective study in Taiwan. Neurobiology of Aging. 2006;27(12):1797–1806. doi: 10.1016/j.neurobiolaging.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Wang PN, Liu HC, Lirng JF, Lin KN, Wu ZA. Accelerated hippocampal atrophy rates in stable and progressive amnestic mild cognitive impairment. Psychiatry Res. 2009;171(3):221–231. doi: 10.1016/j.pscychresns.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Whitwell JL, Jack CR, Jr, Pankratz VS, Parisi JE, Knopman DS, Boeve BF, Petersen RC, Dickson DW, Josephs KA. Rates of brain atrophy over time in autopsy-proven frontotemporal dementia and Alzheimer disease. Neuroimage. 2008a;39(3):1034–1040. doi: 10.1016/j.neuroimage.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Shiung MM, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr MRI patterns of atrophy associated with progression to AD in amnestic mild cognitive impairment. Neurology. 2008b;70(7):512–520. doi: 10.1212/01.wnl.0000280575.77437.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimo A, Winblad B, Aguero-Torres H, von Strauss E. The magnitude of dementia occurrence in the world. Alzheimer Dis Assoc Disord. 2003;17(2):63–67. doi: 10.1097/00002093-200304000-00002. [DOI] [PubMed] [Google Scholar]
- Xu Y, Jack CR, Jr, O'Brien PC, Kokmen E, Smith GE, Ivnik RJ, Boeve BF, Tangalos RG, Petersen RC. Usefulness of MRI measures of entorhinal cortex versus hippocampus in AD. Neurology. 2000;54(9):1760–1767. doi: 10.1212/wnl.54.9.1760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The effect sizes for selected baseline and APC values for the comparison of (a) MCI-C and HC participants, (b) MCI-S and HC participants, (c) AD and MCI-S participants, and (d) AD and MCI-C participants are shown. (n=643*; 143 AD, 57 MCI-C, 253 MCI-S, 190 HC; *30 participants removed due to failed image processing)