Abstract
Troponin T (TnT) binds to tropomyosin (Tm) to anchor the troponin complex in the thin filament, and it thus serves as a vital link in the Ca2+ regulation of striated muscle contraction. Pioneer work three decades ago determined that the T1 and T2 chymotryptic fragments of TnT each contains a Tm-binding site. A more precise localization of the two Tm-binding sites of TnT remains to be determined. In the present study, we tested serial deletion constructs of TnT and carried out monoclonal antibody competition experiments to show that the T1 region Tm-binding site involves mainly a 39 amino acids segment in the N-terminal portion of the conserved middle region of TnT. We further employed another set of TnT fragments to locate the T2 region Tm-binding site to a segment of 25 amino acids near the beginning of the T2 fragment. The localization of the two Tm-binding sites of TnT provided new information for the structure-function relationship of TnT and the anchoring of troponin complex on muscle thin filament.
Keywords: troponin T, tropomyosin, striated muscle, thin filament structure and regulation
The contraction of striated muscle is regulated by Ca2+ via the troponin complex that is composed of three protein subunits: troponin C (TnC1, the calcium-binding subunit), troponin I (TnI, the inhibitory subunit), and troponin T (TnT, the tropomyosin-binding subunit). During the activation of contraction, increased cytosolic Ca2+ binds to TnC and induces conformational changes in the troponin complex, which permits the tropomyosin-actin thin filament to activate myosin ATPase and the production of force [1]. Through binding to tropomyosin (Tm), TnT anchors the troponin complex to the thin filament and transmits the Ca2+-induced allosteric signals from troponin to Tm [2]. The interaction between TnT and Tm is essential to the function of skeletal and cardiac muscles.
Pioneer work three decades ago demonstrated that two TnT fragments generated by limited chymotrypsin or CNBr cleavage, named T1 (CB1) and T2 (amino acids 2–158 and 159–259, respectively, in rabbit fast skeletal muscle TnT), each contains a Tm-binding site [3–6]. The Tm-binding site in the T1 fragment (designated as Site 1 in the present study) was further mapped to the middle region of TnT within the CNBr fragment CB2 (amino acids 71–151 in rabbit fast skeletal muscle TnT) [7]. The T1 region site binds the head-tail junction of Tm [5,8] and the T2 region site (designated as Site 2 in the present study) binds Tm near Cys190 [9,10]. These proteolytic fragment-based mapping data have long served as a foundation for studies on TnT structure and function [2]. However, more precise locations of the two Tm-binding sites of TnT in these partial proteolysis fragments have yet to be determined in order to better understand their function in the Ca2+ regulation of muscle contraction.
Crystallography studies on cardiac and skeletal muscle troponin complex have determined high resolution 3-D structure of the globular domain of troponin including the majority of the T2 region of TnT except for the very C-terminal 14 amino acids [11,12]. The T1 region of TnT was not resolved in the crystal structures. In agreement with the results from previous protein binding studies [2,13–15], the high resolution structure of troponin revealed the TnI and TnC binding sties in the C-terminal T2 region of TnT. To date, no high-resolution structure has been determined for the Tm-binding sites of TnT.
Here we studied the locations of the two Tm-binding sites of TnT using molecular-engineered TnT fragments with precisely defined sequences and site-specific monoclonal antibody (mAb) epitope probes. The results showed that the T1 region Tm-binding Site 1 of TnT involves mainly a 39 amino acids segment in the N-terminal portion of the conserved middle region of TnT and the C-terminal region Tm-binding Site 2 is in a 25 amino acids segment near the beginning of the T2 fragment. The results provided updated information critical to the understanding of the structure-function relationship of TnT and the interaction between troponin and muscle thin filament.
Materials and Methods
Anti-TnT antibodies
The following specific antibodies were used as epitope probes in the present study:
A mouse mAb, CT3, recognizes the exon 9-encoded segment of cardiac TnT with cross-reaction to the corresponding segment of slow TnT (encoded by exon 7) but not fast TnT.
A mouse mAb, 2C8, recognizes a highly conserved epitope in all TnT isoforms in all vertebrate species tested (from hagfish to human) encoded by exon 10 of cardiac TnT, exon 8 of slow TnT and exon 11 of fast TnT genes [16].
A mouse mAb, T12, [17] (a gift from Prof. Jim J.-C. Lin, University of Iowa) recognizes an epitope in the exons 12 and 13-encoded region of fast TnT with cross-reaction to the equivalent region of cardiac TnT (encoded by exons 11 and 12) but not slow TnT [16].
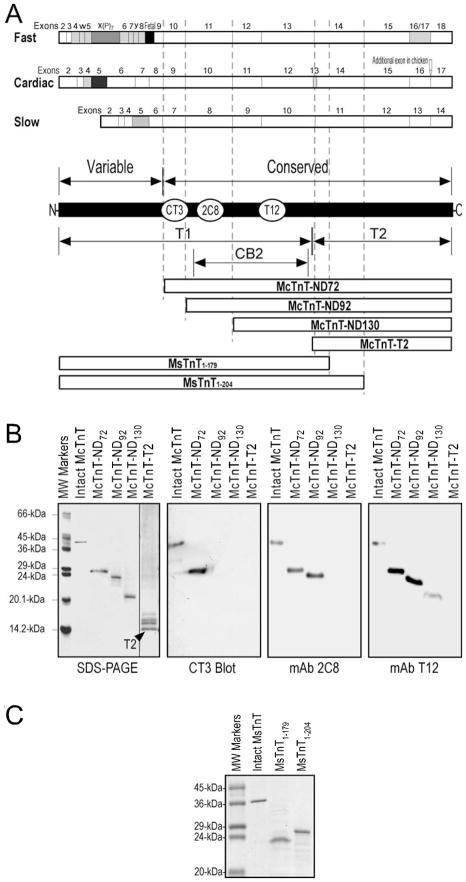
The epitope locations of these anti-TnT mAbs are indicated in Fig. 1A. The mapping of mAb CT3, 2C8 and T12 epitopes using Western blotting on a nested set of N-terminal truncated mouse cardiac TnT fragments is shown in Fig. 1B. It is important to note that the mapping of CT3 epitope using the engineered TnT fragments corrected our previous mapping error using CNBr fragments of bovine cardiac TnT that contained trace amount of partially digested products. The correct location of the CT3 epitope is in the CNBr2 fragment of bovine cardiac TnT, which was not examined in our previous study [18].
Fig. 1. Structural and functional regions of TnT and TnT fragments studied.
(A) The coding regions of the three muscle fiber type isoforms of TnT are aligned. The exon organizations are shown with alternatively spliced exons highlighted (the developmentally regulated fetal exons are in solid black). The structural differences among cardiac, fast and slow skeletal muscle TnT isoforms are mainly in the N-terminal variable region, which result in differences in exon and amino acid numbers in homologous regions of different TnT isoforms. The regions of T1, T2 and CB2 fragments are outlined along with the linear maps of the TnT isoforms and the epitope locations of three anti-TnT mAbs used as structural probes. Three N-terminal truncated cardiac TnTs with deletion of the N-terminal variable region alone (McTnT-ND72) or together with portions of the conserved middle region encoded by exons 9 and 10 (McTnT-ND92 and McTnT-ND130) and the T2 fragment of cardiac TnT (McTnT-T2) were used in the mapping of the CB2 region Tm-binding site. Two C-terminal truncated slow TnT fragments (MsTnT1–179, MsTnT1–204,) were used in the mapping of the T2 region Tm-binding site. The corresponding regions of the TnT fragments are outlined with the dashed lines. (B) The epitope specificities of mAbs CT3, 2C8 and T12 were determined by Western blotting using the N-terminal truncated cardiac TnT fragments. (C) The two representative slow TnT fragments used in the mapping of the T2 region Tm-binding site were expressed in E. coli. The SDS-gel showed intact MsTnT and the MsTnT1–179 and MsTnT1–204 fragments prepared from bacterial culture.
A polyclonal anti-TnT rabbit antiserum, RATnT, was described previously [19].
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting
Protein samples were homogenized in SDS-PAGE sample buffer containing 2% SDS, 0.3% bromphenol blue, 10% glycerol, 50 mM Tris-HCl, pH 8.8 and analyzed on gels in Laemmli buffers as described previously [16]. The resulting gels were stained with Coomassie Blue R250 to reveal the protein bands and proteins from duplicate gels were transferred to nitrocellulose membranes. The membrane was blocked with 1% bovine serum albumin (BSA) in Tris-buffered saline (TBS, 136.9 mM NaCl, 2.7 mM KCl, 25 mM Tris-HCl, pH 7.4) and incubated with anti-TnT antibodies. After high stringency washes with TBS containing 0.5% Triton X-100 and 0.05% SDS, the membranes were incubated with alkaline phosphatase-labeled anti-mouse IgG or anti-rabbit IgG second antibody (Santa Cruz Biotechnology, Santa Cruz, CA), washed again, and developed in 5-bromo-4-chloro-3-indolylphosphate and nitro blue tetrazolium substrate solution as previously described [20].
Construction, expression and purification of a C-terminal truncated mouse slow skeletal muscle TnT fragment
Using a recombinant plasmid encoding full-length mouse slow skeletal muscle TnT as template [21], a shortened cDNA was constructed to encode a C-terminal truncated slow TnT (MsTnT1–204) with the segment encoded by the last three exons (exons 12, 13 and 14) deleted. As described previously [20], the truncated cDNA was generated using PCR mutagenesis, cloned into pAED4 expression plasmid, and verified by DNA sequencing.
MsTnT1–204 protein was expressed in bacteria and purified. BL21(DE3)pLysS E. coli cells were freshly transformed with the expression plasmid and cultured in 2x tryptone-yeast broth (16 g/L Tryptone, 10 g/L yeast extract, 5 g/L NaCl, 1.32 g/L Na2HPO4, pH 7.3) containing 100 μg/mL ampicillin and 25 μg/mL chloramphenicol at 37°C with vigorous shaking. The culture was induced at early log-phase of growth with 0.4 mM isopropyl-1-thiol-β-D-galactoside. After 3 additional hours of culture, the bacterial cells were harvested by centrifugation. Performed at 4°C or on ice, the bacterial pellet was suspended in 2.5 mM EDTA, 50 mM Tris-HCl, pH 8.0, 0.5 mM phenylmethylsulphonyl fluoride (PMSF) and lyzed by three passes through a French Press.
The bacterial lysate was fractionated by ammonium sulfate precipitation between 30–55% saturation, dialyzed and fractionated on a DE52 anion exchange column in 6 M urea at pH 8.0. The MsTnT1–204 peak identified by SDS-PAGE and Western blotting using mAb CT3 was dialyzed and concentrated by lyophilization for further purification on a Sephadex G-75 gel filtration column at pH 7.0 in the presence of 6 M urea, 0.5 M KCl and 0.1 mM EDTA. The MsTnT1–204 peak was identified by SDS-PAGE, dialyzed to remove urea and salt, and lyophilized.
Other TnT and TnT fragments
Intact and N-terminal truncated fragments of mouse cardiac TnT were expressed from cloned cDNA in bacterial cultures and purified as described previously [22]. Among these deletion constructs, fragment McTnT-ND72 had only the N-terminal hypervariable region selectively removed. Fragments McTnT-ND92 and McTnT-ND130 further had portions of the conserved middle region encoded by exons 9 and 10 removed [23]. An engineered T2 fragment of mouse cardiac TnT (McTnT-T2) was also expressed in bacteria and purified as described previously [16].
Intact mouse slow skeletal muscle TnT and its fragment MsTnT1–179 with a C-terminal truncation mimicking the nonsense mutation found in Amish nemaline myopathy were expressed in bacteria and purified as described previously [20].
Microtiter plate protein binding assays
Enzyme-linked immunosorbant assay (ELISA)-based solid phase protein binding experiments [16,19,20,22] were performed to examine the binding of the multiple TnT fragments to Tm. Purified TnT, TnT fragments or BSA control was dissolved in Buffer A (100 mM KCl, 3 mM MgCl2, 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 7.0) at 5 μg/mL to coat 96-well microtiter plates at 100 μL/well at 4°C overnight. Free TnT or TnT fragment was removed and the plate was washed with Buffer T (Buffer A plus 0.05% Tween 20) for three times over a 10 min period to block the plastic surface. Serial dilutions of bovine skeletal muscle Tm purified as previously described [24] in Buffer T containing 0.1% BSA were added to the plates at 100 μL/well and incubated at room temperature for 2 h. The plates were washed three times with Buffer T and the amount of Tm bound to the immobilized TnT in each well was quantified using an anti-Tm mAb CH1 [25] (a gift from Prof. Jim J.-C. Lin, University of Iowa) via standard ELISA procedure using horse radish peroxidase (HRP)-labeled second antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and H2O2-ABTS (2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) substrate reaction. The A415nm values in the linear course of the color development were monitored for each assay well using a BioRab Benchmark automated microplate reader and recorded to construct Tm-binding curves for each of the TnT and TnT fragments. The experiments were done in triplicate wells and repeated.
ELISA competition assay
To examine the spatial relationship between the TnT epitopes recognized by mAbs CT3, 2C8 or T12 and the T1 region Tm-binding site, purified Tm at 2 μg/mL in Buffer A was coated on microtiter plates as above. After washes with Buffer T to remove free Tm, the plate was incubated with mouse cardiac TnT [22] or mouse fast skeletal muscle TnT (α isoform) [16]), both at concentrations predetermined to produce an A415nm value between 0.25–0.50 under the ELISA conditions and mixed with serial dilutions of mAb CT3 (for the cardiac TnT experiments), 2C8 or T12 (for the fast TnT experiments) hybridoma ascites fluid. The bindings of cardiac TnT to immobilized Tm in the presence of mAb CT3 and fast TnT in the presence of mAb 2C8 or T12 and were determined with ELISA as above using polyclonal rabbit anti-TnT antibody RATnT [19] and HRP-labeled anti-rabbit IgG second antibody (Sigma Life Science, St Louise, MO) that had no cross-reaction with the mouse mAb probes. Competitive Tm-binding curves were constructed by normalization to the level of Tm-binding in the absence of blocking mAb. The experiments were done in triplicate wells and repeated.
ELISA mAb affinity titration
The affinity of mAb 2C8 to cardiac, slow and fast skeletal muscle TnT isoforms was compared using ELISA titration. As described previously [16], purified mouse fast, slow and cardiac TnT’s were coated on 96-well microtitering plate in Buffer A and incubated with serial dilutions of mAb 2C8 mouse ascites in Buffer T containing 0.1% BSA. After washes with Buffer T, HRP-anti-mouse IgG second antibody incubation, and H2O2-ABTS substrate reaction, A405nm values were recorded to plot mAb 2C8 titration curves and compare the relative affinities for the three TnT isoforms. The experiments were done in triplicate wells and repeated.
Data analysis
Statistical analyses for the ELISA affinity titration, protein binding and competition assays were performed using Student’s t test. All values are presented as mean ± SD.
Results
Localization of the T1 region Tm-binding site of TnT
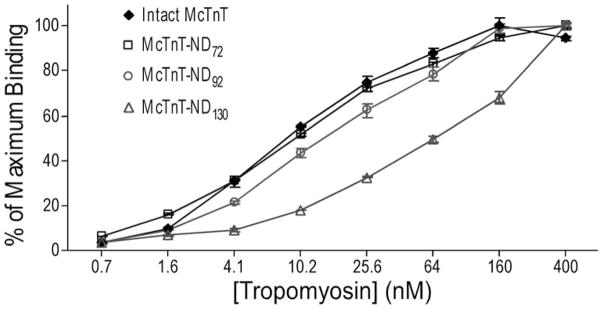
We first mapped the T1 region Tm-binding site by testing a set of N-terminal truncated cardiac TnT fragments (Fig. 1) in ELISA protein binding experiments. The results in Fig. 2 demonstrated that the deletion of only the N-terminal variable region did not significantly affect Tm-binding. Further deletion of the 20 amino acids encoded by exon 9 in the conserved region produced a detectable decrease in the affinity to Tm with statistical significance [22]. The rather moderate destruction is consistent with the previous observation that the T1 region Tm-binding site is in the middle CB2 region (Fig. 1A) [7]. In contrast, deletion of the next 39 amino acids encoded by exon 10 resulted in a major decrease in Tm-binding affinity.
Fig. 2. Binding of N-terminal truncated TnT fragments to Tm.
N-terminal serially truncated cardiac TnT fragments (Fig. 1) were tested for Tm-binding affinity in ELISA protein binding experiments. The results showed that the deletion of the N-terminal variable region (McTnT-ND72) alone did not significantly affect Tm-binding whereas further deletion of 20 amino acids in the conserved middle region (McTnT-ND92) produced a detectable decrease in Tm-binding affinity. Extending the deletion to remove 39 more amino acids encoded by exon 10 (McTnT-ND130) resulted in a major decrease in Tm-binding affinity.
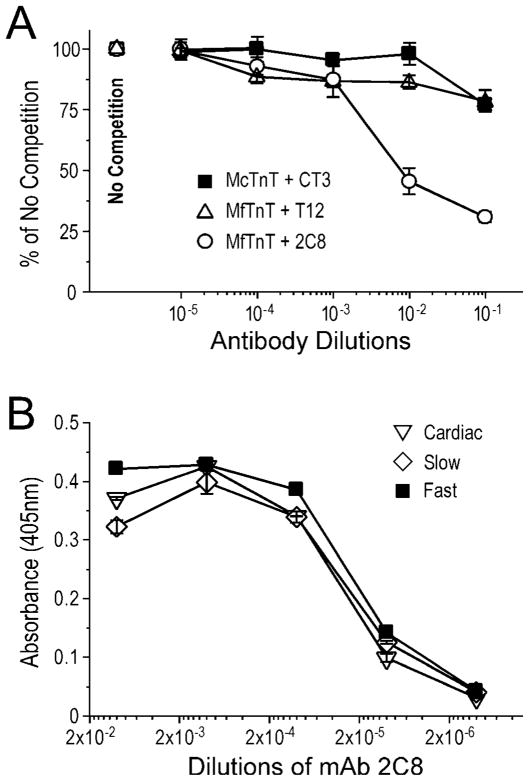
To further map the location of the middle region Tm-binding site of TnT, we applied mAbs that recognize specific regions of TnT as structural probes in competitive Tm-binding experiments. mAb 2C8 recognizing the exon 10-encoded region had a significant blocking effect on Tm-binding (Fig. 3A). In contrast, mAbs CT3 and T12 recognizing the flanking regions (Fig. 1) had only marginally detectable effects. These results are in good agreement with the small decrease in Tm-binding when deleting the exon 9-encoded segment and the large decrease when extending the deletion to remove the exon 10-encoded segment (Fig. 2).
Fig. 3. mAb competition/blocking mapping of the T1 region Tm-binding site of TnT.
(A) The competition curves showed that mAb 2C8 significantly blocked the binding of TnT to Tm (P<0.01). In contrast, mAbs CT3 and T12 recognizing the flanking regions (Fig. 1) had only marginal effects. (B) The ELISA titration curves showed that mAb 2C8 raised against mouse cardiac TnT binds fast, slow and cardiac TnT isoforms with very similar affinities, indicating a highly conserved epitope structure with conserved function. Data are plotted as mean ± SD.
The ELISA titration curves in Fig. 3B demonstrated that the 2C8 epitope is a highly conserved three-dimensional structure among the three diverged TnT isoforms. The results showed that mAb 2C8 raised by immunization with cardiac TnT had nearly identical affinities to fast, slow and cardiac isoforms of TnT. In contrast, previous data showed that mAb CT3 recognizes cardiac TnT and slow TnT but not fast TnT whereas mAb T12 recognizes fast TnT and cardiac TnT but not slow TnT [16]. Therefore, the three-dimensional structure of 2C8 epitope is more conserved than the flanking regions (Fig. 1A), supporting the notion that this segment corresponds to a structure of TnT with evolutionarily conserved function such as the binding to Tm.
Localization of the T2 region Tm-binding site of TnT
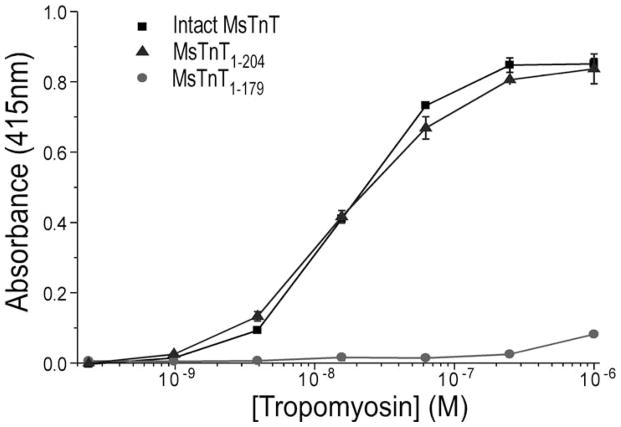
To localize the T2 region Tm-binding site of TnT, we examined another set of TnT fragments in ELISA solid phase protein binding experiments. Fig. 4 shows the Tm-binding curves of intact slow TnT (MsTnT), a mutant slow TnT that causes a recessive nemaline myopathy (MsTnT1–179), and a slow TnT with the exons 12–14 region truncated (MsTnT1–204) (Fig. 1A). In agreement with previous observation [20], MsTnT1–179 containing the T1 region Tm-binding site showed rather low affinity for Tm. This feature allows an easy mapping of the T2 region Tm-binding site using the slow TnT constructs. The results showed that MsTnT1–204 exhibited high affinity Tm-binding, as strong as that of intact MsTnT. The fact that MsTnT1–204 retained the full avidity of Tm binding whereas MsTnT1–179 did not indicates that the T2 region Tm-binding site lies in the segment encoded by exon 11 of slow TnT corresponding to the exon 14-encoded segment of fast and cardiac TnTs [23].
Fig. 4. Mapping of the T2 region Tm-binding site.
Intact MsTnT, MsTnT1–179, and MsTnT1–204 fragments (Fig. 1) were examined for Tm-binding in ELISA protein binding experiments. The binding curves showed that whereas MsTnT1–179, containing the T1 region Tm-binding site had rather weak binding to Tm (P<0.01), MsTnT1–204 exhibited high affinity binding to Tm similar to that of intact MsTnT. Data are plotted as mean ± SD. In contrast to the normalized binding curves of the N-terminal truncated cardiac TnT fragments shown in Fig. 2 based on their similar maximum binding to Tm, the binding curves here were plotted from actual absorbance to demonstrate the significantly lower maximum Tm-binding of MsTnT1–179 than that of intact MsTnT and MsTnT1–204 in the solid phase binding assay.
Highly conserved primary structure of the two Tm-binding sites of TnT
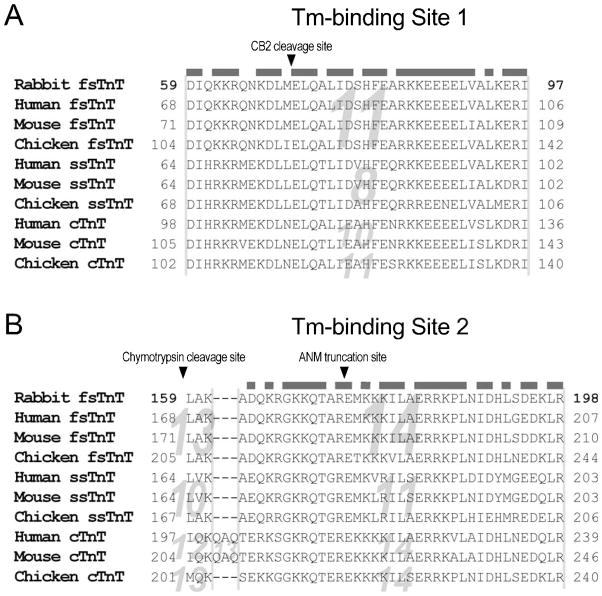
Our data from protein binding and mAb competition experiments determined that the T1 region Tm-binding site mainly depends on the segment encoded by exon 10 of mouse cardiac TnT. Since the hypervariable nature of TnT N-terminal region, the homologous exon encoding this segment is numbered differently in fast TnT (exon 11), slow TnT (exon 8) and chicken cardiac TnT (exons 11) genes (Fig. 1A). This exon encodes amino acids 105–143 of mouse cardiac TnT (92–130 in the adult low molecular weight isoform [22]) corresponding to amino acids 59–97 in rabbit fast skeletal muscle TnT used in the previous mapping of Tm-binding sites [2]. Considering Tm-binding Site 1 was previously mapped in the CB2 fragment of rabbit fast TnT [7], it would mainly consist of amino acids 117–143 in mouse cardiac TnT (amino acids 72–97 in rabbit fast TnT) (Fig. 5A).
Fig. 5. Evolutionarily conserved primary structure of the two Tm-binding sites of TnT.
(A) Amino acid sequences of mammalian and avian fast, slow and cardiac TnT isoforms are aligned to demonstrate the highly conserved primary structure in the T1/CB2 region Tm-binding site (Site 1). The beginning of the CB2 region is indicated with an arrowhead. (B) Amino acid alignment demonstrates the highly conserved primary structure in the T2 region Tm-binding site (Site 2). The beginning of the T2 region and the truncation site of Amish nemaline myopathy slow TnT are indicated with arrowheads. The homologous exons encoding these segments are indicated in the background. Identical or conserved residues encoded by the two Tm-binding sites’ main coding exons are outlined with the grey bars above the sequences. The amino acid numbers of rabbit fast TnT match that in the previous Tm-binding studies [2]. The amino acid numbers for all other TnT isoforms were deduced from the longest splice forms [23]. The variable amino acid start positions and different exon numbers reflect the variation of the N-terminal region of the TnT isoforms (Fig. 1A). Met1 was not counted in this figure to match the earlier studies using rabbit fast skeletal muscle TnT [2].
Also due to the N-terminal hypervariable region, the T2 region Tm-binding site localized in the exon 11-encoded segment of slow TnT (amino acids 179–203, 180–204 when Met1 is included) corresponds to the exon 14-encoded segment of fast TnT or cardiac TnT (Fig. 1A), i.e., amino acids 174–198 in rabbit fast skeletal muscle TnT (Fig. 5B).
Alignment of amino acid sequences of the two regions of fast, slow and cardiac isoforms of TnT from avian and mammalian species demonstrated highly conserved primary structures (Fig. 5). 76.9% of the residues encoded by the main coding exon for Site 1 and 70.3% of the residues encoded by the main coding exon for Site 2 are identical or conserved among the sequences analyzed. The structural conservation during evolution supports conserved fundamental functions in these two regions of TnT, such as the interaction with Tm. The structural conservation also justified our experimental strategy of using fragments of cardiac and slow TnTs as representatives to determine the Tm-binding sites of TnT.
Discussion
Previous studies have determined that TnT is an elongated molecule of which the C-terminal T2 domain interacts with TnI and TnC whereas the middle region and the N-terminal variable region form the asymmetric tail portion of the troponin complex [2,26,27]. This extended molecular conformation of TnT spatially separates the two Tm-binding sites in the C-terminal and middle regions to allow differentiated functions.
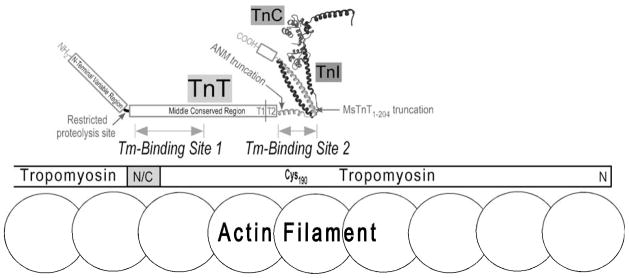
It has been observed that the N-terminal CB3 fragment does not contain any known protein binding sites [2,7] and its deletion would not destroy the function of troponin in vitro [28–30] and in vivo [31]. Consistent with these results, we recently demonstrated that a stress-induced selective proteolysis cleavage of the entire hypervariable region corresponding to the N-terminal 71 amino acids in mouse cardiac TnT [32] did not destruct the middle region Tm-binding site (Fig. 6).
Fig. 6. The two Tm-binding sites of TnT in troponin-tropomyosin assembly.
This proposed model illustrates that the CB2 region Tm-binding site of TnT (Site 1) binds the head-tail (N/C) junction of Tm (indicated by the gray box) in striated muscle thin filament, whereas the T2 region Tm-binding site of TnT (Site 2) binds Tm near Cys190. The high resolution structure of partial troponin complex determined by crystallography is redrawn from published data [11,12]. The boundary between the T1 and T2 regions is indicated with a vertical line. The proteolysis site in cardiac TnT that selectively removes the N-terminal variable region [32], the position of the Amish nemaline myopathy (ANM) nonsense mutation [20], and the position of the MsTnT1–204 truncation (Fig. 1A) are indicated with the arrows.
The N-terminal side boundary of the Tm-binding Site 1 was suggested by the previous observation that the CNBr segment CB2 of rabbit fast TnT bound Tm [7]. To map the C-terminal side boundary of the T1/CB2 region Tm-binding site of TnT, mAb competition experiments demonstrated that mAb 2C8 recognizing the exon 10-encoded segment had a significant blocking effect on Tm-binding whereas mAb T12 recognizing the downstream portion of CB2 segment had only a small effect. Therefore, the data suggest that the Tm-binding Site 1 is located in the N-terminal portion of the CB2 fragment (Fig. 5A). Consistently, a previous study demonstrated that deletion of 25 amino acids in the exon 10-encoded segment resulted in a sharp drop of Tm-binding affinity [33]. Together with the highly conserved amino acid sequence and epitope structure, this 39 amino acids segment encoded by a single exon and predicted with α-helical structure forms the core of the Tm-binding Site 1 of TnT.
A previous study reported that deletion of a C-terminal segment of fast TnT encoded by exon 18, the mutually exclusive exon 17 or 16, plus a part of exon 15 did not abolish Tm-binding [34]. The segment encoded by exon 15 of fast TnT gene (homologous to exon 12 of slow TnT gene) binds TnI in the I-T arm of troponin complex as determined in the crystal structures [11,12]. On the other hand, truncating slow TnT in the middle of the exon 11-encoded segment as that produced by the nonsense mutation in Amish nemaline myopathy abolished high affinity binding to Tm [20]. Therefore, the T2 region Tm-binding Site 2 of TnT would most likely be in the segment encoded by exon 11 of slow TnT corresponding to the exon 14 region of cardiac and fast TnT’s. Our protein binding experiments using exon unit-based deletion constructs of TnT showed that this is indeed the case. While slow TnT with the exons 12–14 region-deleted (MsTnT1–204) retained strong Tm-binding affinity comparable to that of intact TnT, truncation of slow TnT in the middle of exon 11-encoded segment (MsTnT1–179 [20]) severely decreased the binding affinity to Tm and, therefore, removed or destroyed the T2 region Tm-binding site (Fig. 4). Altogether, the data indicate that the 25 amino acids segment (i.e., residues 180–204 in slow TnT, Fig. 5B) in the beginning of the T2 fragment forms a critical portion of Tm-binding Site 2 of TnT.
Deduced from results of the present study and literature data, the proposed locations of the two Tm-binding sites of TnT in muscle thin filament are illustrated in Fig. 6. the Tm-binding Site 2 in the T2 region that binds Tm near Cys190 [9,10] was partially included in the reported crystal structure of troponin [11,12]. These data provide updated information for the anchoring of troponin on the Tm-actin filament in striated muscle and their interactions during Ca2+-regulated contraction and relaxation.
Acknowledgments
We thank Hui Wang for technical assistance and Dr. Jim Lin for the T12 and CH1 mAbs. Stephen Chong participated in the study when this research group was at Northwestern University. This study was supported by grants from the National Institutes of Health (AR-048816 and HL-078773 to J-PJ).
Footnotes
Abbreviations used: TnC, troponin C; BSA, bovine serum albumin; ELISA, enzyme-linked immunosorbant assay; McTnT, mouse cardiac TnT; MsTnT, mouse slow skeletal muscle TnT; ND72, deletion of the N-terminal 71 amino acids; ND91, deletion of the N-terminal 90 amino acids; ND130, deletion of the N-terminal 129 amino acids; SDS-PAGE, SDS-polyacrylamide gel electrophoresis; TBS, Tris-buffered saline; Tm, tropomyosin; TnI, troponin I; TnT, troponin T
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gordon AM, Homsher E, Regnier M. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 2.Perry SV. J Muscle Res Cell Motil. 1998;19:575–602. doi: 10.1023/a:1005397501968. [DOI] [PubMed] [Google Scholar]
- 3.Jackson P, Amphlett GW, Perry SV. Biochem J. 1975;151:85–97. doi: 10.1042/bj1510085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearlstone JR, Smillie LB. Can J Biochem. 1977;55:1032–1038. doi: 10.1139/o77-154. [DOI] [PubMed] [Google Scholar]
- 5.Mak AS, Smillie LB. J Mol Biol. 1981;149:541–550. doi: 10.1016/0022-2836(81)90486-1. [DOI] [PubMed] [Google Scholar]
- 6.Tanokura M, Tawada Y, Ono A, Ohtsuki I. J Biochem. 1983;93:331–337. doi: 10.1093/oxfordjournals.jbchem.a134185. [DOI] [PubMed] [Google Scholar]
- 7.Pearlstone JR, Smillie LB. J Biol Chem. 1982;257:10587–10592. [PubMed] [Google Scholar]
- 8.Pato MD, Mak AS, Smillie LB. J Biol Chem. 1981;256:602–607. [PubMed] [Google Scholar]
- 9.Chong PC, Hodges RS. J Biol Chem. 1982;257:9152–9160. [PubMed] [Google Scholar]
- 10.Morris EP, Lehrer SS. Biochemistry. 1984;23:2214–2220. doi: 10.1021/bi00305a018. [DOI] [PubMed] [Google Scholar]
- 11.Takeda S, Yamashita A, Maeda K, Maeda Y. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 12.Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ. Proc Natl Acad Sci U S A. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heeley DH, Golosinska K, Smillie LB. J Biol Chem. 1987;262:9971–9978. [PubMed] [Google Scholar]
- 14.Ishii Y, Lehrer SS. J Biol Chem. 1991;266:6894–6903. [PubMed] [Google Scholar]
- 15.Schaertl S, Lehrer SS, Geeves MA. Biochemistry. 1995;34:15890–15894. doi: 10.1021/bi00049a003. [DOI] [PubMed] [Google Scholar]
- 16.Chong SM, Jin JP. J Mol Evol. 2009;68:448–460. doi: 10.1007/s00239-009-9202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin JP, Lin JJC. J Biol Chem. 1988;263:7309–7315. [PubMed] [Google Scholar]
- 18.Jin JP, Chen A, Ogut O, Huang QQ. Am J Physiol: Cell Physiol. 2000;279:1067–1077. doi: 10.1152/ajpcell.2000.279.4.C1067. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Jin JP. Biochemistry. 1998;37:14519–14528. doi: 10.1021/bi9812322. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Huang QQ, Breckenridge MT, Chen A, Crawford TO, Morton DH, Jin JP. J Biol Chem. 2005;280:13241–13249. doi: 10.1074/jbc.M413696200. [DOI] [PubMed] [Google Scholar]
- 21.Jin JP, Chen A, Huang QQ. Gene. 1998;214:121–129. doi: 10.1016/s0378-1119(98)00214-5. [DOI] [PubMed] [Google Scholar]
- 22.Biesiadecki BJ, Chong SM, Nosek TM, Jin JP. Biochemistry. 2007;46:1368–1379. doi: 10.1021/bi061949m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin JP, Zhang Z, Bautista JA. Crit Rev Eukar Gene Expr. 2008;18:93–124. doi: 10.1615/critreveukargeneexpr.v18.i2.10. [DOI] [PubMed] [Google Scholar]
- 24.Smillie LB. Methods Enzymol. 1982;85:234–241. doi: 10.1016/0076-6879(82)85023-4. [DOI] [PubMed] [Google Scholar]
- 25.Lin JJC, Chou CS, Lin JLC. Hybridoma. 1985;4:223–242. doi: 10.1089/hyb.1985.4.223. [DOI] [PubMed] [Google Scholar]
- 26.Cabral-Lilly D, Tobacman LS, Mehegan JP, Cohen C. Biophys J. 1997;73:1763–1770. doi: 10.1016/S0006-3495(97)78206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wendt T, Guenebaut V, Leonard KR. J Struct Biol. 1997;118:1–8. doi: 10.1006/jsbi.1996.3834. [DOI] [PubMed] [Google Scholar]
- 28.Pan BS, Gordon AM, Potter JD. J Biol Chem. 1991;266:12432–12438. [PubMed] [Google Scholar]
- 29.Fisher D, Wang G, Tobacman LS. J Biol Chem. 1995;270:25455–25460. doi: 10.1074/jbc.270.43.25455. [DOI] [PubMed] [Google Scholar]
- 30.Chandra M, Montgomery DE, Kim JJ, Solaro RJ. J Mol Cell Cardiol. 1999;31:867–880. doi: 10.1006/jmcc.1999.0928. [DOI] [PubMed] [Google Scholar]
- 31.Feng HZ, Biesiadecki BJ, Yu ZB, Hossain MM, Jin JP. J Physiol (London) 2008;586:3537–3550. doi: 10.1113/jphysiol.2008.153577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Biesiadecki BJ, Jin JP. Biochemistry. 2006;45:11681–11694. doi: 10.1021/bi060273s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinkle A, Goranson A, Butters CA, Tobacman LS. J Biol Chem. 1999;274:7157–7164. doi: 10.1074/jbc.274.11.7157. [DOI] [PubMed] [Google Scholar]
- 34.Jha PK, Leavis PC, Sarkar S. Biochemistry. 1996;35:16573–16580. doi: 10.1021/bi9622433. [DOI] [PubMed] [Google Scholar]