Abstract
Chondroid lipoma, a rare benign adipose tissue tumor, may histologically resemble myxoid liposarcoma or extraskeletal myxoid chondrosarcoma, but is genetically distinct. In the current study, an identical reciprocal translocation, t(11;16)(q13;p13) was identified in three chondroid lipomas, a finding consistent with previous isolated reports. A fluorescence in situ hybridization (FISH)-based positional cloning strategy using a series of bacterial artificial chromosome (BAC) probe combinations designed to narrow the 16p13 breakpoint revealed MKL2 as the candidate gene. Subsequent 5′ RACE studies demonstrated C11orf95 as the MKL2 fusion gene partner. MKL/myocardin-like 2 (MKL2) encodes myocardin-related transcription factor B (MRTF-B) in a megakaryoblastic leukemia gene family, and C11orf95 (chromosome 11 open reading frame 95) is a hypothetical protein. Sequencing analysis of RT-PCR generated transcripts from all three chondroid lipomas defined the fusion as occurring between exons 5 and 9 of C11orf95 and MKL2, respectively. Dual-color breakpoint spanning probe sets custom-designed for recognition of the translocation event in interphase cells confirmed the anticipated rearrangements of the C11orf95 and MKL2 loci in all cases. The FISH and RT-PCR assays developed in this study can serve as diagnostic adjuncts for identification of this novel C11orf95-MKL2 fusion oncogene in chondroid lipoma.
INTRODUCTION
Chondroid lipoma is a benign adipose tissue tumor composed of nests and cords of mature adipocytes and uni- or multivacuolated cells embedded in a prominent myxohyaline matrix (Kindblom et al., 2002). This neoplasm is rare and primarily occurs in the proximal extremities and limb girdles of middle-aged adults, particularly females (Logan et al., 1996). Chondroid lipoma can be mistaken for a sarcoma, especially myxoid liposarcoma or extraskeletal myxoid chondrosarcoma (Meis and Enzinger, 1993; Weiss and Goldblum, 2008). Chondroid lipoma does not recur locally or metastasize (Mentzel and Fletcher, 1998).
Cytogenetic studies of solitary ordinary lipoma are abundant and have revealed rearrangements of 12q13-15 and the underlying chromatin remodeling gene HMGA2 gene as most common (Sandberg, 2004). In striking contrast, karyotypic analyses of chondroid lipoma are few, although a recurrent translocation t(11;16)(q13;p13) has been observed in isolated cases (Gisselsson et al., 1999a; Thomson et al., 1999; Ballaux et al., 2004).
In the current study, cytogenetic analysis of three chondroid lipomas revealed an identical t(11;16)(q13;p13) in all three cases. We hypothesized that this primary translocation generates a fusion gene of central pathogenic importance in chondroid lipoma. The objectives of our study were to: 1) further refine the localization of the 11q13 and 16p13 chromosomal breakpoints; 2) uncover the underlying involved genes; and, 3) establish new molecular diagnostic assays for this rare entity to aid in its distinction from more aggressive, histopathologically similar neoplasms.
MATERIALS AND METHODS
Case History
The clinicohistopathologic features of the patients and corresponding tumors are listed in Table 1. The histopathologic diagnoses were established in accordance with the World Health Organization classification (Kindblom et al., 2002).
Table 1.
Clinicopathologic and Cytogenetic Data
| Case# | Age(years)/Sex | Location (Size in cm) | Karyotype | Reference |
|---|---|---|---|---|
| 1 | 74/F | Left calf (3.3 × 1.9 × 1.2) | 46,XX,t(6;12)(p21;q13),t(11;16)(q13;p13.2)[23]/47,idem,+r, 0-5dmin[8]/46,XX[11] | Present study |
| 2 | 47/F | Left thigh (10.0 × 8.0 × 8.0) | 46,XX,t(11;16)(q13;p13)[13]/46,XX[4] | Present study |
| 3 | 33/M | Left thigh (11.5 × 8.5 × 5.5) | 46,XY,t(11;16)(q13;p13)[5]/46,XY[10] | Present study |
| 4 | 46/M | Left thigh (5.0 × 3.5 × 2.5) | 46,XY,t(11;16)(q13;p12-13) | Thomson et al., 1999 |
| 5 | 21/M | Left calf (7.0 × 7.0 × 7.0) | 46,XY,t(1;2;5)(q32;q37;q31),t(11;16)(q13;p13)[24]/46,XY[1] | Gisselsson et al., 1999a |
| 6 | 72/F | Left paravertebral region (5.0 × 3.1 × 2.5) | 46,XX,t(11;16)(q13;p13)[20]/47-48,XX,idem,+2-3r[cp2] | Ballaux et al., 2004 |
Cytogenetic Analysis
A representative fresh tissue sample from each case was received for conventional cytogenetic analysis. Culturing, harvesting and preparation of slides were performed as previously described (Nishio et al., 2006). Briefly, the tissues were disassociated mechanically and enzymatically and cultured in RPMI 1640 supplemented with 20% fetal bovine serum for 3–8 days. Cultured cells received an overnight exposure to Colcemid (0.02ug/ml). Following hypotonic treatment (0.8% sodium citrate for 20 min), the cells were fixed three times with methanol:glacial acetic acid (3:1). Chromosome analysis was performed on GTG-banded (Giemsa/trypsin) metaphases, and the karyotypes were expressed according to the International System for Human Cytogenetic Nomenclature 2009 (ISCN, 2009).
Fluorescence In Situ Hybridization (FISH)
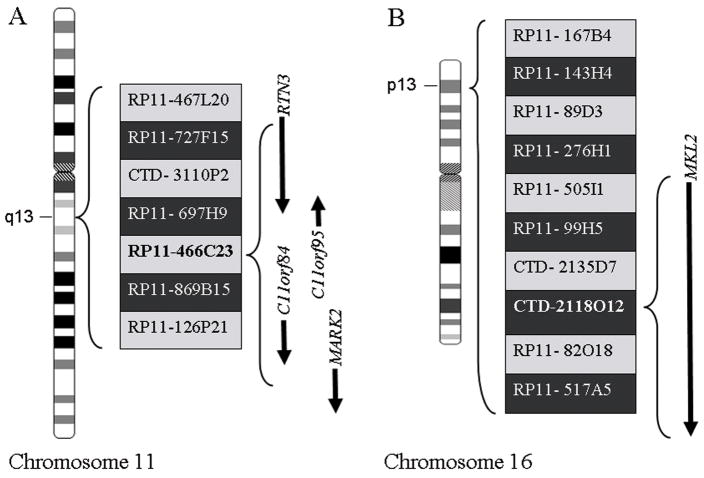
A FISH-based positional cloning strategy using a series of bacterial artificial chromosome (BAC) probe combinations on chondroid lipoma abnormal metaphase cells from case 1 was employed to narrow the derivative chromosomes 11 and 16 breakpoints. All BACs mapped to 11q13 and 16p13 were identified from the NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/mapview) and the Ensembl Genome Browser (http://www.ensembl.org), and were obtained from Invitrogen (Invitrogen, Carlsbad, CA). The 11q13 BACs included: RP11-467L20, RP11-727F15, CTD- 3110P2, RP11-697H9, RP11-466C23, RP11-869B15, and RP11-126P21; and, the 16p13 BACs included: RP11- 167B4, RP11- 143H4, RP11- 89D3, RP11- 276H1, RP11- 505I1, RP11-99H5, CTD- 2135D7, CTD- 2118O12, RP11- 82O18, and RP11- 517A5.
Because the BAC FISH studies revealed that the chromosome 16p13 breakpoint was occurring within clone CTD-2118O12 containing a single candidate gene (MKL2), two long-range PCR products, MKL2/913-25107 (24195 bp) and MKL2/175221-194480 (19279 bp), corresponding to a normal control genomic MKL2 gene 5′ and 3′ region respectively, were constructed by amplification with Expand Long Range, dNTPack (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions to further confirm and define the rearrangement of this gene. MKL2 long-range PCR primer sets included 5′-TGCCCT AACAGACAACCCTCAATCG-3′/5′-CCCTGGAATGAGGAGGACAGCC-3′ for MKL2/913-25107, and 5′-CCAGGAAGTGAACAGCAGCG-3′/5′-TGATTCCCGA CAGCACCTTG-3′ for MKL2/175221-194480. The PCR conditions were as follows: annealing temperature 57°C, elongation at 68°C for 20 min for the first ten cycles and 5 seconds more at each subsequent cycle for a total of 35 cycles. The generated 24 kb and 19 kb genomic fragments were subsequently purified using High Pure PCR Product Purification Kit (Roche Applied Science, Indianapolis, IN) according to the supplier’s instructions.
Each BAC and long-range PCR product probe was directly labeled by nick translation with either Spectrum Green- or Spectrum Orange-dUTP per the manufacturer’s protocol (Abbott Molecular, Inc., Des Plaines, IL). An amount of 3ug of DNA for each probe or 1.5 ug for each of two probes were combined. All nick translation reagents were then multiplied by the total ug of DNA used in the cocktail. Amounts of 200 ng of each probe were hybridized to the target DNA and blocked with approximately 15 fold excess of a combination of Human Cot-I DNA (Invitrogen, Carlsbad, CA) and human placental DNA.
Prior to hybridization, slides were pretreated at 72°C in 2×SSC for 2 min and at 37°C in protease solution [25 mg of protease in 50 ml of protease solution (Abbott Molecular, Inc., Des Plaines, IL)] for 3 min, washed in 1×PBS at room temperature for 5 min, post-fixed in 1% formaldehyde at room temperature for 5 min, and again washed in 1×PBS at room temperature for 5 min. The slides were then dehydrated in gradient ethanol (75, 85, and 100%) at room temperature for 2 min each and air-dried. Following pretreatment, the cells and probes were co-denatured at 75°C for 1 min and incubated overnight at 37°C using the HYBrite™ system (Abbott Molecular, Inc., Des Plaines, IL). Post-hybridization washing was performed in 0.4×SSC/0.3% NP-40 at 72°C for 2 min, followed by 2×SSC/0.1% NP-40 at room temperature for 1 min. The slides were then counterstained with DAPI II (Abbott Molecular, Inc., Des Plaines, IL).
Hybridization signals were assessed in 10 metaphase cells exhibiting the t(11;16)(q13;p13) (case 1) or in 200 interphase nuclei obtained from cytologic touch preparations of lesional tissue (cases 1, 2 and 3) with strong, well-delineated signals by two different individuals. An interphase cell specimen was interpreted as abnormal if a split of the flanking probe or a fusion of the spanning probe signals was detected in >20% of the cells evaluated (more than two standard deviations above the average false positive rate). As additional controls, each probe set was also hybridized to metaphase cell preparations of karyotypically normal peripheral blood lymphocytes to confirm correct mapping, optimal signal strength, and lack of cross-hybridization, before proceeding with analysis of the patient samples. Images were acquired using the Cytovision Image Analysis System (Applied Imaging, Santa Clara, CA, USA).
Rapid Amplification of cDNA Ends (RACE) Analysis and RT-PCR Analysis
Following the identification of the involvement of the MKL2 gene, 5′RACE was employed to determine the MKL2 translocation gene partner. Total RNA was extracted from 100mg of snap frozen tumor of case 1 in the presence of TRIzol®Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and further purified using RNeasy kit (Qiagen, Valencia, CA). The integrity of the RNA was confirmed on the Bioanalyzer 2100 system (Agilent Technologies, Wilmington, DE). 5′ RACE was performed using the SMART™ RACE cDNA Amplification Kit (Clontech Laboratories, Inc., Mountain View, CA) following the protocol provided by the supplier. First strand cDNA was prepared using 1 ug of total RNA in the presence of 5′-CDS primer and SMART II A oligo using Murine Leukemia Virus Reverse Transcriptase to allow for isolation of the complete 5′ sequence of the target fusion transcript. Three 5′-RACE PCR amplifications were achieved by using the Universal primer A Mix (UPM) and three MKL2 gene specific primers MKL2/1509-1485 (5′-ATGATGACACTGCCACGATGCCCCC-3′), MKL2/3130-3109 (5′-GCAGCAAACTGGGTCTCGGAAG-3′), and MKL2/4148-4120 (5′-GGATACCCATTTCCTTACCCCAGCGTAGG-3′). Subsequently, nested PCR was performed using nested UPM and primers MKL2/1509-1485 and MKL2/3130-3109. A diluted initial 5′-RACE PCR (MKL2/4148-4120) product was used as a template. The nested PCR products were gel purified using High Pure PCR Product Purification Kit (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions and then sequenced.
Following identification of C11orf95 as the chromosome 11 translocation partner gene, RT-PCR analysis was performed on all three cases to confirm the presence of the C11orf95-MKL2 fusion transcript. Total RNA was extracted from 40mg of case 2 and 1×105 cultured cells of case 3 using the same method described above. Two sets of C11orf95-MKL2 primers were designed (5′-CCTCGCTCAAGATGAGCACCATCG-3′/5′-ATGATGACACTGCCACGATGCC-3′ and 5′-CGCCTCGGCGGGCCTGTCC-3′/5′-CATTTTCAGCACTTCCACGAGC-3′) to amplify fusion transcript products of 640 bp and 1028 bp in size respectively. PCR was optimized with an initial denaturation step at 94°C for 2 min, followed by the first 10 cycles at 94°C for 15 sec, 63°C for 30 sec, and 72°C for 1 min, then the next 20 cycles at 94°C for 15 sec, 63°C for 30 sec, and 72°C for 1 min with 5 sec increasing elongation time for each successive cycle, and a final elongation at 72°C for 10 min. The PCR products were analyzed on 2% agarose gels and visualized by ethidium bromide staining. The RT-PCR products for case 1 were sequenced following subcloning using the TOPO TA Cloning®Kit for Sequencing (Invitrogen, Carlsbad, CA) and for cases 2 and 3 by direct sequencing of the gel purified products.
RESULTS
Histology
Grossly, all three tumors were well-circumscribed and featured lobulated, gelatinous, yellow cut surfaces. Histologically, each tumor was composed of cords and nests of cells deposited in a variably myxoid or myxochondroid matrix; none of the tumors contained abundant hyaline cartilage (Figure 1A). Individual tumor cells had variable morphology, with some resembling mature adipocytes and some resembling lipoblasts. Many of the cords and nests were composed of smaller cells with eosinophilic granular cytoplasm, while some were reminiscent of physaliferous cells or brown fat cells. The tumor cells were uniformly reactive for vimentin, and variably immunoreactive for S-100 protein, PGP 9.5, Leu 7, and NSE and were negative for AE1/3, EMA, CEA (polyclonal), and neurofilament in case 2. Electron microscopic examination of case 3 revealed lipocytes and lipoblasts with relatively abundant intracytoplasmic lipid and glycogen. An ultrastructural feature previously described in chondroid lipoma, “cytoplasmic knobby protrusions” were also noted (Kindblom et al., 2002). Ultrastructurally, the myxohyaline matrix exhibited cartilage-like features.
Figure 1.
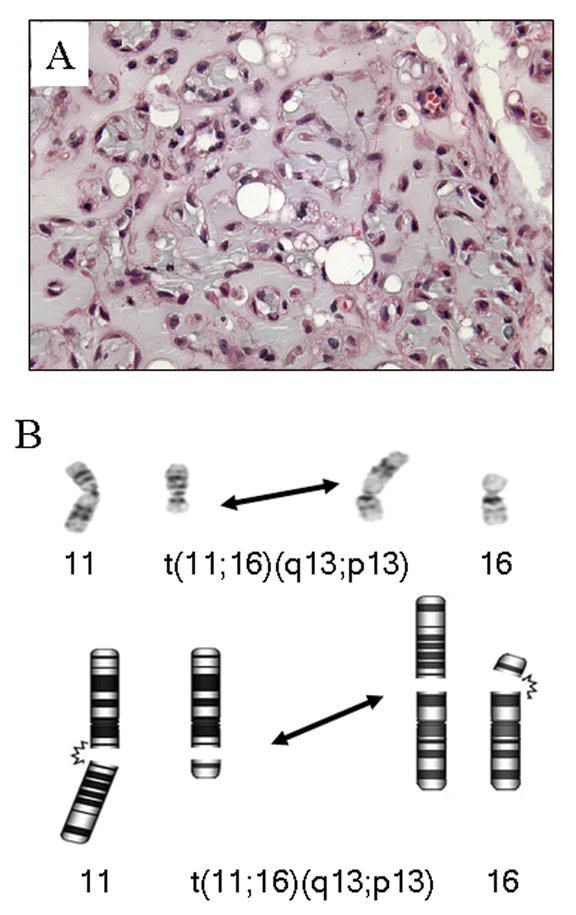
A: Case 1, scattered lipoblasts and cords of cells with eosinophilic cytoplasm deposited in a myxochondroid matrix (H & E; original magnification × 400). B: Partial karyotype and corresponding schematic illustrating the 11;16 translocation recurrent in chondroid lipoma.
Cytogenetic Analysis
Karyotypic analysis revealed a t(11;16)(q13;p13) as the sole cytogenetic aberration in cases 2 and 3 (Figure 1B). In contrast, the t(11;16)(q13;p13) was accompanied by additional cytogenetic abnormalities in case 1. In addition to the current cases, the cytogenetic findings of previously described chondroid lipomas are listed in Table 1.
FISH
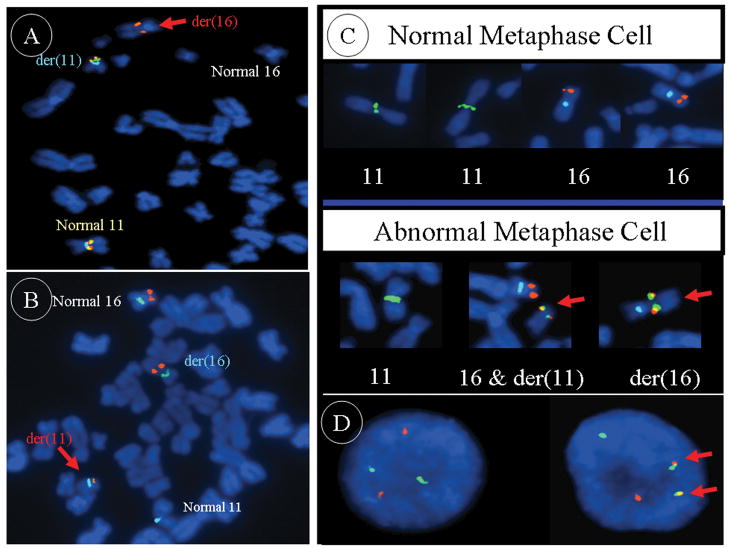
Metaphase FISH analysis revealed that the chromosome 11q13 breakpoint was occurring within a solitary BAC clone (RP11-466C23) (Figures 2A and 3A). It was determined that potentially four candidate genes including RTN3, C11orf95, C11orf84, and MARK2 are mapped to RP11-466C23. In contrast, only a single known gene, MKL2, is localized to the solitary BAC clone (CTD- 2118O12) shown to be disrupted by metaphase FISH analysis of the chromosome 16p13 breakpoint (Figures 2B and 3B). Further evidence of MKL2 involvement was provided by additional FISH analysis using custom-designed MKL2-specific probes generated by long-range PCR. In an effort to develop a dual-color dual-fusion probe set of clinical utility, the following probes were cocktailed: RP11- 697H9 and RP11-466C23 to span the 11q13 breakpoint and CTD-2135D7 and CTD- 2118O12 to span the 16p13 breakpoint. Utilizing this latter probe set, dual fusions indicative of the 11;16 translocation were confirmed on ten abnormal metaphase cells of case 1 (Figure 3C) and in 86% and 94% of the interphase cells analyzed from cases 2 and 3, respectively (Figure 3D). Due to the presence of a ring chromosome and double minutes in a subclone of case 1, standard protocol MDM2 FISH analysis using a commercially available probe (Abbott Molecular, Inc., Des Plaines, IL) was also conducted and was negative for amplification.
Figure 2.
FISH-based positional cloning strategy. A: Schematic illustrating the BAC clones spanning the 11q13 breakpoint region and the four candidate genes located within BAC RP11-466C23; B: Schematic illustrating the BAC clones spanning the 16p13 breakpoint region and the single candidate gene located within BAC CTD-2118O12. A and B: The BAC clones listed in the light gray shaded boxes were labeled in spectrum orange and those in the darker shaded boxes in spectrum green. Arrows show the transcriptional orientation of the individual genes.
Figure 3.
A: The 11q13 breakpoint is shown to lie within the spectrum orange labeled BAC probe RP11-466C23 (BAC probe CTD-3110P2 is labeled in green and CEP11 in aqua). B: The 16p13 breakpoint is shown to lie within the spectrum orange labeled BAC probe CTD-2118O12 (CEP16 is labeled in green and CEP11 in aqua). C: The dual-color dual-fusion probe set (RP11-697H9 and RP11-466C23 cocktail probe labeled in spectrum green, CTD-2135D7 and CTD-2118O12 cocktail probe in orange, and CEP16 in aqua) confirms the presence of the 11;16 translocation on each of the respective derivative chromosomes in case 1 (lower panel) in contrast to single signals on the chromosome 11 and 16 homologues from a normal peripheral blood lymphocyte metaphase cell (upper panel). D: The dual-color dual-fusion probe set in a normal interphase cell (left) and in a translocation positive chondroid lipoma interphase cell (right).
RACE Analyses and RT-PCR
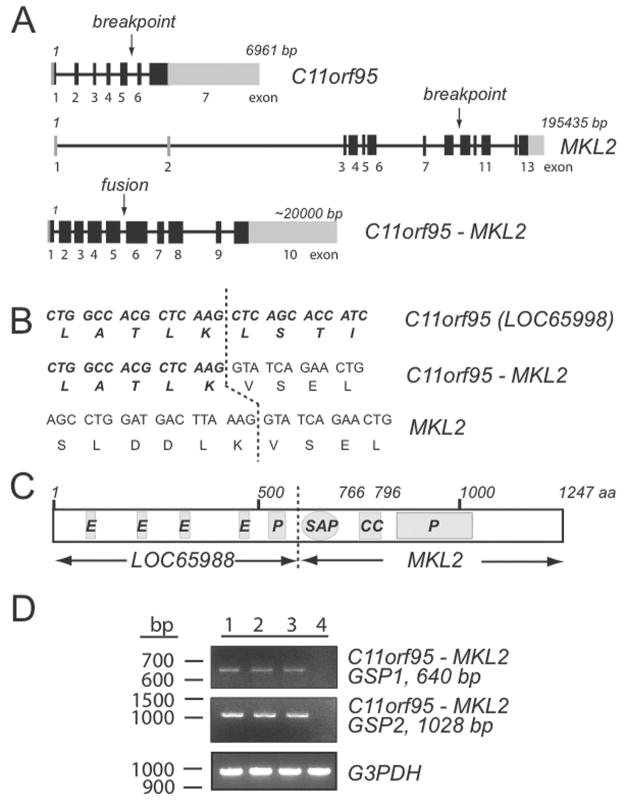
5′ RACE analysis identified C11orf95 as the MKL2 translocation partner gene and sequencing studies of the RT-PCR generated transcripts defined the fusion occurring between exons 5 and 9 of C11orf95 and MKL2, respectively (Figure 4A and B). This C11orf95-MKL2 fusion gene codes for a transcript of 9127 nucleotides with an open reading frame of 3744 nucleotides. The chimeric transcript encodes a protein of 1247 amino acids that retains the SAP domain from MKL2 (Figure 4C). Identical fusion transcripts were confirmed in all three chondroid lipomas (Figure 4D).
Figure 4.
A: Schematic of C11orf95, MKL2 and C11orf95-MKL2 fusion gene, solid bars represent coding exons, gray boxes are non-translated regions. B: Amino acid sequence at the breakpoint. C: Schematic and domain structure of the fusion C11orf95-MKL2 protein; the letters within the bars designate conserved domains; polyglutamic acid region (E), proline rich region (P), DNA-binding SAP domain (SAP), and coiled-coiled region (CC). D: Detection of the presence of C11orf95-MKL2 fusion transcript by RT-PCR analysis.
DISCUSSION
Chondroid lipoma is a rare benign tumor of adipose tissue origin that may mimic myxoid chondrosarcoma and liposarcoma. Recognition of an identical 11;16 translocation in six chondroid lipoma cases [three current cases and three previously reported (Thomson et al., 1999; Gisselsson et al., 1999a; Ballaux et al., 2004)] prompted our efforts in mapping the breakpoints and defining the underlying involving genes. Recurrent chromosomal translocations leading to the formation of chimeric genes have been identified in a number of benign and malignant bone and soft tissue tumors and are believed to be involved in the early development of these tumors (Unni et al., 2005; Ladanyi et al., 2008; Mandahl and Mertens, 2009). The fusion oncogenes found in sarcomas are dominated by the creation of aberrant transcription factors. Rearrangements involving the high mobility group A family of genes (HMGA2 and HMGA1) are common in benign mesenchymal tumors including lipoma and pulmonary chondroid hamartoma among others (Fletcher et al., 1995; Schoenmakers et al., 1995; Ladanyi et al., 2008; Mandahl and Mertens, 2009). The HMGA proteins have an intrinsic flexibility that permit their important role in regulating transcription and chromatin architectural control (Reeves and Beckerbauer, 2001). In ordinary lipoma, HMGA2 is most frequently fused to LPP. Interestingly, Kubo et al. (2006) demonstrated that in addition to its role in adipogenesis, the HMGA2-LPP fusion protein may promote chondrogenesis.
In the current study, a C11orf95-MKL2 fusion was identified in three chondroid lipomas carrying a t(11;16)(q13;p13). C11orf95 encodes a 678 amino acid hypothetical protein of unknown function that exhibits expression in a wide variety of human tissues (Ota et al., 2004). This protein contains four polyglutamic rich and one proline rich region. The MKL2 gene codes for a 1049 amino acid myocardin-like protein and is a member of the myocardin/megakaryoblastic leukemia gene family (Selvaraj and Prywes, 2003). MKL2, an SAP (SAF-A, acinus, PIAS) DNA-binding domain containing protein, has been functionally implicated in chromatin remodeling in addition to serving as a transcriptional co-activator of SRF (serum response factor) (Selvaraj and Prywes, 2003).
In all three chondroid lipoma cases, exons 1–5 of C11orf95 were fused in-frame to the last five exons of MKL2. This chimeric transcript encompasses all putative functional motifs encoded by each gene. The C-terminal portion of the C11orf95-MKL2 chimeric protein contains a SAP DNA-binding domain, a coiled-coiled (CC) domain and a proline-rich region known to be present in transcription factors and oncoproteins. Interestingly, a rearrangement involving another MKL gene family member (MKL1) has been identified in t(1;22)(p13;q13) acute megakaryoblastic leukemia (Ma et al., 2001; Cen et al., 2003) and t(1;22)(p13;q13) acute myeloid leukemia, subtype M1 ( Hsiao et al., 2005). This translocation results in a fusion of RBM15 and MKL1 that also encompasses all putative functional motifs encoded by each gene. Additional studies must be conducted to determine the functionality of the C11orf95-MKL2 fusion oncogene in chondroid lipoma.
Recurrent involvement of the 11q13 region has also been found in another benign adipose tissue tumor, hibernoma, as well as in a common benign renal tumor, oncocytoma (Mertens et al., 1994; Sandberg, 2004; Sukov et al., 2009). The 11q13 rearrangement in renal oncocytoma has been shown to involve CCND1, however, the underlying 11q13 gene involved in hibernoma has not yet been identified (Sukov et al., 2009). Unlike in chondroid lipoma, 11q13 has not been shown to recombine with 16p12-13 in hibernoma. Gisselsson et al. reported five cases of hibernoma with complex rearrangements of chromosome 11, leading to loss of chromosome material in 11q13 (Gisselsson et al., 1999b). The most commonly deleted segment included the MEN1 gene. However, in the chondroid lipoma case described by Gisselsson et al., no deletions were found. With the intention of determining if hibernoma shares the same chromosome 11 breakpoint with chondroid lipoma, we performed FISH analysis using the two-color chromosome 11 breakpoint probe set RP11-697H9 and RP11-466C23 on hibernoma metaphase cells with 11q13 rearrangement. There was no disruption of these probe signals in the abnormal metaphase cells indicating that a gene other than C11orf95 is likely involved (data not shown).
Chondroid lipoma is a rare entity and may be mistaken for several other benign and malignant soft tissue tumors such as extraskeletal chondroma, liposarcoma, extraskeletal myxoid chondrosarcoma, and myoepithelioma/mixed tumor (Kindblom et al., 2002; Weiss and Goldblum, 2008). Unlike chondroid lipoma, myxoid liposarcoma usually exhibits a uniform population of stellate or spindled cells and an arborizing capillary proliferation. Extraskeletal myxoid chondrosarcoma is devoid of a prominent vascular proliferation, but contains uniform oval or spindled cells without cytoplasmic vacuoles, lipoblasts, or mature adipocytes. Both of these sarcomas harbor unique chromosomal abnormalities that can be identified with conventional cytogenetic, molecular cytogenetic, or molecular (e.g. RT-PCR) approaches for diagnostic purposes (Bridge, 2008). Extraskeletal chondroma frequently arises more peripherally than chondroid lipoma and contains abundant hyaline cartilage, a finding not typical of chondroid lipoma. Cytogenetic studies of extraskeletal chondroma or soft part chondroma have not revealed a tumor-specific anomaly although involvement of chromosomal region 12q12~15 appears to be recurrent in a subset of these tumors (Buddingh et al., 2003). Lastly, myoepitheliomas may show a broad spectrum of histologic appearances, but often are composed of spindled and epithelioid cells, some of which may contain vacuoles deposited in a variably chondromyxoid or hyalinized stroma. However in contrast to chondroid lipoma, myoepithelioma lacks adipocytes and lipoblasts and features significantly stronger immunoreactivity to antibodies for epithelial differentiation. Cytogenetic or array comparative genomic hybridization studies of soft tissue myoepitheliomas are rare, however, EWSR1 rearrangements or deletion within 19p13 have been identified as recurrent anomalies in subsets of these neoplasms (van den Berg et al., 2004; Balogh et al., 2008; Brandal et al., 2008; Hallor et al., 2008; Brandal et al., 2009).
In conclusion, chondroid lipoma is characterized by chromosomal translocation t(11;16)(q13;p13) resulting in the fusion of the C11orf95 and MKL2 genes. Novel FISH and RT-PCR assays developed in this study can serve as valuable diagnostic adjuncts for this rare disease entity, particularly when evaluating small samples or when fresh tissue is not available.
Acknowledgments
Supported by: NIH/NCI grant 5 P30 CA036727-2452, Nebraska Department of Health Institutional LB595 Grant for Cancer and Smoking Disease Research, U-10-CA98543-091, University of Nebraska Medical Center Eppley Pediatric Cancer Award, T.Y. supported in part by the Gladys Pearson Fellowship Award.
The authors would like to thank Ms. Kimberly A Christian for her excellent secretarial assistance.
References
- Ballaux F, Debiec-Rychter M, De Wever I, Sciot R. Chondroid lipoma is characterized by t(11;16)(q13;p12–13) Virchows Archiv. 2004;444:208–210. doi: 10.1007/s00428-003-0946-4. [DOI] [PubMed] [Google Scholar]
- Balogh Z, Deák L, Sápi Z. Malignant myoepithelioma of soft tissue: a case report with cytogenetic findings. Cancer Genet Cytogenet. 2008;183:121–124. doi: 10.1016/j.cancergencyto.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Gorunova L, Skjeldal S, Micci F, Heim S. Detection of a t(1;22)(q23;q12) translocation leading to an EWSR1-PBX1 fusion gene in a myoepithelioma. Genes Chromosomes Cancer. 2008;47:558–564. doi: 10.1002/gcc.20559. [DOI] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Heim S. t(19;22)(q13;q12) Translocation leading to the novel fusion gene EWSR1-ZNF444 in soft tissue myoepithelial carcinoma. Genes Chromosomes Cancer. 2009;48:1051–1056. doi: 10.1002/gcc.20706. [DOI] [PubMed] [Google Scholar]
- Bridge JA. Advantages and limitations of cytogenetic, molecular cytogenetic, and molecular diagnostic testing in mesenchymal neoplasms. J Orthop Sci. 2008;13:273–282. doi: 10.1007/s00776-007-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddingh EP, Naumann S, Nelson M, Neffa JR, Birch N, Bridge JA. Cytogenetic findings in benign cartilaginous neoplasms. Cancer Genet Cytogenet. 2003;141:164–168. doi: 10.1016/s0165-4608(02)00726-4. [DOI] [PubMed] [Google Scholar]
- Cen B, Selvaraj A, Burgess RC, Hitzler JK, Ma Z, Morris SW, Prywes R. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol Cell Biol. 2003;23:6597–6608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JA, Longtine J, Wallace K, Mentzer SJ, Sugarbaker DJ. Cytogenetic and histological findings in 17 pulmonary chondroid hamartomas: evidence for a pathogenetic relationship with lipomas and leiomyomas. Genes Chromosomes Cancer. 1995;12:220–223. doi: 10.1002/gcc.2870120310. [DOI] [PubMed] [Google Scholar]
- Gisselsson D, Domanski HA, Hoglund M, Carlen B, Mertens F, Willen H, Mandahl N. Unique cytological features and chromosome aberrations in chondroid lipoma: a case report based on fine-needle aspiration cytology, histopathology, electron microscopy, chromosome banding, and molecular cytogenetics. Am J Surg Pathol. 1999a;23:1300–1304. doi: 10.1097/00000478-199910000-00018. [DOI] [PubMed] [Google Scholar]
- Gisselsson D, Hoglund M, Mertens F, Dal Cin P, Mandahl N. Hibernomas are characterized by homozygous deletions in the multiple endocrine neoplasia type I region. Metaphase fluorescence in situ hybridization reveals complex rearrangements not detected by conventional cytogenetics. Am J Pathol. 1999b;155:61–66. doi: 10.1016/S0002-9440(10)65099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallor KH, Teixeira MR, Fletcher CD, Bizarro S, Staaf J, Domanski HA, von Steyern FV, Panagopoulos I, Mandahl N, Mertens F. Heterogeneous genetic profiles in soft tissue myoepitheliomas. Mod Pathol. 2008;21:1311–1319. doi: 10.1038/modpathol.2008.124. [DOI] [PubMed] [Google Scholar]
- Hsiao HH, Yang MY, Liu YC, Hsiao HP, Tseng SB, Chao MC, Liu TC, Lin SF. RBM15-MKL1 (OTT-MAL) fusion transcript in an adult acute myeloid leukemia patient. Am J Hematol. 2005;79:43–45. doi: 10.1002/ajh.20298. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Slovak ML, Campbell LJ, editors. ISCN. An International System for Human Cytogenetic Nomenclature. Basel: Karger; 2009. [Google Scholar]
- Kindblom LG, Meis-Kindblom JM, Mandahl N. Chondroid lipoma. In: Fletcher CDM, Unni KK, Mertens F, editors. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon: IARC Press; 2002. p. 30. [Google Scholar]
- Kubo T, Matsui Y, Goto T, Yukata K, Yasui N. Overexpression of HMGA2-LPP fusion transcripts promotes expression of the alpha-2 type XI collagen gene. Biochem Biophys Res Commun. 2006;340:476–481. doi: 10.1016/j.bbrc.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Ladanyi M, Antonescu CR, Dal Cin P. Cytogenetic and Molecular Genetic Pathology of Soft Tissue Tumors. In: Weiss SW, Goldblum JR, editors. Soft Tissue Tumors. 5. Philadelphia: Mosby Inc; 2008. pp. 73–102. [Google Scholar]
- Logan PM, Janzen DL, O’Connell JX, Munk PL, Connell DG. Chondroid lipoma: MRI appearances with clinical and histologic correlation. Skeletal Radiol. 1996;25:592–595. doi: 10.1007/s002560050143. [DOI] [PubMed] [Google Scholar]
- Ma ZG, Morris SW, Valentine V, Li M, Herbrick JA, Cui XL, Bouman D, Li Y, Mehta PK, Nizetic D, Kaneko Y, Chan GCF, Chan LC, Squire J, Scherer SW, Hitzler JK. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet. 2001;28:220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- Mandahl N, Mertens F. Soft Tissue Tumors. In: Heim S, Mitelman F, editors. Cancer Cytogenetics. 3. New Jersey: John Wiley & Sons, Inc; 2009. pp. 675–711. [Google Scholar]
- Meis JM, Enzinger FM. Chondroid lipoma. A unique tumor simulating liposarcoma and myxoid chondrosarcoma. Am J Surg Pathol. 1993;17:1103–1112. [PubMed] [Google Scholar]
- Mentzel T, Fletcher CD. Recent advances in soft tissue tumor diagnosis. Am J Clin Pathol. 1998;110:660–670. doi: 10.1093/ajcp/110.5.660. [DOI] [PubMed] [Google Scholar]
- Mertens F, Rydholm A, Brosjo O, Willen H, Mitelman F, Mandahl N. Hibernomas are characterized by rearrangements of chromosome bands 11q13-21. Int J Cancer. 1994;58:503–505. doi: 10.1002/ijc.2910580408. [DOI] [PubMed] [Google Scholar]
- Nishio J, Althof PA, Bailey JM, Zhou M, Neff JR, Barr FG, Parham DM, Teot L, Qualman SJ, Bridge JA. Use of a novel FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of alveolar rhabdomyosarcoma. Lab Invest. 2006;86:547–556. doi: 10.1038/labinvest.3700416. [DOI] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M, Shiratori A, Sudo H, Hosoiri T, Kaku Y, Kodaira H, Kondo H, Sugawara M, Takahashi M, Kanda K, Yokoi T, Furuya T, Kikkawa E, Omura Y, Abe K, Kamihara K, Katsuta N, Sato K, Tanikawa M, Yamazaki M, Ninomiya K, Ishibashi T, Yamashita H, Murakawa K, Fujimori K, Tanai H, Kimata M, Watanabe M, Hiraoka S, Chiba Y, Ishida S, Ono Y, Takiguchi S, Watanabe S, Yosida M, Hotuta T, Kusano J, Kanehori K, Takahashi-Fujii A, Hara H, Tanase TO, Nomura Y, Togiya S, Komai F, Hara R, Takeuchi K, Arita M, Imose N, Musashino K, Yuuki H, Oshima A, Sasaki N, Aotsuka S, Yoshikawa Y, Matsunawa H, Ichihara T, Shiohata N, Sano S, Moriya S, Momiyama H, Satoh N, Takami S, Terashima Y, Suzuki O, Nakagawa S, Senoh A, Mizoguchi H, Goto Y, Shimizu F, Wakebe H, Hishigaki H, Watanabe T, Sugiyama A, Takemoto M, Kawakami B, Yamazaki M, Watanabe K, Kumagai A, Itakura S, Fukuzumi Y, Fujimori Y, Komiyama M, Tashiro H, Tanigami A, Fujiwara T, Ono T, Yamada K, Fujii Y, Ozaki K, Hirao M, Ohmori Y, Kawabata A, Hikiji T, Kobatake N, Inagaki H, Ikema Y, Okamoto S, Okitani R, Kawakami T, Noguchi S, Itoh T, Shigeta K, Senba T, Matsumura K, Nakajima Y, Mizuno T, Morinaga M, Sasaki M, Togashi T, Oyama M, Hata H, Watanabe M, Komatsu T, Mizushima-Sugano J, Satoh T, Shirai Y, Takahashi Y, Nakagawa K, Okumura K, Nagase T, Nomura N, Kikuchi H, Masuho Y, Yamashita R, Nakai K, Yada T, Nakamura Y, Ohara O, Isogai T, Sugano S. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Reeves R, Beckerbauer L. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta. 2001;1519:13–29. doi: 10.1016/s0167-4781(01)00215-9. [DOI] [PubMed] [Google Scholar]
- Sandberg AA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: lipoma. Cancer Genet Cytogenet. 2004;150:93–115. doi: 10.1016/j.cancergencyto.2003.12.018. [DOI] [PubMed] [Google Scholar]
- Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nature Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- Selvaraj A, Prywes R. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J Biol Chem. 2003;278:41977–41987. doi: 10.1074/jbc.M305679200. [DOI] [PubMed] [Google Scholar]
- Sukov WR, Ketterling RP, Lager DJ, Carlson AW, Sinnwell JP, Chow GK, Jenkins RB, Cheville JC. CCND1 rearrangements and cyclin D1 overexpression in renal oncocytomas: frequency, clinicopathologic features, and utility in differentiation from chromophobe renal cell carcinoma. Hum Pathol. 2009;40:1296–1303. doi: 10.1016/j.humpath.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Thomson TA, Horsman D, Bainbridge TC. Cytogenetic and cytologic features of chondroid lipoma of soft tissue. Mod Pathol. 1999;12:88–91. [PubMed] [Google Scholar]
- Unni KK, Inwards CY, Bridge JA, Kindblom LG, Wold LE. Tumors of the Bones and Joints. Maryland: ARP Press; 2005. [Google Scholar]
- van den Berg E, Zorgdrager H, Hoekstra HJ, Suurmeijer AJH. Cytogenetics of a soft tissue malignant myoepithelioma. Cancer Genet Cytogenet. 2004;151:87–89. doi: 10.1016/j.cancergencyto.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Weiss SW, Goldblum JR. Enzinger and Weiss’s Soft Tissue Tumors. 5. Philadelphia: Mosby Elsevier; 2008. pp. 442–444. [Google Scholar]