Abstract
Eukaryotic elongation factor 1A (eEF1A1) is an abundant cytosolic protein in Saccharomyces cerevisiae and is well conserved amongst species. This protein undergoes multiple posttranslational modifications, including the N-methylation of four side chain lysine residues. However, the enzyme(s) responsible for catalyzing these modifications have remained elusive. Here we show by intact protein mass spectrometry that deletion of either of two genes coding for putative methyltransferases results in a loss in mass of eEF1A. Deletion of the YHL039W gene, a member of the SET domain subfamily including cytochrome c and ribosomal protein lysine methyltransferases, results in an eEF1A mass loss corresponding to a single methyl group. Deletion in the YIL064W/SEE1 gene, encoding a well conserved seven beta strand methyltransferase sequence, has been shown previously to affect vesicle transport; in this work we show that deletion results in the loss of two methyl group equivalents from eEF1A. We find that deletion of thirty five other putative and established SET domain and seven beta strand methyltransferases has no effect on the mass of eEF1A. Finally, we show that wild type extracts, but not YIL064W/SEE1 mutant extracts, can catalyze the S-adenosylmethionine-dependent in vitro methylation of hypomethylated eEF1A. We suggest that YHL039W (now designated EFM1 for elongation factor methyltransferase 1) and YIL064W/SEE1 encode distinct eEF1A methyltransferases that respectively monomethylate and dimethylate this protein at lysine residues.
Keywords: eukaryotic elongation factor 1A, protein lysine methylation, posttranslational modification of proteins, methyltransferases, S-adenosylmethionine
1. Introduction
Posttranslational modifications are found throughout eukaryotic cells, playing structural roles and contributing to the regulation of protein function. Great progress has been made recently in the understanding of protein lysine methylation, particularly since the discovery of numerous methyltransferases responsible for these modifications as well as demethylases that can reverse at least some of the modification reactions [1–7]. While the focus to date has largely been on the role of histone lysine methylation in transcriptional control, there are a number of additional non-histone substrates acted upon by lysine methyltransferases [1, 3–6, 8, 9]. For a number of these modified proteins, the identity of the methyltransferase, as well as the function methylation imparts, remains to be discovered.
We have been interested in the methyl modification of proteins involved in translation, and more specifically in the enzymes that catalyze these modifications. There are a number of translation-associated proteins that have been observed to be modified by lysine methylation, including several ribosomal proteins [8, 9]. The functions of these lysine modification reactions remain obscure. One of the most highly methylated proteins at lysine residues is the well conserved translational affiliated protein eEF1A [8, 10–12]. Perhaps best known for its role in escorting tRNA to the ribosomal A-site in a GTP-dependent manner, eEF1A has additionally been described as having multiple moonlighting functions including interactions with the cytoskeleton [13–15]. Phosphorylation has been determined to modify some of the activities of mammalian eEF1A, typically resulting in a stimulatory effect [14, 15]. The impact of methyl modification on the functions of eEF1A is more poorly understood [10, 12, 16]. However, it is known that some methyl modifications of eEF1A are well conserved from simple eukaryotes like Saccharomyces cerevisiaeall the way to humans [11]. The equivalent bacterial GTP-dependent translation factor EF-Tu is lysine trimethylated, though at a residue 12 positions C-terminal to the strictly conserved trimethyl lysine residue of eukaryotes [11, 17]. Nevertheless, the methyltransferase species that are responsible for eEF1A modification have proven elusive; none have been identified to date.
We have focused our efforts on identifying the methyltransferase(s) responsible for eEF1A methylation in S. cerevisiae with the hope that this work can provide a foundation for understanding the functional role of the methylation reactions in this and other eukaryotes. We screened deletion mutants of putative methyltransferases of both the seven beta strand and the SET domain families to identify potential catalysts for eEF1A methyl modification. In the past, we have used in vivo radiolabeling techniques to identify methyltransferase-substrate pairs [18]. However, due to multiple methylated sites, these techniques were not useful in determining the enzymes acting upon eEF1A. In this study we took an approach using intact protein mass spectrometry to analyze protein modifications [19]. We obtained intact mass values for chromatographically purified eEF1A at high enough resolution to observe the 14 Da changes that occur due to loss of methylation in a mutant strain. Using these techniques, we have identified two novel proteins involved in methylating eEF1A in S. cerevisiae.
2. Materials and Methods
2.1 Yeast strains
With the exception noted below, all S. cerevisiae strains were obtained from the Saccharomyces Genome Deletion Project and included the parent “wild type” strains BY4741 and BY4742 as well as the ΔYHL039W and ΔYIL064W/see1 gene deletion strains in both of these backgrounds. The Δset1 gene deletion strain was a gift from Drs. Renee Chosed and Sharon Dent at the MD Anderson Cancer Center (Houston, TX) along with its corresponding parent strain, KT1112. A complete list of strains screened for catalysis of eEF1A methylation is given in Supplemental Table 1.
2.2 Isolation of cytosolic proteins
Cells were grown at 30 °C in YPD media (1% bacto-yeast extract, 2% bacto-peptone, 2% dextrose) to an optical density of 0.5 – 1.0 at 600 nm. The cells were subsequently harvested by centrifugation at 4 °C for 5 min at 5,000 × g. Cell pellets were combined with 1.5 g of baked zirconium glass beads (Biospec Products; Bartlesville, OK) in 3 ml buffer A (20 mM Tris HCl, 15 mM Mg acetate, 60 mM KCl, 1 mM DTT, 1 mM PMSF, and proteinase inhibitors from the Roche Proteinase Inhibitor Cocktail Tablet with 1mM EDTA) and submitted to ten repetitions of one min of vortexing followed by one min at 0 °C. Samples were fractionated as described previously [18]. Briefly, lysates were centrifuged at 4 °C first at 12,000 × g for 5 min and then 20,000 × g for 15 min in a Beckman JA-17 rotor. The final centrifugation was performed at 100,000 × g for 2 hrs at 4 °C in a Beckman Ti-65 rotor. The approximately 4 ml of supernatant containing the cytosolic fraction was stored at −80 °C pending further protein separation.
2.3 Column purification of eEF1A
Isolation of eEF1A was achieved by use of a pair of ion exchange columns in a manner similar to the one described by Lopez-Valenzuela et al. [20]. Specifically, the total volume of each cytosolic sample (approximately 4 ml) was individually loaded onto a 5 ml HiTrap Q HP anion exchange column (GE Healthcare) that had been equilibrated with buffer A (5 mM NaCl, 20 mM HEPES, 5% glycerol, 1 mM DTT, 1 mM EDTA, pH 8) and was then washed with an additional 5 ml of buffer A. The total flow-through containing eEF1A was next loaded at 2 ml/min onto a 5 ml HiTrap SP HP cation exchange column equilibrated in buffer A and the column subsequently washed with buffer A at 5 ml/min for 5 min. To elute eEF1A an increasing salt gradient of 0–50% buffer B (1 M NaCl, 20 mM HEPES, 5% glycerol, 1 mM DTT, 1 mM EDTA, pH 8) run at 5 ml/min over 15 min was used and 1.5 ml fractions collected. All of these steps were performed at 4 °C. Purified eEF1A fractions were identified by the presence of a single 49 kDa polypeptide band on SDS gel electrophoresis and were monitored by UV absorbance at 280 nm.
2.4 Intact mass determination by coupled liquid chromatography-mass spectrometry
The intact mass of eEF1A was analyzed using a PLRP-S polymeric column with pore size of 300Ǻ, bead size of 5 µm, and dimensions of 150 × 1.0 mm (Polymer Laboratories, Amherst, MA) coupled to an Applied Biosystems Q-Star Elite instrument based on the protocol described previously [21]. Here, the column was equilibrated with 5% buffer B (0.05% trifluoroacetic acid in acetonitrile) and 95% buffer A (0.05% trifluoroacetic acid in water) and maintained at 50 °C for the duration of the run. The samples were eluted at 50 µl/min with 5 min at equilibration conditions, followed by a 55 min gradient from 5% to 60% B, and finally a 5 min gradient from 60% to 100% B. Afterwards, the column was returned to equilibration conditions over 5 min. Data was evaluated and intact masses calculated using Analyst software (Applied Biosystems, Foster City, CA). The peak containing eEF1A was found at approximately 42 min.
2.5 In vitro methylation of eEF1A
Cells were grown at 30 °C in YPD media (1% bacto-yeast extract, 2% bacto-peptone, 2% dextrose) to an optical density of 0.5 – 0.8 at 600 nm. The cells were subsequently harvested by centrifugation at 4 °C for 5 min at 5,000 × g. Cell pellets were lysed by combining with 0.5 g glass beads in 1 ml phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, and proteinase inhibitors from the Roche Proteinase Inhibitor Cocktail Tablet with 1mM EDTA, pH 7.4) and submitted to seven rounds of vortexing and ice treatment as described above. After recovering the lysate, the beads were washed with an additional 0.25 ml phosphate-buffered saline which was then combined with the original aliquot. The lysate was centrifuged at 4 °C first at 12,000 × g for 15 min and then 17,000 × g for 10 min, with the supernatants retained. The protein concentration was determined by precipitating an aliquot using 10% trichloroacetic acid by the Lowry method with bovine serum albumin as a standard.
For the in vitro assay, a total of 40 µg of lysate protein (wild type, ΔYHL039W, ΔYIL064W, or a 1:1 mixture of wild type and mutant) was combined in 0.1 M sodium phosphate buffer (pH 7.0) and incubated with 0.66 µM of the radiolabeled methyl donor S-adenosyl-L-[methyl-3H]methionine ([3H]AdoMet; 78.0 Ci/mmol) for 60 min at 30 °C. Reactions were stopped by the addition of an equal volume of 2× SDS running dye (180 mM Tris-HCl, pH 6.8, 4% SDS, 0.1% β-mercaptoethanol, 20% glycerol, and 0.002% bromophenol blue) and subsequently heated at 100 °C for 3 min. Samples were then loaded onto a 12.6% SDS polyacrylamide gel in the Laemmli buffer system [22] and run for 3 h at 30 mA. Afterwards the gels were rocked for 1 hr submerged in Coomassie Brilliant Blue dye and then de-stained overnight to enable visualization of protein bands. The wet gel lanes were chopped into ten 2 mm slices surrounding the 42.7 kDa ovalbumin marker band, and each slice placed in an individual 1.5 ml polypropylene microcentrifuge tube with 1 ml of 30% H2O2. The tubes were then inserted into vials containing 5 ml of Safety-Solve Complete Counting Mixture (Research Products International, Mt. Prospect, IL), caps were loosely put on, and left to incubate 24 h at 37 °C. After incubation, the caps were tightened and vials shaken vigorously to mix the samples so that total radioactivity could be counted by a Beckman LS6500 scintillation counter.
3. Results
3.1 YHL039W is required for monomethylation of eEF1A
It has previously been established that eEF1A is lysine methylated in S. cerevisiae at four sites including two non-conserved monomethylation sites at residues 30 and 390, one conserved dimethylated site at residue 316, and one conserved trimethylated site at residue 79 [11]. Evidence has also been presented for the substoichiometric modification of the C-terminal lysine residue at its alpha-carboxyl group [23]. Therefore, we initiated our screen using deletion mutants of genes encoding known and putative methyltransferases of the S. cerevisiae SET domain family, a family of enzymes that appears to specifically catalyze the methylation of protein lysine residues. Previous work has shown that the S. cerevisiae SET domain family of methyltransferases can be further grouped into two subfamilies [24]. The first subfamily contains the Set1 and Set2 proteins known to methylate histone H3 at lysine residues 4 and 36, respectively [25, 26]. Although no substrates have yet been identified for Set3-Set6, Set3 has been found to be a part of a histone deacetylation complex so is also linked to transcription [27]. The second subfamily contains identified ribosomal lysine methyltransferases Rkm Rkm4 as well as the cytochrome c lysine methyltransferase Ctm1 [9, 24]. Recently, Set1 was shown to have lysine methyltransferase activity for an additional substrate, the kinetochore protein Dam1 [28]. With this study illustrating that SET domain lysine methyltransferases may have multiple substrates, we chose to include all known and putative S. cerevisiae SET domain methyltransferases in our screen for enzymes catalyzing eEF1A lysine methylation.
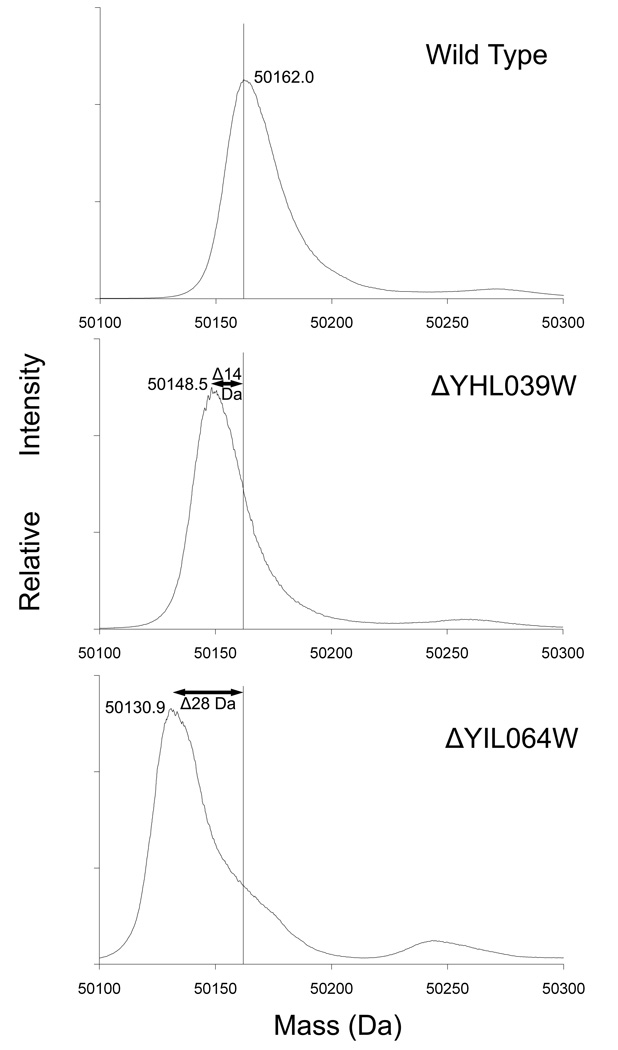
To determine the enzyme(s) responsible for catalyzing the methylation of eEF1A, we purified eEF1A from wild type yeast cells and from deletion mutants in the same background for the twelve known and putative SET domain methyltransferases previously identified [24]. Cells from each strain were lysed, the cytosolic fraction purified by centrifugation, and eEF1A purified by three steps of column chromatography as described in Materials and Methods. The final reverse phase HPLC column was linked directly to the mass spectrometer and samples were injected using an electrospray source, allowing for intact mass measurement. The predicted average mass of the eEF1A polypeptide is 50,032 Da. With the expected N-terminal acetylation and the seven internal methyl groups the expected mass is 50,172 Da. For the wild type sample, we observed a broad peak with an apex indicating the predominant species has a mass of 50,162 Da (Fig. 1, top panel). It is not clear at this point whether the smaller experimental value reflects measurement error (in this case of about 200 ppm) or the presence of some species lacking one or more methyl groups. The C-terminal methyl ester is not observed in our preparations, presumably due to hydrolysis during the purification steps, and its mass is not included in the calculated mass above.
FIGURE 1. Loss of eEF1A methylation in two mutants, ΔYHL039W and ΔYHL064W.
The intact masses of wild type (top panel), ΔYHL039W (middle panel), and ΔYIL064W (lower panel) eEF1A were determined as described in Materials and Methods. Strains shown are in the BY4742 background. Arrows indicate the loss of methyl groups based on the shift of the peak’s apex.
Analysis of eleven of the twelve SET domain family member deletion strains showed mass spectra effectively identical to that of the wild type (data not shown). However, we observed a loss of approximately 14 Da from the total eEF1A mass in the ΔYHL039W deletion strain (Fig. 1, middle panel). This suggests that the gene product encoded by YHL039W i0s responsible for monomethylating eEF1A. These results for the ΔYHL039W deletion strain in the BY4742 background were confirmed in an independently derived ΔYHL039W deletion strain in the BY4741 background (data not shown). Based on the fact that all SET domain methyltransferases to date modify lysine residues, and on the absence of non-lysine stoichiometric modifications of eEF1A, we suggest that the protein product of the YHL039W gene monomethylates eEF1A and designate it elongation factor methyltransferase 1 (Efm1).
3.2 YIL064W is required for dimethylation of eEF1A
While the SET domain family methyltransferases may exclusively act on protein lysine substrates, members of the larger seven beta strand methyltransferase family have also been observed to methylate lysine residues. For example, in S. cerevisiae the seven beta strand methyltransferase Dot1 modifies lysine 79 of histone 3 [29]. With this in mind, we expanded our screen for eEF1A methyltransferases to include putative seven beta strand methyltransferases identified from a recent bioinformatic analysis [30]. Supplemental Table 1 shows the set of nineteen putative seven beta strand methyltransferase deletion strains analyzed, as well as the known seven beta strand lysine methyltransferase Dot1, the known glutamine methyltransferases Mtq1 and Mtq2, and the three known protein arginine methyltransferases Rmt1, Rmt2, and Hsl7.
With one exception, the mass spectra of eEF1A obtained from all of these deletion strains did not deviate significantly from that of the wild type strain (data not shown). However, in the ΔYIL064W/see1 deletion strain, we found a loss of approximately 28 Da from the intact mass of eEF1A as compared to wild type (Fig. 1, bottom panel). This suggests that YIL064W/See1 is a seven beta strand lysine methyltransferase that catalyzes the addition of two methyl groups to eEF1A.
3.3 In vitro methylation of eEF1A
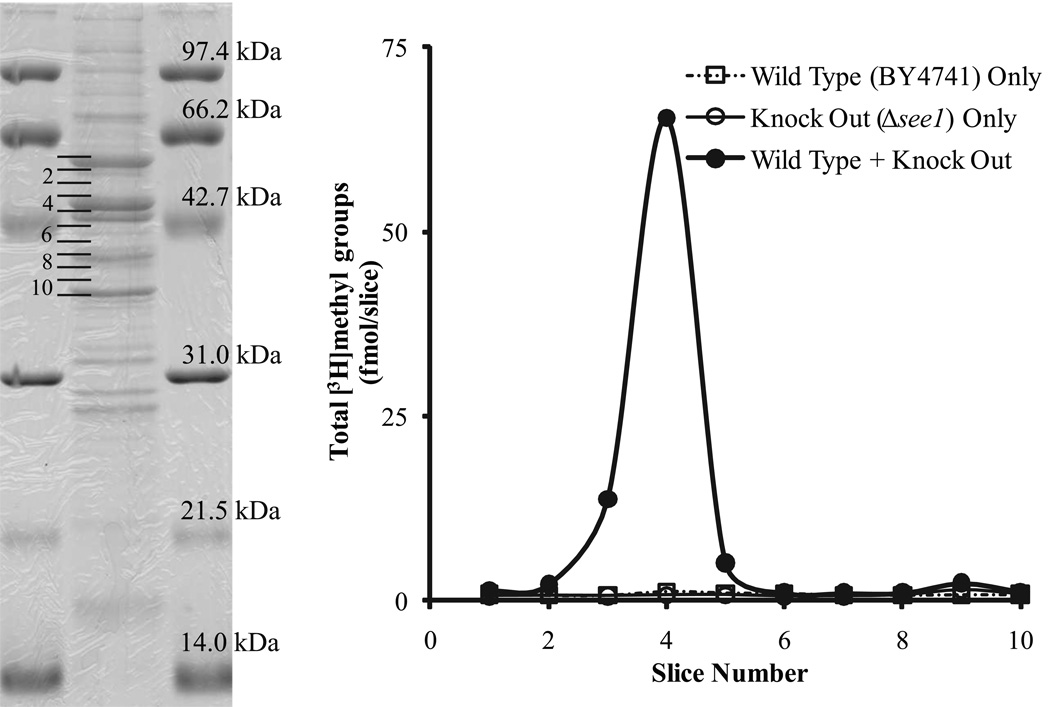
To support these findings, we developed in vitro assays for these enzymes. We measured the transfer of radiolabeled methyl groups from [3H]AdoMet to the eEF1A polypeptide chain by introducing functional enzyme from wild type lysates to the hypomethylated eEF1A obtained from mutant lysates. Accordingly, we combined lysates from the different strains and performed in vitro incubation with [3H]AdoMet followed by the separation of polypeptide chains by SDS gel electrophoresis. The incorporation of radiolabeled methyl groups into eEF1A was monitored by counting gel slices in the region surrounding the eEF1A band as described in Materials and Methods.
In reactions containing only wild type lysate, where eEF1a is presumably fully modified, we observed no significant incorporation of radioactivity at the band corresponding to eEF1A (Fig. 2, open boxes). Likewise, control samples for the mutant-only lysates showed no radiolabel incorporation for eEF1A (Fig. 2, open circles, for ΔYIL064W/see1; data not shown for ΔYHL039W). However, in samples where wild type lysate containing the active protein YIL064w/See1 was combined with lysate from the ΔYIL064W/see1 deletion strain (containing eEF1A lacking the modification dependent upon the YIL064W/See1 protein), we observed an incorporation of the radiolabel consistent with eEF1A methylation (Fig. 2, closed circles). The only difference in the lysates is the presence or absence of YIL064W/See1, further supporting the identity of this protein as an eEF1A methyltransferase. Similar results were obtained using extracts from the independent ΔYIL064W/see1 deletion and wild type strains from the BY4742 background (data not shown).
FIGURE 2. In vitro methylation of eEF1A.
Lysates from the strains of interest were combined and incubated in the presence of [3H]AdoMet as described in Materials and Methods. SDS gel electrophoresis was subsequently used to assist in the isolation of eEF1A, and ten 2mm slices of the gel lanes were collected surrounding the peptide band corresponding to the appropriate molecular weight. The left panel shows the slice location on a representative lane. The right panel displays the total counts obtained after treating gel slices with hydrogen peroxide and counting in a scintillation counter for the BY4741 wild type only lysate (open boxes), the ΔYIL064W only lysate (open circles), and a 1:1 mixture of wild type and ΔYIL064W lysates (closed circles). Methyl groups added were determined from the radioactivity in each slice and the [3H]AdoMet specific radioactivity.
Interestingly, the corresponding lysate combination for YHL039W did not yield in vitro methylated eEF1A (data not shown). Since the folded eEF1A may a poor substrate for this enzyme, we additionally tried in vitro assays using urea-treated lysate from the ΔYHL039W delete strain. This would theoretically denature hypomethylated eEF1A and make the YHL039W site more available to the functional enzyme present in the wild type lysate. However, we were still unable to observe any counts above background (data not shown). It is possible that YHL039W modification occurs co-translationally and that even the denatured mature protein is not a preferred substrate for the methyltransferase.
3.4 Site specificity and amino acid sequence conservation of eEF1A methyltransferases Efm1 and See1
The only known stoichiometric modifications of yeast eEF1A are the monomethylation of lysine 30, the trimethylation of lysine 79, the dimethylation of lysine 316, and the monomethylation of lysine 390 [11]. Since we observed a loss of 14 Da upon deletion of Efm1, this suggests that the site of action of this enzyme is one of the two monomethylated lysine residues of eEF1A. Neither of the monomethylation sites of eEF1A in S. cerevisiae appears to be conserved in higher eukaryotes [11]. It appears as if Efm1 may be an enzyme specific to yeast, as we find closely related species in C. albicans and S. pombe, but not in higher organisms (Fig. 3). In contrast, the dimethylation at lysine 316 is conserved among eukaryotes, although the corresponding residue in higher eukaryotic organisms is found in a trimethylated rather than a dimethylated state [11]. This conservation is reflected in the wide distribution of proteins in eukaryotes with amino acid sequence similarity to YIL064W/See1 (Fig. 4). The apparent ortholog of YIL064W/See1 in humans is METTL10, a protein of as yet unknown function containing a seven beta strand AdoMet binding domain.
FIGURE 3. YHL039W is poorly conserved in nature.
The amino acid sequence of YHL039W was compared to the non-redundant protein database using the BLAST algorithm to obtain expect values. Amino acid sequence alignments determined by CLUSTALW2 are shown for S. cerevisiae (NCBI ID: NP_011824), C. albicans (NCBI ID: XP_716460, expect value 5e-50), and S. pombe (NCBI ID: NP_595446, expect value 7e-9). Not shown: H. sapiens (SETD3, NCBI ID: NP_115609, expect value 2e-04), H. sapiens (SETD4, NCBI ID: NP_059134, expect value 1e-06), A. thaliana (NCBI ID: NP_172856, expect value 2e-5), A. thaliana (NCBI ID: NP_564222, expect value 1e-4), A. thaliana (NCBI ID: NP_001030933, expect value 2e-4), A. thaliana (NCBI ID: NP_191068, expect value 3e-4), C. elegans (NCBI ID: NP_497604, expect value 0.47) and D. melanogaster (NCBI ID: NP_995955, expect value 0.002).
FIGURE 4. YIL064W is well conserved in nature.
The amino acid sequence of YIL064W was compared to the non-redundant protein database using the BLAST algorithm to obtain expect values. Amino acid sequence alignments determined by CLUSTALW2 are shown for S. cerevisiae (NCBI ID: NP_012200), C. albicans (NCBI ID: XP_715004, expect value 2e-49), S. pombe (NCBI ID: NP_595254, expect value 3e-37), D. melanogaster (NCBI ID: NP_608733, expect value 8e-31), C. elegans (NCBI ID: NP_500612, expect value 1e-25), A. thaliana (NCBI ID: NP_176841, expect value 3e-21), and H. sapiens (NCBI ID: NP_997719, expect value 2e-23). Boxes identify the proposed motifs of the conserved AdoMet binding domain.
We compared the monomethylation sites of eEF1A to those of other substrates of SET domain methyltransferases (Table 1). While we found little or no similarity in the sequence surrounding lysine 30, we noted that lysine 390 was preceded by a proline residue that also preceded a methylated lysine residue in the ribosomal proteins Rpl23 and Rpl12 as well as cytochrome c. Furthermore, lysine 390 in eEF1A and the methylated lysine 3 in Rpl12 are both followed by a phenylalanine residue. These considerations suggest that the SET domain methyltransferase Efm1 modifies lysine 390, although this needs to be experimentally confirmed. It is important to note that the overall native structure of at least one substrate protein for SET domain methyltransferases has been observed to be required for substrate recognition and this may be a determining factor rather than, or in addition to, the primary sequence [31].
TABLE 1.
Translation/cytochrome c SET domain subfamily in S. cerevisiae
| Methyltransferase | BLAST expect value |
Substrate | Extent of methylation |
Site | |
|---|---|---|---|---|---|
| Efm1 | YHL039W | - | eEF1A K30 ? | mono | TGHLIY-K-CGGIDK |
| eEF1A K390 ? | mono | KLEDHP-K-FLKSGD | |||
| Rkm1 | YPL208W | 3.00E-174 | Rpl23 K105 | di | GVIANP-K-GEMKGS |
| Rpl23 K109 | NPKGEM-K-GSAITG | ||||
| Rkm2 | YDR198C | 4.00E-86 | Rpl12 K3 | tri | MPP-K-FDPNEV |
| Rkm3 | YBR030W | 7.00E-77 | Rpl42 K40 | mono | SLFAQG-K-RRYDRK |
| Rkm4 | YDR257C | 7.00E-100 | Rpl42 K55 | mono | GFGGQT-K-PVFHKK |
| Ctm1 | YHR109W | 1.00E-19 | Cytochrome c K72 | tri | EYLTNP-K-KYIPGT |
4. Discussion
In this work we have identified two yeast genes required for eEF1A methylation. Previous work found no phenotypic change or in vitro translational defects when the four internal lysine methylation sites of eEF1A were changed to arginine residues [12]. Additionally, strains with deletion mutants in either gene are viable and no phenotype has been observed to date for EFM1 mutants. However, mutant studies of SEE1 have suggested a role for this protein in vesicle trafficking, particularly in early endocytotic transport [32]. With the observation that eEF1A can associate with filamentous actin and microtubules [13–15], we suggest that eEF1A methylation may modulate the cytoskeletal interactions involved in vesicle trafficking.
The ortholog of See1 in humans is METTL10, a protein that has already been annotated in the UniProt database as having a seven beta strand AdoMet binding domain. Additionally, METTL10 has been recently identified as being a target for mitotic phosphorylation at serine 21 in a [pS/pT]-P cyclin-dependent kinase recognition site [33]. The extent of phosphorylation at this site increased 7.3 fold in mitotic arrest, suggesting a regulatory control.
At this time, we have no evidence for the functional role of the Efm1 methyltransferase. Unlike See1, the distribution of Efm1 in nature is narrow and appears to be limited to fungal species. The two best BLAST matches of Efm1 with human proteins are putative SET domain family proteins SETD3 and SETD4 (Fig. 3). However, these species are unlikely to have the same function because BLAST searches with these latter proteins demonstrate comparable similarity to yeast ribosomal methyltransferases. Additionally, human SETD4 has significantly better matches to S. pombe (NCBI ID: NP_588349) and C. albicans (NCBI ID: XP_722076) proteins that are distinct from the Efm1 orthologs shown in Fig. 3. Interestingly, the identification of Efm1 now completes the yeast translation/cytochrome c SET domain subfamily, including members that modify ribosomal proteins and cytochrome c [9, 24, 31, 34].
Supplementary Material
Acknowledgments
This work was funded by NIH Grant GM026020. R.S.L. was supported by the UCLA Cellular and Molecular Biology Training Program funded by NIH Grant GM007185. Mass spectrometry was performed in the UCLA Molecular Instrumentation Center supported by Grant S10RR024605 from the National Center for Research Resources. We thank Drs. Renee Chosed and Sharon Dent for providing yeast strains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used: eEF1A, eukaryotic elongation factor 1A; [3H]AdoMet, S-adenosyl-L-[methyl-3H]methionine; AdoMet, S-adenosyl-L-methionine.
References
- 1.Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 2.Secombe J, Eisenman RN. The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection. Cell Cycle. 2007;6:1324–1328. doi: 10.4161/cc.6.11.4269. [DOI] [PubMed] [Google Scholar]
- 3.Couture JF, Dirk LM, Brunzelle JS, Houtz RL, Trievel RC. Structural origins for the product specificity of SET domain protein methyltransferases. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20659–20664. doi: 10.1073/pnas.0806712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr. Opin. Genet. Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Rathert P, Dhayalan A, Ma H, Jeltsch A. Specificity of protein lysine methyltransferases and methods for detection of lysine methylation of non-histone proteins. Mol. Biosyst. 2008;4:1186–1190. doi: 10.1039/b811673c. [DOI] [PubMed] [Google Scholar]
- 6.Lan F, Shi Y. Epigenetic regulation: methylation of histone and non-histone proteins. Sci. China C. Life Sci. 2009;52:311–322. doi: 10.1007/s11427-009-0054-z. [DOI] [PubMed] [Google Scholar]
- 7.Ng SS, Yue WW, Oppermann U, Klose RJ. Dynamic protein methylation in chromatin biology. Cell Mol. Life Sci. 2009;66:407–422. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polevoda B, Sherman F. Methylation of proteins involved in translation. Mol. Microbiol. 2007;65:590–606. doi: 10.1111/j.1365-2958.2007.05831.x. [DOI] [PubMed] [Google Scholar]
- 9.Webb KJ, Laganowsky A, Whitelegge JP, Clarke SG. Identification of two SET domain proteins required for methylation of lysine residues in yeast ribosomal protein Rpl42ab. J. Biol. Chem. 2008;283:35561–35568. doi: 10.1074/jbc.M806006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiatt WR, Garcia R, Merrick WC, Sypherd PS. Methylation of elongation factor 1 alpha from the fungus Mucor. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3433–3437. doi: 10.1073/pnas.79.11.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavallius J, Zoll W, Chakraburtty K, Merrick WC. Characterization of yeast EF-1 alpha: non-conservation of post-translational modifications. Biochim. Biophys. Acta. 1993;1163:75–80. doi: 10.1016/0167-4838(93)90281-u. [DOI] [PubMed] [Google Scholar]
- 12.Cavallius J, Popkie AP, Merrick WC. Site-directed mutants of post-translationally modified sites of yeast eEF1A using a shuttle vector containing a chromogenic switch. Biochim. Biophys. Acta. 1997;1350:345–358. doi: 10.1016/s0167-4781(96)00181-9. [DOI] [PubMed] [Google Scholar]
- 13.Moore RC, Cyr RJ. Association between elongation factor-1alpha and microtubules in vivo is domain dependent and conditional. Cell Motil. Cytoskeleton. 2000;45:279–292. doi: 10.1002/(SICI)1097-0169(200004)45:4<279::AID-CM4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Ejiri S. Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Biosci. Biotechnol. Biochem. 2002;66:1–21. doi: 10.1271/bbb.66.1. [DOI] [PubMed] [Google Scholar]
- 15.Lamberti A, Caraglia M, Longo O, Marra M, Abbruzzese A, Arcari P. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: review article. Amino Acids. 2004;26:443–448. doi: 10.1007/s00726-004-0088-2. [DOI] [PubMed] [Google Scholar]
- 16.Sherman M, Sypherd PS. Role of lysine methylation in the activities of elongation factor 1 alpha. Arch. Biochem. Biophys. 1989;275:371–378. doi: 10.1016/0003-9861(89)90384-6. [DOI] [PubMed] [Google Scholar]
- 17.Van Noort JM, Kraal B, Sinjorgo KMC, Persoon NLM, Johanns ESD, Bosch L. Methylation in vivo of elongation factor EF-Tu at lysine-56 decreases the rate of tRNA-dependent GTP hydrolysis. Eur. J. Biochem. 1986;160:557–561. doi: 10.1111/j.1432-1033.1986.tb10074.x. [DOI] [PubMed] [Google Scholar]
- 18.Porras-Yakushi TR, Whitelegge JP, Miranda TB, Clarke S. A novel SET domain methyltransferase modifies ribosomal protein Rpl23ab in yeast. J. Biol. Chem. 2005;280:34590–34598. doi: 10.1074/jbc.M507672200. [DOI] [PubMed] [Google Scholar]
- 19.Lee SW, Berger SJ, Martinović S, Pasa-Tolić L, Anderson GA, Shen Y, Zhao R, Smith RD. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5942–5947. doi: 10.1073/pnas.082119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Valenzuela JA, Gibbon BC, Hughes PA, Dreher TW, Larkins BA. eEF1A isoforms change in abundance and actin-binding activity during maize endosperm development. Plant Physiol. 2003;133:1285–1295. doi: 10.1104/pp.103.027854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipson RS, Webb KJ, Clarke SG. Rmt1 catalyzes zinc-finger independent arginine methylation of ribosomal protein Rps2 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2010;391:1658–1662. doi: 10.1016/j.bbrc.2009.12.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miranda TB, Lowenson JD, Clarke S. A new type of protein methylation activated by tyrphostin A25 and vanadate. FEBS Let. 2004;577:181–186. doi: 10.1016/j.febslet.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 23.Zobel-Thropp P, Yang MC, Machado L, Clarke S. A novel post-translational modification of yeast elongation factor 1A. Methylesterification at the C terminus. J. Biol. Chem. 2000;275:37150–37158. doi: 10.1074/jbc.M001005200. [DOI] [PubMed] [Google Scholar]
- 24.Porras-Yakushi TR, Whitelegge JP, Clarke S. A novel SET domain methyltransferase in yeast: Rkm2-dependent trimethylation of ribosomal protein L12ab at lysine 10. J. Biol. Chem. 2006;281:35835–35845. doi: 10.1074/jbc.M606578200. [DOI] [PubMed] [Google Scholar]
- 25.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Séraphin B, Aasland R, Stewart AF. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, Tatchell K, Hawke DH, Kobayashi R, Dent SY. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 30.Petrossian T, Clarke S. Bioinformatic identification of novel methyltransferases. Epigenomics. 2009;1:163–175. doi: 10.2217/epi.09.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takakura H, Yamamoto T, Sherman F. Sequence requirement for trimethylation of yeast cytochrome c. Biochem. 1997;36:2642–2648. doi: 10.1021/bi962245n. [DOI] [PubMed] [Google Scholar]
- 32.Martín-Granados C, Riechers SP, Stahl U, Lang C. Absence of See1p, a widely conserved Saccharomyces cerevisiae protein, confers both deficient heterologous protein production and endocytosis. Yeast. 2008;25:871–877. doi: 10.1002/yea.1641. [DOI] [PubMed] [Google Scholar]
- 33.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci U. S. A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polevoda B, Martzen MR, Das B, Phizicky EM, Sherman F. Cytochrome c methyltransferase, Ctm1p, of yeast. J. Biol. Chem. 2000;275:20508–20513. doi: 10.1074/jbc.M001891200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.