Summary
Caspase-9 activation is critical for intrinsic cell death. The activity of caspase-9 is increased dramatically upon association with the apoptosome and the apoptosome bound caspase-9 is the caspase-9 holoenzyme (C9Holo). In this study, we use quantitative enzymatic assays to fully characterize C9Holo and a leucine-zipper linked dimeric caspase-9 (LZ-C9). We surprisingly show that LZ-C9 is more active than C9Holo for the optimal caspase-9 peptide substrate LEHD-AFC, but is much less active than C9Holo for the physiological substrate pro-caspase-3. The measured Km values of C9Holo and LZ-C9 for LEHD-AFC are similar, demonstrating that dimerization is sufficient for catalytic activation of caspase-9. The lower activity of C9Holo against LEHD-AFC may be attributed to incomplete C9Holo assembly. However, the measured Km of C9Holo for pro-caspase-3 is much lower than that of LZ-C9. Therefore, in addition to dimerization, the apoptosome activates caspase-9 by enhancing its affinity for pro-caspase-3, which is important for pro-caspase-3 activation at the physiological concentration.
Introduction
Caspase activation is a hallmark of apoptotic cell death (Riedl and Shi, 2004; Salvesen, 2002). According to their sequence of activation, caspases may be divided into two groups: initiator caspases such as caspase-8, 9 and 10, and effector caspases such as caspase-3 and 7 (Salvesen, 2002; Shi, 2002). In the classical paradigm of apoptosis, there are two general pathways for caspase activation and apoptosis induction (Danial and Korsmeyer, 2004). In the extrinsic apoptosis pathway, death ligands interact with and activate death receptors, leading to the assembly of the death inducing signaling complex (DISC) that activates caspase-8 and caspase-10. In the intrinsic apoptotic pathway, cellular insults lead to mitochondrial alteration and cytochrome c release. In the presence of cytochrome c and dATP or ATP, the adapter protein Apaf-1 assembles into an oligomeric structure known as the apoptosome, which recruits and activates caspase-9 (Li et al., 1997; Zou et al., 1997). Caspase-8, 9, and 10 then activate effector caspases such as caspase-3 and 7, which cleave a wide range of substrates to execute apoptosis.
Caspases are synthesized as single chain pro-caspases, which undergo intra-chain cleavage to generate the large and small subunits. The molecular requirements for caspases to be catalytically active have been revealed by previous elegant structural studies on active and inactive caspases (Chai et al., 2001; Riedl et al., 2001; Wei et al., 2000). First, a caspase needs to be dimeric to be active because regions from both its own chain and the neighboring chain form the caspase active sites. Specifically, these regions include the L1 and L2 loops of the large subunit and the L3 and L4 loops of the small subunit of one chain and the L2′ loop of the small subunit of the neighboring chain. Second, intra-chain cleavage facilitates formation of the active site because the L2 and L2′ loops are released upon cleavage to become part of the loop-bundle that supports the active site. This intra-chain cleavage may also stabilize dimerization of some caspases (Boatright et al., 2003; Donepudi et al., 2003). The requirement for cleavage differs among caspases in a manner that appears to depend on the length of the L2-L2′ linker region.
Because effector caspases are constitutive dimers, their activation is strictly a consequence of intra-chain cleavage by initiator caspases. In contrast to effector caspase activation, intra-chain cleavage does not appear to be the crucial factor for initiator caspase activation (Boatright et al., 2003; Donepudi et al., 2003; Srinivasula et al., 2001; Stennicke et al., 1999). For example, caspase-9 can be activated without proteolytic processing. Processed caspase-9 alone is not much more active than pro-caspase-9. A small population of dimerized uncleavable caspase-8 is as active as processed caspase-8.
An induced proximity model for initiator caspase activation was initially proposed from observations that initiator caspases fused to heterologous dimerizing partners underwent processing and induced cell death (MacCorkle et al., 1998; Muzio et al., 1998; Srinivasula et al., 1998; Yang et al., 1998a; Yang et al., 1998b). This model is consistent with the presence of prodomains in initiator caspases that participate in the assembly of oligomeric complexes in an apoptotic signal dependent fashion. In this model, the pro-form of an initiator caspase has low activity that is sufficient for autocatalytic processing when brought into close proximity in these complexes. More recent biochemical studies have led to a revised proximity-induced dimerization model for initiator caspase activation because caspase-8 and 9 are not constitutive dimers in solution and specific homo-dimerization appears to be crucial for their activation (Boatright et al., 2003; Donepudi et al., 2003; Renatus et al., 2001).
For caspase-9, its activity is increased dramatically (~103 fold) upon association with the apoptosome and the apoptosome bound caspase-9 is the caspase-9 holoenzyme (C9Holo) (Jiang and Wang, 2000; Rodriguez and Lazebnik, 1999; Srinivasula et al., 2001). Based on the proximity-induced dimerization model, the function of the apoptosome is to promote homo-dimerization of caspase-9 and therefore its activation. In the context of the cryo-EM structure of the heptameric apoptosome, this homo-dimerization could occur among caspase-9 chains bound on the apoptosome or the apoptosome-bound caspase-9 may dimerize with additional caspase-9 monomers from the solution (Acehan et al., 2002; Shi, 2004; Yu et al., 2005).
Although dimerization does enhance caspase-9 activity, it is not known whether dimerization is sufficient for caspase-9 activation. To address this question, a dimeric form of caspase-9 needs to be constructed and its activity needs to be quantitatively compared with C9Holo. In an effort to answer this question, a constitutive caspase-9 dimer was engineered by replacing the dimerization residues of caspase-9 with those from caspase-3 (Chao et al., 2005). This engineered caspase-9 dimer is more active than caspase-9, but much less active than C9Holo. Therefore, it was argued that dimerization of caspase-9 is not sufficient for its activation (Chao et al., 2005).
Because replacing residues at the dimerization interface may have compromised the catalytic activity of the engineered caspase-9 dimer, we took an alternative approach to address whether dimerization is sufficient for caspase-9 activation. We replaced the pro-domain of caspase-9 (caspase recruitment domain, CARD) with the leucine-zipper dimerization domain of GCN4 (Hu et al., 1990) to promote caspase-9 homo-dimerization through its intrinsic weak dimerization interface (LZ-C9). We surprisingly found that LZ-C9 is not less active, but more active than C9Holo in cleaving the tetrapeptide substrate LEHD-AFC. The measured Km values of LZ-C9 and C9Holo for LEHD-AFC are similar. The lower activity of C9Holo against LEHD-AFC may be attributed to incomplete C9Holo assembly. This establishes that dimerization is sufficient for catalytic activation of caspase-9. In contrast, LZ-C9 activity for pro-caspase-3, the physiological substrate of caspase-9, is much lower than C9Holo. This inverse activity of LZ-C9 and C9Holo for LEHD-AFC and pro-caspase-3 is entirely unexpected. It is not due to feedback cleavage of caspase-9 by caspase-3 or promotion of processed caspase-3 activity by C9Holo. Instead, Km determination reveals that the Km of C9Holo for pro-caspase-3 is much lower than that of LZ-C9, suggesting that apoptosome activates caspase-9 by enhancing its affinity to pro-caspase-3. This affinity enhancement is important for efficient pro-caspase-3 activation at the low physiological concentration and makes C9Holo a specific and optimal pro-caspase-3 processing machine. Collectively, these studies provide unforeseen insights into the molecular basis of caspase-9 activation by the apoptosome.
Results
Engineering of LZ-C9, a dimeric caspase-9
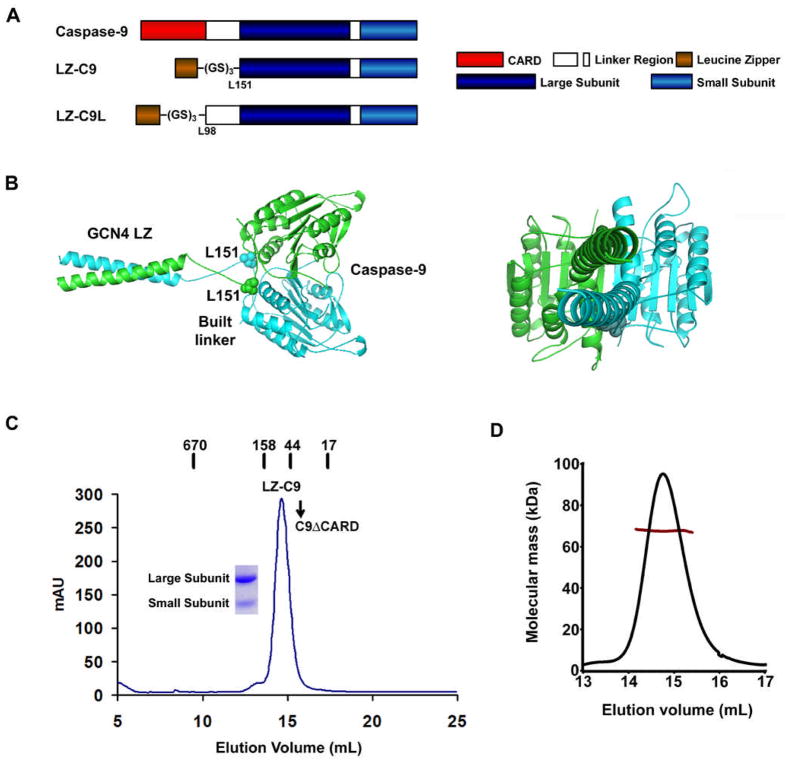
To determine whether caspase-9 dimerization is sufficient for its activation, we removed the CARD of pro-caspase-9 and replaced it with the leucine zipper dimerization domain of the transcription factor GCN4. A six residue linker was added in between the leucine zipper and the beginning of the catalytic domain of caspase-9 (LZ-C9, residues 151–416, Figure 1A). In the crystal structure of dimeric CARD-deleted caspase-9 (Renatus et al., 2001), residues L151 at the beginning of the two large subunits in the caspase-9 dimer reside at the same surface of the molecule with a Cα distance of 21 Å. In the crystal structure of the GCN4 leucine zipper, the last two residues are disordered and the distance between the last ordered residue and its symmetry mate is 13 Å (O’Shea et al., 1991). A minimal distance of 4 Å is required to bridge each end of the parallel leucine zipper to L151 of caspase-9. Because the two disordered leucine zipper residues and a linker of 6 residues could stretch to approximately 30Å, the LZ-C9 construct should be sufficient to allow intrinsic caspase-9 dimerization to occur without any constraints, as also shown in a molecular model of LZ-C9 we constructed (Figure 1B).
Figure 1.
Engineering of LZ-C9, a dimeric caspase-9. (A) Schematic diagram of caspase-9, LZ-C9 and LZ-C9L. (B) A molecular model of LZ-C9 shown as a ribbon diagram. L151 residues of dimeric caspase-9 are shown as space filling models. Two orientations are show, left panel with the twofold axis horizontal and right panel with the twofold axis into the page. (C) Gel filtration profile of LZ-C9. Locations of molecular weight standards and C9ΔCARD are marked. SDS-PAGE of the peak fraction is shown. (D) Multiangle light scattering of LZ-C9. The trace in black corresponds to light scattering signals at 90°. The trace in dark red shows the variation in molecular mass determination as a function of peak position or elution volume.
The calculated molecular weight of LZ-C9 is 34.7 kD. It elutes as a single peak at 14.6 ml in a Superdex 200 HR 10/300 gel filtration column, suggesting that it is a dimer (Figure 1C). Higher order oligomerization was not observed. In contrast, CARD deleted caspase-9 (C9ΔCARD) with a calculated molecular weight of 32.7 kD elutes at 15.5 ml in the same gel filtration column, which is consistent with the monomeric state of caspase-9. Since gel filtration column elution positions may be influenced by both mass and shape, we used static multiangle light scattering (MALS) in conjunction with refractive index measurements to determine the molecular mass of intact LZ-C9. Consistent with the gel filtration data, MALS measurements gave a molecular mass of 67.6 kD (0.7% fitting error) with a polydispersity of 1.000, confirming the dimeric state of LZ-C9 (Figure 1D). In contrast, MALS analysis showed that C9ΔCARD is a monomer with a measured molecular mass of 35.6 kD (0.7% fitting error) and a polydispersity of 1.000.
LZ-C9 is more active than C9Holo for the tetrapeptide substrate LEHD-AFC but much less active than C9Holo for the physiological substrate pro-caspase-3
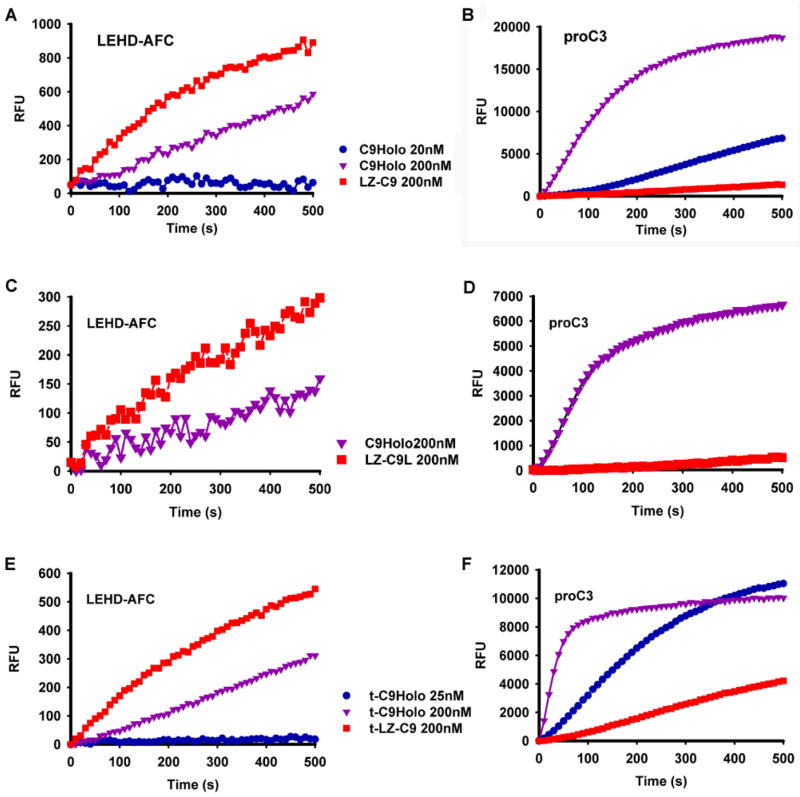
We used the fluorogenic peptide substrate LEHD-AFC, which is an optimal peptide substrate for caspase-9 (Thornberry et al., 1997), to compare the activity of LZ-C9 with C9Holo. Because intra-chain cleavage does not play a significant role in caspase-9 activation, both caspase-9 and LZ-C9 we used are the processed forms. Unexpectedly, at the same caspase-9 concentrations, LZ-C9 gave a significantly faster rate of substrate cleavage than C9Holo (Figure 2A).
Figure 2.
LZ-C9 and C9Holo have inverted activity for LEHD-AFC and pro-caspase-3 substrates. (A, C, E) Reaction progress curves of tetrapeptide substrate LEHD-AFC cleavage by LZ-C9, LZ-C9L or t-LZ-C9, respectively, and C9Holo at indicated concentrations. (B, D, F) Reaction progress curves of DEVD-AMC cleavage by caspase-3 activated by LZ-C9, LZ-C9L or t-LZ-C9, respectively, and C9Holo at the same concentrations as in A, C and E.
Because the physiological substrate of caspase-9 is pro-caspase-3, we measured the activity of LZ-C9 and C9Holo for pro-caspase-3 activation. An indirect fluorogenic assay was used, in which cleavage of the caspase-3 substrate DEVD-AMC by caspase-9 activated caspase-3 was measured. In contrast to the higher activity of LZ-C9 for the LEHD-AFC peptide substrate, C9Holo is much more efficient in activating pro-caspase-3 than LZ-C9 (Figure 2B).
Since there are approximately 50 additional residues between the CARD and the beginning of the catalytic domain, we also constructed a longer LZ-C9 construct that includes this additional region (LZ-C9L, residues 98–416). MALS analysis showed that LZ-C9L, with a calculated molecular weight of 40.3 kD, is also a dimer with a measured molecular mass of 79.1 kD (2.0% fitting error) and a polydispersity of 1.000. We tested the activity of LZ-C9L against LEHD-AFC and pro-caspase-3. In comparison with C9Holo, LZ-C9L is more effective in cleaving LEHD-AFC, but is much less active against the physiological substrate pro-caspase-3 (Figure 2C, 2D). Therefore, LZ-C9 (or LZ-C9L) and C9Holo exhibit an inverse capability in cleaving the LEHD-AFC substrate and the physiological substrate pro-caspase-3.
Feedback cleavage of caspase-9 by caspase-3 does not contribute to the inverse activity of LZ-C9 and C9Holo on LEHD-AFC and pro-caspase-3
This inverse activity of LZ-C9 and C9Holo for LEHD-AFC and pro-caspase-3 is intriguing. The human caspase-9 self-mediated intra-chain cleavage occurs at Asp 315 (sequence 312PEPD315) to generate the large (p35) and small (p12) caspase-9 subunits. However, it has also been shown that activated caspase-3 can further feedback cleave the N-terminus of the small subunit of caspase-9 at Asp330 (sequence 327DQLD330) and enhance its activity (Zou et al., 2003). Therefore, we wondered if this feedback cleavage could contribute to the observed difference between C9Holo and LZ-C9 towards the two different substrates.
We mixed the cell pellets of caspase-9 and LZ-C9 with those of non-tagged caspase-3 and purified the truncated caspase-9 forms (t-caspase-9 and t-LZ-C9) that have their p12 converted to p10 by the additional cleavage at Asp330. We then repeated the above experiments to determine whether our observations still hold true for these truncated caspase-9 forms (t-C9Holo and t-LZ-C9). Measurement of LEHD-AFC cleavage in the direct fluorogenic assay and pro-caspase-3 activation in the indirect fluorogenic assay showed that the inverse activity of t-LZ-C9 and t-C9Holo for these substrates is preserved (Figure 2E, 2F) Therefore, caspase-3 mediated cleavage of caspase-9 does not explain this discrepancy in capability between LZ-C9 and C9Holo for LEHD-AFC and pro-caspase-3.
Association of processed caspase-3 with C9Holo does not increase caspase-3 activity
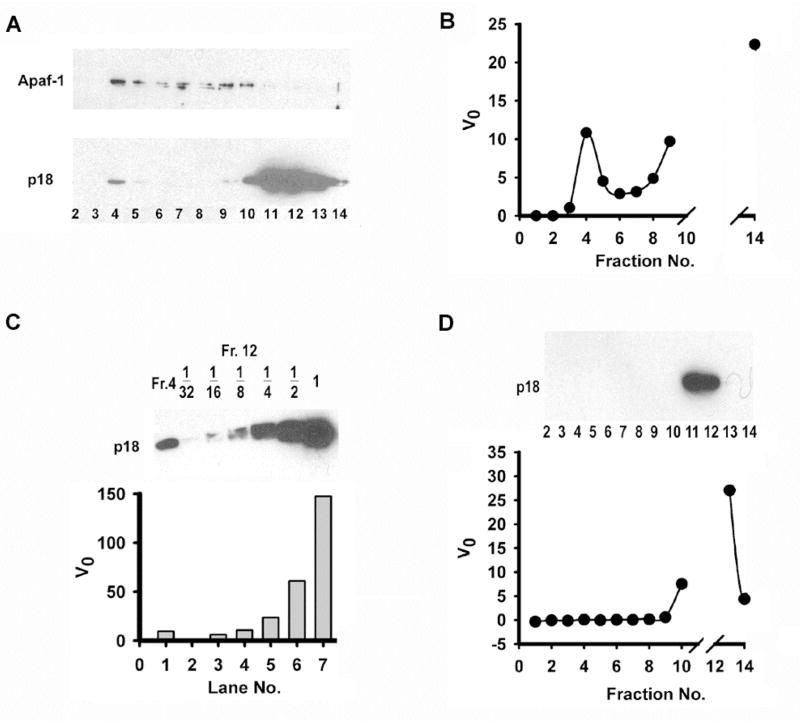
We then examined whether the high efficiency of pro-caspase-3 activation by C9Holo is due to potential promotion of processed caspase-3 catalytic activity, making C9Holo processed caspase-3 a caspase-3 holoenzyme. To determine this, we first examined whether caspase-3 can stay associated with C9Holo after processing. We mixed Apaf-1, cytochrome c, dATP, full-length caspase-9 and excess pro-caspase-3 and subject the mixture to gel filtration chromatography. All pro-caspase-3 was cleaved. Indeed, some processed caspase-3 was associated with C9Holo in the early elution fractions (Figure 3A).
Figure 3.
Association of C9Holo with processed caspase-3 does not enhance caspase-3 activity. (A) Excess pro-caspase-3 was incubated with C9Holo and the mixture was subjected to gel-filtration. Aliquots of fractions were resolved by 15% SDS-PAGE followed by western blotting using antibodies against Apaf-1 (upper panel) and caspase-3 (lower panel). (B) Fractions were also assayed for their cleavage activity against DEVD-AMC. The initial velocities were plotted as a function of fraction numbers. Activities of fractions 10–13 were too high for initial velocities to be determined precisely and they were not shown. (C) Fraction 12 was serial diluted in 2-fold steps. Aliquots of each dilution were analyzed by 15% SDS-PAGE along with an aliquot of fraction 4 and blotted against caspase-3 (upper panel). They were also assayed for cleavage activity against DEVD-AMC. The initial velocities were plotted (lower panel). (D) Excess pro-caspase-3 was incubated with LZ-C9 under same conditions as in (A) and the mixture was subjected to gel-filtration. Fractions were resolved by 15% SDS-PAGE and blotted against caspase-3 (upper panel). Aliquots of fractions were assayed for cleavage activity against DEVD-AMC. The initial velocities were plotted as a function of fraction numbers (lower panel). Activities of fractions 11 and 12 were too high for initial velocities to be determined precisely and they were not shown.
We then measured the DEVD-AMC cleavage activities of caspase-3 that was either co-eluted with C9Holo or eluted alone to determine whether C9Holo-associated caspase-3 exhibits higher activity. The experiment showed that similar protein levels of caspase-3 in either fraction gave similar catalytic activities (Figure 3B, compare fractions 4 and 9). Serial dilution of a high concentration caspase-3 fraction to match the caspase-3 level in the C9Holo fraction confirmed the same observation (Figure 3C, compare fraction 4 with 4-fold and 8-fold diluted fraction 12). Similar experiments replacing C9Holo with LZ-C9 showed the absence of caspase-3 protein or activity in the early elution fractions (Figure 3D). These experiments demonstrate that C9Holo-associated caspase-3 does not exhibit higher catalytic activity to cause the observed difference in LZ-C9 and C9Holo against LEHD-AFC and pro-caspase-3.
LZ-C9 and C9Holo have similar Km values for LEHD-AFC and dimerization is sufficient for catalytic activation of caspase-9
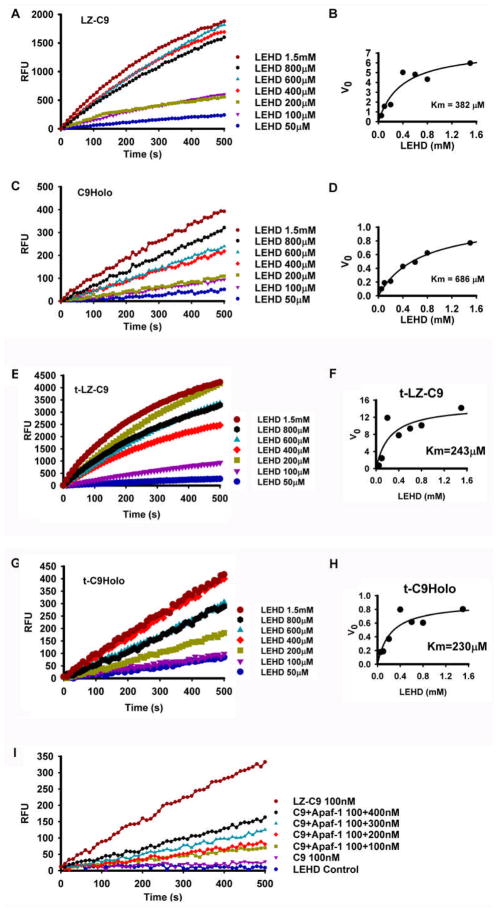
To further characterize the catalytic parameters of LZ-C9 and C9Holo, we measured the initial rates of substrate cleavage at different concentrations of LEHD-AFC, from which the Km values of LZ-C9 and C9Holo for the peptide substrate were determined. The Km values of LZ-C9 and C9Holo for LEHD-AFC are similar, 465.7 ± 118.4 μM and 686.2 ± 136.9 μM, respectively (Figure 4A–D, Table 1). The Km values for the feedback cleaved t-LZ-C9 and t-C9Holo are 260.0 ± 22.84 μM and 276.5 ± 65.48μM, respectively (Figure 4E–H, Table 1). They are high in comparison with the Km values of other caspases for their peptide substrates (Zhou et al., 1998).
Figure 4.
LZ-C9 and C9Holo have similar Km values for tetrapeptide substrate LEHD-AFC. (A, C, E, G) Reaction progress curves of LEHD-AFC cleavage at different concentrations by LZ-C9 (100 nM), C9Holo (100 nM full length caspase-9, 100 nM Apaf-1, 10 μM dATP and 500 nM cytochrome c), t-LZ-C9 (100 nM) and t-C9Holo (100 nM full length truncated caspase-9, 100 nM Apaf-1, 10 μM dATP and 500 nM cytochrome c), respectively. (B, D, F, H) Plots of initial velocities as a function of LEHD-AFC concentrations for LZ-C9, C9Holo, t-LZ-C9 and t-C9Holo, respectively. (I) Reaction progress curves of LEHD-AFC cleavage by caspase-9 at different Apaf-1 concentrations.
Table 1.
Measured Km values of C9Holo, t-C9Holo, LZ-C9 and t-LZ-C9 for LEHD-AFC and pro-caspase-3.
| Km for LEHD-AFC | Km for pro-caspase-3 | |
|---|---|---|
| C9Holo | 686.2 ± 136.9 μM† | Average 139.3 ± 23.19 nM#, & |
| t-C9Holo | Average 276.5 ± 65.48 μM#, $ | 79.8 ± 18.5 nM† |
| LZ-C9 | Average 465.7 ± 118.4 μM#, & | High@ |
| t-LZ-C9 | Average 260.0 ± 22.84 μM#, $ | High@ |
The errors in individual Km measurements represent the scatter of the data points from the hyperbolic fittings.
When two measurements are available, the average Km values are shown along with the individual measurements. The errors in average Km values are standard deviations from the two measurements.
One protein preparation was used in the two individual measurements for obtaining the average Km values.
Two different protein preparations were used in the two individual measurements for obtaining the average Km values.
Km values could not be determined from the current data.
Since caspase-9 and Apaf-1 need to be assembled into the large multimeric caspase-9 holoenzyme for caspase-9 activation, we asked whether the observed lower activity of C9Holo for LEHD-AFC is due to incomplete apoptosome and C9Holo assembly and hence lower effective concentration of C9Holo. Alternatively, a lower intrinsic turnover rate, kcat, of C9Holo, can also contribute to the observed lower activity. To distinguish these two possibilities, we titrated excess Apaf-1 into caspase-9 to facilitate assembly of C9Holo. This titration increased the rate of LEHD-AFC cleavage when caspase-9 concentration remained constant (Figure 4I). At the highest Apaf-1 concentration we could provide, the activity of C9Holo did not catch up with the activity of LZ-C9. These data suggest that holoenzyme assembly is a limiting factor contributing to the lower activity of C9Holo to the LEHD-AFC substrate. The data also suggest that the observed striking difference between C9Holo and LZ-C9 in activating pro-caspase-3 (Figure 2B) is still an underestimation because of incomplete holoenzyme assembly. On the other hand, it is likely that the catalytic activities of C9Holo and LZ-C9, exemplified in Km and kcat for LEHD-AFC, are equivalent, Thus dimerization appears to be sufficient for catalytic activation of caspase-9.
C9Holo has dramatically higher substrate affinity for pro-caspase-3 than LZ-C9
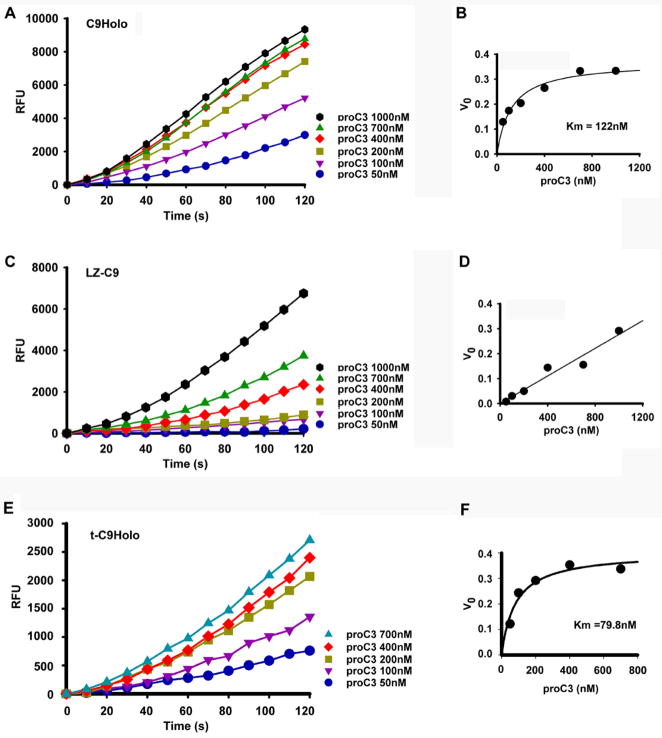
Then why does C9Holo activate caspase-3 much more efficiently than LZ-C9? Is it possible that C9Holo possesses a higher affinity for pro-caspase-3 than LZ-C9? To examine this, we measured the rates of substrate cleavage at different concentrations of pro-caspase-3 to determine the Km values of C9Holo and LZ-C9 for pro-caspase-3. We used the indirect fluorogenic assay, in which cleavage of the caspase-3 substrate DEVD-AMC was measured. The slope of the DEVD-AMC cleavage at a given time point is indicative of the active casapse-3 concentration at that time point and was used to derive Km values for the pro-caspase-3 cleavage reaction. Surprisingly, the Km of C9Holo for pro-caspase-3 is 139.3 ± 23.19 nM (Figure 5A–B, Table 1), about three orders of magnitude lower than that for LEHD-AFC. In contrast, the Km of LZ-C9 for pro-caspase-3 was estimated to be much higher; at the highest possible concentration of pro-caspase-3 we could supply, the reaction did not show any sign of saturation (Figure 5C-D). This difference in Km values between C9Holo and LZ-C9 is also exemplified in the much higher effect of increasing pro-caspase-3 concentration on the ability of LZ-C9 than on the ability of C9Holo to activate pro-caspase-3 (Figure 5A, 5C).
Figure 5.
C9Holo has much reduced Km for pro-caspase-3. (A, C, E) Reaction progress curves of pro-caspase-3 activation at different pro-caspase-3 concentrations by C9Holo (25 nM), LZ-C9 (200 nM) and t-C9Holo (25 nM), respectively. These were measured indirectly by the cleavage of the caspase-3 substrate DEVD-AMC. (B, D, F) Plots of initial velocities as a function of pro-caspase-3 concentrations for C9Holo, LZ-C9 and t-C9Holo, respectively.
Similarly, we showed that the Km value of t-C9Holo for pro-caspase-3 is 79.8 ± 18.5 nM (Figure 5E–F, Table 1), similar to its non-truncated counterpart. These data demonstrate that both C9Holo and t-C9Holo have high affinity to pro-caspase-3. Therefore, we show unexpectedly that one important mechanism of caspase-9 activation by the apoptosome is by enhancing its affinity to the physiological substrate pro-caspase-3.
Discussion
Dimerization is sufficient for catalytic activation of caspase-9
Caspase-9 is unusual in that it is only active when bound to the apoptosome. Such activation is so dramatic that the apoptosome-bound caspase-9 is known as the caspase-9 holoenzyme (Rodriguez and Lazebnik, 1999; Srinivasula et al., 2001). Other caspases, including caspase-8, an initiator caspase in the extrinsic apoptosis pathway, can directly activate their substrates without the requirement to be bound to an accelerator. Our data on the catalytic properties of LZ-C9 and C9Holo show that caspase-9 dimerized by a leucine zipper is not less active, but more active than the caspase-9 holoenzyme. This demonstrates that dimerization is sufficient for catalytic activation of caspase-9.
The measured Km values of caspase-9 holoenzyme and LZ-C9 for LEHD-AFC are similar. They are also similar to the previously reported Km values of caspase-9 for LEHD-AFC (Renatus et al., 2001; Zou et al., 2003). How about the intrinsic turnover rate of caspase-9 holoenzyme in comparison with LZ-C9? The similar Km but lower activity of the caspase-9 holoenzyme could indicate that it possesses a lower kcat than LZ-C9 for LEHD-AFC. However, because the apoptosome might also mediate caspase-9 dimerization, it is unlikely that its kcat is lower than that of leucine-zipper dimerized caspase-9. Since caspase-9 holoenzyme requires assembly, a more plausible explanation to the lower activity is that the effective concentration of C9Holo is lower than its apparent concentration. This is supported by the increase of activity upon titration of more Apaf-1. Therefore, we conclude that the catalytic activity of LZ-C9 is equivalent to that of caspase-9 holoenzyme for peptide substrate LEHD-AFC. Our data contradict the observation from an engineered dimeric caspase-9 made by replacing the weak dimerization residues of caspase-9 with the strong dimerization residues of caspase-3 (Chao et al., 2005). The difference may be that LZ-C9 likely promotes dimerization through its intrinsic dimerization interface, while the altered dimerization interface in the engineered dimer may have affected its catalytic ability, even against LEHD-AFC.
Enhanced affinity of C9Holo for pro-caspase-3 is important for efficient pro-caspase-3 activation at the physiological concentration
Although dimerization appears to be sufficient for establishing the catalytic competency of caspase-9, it is not sufficient for the high efficiency of caspase-9 holoenzyme to support pro-caspase-3 activation because LZ-C9 is much less efficient in pro-caspase-3 activation. Our data indicate that pro-caspase-3 is a much better substrate for the caspase-9 holoenzyme with dramatically increased affinity. Therefore, apoptosome activates caspase-9 by both dimerization and enhanced affinity to its physiological substrate, pro-caspase-3.
This affinity enhancement is important if the endogenous concentration of pro-caspase-3 in cells is low, for example, around the Km of C9Holo, but much below the Km of LZ-C9. Indeed, previous reports have shown this to be the case. Using quantitative Western analysis, the approximate concentration of pro-caspase-3 in 293 cells was found to be around 100 nM (Stennicke et al., 1998), which is similar to the Km of C9Holo for pro-caspase-3. Independently, caspase-3 concentration in Jurkat T cells was estimated to be below 100 nM (Pop et al., 2001; Saunders et al., 2000). At this pro-caspase-3 concentration, only C9Holo, but not dimerized caspase-9, can efficiently cleave pro-caspase-3 substrate. The small yet likely significant reduction in the Km of C9Holo for pro-caspase-3 by the feedback cleavage of caspase-9 (Table 1) may further enhance the ability of C9Holo to process pro-caspase-3 at the physiological concentration.
C9Holo is a specific pro-caspase-3 processing machine
In the intrinsic apoptosis pathway, C9Holo appears to have a strict specificity for pro-caspase-3 and its relative pro-caspase-7. Other caspase-9 substrates have not been reported. This situation differs from the initiator caspase in the extrinsic apoptosis pathway, caspase-8, which cleaves pro-caspase-3 as well as BID, a BH3 only Bcl-2 family member (Li et al., 1998; Luo et al., 1998).
This strict specificity may be conferred by association of caspase-9 with the apoptosome in the form of C9Holo. In contrast, caspase-8 gets released upon activation from the death inducing signaling complex (Peter and Krammer, 2003). The intrinsic substrate specificity of caspase-9, as determined by its specificity for four-residue peptide sequences, is not stricter than caspase-8 or any other caspases (Thornberry et al., 1997). However, because the Km of caspase-9 or C9Holo for these peptides are unusually high, caspase-9 is essentially inactive against potential substrates. Only in the form of C9Holo, caspase-9 has an enhanced affinity specifically for pro-caspase-3, which makes C9Holo a specific processing machine for pro-caspase-3. Therefore, in addition to providing higher affinity for pro-caspase-3, association of caspase-9 with the apoptosome might also ensure its specificity for pro-caspase-3.
Potential molecular mechanisms of affinity enhancement of C9Holo for pro-caspase-3
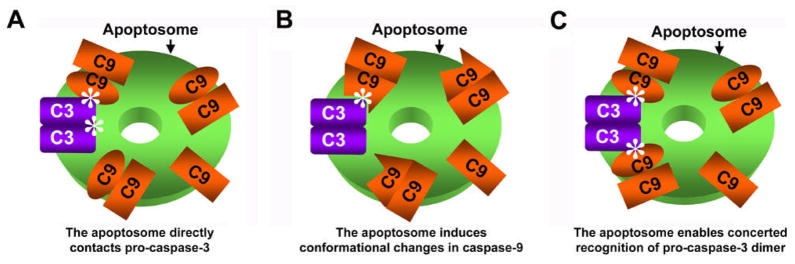
So what are the possible mechanisms for the enhanced affinity of caspase-9 holoenzyme for pro-caspase-3? First, the apoptosome may directly interact with pro-caspase-3 and therefore enhances the affinity of bound caspase-9 to pro-caspase-3 (Figure 6A). In this model, the apoptosome plays a direct contact role in pro-caspase-3 recruitment. Alternatively, the apoptosome may induce conformational changes in bound caspase-9 and therefore increases its affinity to pro-caspase-3 (Figure 6B). The conformational changes may possibly lie outside the immediate active site, as it does not affect the cleavage of LEHD-AFC.
Figure 6.
Potential molecular mechanisms of affinity enhancement of C9Holo for pro-caspase-3. The apoptosome, caspase-9 (C9) and pro-caspase-3 (C3) are shown in green, orange and purple, respectively. The inactive caspase-9 subunits are shown as rectangles. Asterisks denote contacts of pro-caspase-3 with either the apoptosome or caspase-9. For simplicity, only one pro-caspase-3 dimer is shown.
It is also possible that this enhanced affinity may be due to avidity by a simultaneous recognition of two cleavage sites from two adjacent caspase-9 dimers (Figure 6C). The rationale is that in the platform of the oligomeric apoptosome, C9Holo may simultaneously interact with both cleavage sites on pro-caspase-3, leading to enhanced affinity via avidity. In contrast, LZ-C9, with its single productive active site (Chao et al., 2005; Renatus et al., 2001), cannot gain affinity through this avidity. We have tried to test this hypothesis by linking two active site mutant (C163A) of pro-caspase-3 sequences in tandem via a flexible covalent linker [1-277- (GS)7-1-277]. This single-chain pro-caspase-3-C163A construct harbors the entire heterotetrameric sequence of pro-caspase-3 and contains two caspase-9 cleavage sites. We then mutated the second caspase-9 cleavage site in this single chain pro-caspase-3 construct (D175A) to generate a pro-caspase-3 dimer with a single caspase-9 recognition sequence. However, the cleavage activities of C9Holo against the single chain wildtype or D175 mutant did not show dramatic differences (data not shown). It is possible that the lack of dramatic differences may also be due to the incomplete destruction of the second caspase-9 recognition site by the single D175A mutation. A more stringent test on this hypothesis is to generate monomeric pro-caspase-3. But previous (Bose and Clark, 2001) as well as our own experiments (data not shown) have indicated that monomeric pro-caspase-3 mutant cannot be generated without greatly altering the structure of pro-caspase-3. Therefore, it remains to be seen whether the potential concerted recognition of two cleavage sites by C9Holo contributes to this high affinity.
Regardless of the exact mechanism, it is clear that C9Holo has dramatically increased affinity to pro-caspase-3 and therefore exhibits dramatically increased activity and specificity for this physiological substrate. This characteristic places caspase-9 as a single unique caspase in the entire caspase superfamily in that it defies the otherwise universal activation mechanism of cleavage and/or dimerization. While dimerization of caspase-9 transforms it into an active enzyme when using its optimal peptide as the substrate, the Km of the enzyme for the substrate is high (~10−3 – 10−4 M) compared to those of other caspases (~10−4 – 10−5 M) (Zhou et al., 1998). Therefore, caspase-9 requires the additional step of activation, the association into C9Holo, to be fully activated for processing its physiological substrate at the low physiological concentration (~100 nM), ensuring a precise and versatile regulation. In summary, our quantitative characterization of C9Holo and LZ-C9 provided unexpected insights into the molecular mechanism of caspase-9 activation by the apoptosome and adds to our understandings of caspase activation in general.
Experimental Procedures
Protein Expression and Purification
His-tagged full length human caspase-9, leucine-zippered caspase-9 (LZ-C9 and LZ-C9L) and CARD-deleted caspase-9 (C9ΔCARD, residues 139–416) were expressed in E. coli. LZ-C9 contains the initiation Met, leucine zipper sequence from GCN4 (251–281), a linker (GSGSGS) and the C-terminally His-tagged catalytic domain of human caspase-9 (151–416). LZ-C9L is similar to LZ-C9 except that it includes the additional region between the CARD and the catalytic domain of caspase-9 (98–416). Both LZ-C9 and LZ-C9L were purified by Ni-NTA affinity chromatography (Qiagen), anion exchange at pH 8.0 with HiTrap Q (GE Healthcare) and gel filtration chromatography with Superdex 200 HR 10/300 (GE Healthcare). The buffer for gel filtration contains 20 mM Tris at pH 7.5, 150 mM NaCl and 5mM DTT. To generate caspase-3 feedback-cleaved full-length caspase-9 and LZ-C9, their respective cell pellets were mixed with those of non-tagged caspase-3 and purified similarly. His-tagged Apaf-1 and pro-caspase-3 were expressed in the baculovirus-mediated insect cell expression system. They were purified by Ni-NTA affinity chromatography followed by gel filtration. Caspase concentrations were determined by both the Bradford Assay and active site titration as described previously (Stennicke and Salvesen, 2000; Zou et al., 2003), which agree well with each other.
Modeling of LZ-C9
A molecular model of LZ-C9 was constructed using the GCN4 leucine zipper coordinates (PDB ID 2ZTA) and a caspase-9 coordinates (PDB ID 2AR9). The eight residue linker comprising the two disordered residues in GCN4 and the GSGSGS sequence was built from an eight residue loop in PDB ID 1Z3G containing a similar sequence. Model building was done in O (Jones et al., 1991) and the ribbon diagram was produced in PyMOL (DeLano Scientific).
Caspase Activity Assay
C9Holo and LZ-C9 activities were monitored by turnover of fluorogenic tetrapeptide substrates in both direct and indirect assays. Fluorescence measurement upon substrate cleavage was performed on SpectraMax M2 plate reader (Molecular Devices) using 384-well plates (Corning). All measurements were performed in 20 μL reactions in Buffer A (20 mM Hepes pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA and 1 mM DTT) at 37 °C. Reaction mixtures also contain 10 μM dATP (Sigma) and 500 nM bovine heart cytochrome c (1 μM if Apaf-1 concentration is higher than 200 nM, Sigma) when measuring C9Holo activity. In direct assay, C9Holo or LZC9 of indicated concentration was incubated at 37 °C for 5 min. The reactions were initiated by adding the substrate Ac-LEHD-AFC (100 μM, Calbiochem). Release of the AFC moiety was monitored at emission wavelength of 505 nm with the excitation wavelength of 400 nm. In indirect assay, C9Holo or LZ-C9 was incubated together with Ac-DEVD-AMC (15 μM) at 37°C for 5 min before pro-caspase-3 was added to initiate the reactions. The reactions were then followed at emission wavelength of 465 nm with excitation wavelength of 360 nm. Reactions were monitored at 10-second intervals. V0 was drawn from the initial phase of the progress curve. Km values were determined using nonlinear regression method to fit Michaelis-Menton equation.
Multiangle light scattering
Molar mass of purified LZ-C9, LZ-C9L and C9ΔCARD was determined by static multiangle light scattering (MALS). Protein was injected onto a Superdex 200 HR 10/300 gel filtration column equilibrated in a buffer containing 20mM Tris at pH 7.5, 150mM NaCl and 0.2mM DTT. The chromatography system was coupled to a three angle light scattering detector (mini-DAWNEOS) and refractive index detector (Optilab DSP) (Wyatt Technology). Data were collected every 0.5 s at a flow rate of 0.2 ml/min. Data analysis was carried out using the program ASTRA, yielding the molar mass and mass distribution (polydispersity) of the sample.
Fractionation of Pro-caspase-3 and Apoptosome Mixture
Excess pro-caspase-3 (2μM) was incubated with 1 μM Apaf-1, 2 μM cytochrome c, 10 μM dATP and 1 μM caspase-9 at 30°C for 30 min in a total volume of 600 μL. Reaction mixture was then fractionated on a Superdex 200 column pre-equilibrated with Buffer A at 4 °C. The column was eluted with Buffer A and fractions of 1 ml were collected. Aliquots of 2 μL of each fraction were assayed for their cleavage activity against DEVD-AMC as described. Aliquots of 10 μL of each fraction were resolved by 15 % SDS-PAGE followed by western blot analysis (see below). Alternatively, excess pro-caspase-3 was incubated with 1 μM LZ-C9 for 30 min at 30 °C. The mixture underwent the same fractionation and analyses as described above.
Western Blot Analysis
Proteins were transferred from SDS-PAGE gel to PVDF membrane using Trans-Blot SD (Bio-rad). After transfer, membrane was probed with either mouse monoclonal anti-caspase-3 (Cell Signaling) (1:1000) or polyclonal anti-Apaf-1 (1:1000). Washed blot was then incubated with horseradish peroxidase-conjugated horse anti-mouse (caspase-3) or goat anti-rabbit (Apaf-1) IgG. Reactive bands were visualized using SuperSignal West Pico Kit (Pierce) as instructed.
Acknowledgments
The work was supported by the National Institute of Health (R01 AI-50872). S.C.L and Y.C.L are Cancer Research Institute postdoctoral fellows..
References
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Bose K, Clark AC. Dimeric procaspase-3 unfolds via a four-state equilibrium process. Biochemistry. 2001;40:14236–14242. doi: 10.1021/bi0110387. [DOI] [PubMed] [Google Scholar]
- Chai J, Wu Q, Shiozaki E, Srinivasula SM, Alnemri ES, Shi Y. Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- Chao Y, Shiozaki EN, Srinivasula SM, Rigotti DJ, Fairman R, Shi Y. Engineering a dimeric caspase-9: a re-evaluation of the induced proximity model for caspase activation. PLoS Biol. 2005;3:1079. doi: 10.1371/journal.pbio.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Donepudi M, Mac Sweeney A, Briand C, Grutter MG. Insights into the regulatory mechanism for caspase-8 activation. Mol Cell. 2003;11:543–549. doi: 10.1016/s1097-2765(03)00059-5. [DOI] [PubMed] [Google Scholar]
- Hu JC, O’Shea EK, Kim PS, Sauer RT. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang X. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J Biol Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building models in electron density maps and the location of errors in those models. Acta Crystallgr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- MacCorkle RA, Freeman KW, Spencer DM. Synthetic activation of caspases: artificial death switches. Proc Natl Acad Sci U S A. 1998;95:3655–3660. doi: 10.1073/pnas.95.7.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. JBC. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- O’Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 2003;10:26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- Pop C, Chen YR, Smith B, Bose K, Bobay B, Tripathy A, Franzen S, Clark AC. Removal of the pro-domain does not affect the conformation of the procaspase-3 dimer. Biochemistry. 2001;40:14224–14235. doi: 10.1021/bi011037e. [DOI] [PubMed] [Google Scholar]
- Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS. Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci U S A. 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Fuentes-Prior P, Renatus M, Kairies N, Krapp S, Huber R, Salvesen GS, Bode W. Structural basis for the activation of human procaspase-7. Proc Natl Acad Sci U S A. 2001;98:14790–14795. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Lazebnik Y. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 1999;13:3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen GS. Caspases and apoptosis. Essays Biochem. 2002;38:9–19. doi: 10.1042/bse0380009. [DOI] [PubMed] [Google Scholar]
- Saunders PA, Cooper JA, Roodell MM, Schroeder DA, Borchert CJ, Isaacson AL, Schendel MJ, Godfrey KG, Cahill DR, Walz AM, et al. Quantification of active caspase 3 in apoptotic cells. Anal Biochem. 2000;284:114–124. doi: 10.1006/abio.2000.4690. [DOI] [PubMed] [Google Scholar]
- Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- Shi Y. Caspase activation: revisiting the induced proximity model. Cell. 2004;117:855–858. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, Alnemri ES. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS. Caspase-9 can be activated without proteolytic processing. J Biol Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Jurgensmeier JM, Shin H, Deveraux Q, Wolf BB, Yang X, Zhou Q, Ellerby HM, Ellerby LM, Bredesen D, et al. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem. 1998;273:27084–27090. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS. Caspase assays. Methods Enzymol. 2000;322:91–100. doi: 10.1016/s0076-6879(00)22010-7. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- Wei Y, Fox T, Chambers SP, Sintchak J, Coll JT, Golec JM, Swenson L, Wilson KP, Charifson PS. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem Biol. 2000;7:423–432. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
- Yang X, Chang HY, Baltimore D. Autoproteolytic activation of pro-caspases by oligomerization. Mol Cell. 1998a;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- Yang X, Chang HY, Baltimore D. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science. 1998b;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW. A structure of the human apoptosome at 12.8 A resolution provides insights into this cell death platform. Structure (Camb) 2005;13:1725–1735. doi: 10.1016/j.str.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Krebs JF, Snipas SJ, Price A, Alnemri ES, Tomaselli KJ, Salvesen GS. Interaction of the baculovirus anti-apoptotic protein p35 with caspases. Specificity, kinetics, and characterization of the caspase/p35 complex. Biochemistry. 1998;37:10757–10765. doi: 10.1021/bi980893w. [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- Zou H, Yang R, Hao J, Wang J, Sun C, Fesik SW, Wu JC, Tomaselli KJ, Armstrong RC. Regulation of the Apaf-1/caspase-9 apoptosome by caspase-3 and XIAP. J Biol Chem. 2003;278:8091–8098. doi: 10.1074/jbc.M204783200. [DOI] [PubMed] [Google Scholar]