Abstract
Poliovirus is the most extensively studied member of the order Picornavirales, which contains numerous medical, veterinary and agricultural pathogens. The picornavirus genome encodes a single polyprotein that is divided into three regions: P1, P2 and P3. P3 proteins are known to participate more directly in genome replication, for example by containing the viral RNA-dependent RNA polymerase (RdRp or 3Dpol), among several other proteins and enzymes. We will review recent data that provide new insight into the structure, function and mechanism of P3 proteins and their complexes, which are required for initiation of genome replication. Replication of poliovirus genomes occurs within macromolecular complexes, containing viral RNA, viral proteins and host-cell membranes, collectively referred to as replication complexes. P2 proteins clearly contribute to interactions with the host cell that are required for virus multiplication, including formation of replication complexes. We will discuss recent data that suggest a role for P3 proteins in formation of replication complexes. Among the least understood steps of the poliovirus lifecycle is encapsidation of genomic RNA. We will also describe data that suggest a role for P3 proteins in this step.
Keywords: encapsidation, genome replication, nonstructural proteins, picornaviruses, poliovirus, protein-nucleic acid interaction, protein-protein interaction, secretory or transport vesicles, virus-host interaction
Poliovirus genome, encoded proteins & lifecycle
Poliovirus (PV) is the type virus of the Enterovirus genus in the family Picornaviridae [1]. This family of viruses is now a part of a larger order that has been termed Picornavirales [2]. The order Picornavirales contains numerous existing and emerging pathogens of medical, veterinary and agricultural importance [2]. PV is the most extensively studied virus in this order, at least in terms of our collective understanding of its molecular and cellular biology, biochemistry, pathogenesis and evolution, and is therefore an important model system [3].
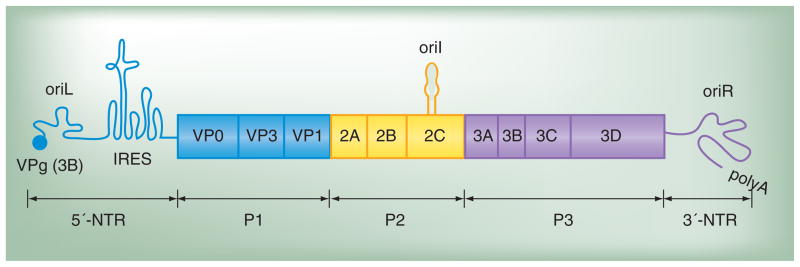
The PV genome organization is shown in Figure 1. The genome is single-stranded RNA of positive polarity – that is, it is a functional mRNA. The 5′-end of the genome contains a covalently linked 22 amino acid peptide, termed virion protein genome linked (VPg). The 5′-nontranslated region (5′-NTR) contains a cis-acting genome replication element (CRE) termed oriL (left). oriL forms a cloverleaf structure. Also present in the 5′-NTR is the internal ribosome entry site (IRES), a complex RNA structure that permits translation initiation to occur in the absence of a 5′-m7G cap. The 3′-NTR contains another CRE, termed oriR (right), that forms a complex RNA structure that includes a pseudoknot. The genome is terminated at its 3′-end by a poly(rA) tail. These aspects of genome organization are highly conserved in position and function in all picornaviruses that have been analyzed [1]. However, a third CRE is also present in the genome. This CRE is termed oriI (internal) and is the template for initiation of minus-strand RNA synthesis. Unlike the other CREs, oriI can function independently of position and can therefore be located at different positions in the genome of other picornaviruses relative to that observed for PV [4].
Figure 1. Poliovirus genome. The 5′-end of the genome is covalently linked to a peptide (VPg) encoded by the 3B region of the genome. The 3′-end contains a poly(rA) tail.
Three cis-acting replication elements are known. oriL is located in the 5′-NTR. oriR is located in the 3′-NTR and is not absolutely required for poliovirus genome replication [101,102]. oriI is located in the 2C-coding sequence for poliovirus and is also referred to as 2C-cis-acting genome replication element; the position of this element is virus dependent. oriI is required for initiation of minus-strand RNA synthesis. Translation initiation employs an IRES. The single open reading frame encodes a polyprotein. P1 encodes virion structural proteins as indicated. P2 encodes proteins thought to participate in virus–host interactions required for genome replication. The 2A protein is a protease that releases the structural protein precursor from the polyprotein. This protein also cleaves cellular proteins to enhance virus multiplication. Protein 2B, 2C and 2BC interact with membranes and contribute to the formation of replication complexes. The 2C protein is ATPase; this activity is required for 2C to contribute to replication complex formation. P3 encodes proteins thought to participate directly in genome replication. Polyprotein processing is mediated by protease activity residing in 2A, 3C and/or 3CD proteins.
IRES: Internal ribosome entry site; NTR: Nontranslated region; VPg: Virion protein genome linked.
The PV genome encodes a single polyprotein, approximately 3000 amino acids in length (Figure 1). The polyprotein can be divided into three functional regions: P1, P2 and P3, and is cleaved co- and post-translationally by viral 2A and 3C protease activities. P1 proteins are capsid proteins. Four capsid proteins exist: VP1–4. VP2 and VP4 are produced in assembled virus particles that contain genomic RNA by autocatalytic cleavage of VP0 [5, 6].
P2 proteins are known to participate in processes that convert the host cell into an environment that permits robust genome replication. The 2A protein is a protease that cleaves at the VP1–2A boundary to release P1 from the rest of the polyprotein. However, this protease also cleaves cellular proteins, inhibiting cellular processes such as cap-dependent translation [7,8]. The 2B, 2C and 2BC proteins interact with cellular membranes and have been shown to induce vesicles in cells that appear similar to vesicles induced during PV infection, which are the sites of genome replication; the so-called replication complexes (RCs) [9].
P3 proteins represent the primary focus of this article. These proteins have the most direct roles in genome transactions required for genome replication (Figure 2) [3]. The P3 polyprotein is processed by two pathways: a major pathway (Figure 2A) and a minor pathway (Figure 2B) [10,11]. The major pathway produces the 3AB and 3CD proteins. Both of these proteins bind to one or more CREs and interact with each other [12,13]. 3AB protein has recently been demonstrated to remodel RNA as a result of its helix-destabilizing activity [14]. 3AB can also stimulate the activity of the RNA-dependent RNA polymerase (RdRp), 3Dpol, encoded by the 3D region of the genome [15]. 3CD protein is a protease, the active site of which is located in the 3C protein domain. The 3C-dependent protease activity of 3CD, and perhaps other 3C-containing precursors, is responsible for the majority of polyprotein cleavage events. In spite of the presence of the 3D protein domain, 3CD protein does not exhibit RdRp activity [16]. New roles for 3CD protein before and after genome replication are beginning to emerge and will be discussed.
Figure 2. P3 precursor processing pathways and functions of proteins encoded in the P3 region.
Processing of the P3 precursor occurs by two independent pathways, major (A) and minor (B). In the major pathway (A), processing between 3B and 3C yields 3AB and 3CD. In the minor pathway (B), processing between 3A and 3B yields 3A and 3BCD. 3BCD processing yields 3BC and 3D; 3BC processing yields 3B and 3C. Known functions for each of the P3 proteins are listed; those colored red are described in the text.
RC: Replication complex; RdRp: RNA-dependent RNA polymerase.
The minor pathway produces 3A and 3BCD proteins. 3BCD protein can be cleaved further to produce 3BC and 3Dpol; 3BC can be cleaved further to produce 3B and 3C. The 3B region of the genome encodes the 22 amino acid peptide (VPg) that is linked to the 5′ end of the genome and the 5′-end of its replication intermediate, minus-strand RNA. 3B and VPg are therefore used interchangeably. The 3A protein interacts with membranes and inhibits secretion [17,18]. The molecular mechanism for this activity of 3A is becoming clearer and will be discussed. As described previously, 3C is the viral protease and 3Dpol is the viral RdRp. 3C protein also has RNA-binding activity and is capable of interacting with 3Dpol [19–21]. Roles for proteins 3BCD and 3BC in genome replication and the virus lifecycle remain unclear and controversial. These proteins do not accumulate to significant levels, if at all, in picornavirus-infected cells [10]. However, both of these proteins can substitute for the VPg peptide in reactions performed in vitro that mimic initiation of genome replication [10]. This observation has led to the proposal that 3BC(D) serves as the primer for initiation of genome replication in vivo, while more traditional models favor the use of the VPg peptide produced by proteolysis of the 3AB protein [3]. Aspects of this controversial model for initiation of genome replication will be considered in detail here.
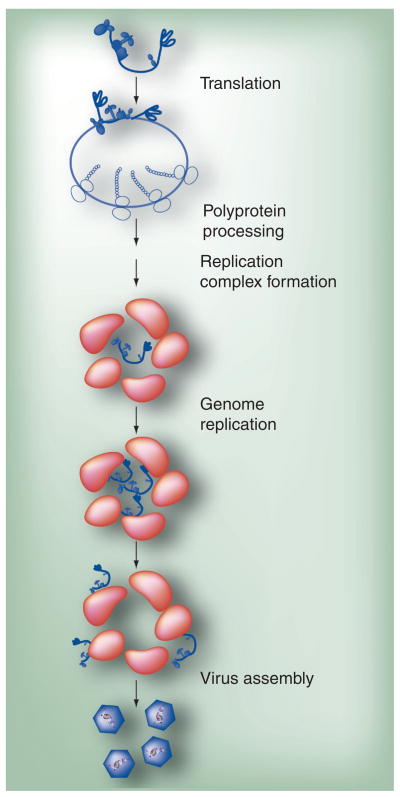
The PV lifecycle is typical relative to most viruses (Figure 3). The virus enters the cell by a receptor (CD155)-facilitated process. A great deal of very detailed structural and mechanistic information is available about the PV virion, its receptor, the virion–receptor interaction and the consequences of receptor binding for virion remodeling [6, 22–26]. However, much less is known about the postattachment steps that ultimately lead to the release of the genome into the host cell cytoplasm [27,28]. Once in the cytoplasm, VPg is removed by a host enzyme [29], the genome is translated, polyproteins are produced, genome replication ensues and progeny virions are produced. The pathway for assembly of PV particles has been well studied, with numerous intermediates identified and characterized [30–36]. However, very little is known about genome encapsidation. Most RNA viruses have packaging signals. The determinants of picornavirus genome encapsidation are unknown. There have been some genetic data that suggest coupling of genome replication and genome encapsidation [37]. If this is the case, then it is likely that sites for genome replication and genome encapsidation are the same. New data suggest that genome encapsidation may occur as a distinct step after genome replication that requires the cooperation of P2 and P3 proteins. These new data will be discussed.
Figure 3. Poliovirus multiplication cycle.
Genomic RNA is first translated to produce the poliovirus polyprotein. The polyprotein is co- and post-translationally processed to produce the various precursors and processed proteins that are needed for poliovirus multiplication. Replication complexes are formed, followed by genome replication. Replicated RNA enters either the translation–replication cycle or viral particle assembly step. For virus assembly, RNA encapsidation and virus particle maturation must occur.
Adapted from [78].
P3 proteins: structure does not always inform function
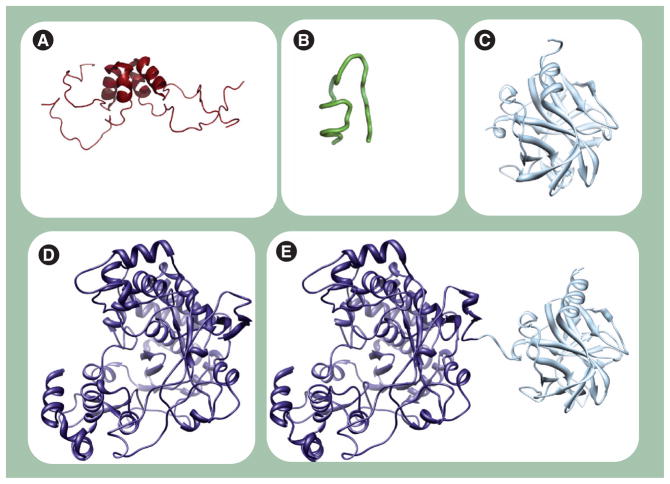
Any discussion of PV P3 proteins must include some discussion of structure, as structural information is available for at least a portion of all of the fully processed P3 proteins and one intermediate (Figure 4) [21,38–41]. It is now generally expected that structural information will provide insight into function. However, as discussed below, the available structures of P3 proteins have created far more questions than insight.
Figure 4. Structures of the poliovirus proteins.
Poliovirus proteins (A) 3A (PDB 1NG7 [40]), (B) 3B (VPg)(2BBP [39]), (C) 3Cpro (1L1N [38]), (D) 3Dpol (1RA6 [41]) and (E) 3CD (2IJD [21]) are shown. The 3A (soluble domain, residues 1–59) and 3B structures were determined by nuclear magnetic resonance; the other three are crystallographic structures.
VPg: Virion protein genome linked.
3A
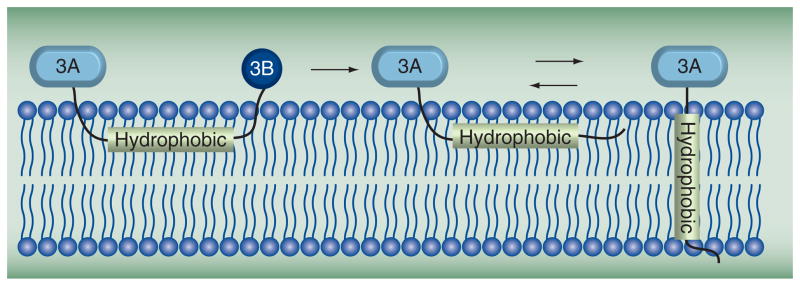
3A is a small protein (87 residues) in which the amino-terminal domain (58 residues) is soluble. The carboxy-terminal domain is hydrophobic and functions to anchor the protein to the surface of the membranous vesicles. The structure of the soluble amino-terminal domain was determined by nuclear magnetic resonance (NMR) [40]. The solution structure (Figure 4A) revealed a symmetric homodimer, with each monomer consisting of an α-helical hairpin. The structure is unique, but provided little information on function. The amino- and carboxy-terminal residues (1–22 and 42–58, respectively) were disordered. The carboxy-terminal region could be merely a linker to the hydrophobic domain; however, it is unclear why the amino-terminal region is unstructured. In fact, this region includes conserved residues that when mutated adversely affected 3A and 3AB activities. Perhaps the disordered amino terminus becomes structured under physiological conditions and/or in the presence of its binding partners. The conserved hydrophobic residues (Ile22, Leu25, Leu26, Val29, Val34, Tyr37, Cys38 and Trp43) in the structured region form the dimer interface. It has been suggested that attachment of the 3B protein domain to 3A to form 3AB causes a change in the organization of the hydrophobic carboxy terminus of 3A (Figure 5) [18]. This effect of 3B on 3A may reflect a change in 3AB quaternary structure relative to 3A as the hydrophobic carboxy terminus may contribute to 3A dimerization.
Figure 5. Model for 3AB and 3A topologies on membranes.
The hydrophobic domain of 3A interacts with membranes in the context of 3AB. Once 3B is cleaved and released, 3A equilibrates to both trans- and nontrans-membrane forms.
Adapted from [18].
3B (VPg)
The solution structure of the 22-residue VPg peptide from PV was determined by NMR [39]. The structure consists of a long loop, encompassing residues 1–14, and a short C-terminal helix (residues 18–21) (Figure 4B). Structures of the VPg peptide from foot-and-mouth disease virus (FMDV) and coxsackievirus B3 (CVB3) have been determined in complex with the cognate RdRp [42,43]. In both of these cases, the VPg peptide adopted an extended conformation. However, only residues 1–15 and 7–15 were resolved for the VPg peptides from FMDV and CVB3, respectively. The PV VPg structure was determined in the presence of trimethylamine N-oxide, which stabilizes helical structures in proteins. The use of trimethylamine N-oxide may explain these differences in VPg structures.
3C
The crystal structure of 3C protein [38] resembles that of chemotrypsin-like serine proteinases (Figure 4C) [44]. 3C protein crystallized as a dimer; however, a monomer is likely sufficient for protease activity. Each subunit consists of two six-stranded antiparallel β-barrel domains (Figure 4C). The amino and carboxy termini form two α-helices packing against the β-barrels. A long loop extends to link the two β-barrels, with a short helix formed in the middle of the loop that is sandwiched between the N- and C-terminal helices. The dimer is stabilized by extensive H bonding and electrostatic interactions between the monomers. Indeed, dimer formation provides an extended, positively charged surface for RNA binding, which is located opposite to the protease active sites of the molecule [45]. Whether or not this binding surface would contribute to specific binding is unclear. How the 3C domain recognizes the diverse CREs of the virus is not obvious from studying by the structure.
3Dpol
The structure of PV 3Dpol [41] is representative of all picornaviral RdRps (Figure 4D) [46–48]. The 3Dpol structure resembles the cupped-right hand that was first described for the Klenow fragment of DNA polymerase I, with fingers, palm and thumb subdomains [49]. The strong interaction between the fingers and thumb subdomains is responsible for the closed polymerase structure. The fingers subdomain comprises residues 1–68, 96–190 and 269–286; they are interspersed with the palm residues 69–95 and 191–268 containing motif A. The rest of the palm comprises residues 287–380, which harbor the four conserved structural motifs B, C, D and E. Residues 1–68 (index finger) at the N-terminus extend to reach the thumb before folding back toward the palm. Residues 269–286 (middle finger) are part of an antiparallel β-sheet; also part of this sheet are the first eight residues at the amino terminus. Residues 150–179 (ring finger) traverse the active site under the index finger and form a short α-helix protruding to the surface of the polymerase. The ring finger represents the top of the nucleoside triphosphate entry channel; it harbors motif F, which is critical for interaction with the triphosphate. Residues 96–149 and 180–190 (pinky finger) form a relatively large structure separated from the other fingers. This finger contains structural motif G, which may play a role in RNA recognition. Finally, the carboxy-terminal residues 381–461 form a bundle of α-helices that constitute the thumb subdomain. The palm has a fairly compact structure with more helical contents; the structural elements are connected by long loops. Unlike the palm and thumb, the fingers subdomain is more disordered with far less structured contents.
3CD
The picornaviral 3CD protein is among the most interesting P3-processing intermediates. This protein is thought to be the major protease and the major CRE-binding protein. Both of these activities require the 3C protein domain. The specificity of 3CD differs from 3C [50,51]. In addition, although the 3CD protein contains the 3D domain, 3CD lacks RdRp activity [16,52]. How these functional differences are caused by the fusion of the 3C and 3D domains is unclear. There has been some suggestion that the fingers domain of 3D is a ‘chameleon’ structure that folds into a different conformation when fused to 3C. As shown in Figure 4E, PV 3CD appears to be a composite of the 3C and 3D proteins [21]. So, again, structure has not been able to explain completely the observed functional differences.
P3 proteins & the vesicular trafficking machinery
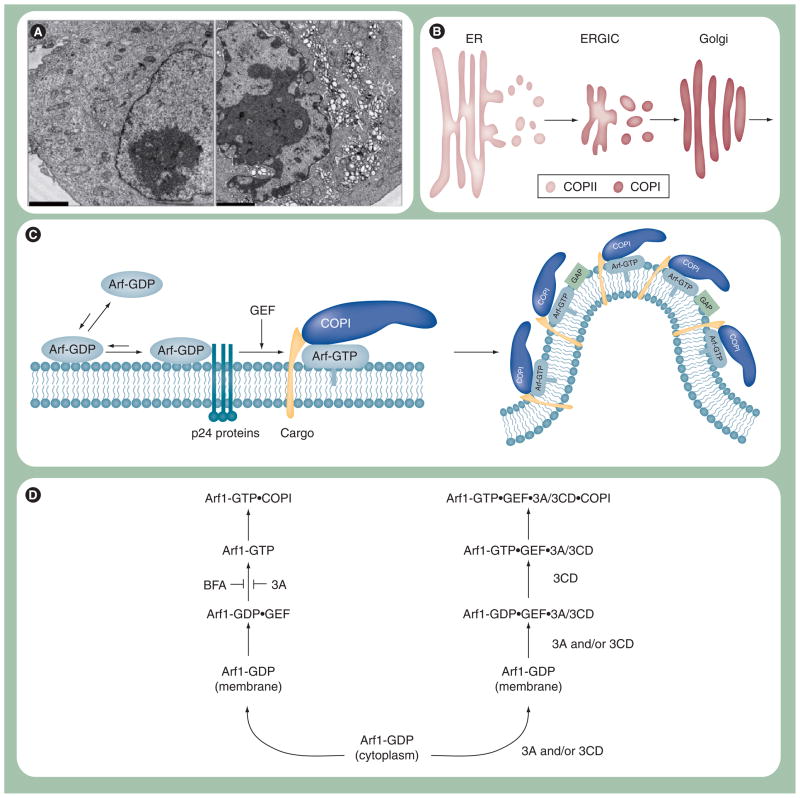
PV infection, like all picornavirus infections, induces vesicles in the infected cell (Figure 6A) [53,54]. These vesicles localize in the perinuclear region of the cell and are thought to be the sites of RNA replication because viral RNA can be detected in these clusters of vesicles [55]. As a result, these vesicle clusters have been referred to as RCs. The origin of the membranes that form the RCs is not known, but it is generally accepted that RCs derive from cellular membranes hijacked by viral proteins [9,56–58]. Evidence exists for hijacking of membranes participating in endoplasmic reticulum (ER)-to-Golgi trafficking [9] and/or autophagy [58,59]. It is worth noting that the size of vesicles induced by infection is larger than normal transport vesicles, suggesting that viral proteins may also promote vesicle fusion [53,57].
Figure 6. Proposed mechanisms for poliovirus replication complex formation.
(A) Poliovirus (PV)-induced vesicles. Electron micrograph of uninfected (left panel) and PV-infected (right panel) HeLa cells. Uninfected HeLa cells show normal intracellular membrane structures. PV-infected HeLa cells show PV-induced vesicles that cluster perinuclearly. HeLa cells were infected with PV at MOI of 10, fixed 7 h postinfection and visualized by electron microscopy. Scale bar: 2 μm. (B) Model for COP-coated vesicular trafficking from ER to Golgi. COPII vesicles originate from the ER and transport and fuse to ERGIC. COPI vesicles originate from ERGIC and transport and fuse to the Golgi. (C) Proposed mechanism for Arf-mediated vesicle formation. The GTPase-inactive form of Arf, Arf-GDP, prefers to be soluble in the cytosol. Once Arf-GDP binds to the membrane, its membrane association is stabilized by the Arf–p24 protein–protein interaction. Arf-GDP is then converted to the active form of Arf, Arf-GTP, by a GEF. Arf-GTP recruits cargo and coatomer proteins, such as COPI, and curvature is induced to form a vesicle. (D) Proposed mechanism for 3A and 3CD regulation of Arf-mediated vesicle formation. 3A and BFA inhibit the normal, cellular pathway required for Arf activation by preventing GEF function (left). In this case the GEF is GBF1. Poliovirus may have evolved a virus-specific mechanism for Arf activation. For example, 3A and/or 3CD may bind to Arf-GDP–GEF complex, causing activation of Arf and COPI recruitment by a mechanism that may no longer be sensitive to GAPs. It is also possible that COPI is not recruited at all.
ER: Endoplasmic reticulum; ERGIC: ER–Golgi intermediate compartment; GAP: GTPase-activating protein; GEF: Guanine nucleotide exchange factor.
COPII coats initiate budding at ER exit sites (Figure 6B) [60]. These vesicles undergo homotypic fusion to create the ER-Golgi intermediate compartment (ERGIC) (Figure 6B) [61,62]. COPI coats initiate budding from ERGIC for transit to the Golgi (Figure 6B) [61]. Association of coats at sites of budding depend on GTPases: SarI (COPII) and ArfI (COPI) (Figure 6C) [63]. These GTPases are usually found in a GDP-bound form (Figure 6C). Guanine nucleotide exchange factors (GEFs) are used to catalyze formation of the active, membrane-associated GTP-bound form (Figure 6C) [64]. This form of the protein then recruits the coat. GTPase-activating proteins (GAPs) stimulate GTP hydrolysis to produce the GDP-bound form, which leads to coat dissociation (Figure 6C) [65].
The first hint that ER–Golgi trafficking was involved in RC formation came from studies of the effect of brefeldin A (BFA) on PV multiplication [66]. BFA inhibits PV replication. BFA targets the Arf1 GEFs: GBF1, BIG1 and BIG2, preventing COPI coat assembly (Figure 6D) [67,68]. Was it the flow of membranes from ERGIC to Golgi or within Golgi that was inhibited? ERGIC-to-Golgi trafficking was a possibility as PV infection inhibits secretion at the level of Golgi that is mediated by the viral 3A protein [69]. It is now clear that 3A binds to GBF1, preventing Arf1 activation without preventing membrane association [67,68]. This observation could be interpreted to mean that GBF1 is not the cause of the inhibitory effect of BFA on PV multiplication. Binding of BFA to GBF1 was recently shown unambiguously to be the cause of inhibition of PV multiplication [67]. A BFA-resistant form of GBF1 supports PV multiplication in the presence of BFA [67]. It is now thought that there is a 3A–GBF1 interaction that is required for replication [67]. This interaction may not occur or may not be functional in the presence of BFA. It should be noted that not all viruses in the family Picornaviridae are sensitive to BFA, suggesting that the 3A–GBF1 interaction may not be required for BFA-insensitive viruses.
In addition to GBF1, 3A also binds to LIS1 [70]. LIS1 is a component of a dynein motor complex that is required for integrity of the Golgi apparatus [71–73]. Studies of LIS1 mutants have revealed a role for this protein in other aspects of vesicular trafficking [70]. Therefore, the 3A–LIS1 interaction may also contribute to changes in secretion and to formation of replication complexes.
Translation of PV polyprotein leads to association of the P2–P3 region with ER exit sites, locations of ER vesicle budding, and perhaps COPII-coated vesicles [9]. COPII-coated vesicles have two fates: homotypic fusion to form vesicular tubular clusters (VTCs) or fusion with existing VTCs. This VTC compartment is often referred to as ERGIC. Inhibition of ER-to-Golgi trafficking would appear to suggest that ERGIC may be the site of PV multiplication [74,75]. However, this may not be the case because PV proteins and ERGIC53, a marker for ERGIC, do not co-localize [Oh HS, Cameron CE, Unpublished Data].
Poliovirus infection inhibits anterograde trafficking at the level of ERGIC, without inhibiting retrograde trafficking, causing dissolution of the Golgi [75]. Both anterograde and retrograde trafficking require COPI, but use different GEFs. Recently, it was suggested that COPI is required for PV replication [56]. It is possible that this is a requirement for retrograde trafficking for induction of vesicles by PV. Alternatively, COPI vesicles may assemble and bind in the absence of an ‘active’ GBF1. Some BFA-sensitive picornaviruses do have vesicles that colocalize with COPI [76]. Such colocalization would imply the stable presence of the coat. Stabilization of the coat would imply a failure to convert Arf-GTP to Arf-GDP or use of an Arf-GTP-independent mechanism for COPI coat assembly.
How would COPI-coated vesicles form in the absence of GBF1? GBF1 is required for production of Arf1-GTP, which is essential for association of the coat. Interestingly, during PV infection Arf1-GTP is not reduced; in fact, it is elevated over time [56]. It appears that 3CD is not only capable of causing the relocalization of Arf1 to membranes but may also induce the activated state (Figure 6D) [56]. Proteins 3C and 3BCD do not have this activity [77]. Is this activated state mediated by Arf1-GTP or just Arf1-GTP-like? There is no evidence of GEF activity of PV proteins, although the signal:noise for this experiment is unclear [56]. The ability of the virus to bypass the normal COPI assembly process could prevent vesicles associated with replication complexes from being lost to downstream, default transactions of COPI-coated vesicles that would prevent RC accumulation. Therefore, PV, and other picornaviruses, may have evolved a unique strategy to induce vesicles that contain a modified COPI coat or function without coat (Figure 6D). Also consistent with a unique role of 3CD in replication complex formation is the observation that delaying the processing between 3B and 3C leads to accumulation of 3BCD and a possible defect to RC formation [78].
P3 proteins & genome replication
Interactions between processed forms of P3 proteins and between these proteins and viral RNA are required for genome replication. Many interactions between P3 proteins have been documented, including 3AB–3CD [13], 3AB–3D [15,79,80], 3B–3D [42,43,79], 3C–3C [20,21,38], 3CD–3CD [21,81], 3D–3D [21,82], 3CD–3D [21] and 3C–3D [20,21]. It is becoming increasingly clear that the processed proteins can interact with each other in many different ways. Some P3 proteins interact with more than one of the sequence and structurally distinct CREs [12,81]. The diversity of protein–protein and protein–RNA interactions that have been observed in PV and other picornaviral systems has led to controversies that have yet to be resolved.
Protein–protein interactions
Studies of protein–protein interactions have turned out to be much more complicated than first envisioned, especially as it relates to determining the universality of interactions amongst picornaviruses and the step(s) of the lifecycle in which the interactions function. A few anecdotes will be described that will illustrate the complexity.
It is well known that the VPg peptide can be used by the RdRp as a primer for RNA synthesis [83]. It seems reasonable that 3B would bind in the active site. However, genetic studies suggested that the 3B-binding site (at least in the context of 3AB) was on the surface of the polymerase near motif E of the palm, remote from the catalytic center [80]. This orientation was supported by computational docking experiments [84]. RdRp derivatives containing changes in the proposed 3AB-binding site were less active for VPg priming than RNA elongation [80]. Therefore, if this site were used for binding of the VPg used to prime RNA synthesis, then this RdRp-bound VPg would be extended by a second RdRp molecule.
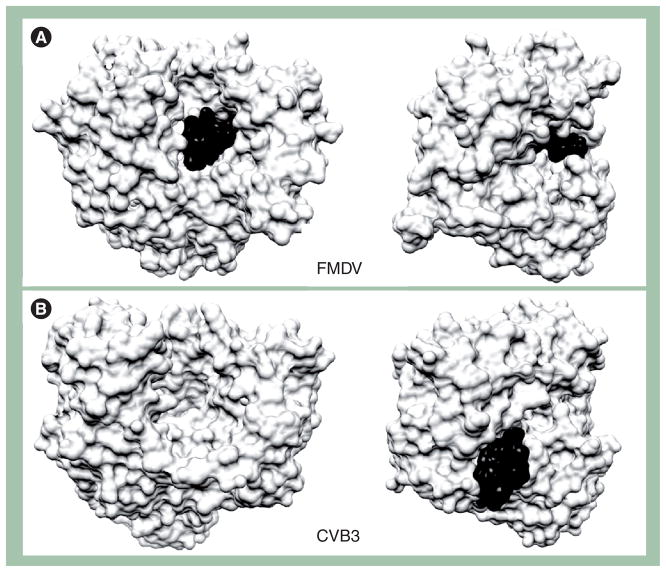
A completely different interpretation was made by investigators studying rhinovirus RdRp [85]. These investigators predicted a ‘front-loading’ model for binding and utilization of VPg. This prediction was confirmed by a crystal structure of FMDV RdRp and VPg (Figure 7A) [42]. The story was brought full circle recently with the solution of a crystal structure for CVB3 RdRp and VPg (Figure 7B) [43]. In this system, VPg does not bind to the active site, consistent with the genetic, biochemical and computational studies of the PV system. Do unique, virus-specific solutions exist for the RdRp–VPg interaction or are all possibilities real and important?
Figure 7. Two binding sites for VPg(3B) in 3Dpol/VPg(3B) complexes.
The complex structure of 3Dpol/VPg(3B) from FMDV is from PDB 2F8E [42] (A) and that of CVB3 is from PDB 3CDW [43] (B). VPg(3B) is shown in black, and 3Dpol is shown in white. Two different views are shown to emphasize the different binding sites for VPg(3B) in the two complexes. The views on the right can be obtained by rotating the left view 90° counterclockwise. In FMDV, the VPg(3B) peptide occupied the active site of the polymerase with the tyrosyl residue (Tyr3) positioned at the catalytic center. In the CVB3 complex, the VPg(3B) peptide binds to the back of the thumb of the polymerase.
CVB3: Coxsackievirus B3; FMDV: Foot-and-mouth disease virus; VPg: Virion protein genome linked.
Another example of complications associated with studying interactions between P3 proteins began with the solution of the crystal structure for PV RdRp [86]. This was the first structure for an RdRp, so the observations made were quite influential. It is worth noting that a significant amount of the electron density map for the fingers subdomain was missing. The most fascinating observation was that RdRp molecules interacted in a head-to-tail fashion, forming fibers that were also capable of interacting. The PV RdRp was found to form a lattice [82]. The implication was a unique, 2D mechanism for RNA synthesis [82,87]. RNA would be elongated by moving from one active site to another in the lattice instead of having the RdRp tracking along the RNA.
The head-to-tail fiber was formed by an interaction interface (termed interface I) that consisted of the back of the thumb of one molecule interacting with the back of the palm of another [86]. Mutations of the back of the thumb that disrupted the interactions were lethal to replication and virus viability, consistent with an important role for fibers in genome replication [87,88]. Importantly, it was even possible to obtain evidence for sheets of polymerase forming in cells [82].
A new way of thinking about RNA virus genome replication was beginning to emerge, but then a variety of confounding observations were reported. One RdRp structure after another began to appear. None had a lattice similar to that observed for PV. Another significant difference was that all of these other structures showed an intramolecular interaction between the fingertips and thumb that was absent in the first structure. If this intramolecular interaction occurs, then the lattice will be lost. Modeling of PV polymerase suggested that the intramolecular interaction between fingertips and thumb could occur [89]. This prediction was confirmed experimentally when complete structures of PV RdRp were obtained [41]. Solution of this structure required use of a polymerase engineered to lack interface I.
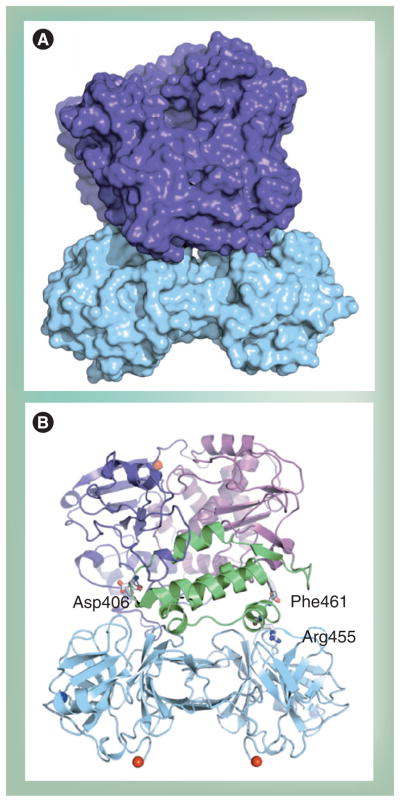
Another confounding observation was that mutations that disrupted interface I exhibited different phenotypes in the context of the virus, depending on the side on which amino acid substitutions were introduced [90]. The back of the thumb was more sensitive to substitutions than the back of the palm. It is now clear that the back of the thumb can interact with the top of a 3C dimer (Figure 8) [20]. This interaction is essential for VPg uridylylation in vitro and genome replication in cells. Similar structures are predicted for other precursors and are supported by genetic data [78]. The 3C–3D interaction may explain more complex interactions between P3 proteins bound at various locations in the genome. Does a unique or virus-specific solution exist to explain the interaction of the back of the thumb of the RdRp with itself or other proteins?
Figure 8. Model of 3C2–3D complex.
(A) Surfaces of 3C dimer and 3Dpol are shown in cyan and purple, respectively. (B) Ribbon diagram of the model shown in (A). The polymerase subdomains are depicted in different colors: fingers (purple), palm (magenta) and thumb (green). 3Dpol residues Asp406, Arg455 and Phe461, known to be required for uridylylation, are highlighted.
Adapted from [20].
The final example relates to studies of the PV 3CD protein. Crystal structures for PV 3C revealed 3C dimers and, as discussed previously, crystal structures of PV 3D revealed a complex lattice [86]. Such interactions could clearly complicate the solution properties of 3CD, a fusion between 3C and 3D. In order to study 3CD structure, all known intermolecular interactions were removed by site-directed mutagenesis [21]. This 3CD derivative crystallized [21]. Suprisingly, quite extensive and potentially conserved 3C–3C, 3C–3D and 3D–3D interactions were observed. None of these interactions had been observed before. Of note was the 3C–3D interaction, which did not resemble the interaction discussed previously. These aforementioned observations pose the question: Do protein–protein interactions observed crystallographically for P3 proteins matter at all?
We would like to suggest that all of the interactions observed to date for P3 proteins are important. PV, indeed all RNA viruses, have limited coding capacity. Each encoded protein may need to serve many unique functions in the lifecycle. These functions may require cooperation with the same or different proteins, leading to numerous unique macromolecular assemblies. The distribution of determinants that direct protein–protein interactions need not be evolutionarily conserved. For example, a heterotrimer that requires six determinants for stability could achieve stability in many ways (Figure 9). Distribution of the determinants for interaction will dictate the nature of the subassemblies that will exist in solution. In the case of an RNA virus, one of these components could be viral RNA. Some heterodimers may be seen in all systems; some heterodimers may only be observed in a single system (Figure 9). This line of reasoning would also apply to homodimeric interactions. This perspective provides an explanation for the apparent inconsistencies described. Therefore, when between-system inconsistencies arise, the first question really should be ‘what’s missing?’
Figure 9. The same heteromeric complex from different viruses may form different subassemblies.
A cartoon is shown of three heterotrimeric complexes. These complexes are identical in structure. However, the interactions used to stabilize the complex (lines between subunits of the complex) are distributed differently. As a result, the stable subassemblies will differ as indicated.
Protein–RNA interactions
As indicated previously, viral RNA represents a key component of the macromolecular assemblies that carry out genome replication. Three CREs exist; they are all different but are bound by the same P3-encoded proteins. The 3C domain (alone and in combination with other domains) is the major determinant of P3-encoded RNA-binding activity [10]. 3AB may also contribute to CRE recognition [13].
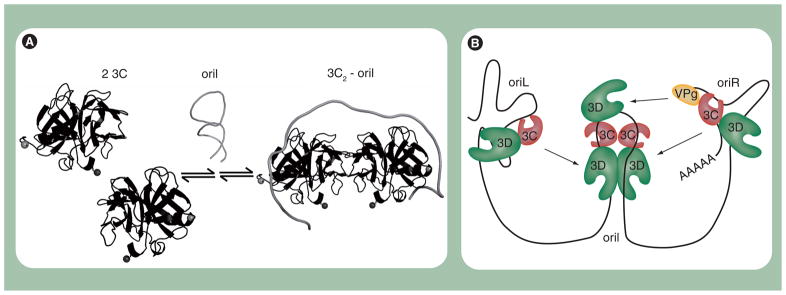
Nuclear magnetic resonance spectroscopy is now being used to understand how the 3C domain interacts with different RNA elements [45,91]. It is clear that 3C is capable of specific recognition of single-stranded and double-stranded RNA [81]. Formation of the VPg uridylylation complex has been suggested to require a dimer and melting of the oriI element for interaction of each 3C molecule with a single-stranded portion of the stem (Figure 10A). Binding to stem-loop d of oriL only requires a monomer binding to an intact double-stranded stem [91]. This initial structural information makes it clear that some 3C residues are probably used for binding to all RNAs and some 3C residues are probably only used for a specific RNA. The next decade promises to construct the cipher for the complex code employed by 3C for recognition.
Figure 10. Assembly and organization of the picornavirus 3B (VPg) uridylylation complex.
(A) Model for organization of 3C2–oriI complex. Protein 3C exists in a monomer–dimer equilibrium. OriI contains two 3C-binding sites. Binding of oriI to 3C shifts the equilibrium in favor of a 3C dimer (3C2). A consequence of 3C binding to oriI is destabilization of the secondary structure associated with unbound oriI, thus extending the loop. (B) Model for initiation of negative-strand RNA synthesis. 3(B)CD proteins bound to oriL and oriR interact with 3D and/or dimerized 3CDs bound to oriI. Multiple interactions among 3CDs on oriL, oriI and oriR facilitate circularization of the poliovirus genome and recruit both the 5′- and 3′-ends to oriI.
We suggest that the combination of protein–protein and protein–RNA interactions occurring at oriL, oriR and oriI facilitate formation of a single, functional ribonucleoprotein complex (Figure 10B). The existence of such a complex would explain the requirement of all cis-acting elements for genome replication, especially minus-strand synthesis.
P3 proteins & encapsidation
Encapsidation is the least understood aspect of picornavirology. In spite of the longstanding appreciation that replicons can be packaged in trans [92], there has been very little use of this system to identify cis-acting elements required for encapsidation. It has been suggested that genome replication and encapsidation are coupled [37]. It is now clear that replication can occur completely prior to the initiation of encapsidation [78]. 5-(3,4-dichlorophenyl)-methylhydantoin (hydantoin), which targets the 2C protein, is a reversible inhibitor of virion assembly that does not affect genome replication. Removal of hydantoin after genome replication is complete and in the presence of a replication inhibitor leads to production of normal levels of infectious virus [78].
Hydantoin blockage of encapsidation is consistent with other studies that suggest a role for 2C protein in this process [93–96]. Importantly, processed 2C need not be the target; rather, a 2C-containing precursor could be the target. Recently, studies using a cell-free system for virus production have shown that viral 3CD protein stimulates virus production at a step after RNA synthesis [97]. This activity does not require 3CD protease activity, so it is not related to capsid precursor maturation. 3CD must be intact, must contain a 3C capable of RNA binding and must contain residues involved in interface I [97]. The activity is inhibited by addition of 3C but not by addition of 3CD. Further analysis suggested that 3CD functions in a late assembly step and increases the specific infectivity of the virus particle. All of these observations point to a possible role of 3CD in encapsidation. To date, the only other P3-encoded protein that has been implicated in egress is VPg. VPg has been suggested to be a determinant of encapsidation [98,99]. Consistent with this possibility, only VPg-linked RNA exhibited 3CD stimulation [100]. The potential role of 3CD and other P3 proteins in encapsidation merits additional attention.
Conclusion & future perspective
As always, picornavirology is thriving and studies of PV continue to lead the way. Formation and function of replication complexes are now dependent on both P2 and P3 proteins, thus expanding the role of P3 proteins in the picornavirus lifecycle. The substantial functional versatility of P3 proteins is remarkable and not explained completely by structure. Separating these functions genetically and development of tools to study viruses defective in individual functions represent major challenges to our understanding of the physical basis for P3 protein versatility. Moreover, movement in this direction will help to clarify whether or not determinants of P3 protein versatility are conserved and define a rule rather than a PV-specific phenomenon.
Executive summary.
P3 proteins: structure & function
At least partial structures are available for all P3 proteins.
Structural information explains expected functions but not the unanticipated functions.
Extent of processing of P3 proteins dictates function and functional interactions with viral and host factors, including membranes.
P3 proteins & the vesicular trafficking machinery
Genome replication occurs on membranes hijacked from the cell and are referred to as replication complexes.
Poliovirus (perhaps most picornaviruses) has developed strategies to redirect cellular membranes.
Hijacking of cellular membranes is now known to require the viral 3A and 3CD proteins in addition to 2B(C).
P3 proteins & genome replication
The same P3 proteins from different picornaviruses can exhibit different interactions in vitro, complicating assignment of function.
All interactions observed in vitro likely occur in cells.
Interactions that appear to be (mutually) exclusive in vitro may not be in vivo.
P3 proteins & encapsidation
Genome replication and encapsidation are not necessarily coupled.
Protein 3CD functions at a step after RNA synthesis that may be encapsidation.
Acknowledgments
The authors thank Jamie Arnold for his assistance in preparation of the manuscript and thank Jamie Arnold, Eric Smidansky and Spencer Weeks for helpful comments on the manuscript.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
Financial support for our work in this area was provided by the NIH (AI053531 to Craig E Cameron). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Racaniello VR. Picornaviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. pp. 765–838. [Google Scholar]
- 2.Le Gall O, Christian P, Fauquet CM, et al. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch Virol. 2008;153(4):715–727. doi: 10.1007/s00705-008-0041-x. [DOI] [PubMed] [Google Scholar]
- 3.Paul AV, Belov GA, Ehrenfeld E, Wimmer E. Model of picornavirus RNA replication. In: Cameron CE, Götte M, Raney KD, editors. Viral Genome Replication. 1. Vol. 3. Springer; Philadelphia, PA: 2009. p. 23. [Google Scholar]
- 4.Steil BP, Barton DJ. Cis-active RNA elements (CREs) and picornavirus RNA replication. Virus Res. 2009;139(2):240–252. doi: 10.1016/j.virusres.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold E, Luo M, Vriend G, et al. Implications of the picornavirus capsid structure for polyprotein processing. Proc Natl Acad Sci USA. 1987;84(1):21–25. doi: 10.1073/pnas.84.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogle JM. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu Rev Microbiol. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventoso I, MacMillan SE, Hershey JW, Carrasco L. Poliovirus 2A proteinase cleaves directly the eIF-4G subunit of eIF-4F complex. FEBS Lett. 1998;435(1):79–83. doi: 10.1016/s0014-5793(98)01027-8. [DOI] [PubMed] [Google Scholar]
- 8.Gradi A, Svitkin YV, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95(19):11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rust RC, Landmann L, Gosert R, et al. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J Virol. 2001;75(20):9808–9818. doi: 10.1128/JVI.75.20.9808-9818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10▪.Pathak HB, Oh HS, Goodfellow IG, Arnold JJ, Cameron CE. Picornavirus genome replication: roles of precursor proteins and rate-limiting steps in oriI-dependent VPg uridylylation. J Biol Chem. 2008;283(45):30677–30688. doi: 10.1074/jbc.M806101200. Suggests that precursor forms of VPg may be used for initiation of genome replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawson MA, Semler BL. Alternate poliovirus nonstructural protein processing cascades generated by primary sites of 3C proteinase cleavage. Virology. 1992;191(1):309–320. doi: 10.1016/0042-6822(92)90193-s. [DOI] [PubMed] [Google Scholar]
- 12.Harris KS, Xiang W, Alexander L, Lane WS, Paul AV, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269(43):27004–27014. [PubMed] [Google Scholar]
- 13.Xiang W, Harris KS, Alexander L, Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol. 1995;69(6):3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14▪.DeStefano JJ, Titilope O. Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities. J Virol. 2006;80(4):1662–1671. doi: 10.1128/JVI.80.4.1662-1671.2006. First report of the nucleic acid chaperone activity of a picornaviral 3AB protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plotch SJ, Palant O. Poliovirus protein 3AB forms a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J Virol. 1995;69(11):7169–7179. doi: 10.1128/jvi.69.11.7169-7179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris KS, Reddigari SR, Nicklin MJ, Hammerle T, Wimmer E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J Virol. 1992;66(12):7481–7489. doi: 10.1128/jvi.66.12.7481-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe SS, Dodd DA, Kirkegaard K. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology. 2005;337(1):18–29. doi: 10.1016/j.virol.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 18▪▪.Fujita K, Krishnakumar SS, Franco D, Paul AV, London E, Wimmer E. Membrane topography of the hydrophobic anchor sequence of poliovirus 3A and 3AB proteins and the functional effect of 3A/3AB membrane association upon RNA replication. Biochemistry. 2007;46(17):5185–5199. doi: 10.1021/bi6024758. Very elegant, biophysical demonstration of the different conformations adopted by 3A and 3AB when interacting with membranes. This approach can be used to study interactions of other viral proteins with membranes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blair WS, Parsley TB, Bogerd HP, Towner JS, Semler BL, Cullen BR. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA. 1998;4(2):215–225. [PMC free article] [PubMed] [Google Scholar]
- 20▪▪.Shen M, Reitman ZJ, Zhao Y, et al. Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2–3Dpol complex. J Biol Chem. 2008;283(2):875–888. doi: 10.1074/jbc.M707907200. Genetic and biochemical data are used to delimit possible structures of the oriI ribonucleoprotein complex. This study represents the highest resolution hypothesis for this complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21▪.Marcotte LL, Wass AB, Gohara DW, et al. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J Virol. 2007;81(7):3583–3596. doi: 10.1128/JVI.02306-06. First crystal structure of a picornaviral 3CD protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basavappa R, Gomez-Yafal A, Hogle JM. The poliovirus empty capsid specifically recognizes the poliovirus receptor and undergoes some, but not all, of the transitions associated with cell entry. J Virol. 1998;72(9):7551–7556. doi: 10.1128/jvi.72.9.7551-7556.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arita M, Koike S, Aoki J, Horie H, Nomoto A. Interaction of poliovirus with its purified receptor and conformational alteration in the virion. J Virol. 1998;72(5):3578–3586. doi: 10.1128/jvi.72.5.3578-3586.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belnap DM, McDermott BM, Jr, Filman DJ, et al. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc Natl Acad Sci USA. 2000;97(1):73–78. doi: 10.1073/pnas.97.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuthill TJ, Bubeck D, Rowlands DJ, Hogle JM. Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J Virol. 2006;80(1):172–180. doi: 10.1128/JVI.80.1.172-180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J Virol. 2005;79(12):7745–7755. doi: 10.1128/JVI.79.12.7745-7755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergelson JM. New (fluorescent) light on poliovirus entry. Trends Microbiol. 2008;16(2):44–47. doi: 10.1016/j.tim.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. Imaging poliovirus entry in live cells. PLoS Biol. 2007;5(7):e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambros V, Pettersson RF, Baltimore D. An enzymatic activity in uninfected cells that cleaves the linkage between poliovirion RNA and the 5′ terminal protein. Cell. 1978;15(4):1439–1446. doi: 10.1016/0092-8674(78)90067-3. [DOI] [PubMed] [Google Scholar]
- 30.Nugent CI, Kirkegaard K. RNA binding properties of poliovirus subviral particles. J Virol. 1995;69(1):13–22. doi: 10.1128/jvi.69.1.13-22.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Tomas CB, Baltimore D. Morphogenesis of poliovirus. II. Demonstration of a new intermediate, the proviron. J Virol. 1973;12(5):1122–1130. doi: 10.1128/jvi.12.5.1122-1130.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobson MF, Baltimore D. Morphogenesis of poliovirus. I. Association of the viral RNA with coat protein. J Mol Biol. 1968;33(2):369–378. doi: 10.1016/0022-2836(68)90195-2. [DOI] [PubMed] [Google Scholar]
- 33.Guttman N, Baltimore D. Morphogenesis of poliovirus. IV. existence of particles sedimenting at 150S and having the properties of provirion. J Virol. 1977;23(2):363–367. doi: 10.1128/jvi.23.2.363-367.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ansardi DC, Porter DC, Morrow CD. Myristylation of poliovirus capsid precursor P1 is required for assembly of subviral particles. J Virol. 1992;66(7):4556–4563. doi: 10.1128/jvi.66.7.4556-4563.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verlinden Y, Cuconati A, Wimmer E, Rombaut B. Cell-free synthesis of poliovirus: 14S subunits are the key intermediates in the encapsidation of poliovirus RNA. J Gen Virol. 2000;81(Pt 11):2751–2754. doi: 10.1099/0022-1317-81-11-2751. [DOI] [PubMed] [Google Scholar]
- 36.Rombaut B, Vrijsen R, Boeye A. New evidence for the precursor role of 14 S subunits in poliovirus morphogenesis. Virology. 1990;177(1):411–414. doi: 10.1016/0042-6822(90)90502-i. [DOI] [PubMed] [Google Scholar]
- 37.Nugent CI, Johnson KL, Sarnow P, Kirkegaard K. Functional coupling between replication and packaging of poliovirus replicon RNA. J Virol. 1999;73(1):427–435. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosimann SC, Cherney MM, Sia S, Plotch S, James MN. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J Mol Biol. 1997;273(5):1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 39.Schein CH, Oezguen N, Volk DE, Garimella R, Paul A, Braun W. NMR structure of the viral peptide linked to the genome (VPg) of poliovirus. Peptides. 2006;27(7):1676–1684. doi: 10.1016/j.peptides.2006.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauss DM, Glustrom LW, Wuttke DS. Towards an understanding of the poliovirus replication complex: the solution structure of the soluble domain of the poliovirus 3A protein. J Mol Biol. 2003;330(2):225–234. doi: 10.1016/s0022-2836(03)00577-1. [DOI] [PubMed] [Google Scholar]
- 41.Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23(17):3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42▪.Ferrer-Orta C, Arias A, Agudo R, et al. The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J. 2006;25(4):880–888. doi: 10.1038/sj.emboj.7600971. The first structure of a picornaviral VPg–3Dpol complex. This structure shows binding of VPg in an extended conformation at the active site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43▪.Gruez A, Selisko B, Roberts M, et al. The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases. J Virol. 2008;82(19):9577–9590. doi: 10.1128/JVI.00631-08. The second structure of a picornaviral VPg–3Dpol complex. This structure shows binding of VPg remote from the active site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4(3):337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45▪.Amero CD, Arnold JJ, Moustafa IM, Cameron CE, Foster MP. Identification of the oriI-binding site of poliovirus 3C protein by nuclear magnetic resonance spectroscopy. J Virol. 2008;82(9):4363–4370. doi: 10.1128/JVI.02087-07. The first report of residues of 3C that change in response to oriI binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campagnola G, Weygandt M, Scoggin K, Peersen O. Crystal structure of coxsackievirus B3 3Dpol highlights the functional importance of residue 5 in picornavirus polymerases. J Virol. 2008;82(19):9458–9464. doi: 10.1128/JVI.00647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J Biol Chem. 2004;279(45):47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- 48.Love RA, Maegley KA, Yu X, et al. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure. 2004;12(8):1533–1544. doi: 10.1016/j.str.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 49.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 50.Parsley TB, Cornell CT, Semler BL. Modulation of the RNA binding and protein processing activities of poliovirus polypeptide 3CD by the viral RNA polymerase domain. J Biol Chem. 1999;274(18):12867–12876. doi: 10.1074/jbc.274.18.12867. [DOI] [PubMed] [Google Scholar]
- 51.Ypma-Wong MF, Dewalt PG, Johnson VH, Lamb JG, Semler BL. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166(1):265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- 52.Flanegan JB, Van Dyke TA. Isolation of a soluble and template-dependent poliovirus RNA polymerase that copies virion RNA in vitro. J Virol. 1979;32(1):155–161. doi: 10.1128/jvi.32.1.155-161.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dales S, Eggers HJ, Tamm I, Palade GE. Electron microscopic study of the formation of poliovirus. Virology. 1965;26:379–389. doi: 10.1016/0042-6822(65)90001-2. [DOI] [PubMed] [Google Scholar]
- 54.Bienz K, Egger D, Rasser Y, Bossart W. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology. 1983;131(1):39–48. doi: 10.1016/0042-6822(83)90531-7. [DOI] [PubMed] [Google Scholar]
- 55.Egger D, Bienz K. Recombination of poliovirus RNA proceeds in mixed replication complexes originating from distinct replication start sites. J Virol. 2002;76(21):10960–10971. doi: 10.1128/JVI.76.21.10960-10971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56▪▪.Belov GA, Altan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, Ehrenfeld E. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J Virol. 2007;81(2):558–567. doi: 10.1128/JVI.01820-06. Suggests Arf guanine nucleotide exchange factors as the target for brefeldin A required for poliovirus replication. First hint that 3CD protein alters localization of factors other than Arf proteins required for vesicular trafficking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho MW, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202(1):129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 58.Suhy DA, Giddings TH, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74(19):8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor MP, Kirkegaard K. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy. 2008;4(3):286–289. doi: 10.4161/auto.5377. [DOI] [PubMed] [Google Scholar]
- 60.Hughes H, Stephens DJ. Assembly, organization, and function of the COPII coat. Histochem Cell Biol. 2008;129(2):129–151. doi: 10.1007/s00418-007-0363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Appenzeller-Herzog C, Hauri HP. The ER–Golgi intermediate compartment (ERGIC): in search of its identity and function. J Cell Sci. 2006;119(Pt 11):2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- 62.Xu D, Hay JC. Reconstitution of COPII vesicle fusion to generate a pre-Golgi intermediate compartment. J Cell Biol. 2004;167(6):997–1003. doi: 10.1083/jcb.200408135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 64.Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8(11):1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 65.Inoue H, Randazzo PA. Arf GAPs and their interacting proteins. Traffic. 2007;8(11):1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- 66.Maynell LA, Kirkegaard K, Klymkowsky MW. Inhibition of poliovirus RNA synthesis by brefeldin A. J Virol. 1992;66(4):1985–1994. doi: 10.1128/jvi.66.4.1985-1994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67▪▪.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 2008;4(11):e1000216. doi: 10.1371/journal.ppat.1000216. Establishes definitively that GBF1 is the target for brefeldin A required for poliovirus replication using a very elegant approach, providing a nice example of how similar questions can be addressed in other viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wessels E, Duijsings D, Lanke KH, Melchers WJ, Jackson CL, van Kuppeveld FJ. Molecular determinants of the interaction between coxsackievirus protein 3A and guanine nucleotide exchange factor GBF1. J Virol. 2007;81(10):5238–5245. doi: 10.1128/JVI.02680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deitz SB, Dodd DA, Cooper S, Parham P, Kirkegaard K. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc Natl Acad Sci USA. 2000;97(25):13790–13795. doi: 10.1073/pnas.250483097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kondratova AA, Neznanov N, Kondratov RV, Gudkov AV. Poliovirus protein 3A binds and inactivates LIS1, causing block of membrane protein trafficking and deregulation of cell division. Cell Cycle. 2005;4(10):1403–1410. doi: 10.4161/cc.4.10.2041. [DOI] [PubMed] [Google Scholar]
- 71.Karki S, Holzbaur EL. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr Opin Cell Biol. 1999;11(1):45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 72.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279(5350):519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 73.Harada A, Takei Y, Kanai Y, Tanaka Y, Nonaka S, Hirokawa N. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol. 1998;141(1):51–59. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doedens JR, Giddings TH, Jr, Kirkegaard K. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J Virol. 1997;71(12):9054–9064. doi: 10.1128/jvi.71.12.9054-9064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beske O, Reichelt M, Taylor MP, Kirkegaard K, Andino R. Poliovirus infection blocks ERGIC-to-Golgi trafficking and induces microtubule-dependent disruption of the Golgi complex. J Cell Sci. 2007;120(Pt 18):3207–3218. doi: 10.1242/jcs.03483. [DOI] [PubMed] [Google Scholar]
- 76.Gazina EV, Mackenzie JM, Gorrell RJ, Anderson DA. Differential requirements for COPI coats in formation of replication complexes among three genera of Picornaviridae. J Virol. 2002;76(21):11113–11122. doi: 10.1128/JVI.76.21.11113-11122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belov GA, Fogg MH, Ehrenfeld E. Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor. J Virol. 2005;79(11):7207–7216. doi: 10.1128/JVI.79.11.7207-7216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78▪▪.Oh HS, Pathak HB, Goodfellow IG, Arnold JJ, Cameron CE. Insight into poliovirus genome replication and encapsidation obtained from studies of 3B–3C cleavage site mutants. J Virol. 2009;83(18):9370–9387. doi: 10.1128/JVI.02076-08. Reports the first viable P3-processing-site mutant poliovirus. This mutant reveals a role for 3CD before and after genome replication that has not been defined. The use of processing site mutants with subtle changes in efficiency may be useful for uncovering new roles for other proteins in the picornaviral lifecycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiang W, Cuconati A, Hope D, Kirkegaard K, Wimmer E. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J Virol. 1998;72(8):6732–6741. doi: 10.1128/jvi.72.8.6732-6741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lyle JM, Clewell A, Richmond K, et al. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J Biol Chem. 2002;277(18):16324–16331. doi: 10.1074/jbc.M112429200. [DOI] [PubMed] [Google Scholar]
- 81.Pathak HB, Arnold JJ, Wiegand PN, Hargittai MR, Cameron CE. Picornavirus genome replication: assembly and organization of the VPg uridylylation ribonucleoprotein (initiation) complex. J Biol Chem. 2007;282(22):16202–16213. doi: 10.1074/jbc.M610608200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lyle JM, Bullitt E, Bienz K, Kirkegaard K. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science. 2002;296(5576):2218–2222. doi: 10.1126/science.1070585. [DOI] [PubMed] [Google Scholar]
- 83.Paul AV, van Boom JH, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393(6682):280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 84▪.Tellez AB, Crowder S, Spagnolo JF, et al. Nucleotide channel of RNA-dependent RNA polymerase used for intermolecular uridylylation of protein primer. J Mol Biol. 2006;357(2):665–675. doi: 10.1016/j.jmb.2005.12.044. Prediction of a site on the back of the polymerase that binds VPg and may be used for initiation of genome replication. [DOI] [PubMed] [Google Scholar]
- 85.Appleby TC, Luecke H, Shim JH, et al. Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer. J Virol. 2005;79(1):277–288. doi: 10.1128/JVI.79.1.277-288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hansen JL, Long AM, Schultz SC. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5(8):1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 87.Hobson SD, Rosenblum ES, Richards OC, Richmond K, Kirkegaard K, Schultz SC. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 2001;20(5):1153–1163. doi: 10.1093/emboj/20.5.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Diamond SE, Kirkegaard K. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J Virol. 1994;68(2):863–876. doi: 10.1128/jvi.68.2.863-876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gohara DW, Crotty S, Arnold JJ, Yoder JD, Andino R, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J Biol Chem. 2000;275(33):25523–25532. doi: 10.1074/jbc.M002671200. [DOI] [PubMed] [Google Scholar]
- 90.Pathak HB, Ghosh SK, Roberts AW, et al. Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol) A surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J Biol Chem. 2002;277(35):31551–31562. doi: 10.1074/jbc.M204408200. [DOI] [PubMed] [Google Scholar]
- 91.Claridge JK, Headey SJ, Chow JY, et al. A picornaviral loop-to-loop replication complex. J Struct Biol. 2009;166(3):251–262. doi: 10.1016/j.jsb.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Porter DC, Ansardi DC, Wang J, McPherson S, Moldoveanu Z, Morrow CD. Demonstration of the specificity of poliovirus encapsidation using a novel replicon which encodes enzymatically active firefly luciferase. Virology. 1998;243(1):1–11. doi: 10.1006/viro.1998.9046. [DOI] [PubMed] [Google Scholar]
- 93.Verlinden Y, Cuconati A, Wimmer E, Rombaut B. The antiviral compound 5-(3,4-dichlorophenyl) methylhydantoin inhibits the post-synthetic cleavages and the assembly of poliovirus in a cell-free system. Antiviral Res. 2000;48(1):61–69. doi: 10.1016/s0166-3542(00)00119-4. [DOI] [PubMed] [Google Scholar]
- 94.Vance LM, Moscufo N, Chow M, Heinz BA. Poliovirus 2C region functions during encapsidation of viral RNA. J Virol. 1997;71(11):8759–8765. doi: 10.1128/jvi.71.11.8759-8765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dove AW, Racaniello VR. Cold-adapted poliovirus mutants bypass a postentry replication block. J Virol. 1997;71(6):4728–4735. doi: 10.1128/jvi.71.6.4728-4735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li JP, Baltimore D. An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J Virol. 1990;64(3):1102–1107. doi: 10.1128/jvi.64.3.1102-1107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Franco D, Pathak HB, Cameron CE, Rombaut B, Wimmer E, Paul AV. Stimulation of poliovirus synthesis in a HeLa cell-free in vitro translation–RNA replication system by viral protein 3CDpro. J Virol. 2005;79(10):6358–6367. doi: 10.1128/JVI.79.10.6358-6367.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nomoto A, Kitamura N, Golini F, Wimmer E. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc Natl Acad Sci USA. 1977;74(12):5345–5349. doi: 10.1073/pnas.74.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reuer Q, Kuhn RJ, Wimmer E. Characterization of poliovirus clones containing lethal and nonlethal mutations in the genome-linked protein VPg. J Virol. 1990;64(6):2967–2975. doi: 10.1128/jvi.64.6.2967-2975.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100▪.Franco D, Pathak HB, Cameron CE, Rombaut B, Wimmer E, Paul AV. Stimulation of poliovirus RNA synthesis and virus maturation in a HeLa cell-free in vitro translation-RNA replication system by viral protein 3CDpro. Virol J. 2005;2(86):86. doi: 10.1186/1743-422X-2-86. Provides the first suggestion that 3CD protein is required in a step after genome replication that is related to virion assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brown DM, Cornell CT, Tran GP, Nguyen JH, Semler BL. An authentic 3′ noncoding region is necessary for efficient poliovirus replication. J Virol. 2005;79(18):11962–11973. doi: 10.1128/JVI.79.18.11962-11973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Todd S, Towner JS, Brown DM, Semler BL. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J Virol. 1997;71(11):8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol Cell. 2001;7(3):581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barton DJ, O’Donnell BJ, Flanegan JB. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20(6):1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]