Abstract
In addition to the heptahelical transmembrane domain shared by all G protein-coupled receptors (GPCRs), many class A GPCRs adopt a helical domain, termed helix 8, in the membrane-proximal region of the C terminus. We investigated the role of residues in the hydrophobic and hydrophilic faces of amphiphilic helix 8 of human cannabinoid receptor 1 (CB1). To differentiate between a role for specific residues and global features, we made two key mutants: one involving replacement of the highly hydrophobic groups, Leu404, Phe408, and Phe412, all with alanine and the second involving substitution of the basic residues, Lys402, Arg405, and Arg409, all with the neutral glutamine. The former showed a very low Bmax based on binding isotherms, a minimal Emax based on GTPγS binding analysis, and defective localization relative to the wild-type CB1 receptor as revealed by confocal microscopy. However, the latter mutant and the wild-type receptors were indistinguishable. Circular dichroism spectroscopy of purified peptides with corresponding sequences indicated that the highly hydrophobic residues are critical for maintaining a strong helical structure in detergent, whereas the positively charged residues are not. Further investigation of mutant receptors revealed that CB1 localization requires a threshold level of hydrophobicity but not specific amino acids. Moreover, mutant receptors carrying two- to six-residue insertions amino-terminal to helix 8 revealed a graded decrease in Bmax values. Our results identify the key helix 8 components (including hydrophobicity of specific residues, structure, and location relative to TM7) determinant for receptor localization leading to robust ligand binding and G protein activation.
Activation of G protein-coupled receptors (GPCRs)1 induces conformational changes that promote heterotrimeric guanine nucleotide binding protein (G protein) activation and the subsequent stimulation of downstream effector proteins. Cannabinoid receptor 1 (CB1), a member of the rhodopsin-like GPCR family, primarily activates pertussis toxin (PTX) sensitive inhibitory G protein leading to the inhibition of adenylyl cyclase and N- and P/Q-type calcium channels and the activation of D-type outward potassium channels and mitogen-activated protein kinase (for a review, see ref 1). However, CB1 also couples to Gs or Gq proteins under certain conditions (2, 3). CB1 is primarily expressed in brain, including the hippocampus, cerebral cortex, basal ganglia, and cerebellum, whereas its variant, CB2, resides mainly in immune cells. As the most abundant G protein-coupled receptor in human brain, CB1 is thought to play critical physiological roles, including appetite stimulation, pain reduction, learning and memory, drug reward, and motor regulation (4–8).
Ligand-activated GPCRs are rapidly desensitized and lost from the cell surface by a mechanism that includes phosphorylation and arrestin binding. Phosphorylation predominantly occurs on serine and threonine residues within intracellular domains, including intracellular loops and the carboxyl terminus (C terminus). Once phosphorylated, activated GPCRs bind arrestins, resulting in key conformational changes that facilitate their interaction with endocytic machinery proteins such as clathrin and AP-2. After localization to endosomes, GPCRs are either dephosphorylated and recycled to the plasma membrane or targeted to lysosomes for degradation (for a review, see ref 9). Prior studies demonstrated that the CB1 receptor behaves similarly, undergoing agonist-induced desensitization and subsequent endocytosis (10, 11). However, CB1 also displays relatively high basal activity and constitutive endocytosis with 85% of the receptor localized to the endosome in the absence of ligand (12–14). Recently, the C terminus of CB1 has been a focal point for investigating receptor phosphorylation, desensitization, and endocytosis (15), yet an understanding of the possible structure–function correlations involving this region of the receptor remains obscure.
In previous studies, we purified the full-length C terminus of the CB1 receptor and, using spectroscopic techniques, demonstrated its strong helical properties induced by negatively charged and zwitterionic detergents. We further defined two amphipathic helical domains within the C terminus of the human CB1 receptor, helix 8 and helix 9 (16). Helix 8 has been identified as a highly conserved structural motif among the rhodopsin-like class A GPCRs. Moreover, this helical motif has also been found in some class B GPCRs (17). Helix 9, a helical domain in the distal region of the C terminus and newly identified in CB1, may be involved in intracellular signaling and constitutive activity.
The recently determined crystal structure of squid rhodopsin offers insight into the importance of the helix 8 domain (18). Like the corresponding domain identified in the crystal structure of the bovine rhodopsin and β-adrenergic receptors, helix 8 of squid rhodopsin is tethered to the membrane surface by TM7 and palmitoylation at Cys337. This helix 8 interacts with an additional helical domain in the distal part of the C terminus, constraining its movement and poising it for interaction with the third intracellular loop (IC3) that is critical for signaling. In addition, the residues in the hydrophobic and hydrophilic faces of helix 8 have been examined for their potential involvement in receptor function. The basic residues in the hydrophilic face, in particular, have been suggested to contribute to the intracellular signaling of many class A GPCRs, including the β1-adrenergic receptor (19), the oxytocin (OT) receptor (20), and rat melanin-concentrating hormone (MCH) receptor 1 (21). Swift and coworkers have reported that the network of noncovalent interactions among the protease-activated receptor helix 8, TM7, and intracellular loop 1 (IC1) contributes to receptor signaling (22). Other studies suggest the hydrophobic residues in helix 8 play a role in receptor biosynthesis, folding, and trafficking (23–25). However, since few studies combine structural and functional analysis of helix 8, it remains difficult to distinguish the extent to which the specific residues versus the overall structure of this domain are critical for the receptor life cycle.
In this study, we show for the first time a direct correlation between the presence of an inducible helical motif and its location in the C terminus of CB1 with particular biological properties exhibited by the receptor. Circular dichroism spectroscopy suggests that the highly hydrophobic face of the helix in the membrane-proximal region of the C terminus is critical for its formation and that the helical unit, rather than particular amino acids, is responsible for receptor trafficking leading to robust ligand binding and G protein coupling. The combination of secondary structural analysis, pharmacological evaluation, and global mutagenesis employed here has led to insights that should help clarify the wide variety of data reported for the role of helix 8 in other GPCRs.
EXPERIMENTAL PROCEDURES
Plasmid Constructions
For mammalian cell expression, all mutant receptors were generated by site-directed mutagenesis (QuickChange, Stratagene, La Jolla, CA) using the human CB1 cDNA cloned into pcDNA3.1 as a template, according to the manufacturer’s instructions. The corresponding green fluorescent protein (GFP)-tagged receptors were generated by subcloning the GFP sequence from the pEGFP-N1 plasmid (BD Biosciences, Franklin Lakes, NJ) into the pcDNA3.1 vector immediately downstream of the CB1 receptor coding region. For bacterial expression and purification of the receptor C terminus, the cDNA containing the coding region for the human CB1 C terminus (73 residues) was subcloned into pGEX-6P-1 (GE Healthcare, Piscataway, NJ). The DNA bases encoding a six-histidine tag (His tag) and two lysine residues to increase solubility during purification were inserted between the last residue of the C terminus and a stop codon as previously described (16). All constructs were confirmed by sequencing analysis.
Peptide Expression and Purification
CB1 C-terminal peptides were prepared as previously described (16). Briefly, Escherichia coli strain BL21(DE3) (Invitrogen), transformed with the wild-type and mutant C-terminal constructs, was inoculated into M9 minimal medium and grown at 37 °C until the OD600 reached 0.5. The culture was then induced with 0.5 mM isopropyl β-thiogalactopyranoside (IPTG) for 6 h at 30 °C. The cells were harvested, and the pellet was resuspended in lysis buffer [50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole (pH 8.0), and 0.1mM phenylmethanesulfonyl fluoride]. The cells were lysed by sonication for 3 min at a power setting of 5 and a duty cycle of 50%. Triton X-100 was added to a final concentration of 1%(v/v), and the lysate was stirred at 4 °C for 30 min and then centrifuged for 30 min at 22500g. The peptide samples were purified from the supernatant using Ni-NTA agarose affinity resin (Qiagen, Valencia, CA) and glutathione-Sepharose 4B (GE Healthcare), followed by PreScission protease (GE Healthcare) cleavage for 18 h at 4 °C. The sample was then desalted and concentrated using a Centricon filter device (Millipore, Billerica, MA). The purity of the peptide was verified by SDS–PAGE and its identity confirmed via Western blot using antisera to the CB1 carboxyl terminus and separately the His tag.
Circular Dichroism Spectroscopy
The CD spectra were recorded on a Jasco (Tokyo, Japan) J-715 CD spectrometer as previously described (16). Peptide concentrations were determined by amino acid analysis (Yale University Keck Biotechnology Resource Laboratory, New Haven, CT). The peptide concentration used was 50 μM in 10 mM phosphate buffer (pH 7.3). All spectra were corrected by subtraction of spectra for reference samples containing only the buffer, and detergents as appropriate. The spectra were recorded using a 0.1 cm path length quartz cuvette from250 to 190 nm at 20 °C at a scan speed of 50 nm/min and a response time of 0.5 s. Four spectra were recorded and averaged per sample. CD intensities were converted to mean residue molar ellipticity [θ], using the equation [θ] = θobs/(10lcn) (in degrees square centimeters per decimole), where θobs is the observed ellipticity in millidegrees, l is the path length in centimeters, c is the final concentration of the peptides in molarity, and n is number of amino acid residues. The CD spectra were analyzed to produce quantitative estimations of the secondary structure using three different methods, CDSSTR, CONTINLL, and SELCON3 (CDPRO). CONTINLL gave a best fit with the lowest root-mean-square deviation (rmsd) for the observed sample spectra and the derived combination of basis spectra and is the method used for values reported in the text.
CB1 Expression and Membrane Preparation
HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 3.5 mg/mL glucose at 37 °C in 5%CO2. For transient transfection, HEK293 cells were seeded at a density of 8 × 105 cells/100 mm dish on the day prior to transfection. Transfection was conducted using the calcium phosphate precipitation method (26) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Membranes of transfected cells expressing the wild-type or mutant receptors were prepared as previously described (27).
Saturation Radioligand Binding
Saturation binding assays were performed as previously described (28). Approximately 30–40 μg of membranes was incubated at 30 °C for 90 min in 200 μL of TME buffer (pH 7.4) containing 0.1% fatty acid-free bovine serum albumin using [3H]CP55940 (139.6 Ci/mmol) (PerkinElmer Life Sciences, Boston, MA). At least nine radio-labeled ligand concentrations (ranging from 0.24 to 37.60 nM) were used to determine the Kd values of the receptors. Nonspecific binding was assessed in the presence of 1 μM unlabeled ligand. Reactions were terminated via addition of 250 μL of TME containing 5% BSA followed by filtration with a Brandel cell harvester through Whatman GF/C filter paper.
GTPγS Binding Assay
Fifteen micrograms of membranes was incubated for 60 min at 30 °C in GTPγS binding assay buffer [50 mM Tris-HCl (pH 7.4), 3 mM MgCl2, 0.2 mM EGTA, and 100 mM NaCl] with unlabeled ligand (at least nine different concentrations were used ranging between 100 pM and 100 μM), 0.1 nM[35S]GTPγS (1250 Ci/mmol) (PerkinElmer Life Sciences), 10 μM GDP, and 0.1% (w/v) BSA. Nonspecific binding was assessed with 10 μM unlabeled GTPγS (Sigma, St. Louis, MO). The reaction was terminated by rapid filtration through Whatman GF/C filters. The radioactivity trapped in filters was determined by liquid scintillation counting.
Confocal Microscopy
HEK293 cells expressing CB1 receptors C-terminally fused with GFP were seeded onto 35 mm glass-bottomed dishes (Matek, MA) precoated with poly-D-lysine. Cells were washed three times with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. For colocalization studies, the cells were permeabilized with 0.1% Triton X-100 in DME containing 5% normal goat serum (pH 7.6). After incubation with blocking solution (5% normal goat serum in DME) for 30 min at room temperature, the cells were incubated with the late endosome/lysosomal marker, lysosomal-associated membrane protein 1 (LAMP-1) (H4A3) (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), diluted in DME containing 5% normal goat serum. For early endosome and endoplasmic reticulum (ER) staining, anti-early endosome antigen antibody (anti-EEA1) (BD Biosciences, San Jose, CA) and mouse anti-protein disulfide isomerase (anti-PDI) (Affinity BioReagents, Golden, CO) were used, respectively. After being washed with PBS, cells were incubated with Cy3-labeled donkey anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories) diluted 1:200 for 1 h at room temperature. Cells were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and visualized using a Leica TCS SP2 confocal microscope (Leica Microsystems, Wetzler, Germany). Images were collected from at least three independently transfected cell dishes and processed for presentation in the figures using Adobe Photoshop version 6.0 (Adobe Systems, San Jose, CA). Quantification of colocalization was performed using ImageJ (National Institutes of Health, Bethesda, MD) with the JACoP plugin (29). For the analysis, images of approximately 10–15 cells were taken. Images for the GFP-bound receptor (green) and subcellular marker (red) were processed in parallel; the background was subtracted, and the images were converted into 8-bit format for analysis. The extent of colocalization was quantified using the intensity correlation analysis. Pearson’s correlation coefficient (PCC) r, which provides an estimate of the goodness of colocalization, was determined (−1 means a negative correlation, 0 means no correlation, and 1 means a positive correlation). PCC (r) was presented as the mean ± the standard deviation (SD).
Data Analysis
All ligand binding assays and functional assays were conducted in duplicate. Data are presented as the mean ± the standard error of the mean or the median with the corresponding 95% confidence limits from at least three independent experiments. The Kd and Bmax values were calculated by nonlinear regression (fitted to a one-site binding model) using GraphPad Prism (GraphPad Software Inc., San Diego, CA). EC50 values for GTPγS binding were calculated using a sigmoidal dose–response relationship. Kd and EC50 values for the wild-type and mutant receptors were compared using analysis of variance (ANOVA) followed by Dunn’s post hoc test for significance. p values of <0.05 were considered to be statistically significant.
RESULTS
Highly Hydrophobic Residues Are Critical for Helix 8 Formation in Membrane Mimetics
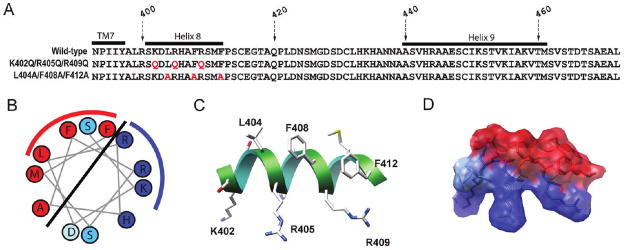
Since the existence of helix 8 was revealed from the crystal structure of bovine rhodopsin, a comparable helix has been identified in many receptors of the rhodopsin-like class A GPCR family. This helical domain has a strong amphipathic nature consistent with its location parallel to the surface of the membrane and at the interface with the cytosolic milieu. Figure 1A shows the sequence of the entire 73-residue C terminus of CB1 following the highly conserved NPxxY motif of TM7. Representation of the 12 residues from S401 to F412 on a helical wheel diagram (Figure 1B) and in molecular models (Figure 1C,D) consistent with our previous NMR study (16) reveals the helix 8 domain of CB1 is also amphipathic with clearly separated hydrophobic and hydrophilic faces.
Figure 1.
CB1 C-terminal sequences and structural representations of helix 8. (A) C-Terminal sequences of the wild-type and two triple mutant receptors; K402Q/R405Q/R409Q and L404A/F408A/F412A. The amino acid numbers corresponding to the sequence of the full-length human CB1 receptor are indicated. The locations of the C terminus of TM7, helix 8, and helix 9 are marked with black bars. The residues mutated to alanine or glutamine are highlighted in red. (B) Helical wheel projection of CB1 helix 8. Hydrophobic and positively charged residues are colored red and blue, respectively. Two serines and an aspartic acid are colored turquoise and light blue, respectively. The hydrophilic and hydrophobic faces of the helix are separated by the solid line, and the residues on which we focused in this study are further indicated by the red and blue arcs, respectively. (C) Schematic view of the amphipathic nature of helix 8 of the C terminus of the human CB1 receptor. The illustration shown here is generated on the basis of our previous structural study of the CB1 C terminus (16). Helix 8 (residues 401–412) is shown as a ribbon. Some key residues are labeled with the amino acid numbers corresponding to the human CB1 sequence. (D) Transparent surface of helix 8 with the side chains highlighted. Hydrophobic and positively charged residues are colored red and blue, respectively.
To investigate the significance of the residues of helix 8, two mutant receptors were generated in the full-length CB1, each involving triple substitutions on one face of the α-helix. These are the K402Q/R405Q/R409Q and L404A/F408A/F412A receptors with substitutions in the hydrophilic and hydrophobic faces, respectively (Figure 1A). Separately, to determine whether these mutations influence secondary structure, the same sequences were generated as a peptide corresponding to the C terminus (R400–L472; see Experimental Procedures), purified, and analyzed using CD spectroscopy. Typically, 50 μM peptide was used to examine its secondary structural characteristics in aqueous solution and in different detergents. Dodecylphosphocholine (DPC), a zwitterionic detergent, and sodium dodecyl sulfate (SDS), a negatively charged detergent, were used in this study since the wild-type C-terminal peptide exhibited helical structure formation with these detergents (Figure 2A,D), whereas no significant structure was evident from the spectra for the peptide in the nonionic detergent, n-dodecyl β-D-maltoside (DDM) (data not shown). In the absence of detergent, all C-terminal peptides, including the wild-type, K402Q/R405Q/R409Q, and L404A/F408A/F412Asequences, exhibited spectra with a single negative minimum at ~200 nm consistent with an unordered structure (43–49%, with small amounts of α-helix, β-sheet, and other structure). In the presence of DPC, the wild-type and K402Q/R405Q/R409Q peptides (Figure 2A,B) display similar CD spectral patterns with signatures of unordered structure below the CMC (1.1 mM) and high helical content above the CMC (36.4 and 38.0% helix at 50 mM DPC for the wild-type and the K402Q/R405Q/R409Qpeptide, respectively). These spectra were characterized by a positive peak in the 190–195 nm range, a negative peak at ~208, and a second broader negative peak at ~222 nm. In marked contrast, the L404A/F408A/F412A peptide (Figure 2C) exhibited an only very small increase in helicity in the presence of the highest concentration of DPC employed (9.9 and 14.4% helical content in aqueous solution and 50 mM DPC, respectively) (Figure 2C and Table 1). When increases in helix content were observed, it was largely at the expense of random coil, and no significant changes in the fraction of other structural motifs were noted. While the negatively charged detergent SDS promotes higher levels of helical structure, the same hierarchical pattern distinguishes the wild-type and K402Q/R405Q/R409Q peptides (Figure 2D,E) relative to the L404A/F408A/F412A peptide (Figure 2F). The spectra indicate that the former adopt 50.6 and 49.2%helix content in 50 mM SDS, respectively, while the latter exhibited significantly lower levels (32.2% helix in 50 mM SDS). Taken together, these data indicate that the wild-type and K402Q/R405Q/R409Q peptides adopt substantial and comparable helical content induced by the presence of detergent while the helical content of the L404A/F408A/F412A peptide is considerably reduced. This clearly demonstrates that while the positive charges are not required, the highly hydrophobic residues of the wild-type sequence are critical for α-helix formation in a membrane mimetic environment.
Figure 2.
CD spectra of the CB1 C-terminal peptides in various detergents. The peptide concentration was 50 μM in 10 mM phosphate buffer (pH 7.3). CD spectra of (A) the wild-type, (B) K402Q/R405Q/R409Q, and (C) L404A/F408A/F412Apeptides in the absence (■) and presence of 0.5 (▲, red), 5 (▼, green), or 50 mM DPC (◆, blue). CD spectra of (D) the wild-type, (E) K402Q/R405Q/R409Q, and (F) L404A/F408A/F412A peptides in the absence (■) and presence of 2 (▲, red), 10 (▼, green), or 50 mM SDS (◆, blue). Data represent the averages of four experiments.
Table 1.
Impact of CB1 Helix 8 Amino Acid Mutation on Ligand Binding, Activation, and Helical Contenta
| receptor | CP55940b |
GTPγSc |
percent helixd | ||
|---|---|---|---|---|---|
| Kd (nM) | Bmax (fmol/mg) | EC50 (nM) | Emax (% WT) | ||
| wild-type | 3.4 ± 0.4 | 3499 ± 129 | 5.5 (3.0–10.2) | 100.0 | 36.4 |
| K402Q/R405Q/R409Q | 3.7 ± 0.6 | 3118 ± 142 | 3.8 (2.4–6.0) | 80.6 ± 2.0 | 38.0 |
| L404A/F408A/F412A | 3.8 ± 0.8 | 867 ± 80 | not available | 14.4 ± 1.9 | 14.4 |
Data are the median and corresponding 95% confidence limits or the mean ± the standard error of the mean of three independent experiments performed in duplicate (n=3 determinations).
Kd and Bmax values were determined from saturation binding assays using [3H]CP55940.
EC50 and Emax values were determined from the CP55940 dose–response model.
Percentage of helical content calculated from spectra of 50 μM peptide at 50 mM DPC using CONTINLL.
CB1 Agonist Binding, Agonist-Stimulated Efficacy, and Localization Correlate with the Helical Propensity of Helix 8
To evaluate the influence of the residues on each face of helix 8 on the ligand binding parameters, saturation binding analysis with [3H]CP55940 was performed with the membrane prepared from HEK293 cell lines expressing the wild-type, K402Q/R405Q/R409Q, and L404A/F408A/F412A receptors at their full length. As shown in Figure 3A, the wild-type and K402Q/R405Q/R409Q receptors produced similar binding isotherms with Kd values of 3.4 ± 0.4 and 3.7 ± 0.6 nM and Bmax values of 3499 ± 129 and 3118 ± 142 fmol/mg, respectively. The L404A/F408A/F412A receptor also displayed a high affinity for CP55940 with a Kd value of 3.8 ± 0.8 nM, comparable to those of the wild-type and K402Q/R405Q/R409Q receptors, suggesting that at least a population of the receptor retains the ability to bind ligand and, therefore, likely folds properly. However, a 4-fold reduced Bmax value was obtained for the membrane with the L404A/F408A/F412A receptor (867 ± 80 fmol/mg), indicating that the cells expressing this mutant receptor have substantially fewer functional receptors with wild-type-like ligand binding affinity compared to the cells expressing the wild-type receptor (Table 1).
Figure 3.

CP55940 saturation binding and stimulation of binding of GTPγS to membranes of HEK293 cells expressing the CB1 wild-type and mutant receptors. (A) Saturation binding with [3H]CP55940 to a membrane prepared fromHEK293 cells expressing the wild-type (■) and mutant receptors, K402Q/R405Q/R409Q (△) and L404A/F408A/F412A (●). Each data point represents the mean ± the standard error of the mean of at least three independent experiments performed in duplicate. (B) Stimulation of [35S]GTPγS binding by CP55940 in membrane preparations from HEK293 cells expressing wild-type (■), K402Q/R405Q/R409Q (△), and L404A/F408A/F412A (●) receptors. Data are presented as specific binding of GTPγS to the membranes. Nonspecific binding was assessed in the presence of 10 μM unlabeled GTPγS. Each data point represents the mean ± the standard error of the mean of at least three independent experiments performed in duplicate.
The relationship between the ligand binding parameters and the functional properties of each receptor was assessed by agonist-stimulated [35S]GTPγS binding (Figure 3B). The EC50 value of the wild-type receptor for agonist, CP55940, was 5.5 (3.0–10.2) nM. The K402Q/R405Q/R409Q receptor displayed a similar EC50 value of 3.8 (2.4–6.0) nM and an Emax value that was 80.6 ± 2.0% of the wild-type value (Table 1). In contrast, the L404A/F408A/F412A receptor displayed virtually no [35S]GTPγS binding as a result of CP55940 treatment (Figure 3B). These data parallel the ligand binding parameters and indicate that the low helical propensity of the L404A/F408A/F412A receptor impacts the availability of the functional receptor to bind ligand and G protein effectively (Table 1).
Confocal microscopy of cells expressing GFP-tagged receptors was used to probe whether the observed reduction in the level of functional receptor expression and activation of the L404A/F408A/F412A receptor is reflected in its subcellular localization pattern. The wild-type CB1-GFP receptor was found mainly in intracellular vesicles as previously reported (13, 30) and was colocalized with the late endosome/lysosome marker, LAMP-1 (Figure 4, top row) with a PCC r of 0.868 ± 0.041 (n = 11) as described in Experimental Procedures. The K402Q/R405Q/R409Q-GFP receptor displayed a localization pattern very similar to that of the wild-type CB1-GFP receptor (Figure 4, middle row) with a PCC r of 0.792 ± 0.111 (n=10). Consistent with these findings, the wild-type GFP-bound receptor and the K402Q/R405Q/R409Q-GFP receptor did not show any significant colocalization with either the early endosome marker, EEA1, or the ER marker, PDI (data not shown). In contrast, the cells expressing the L404A/F408A/F412A-GFP receptor showed a more diffuse pattern of fluorescence and showed little colocalization with LAMP-1 (Figure 4, bottom row) with a PCC r of 0.185 ± 0.045 (n = 11). Interestingly, this mutant receptor exhibited a significant colocalization with the ER marker (data not shown). Thus, this mutant receptor appears to have a trafficking defect that accompanies diminished ligand binding relative to the wild-type and K402Q/R405Q/R409Q receptors.
Figure 4.
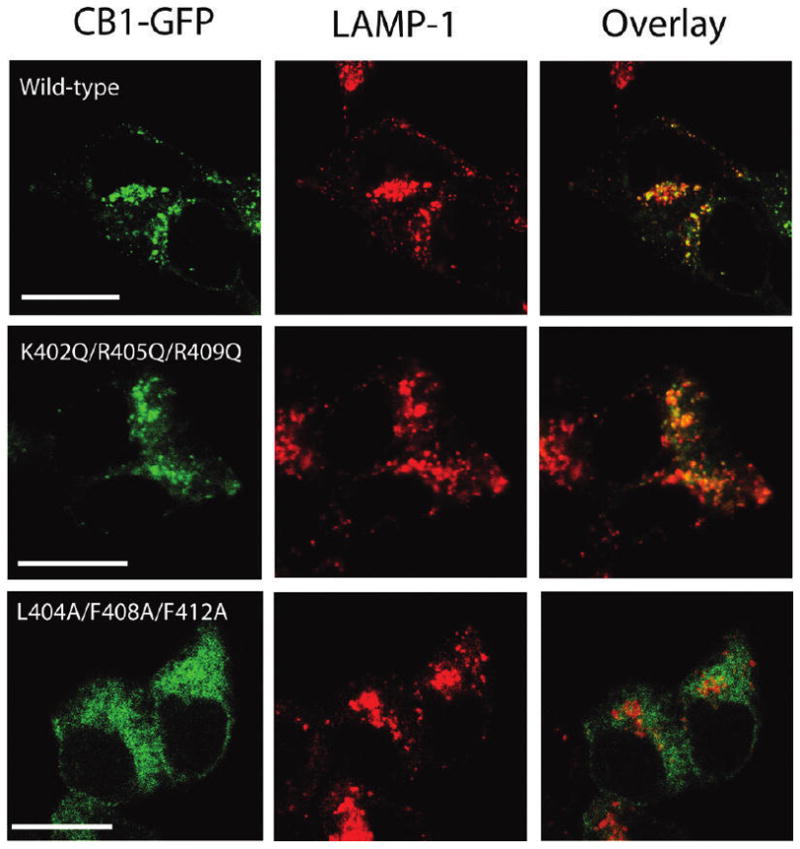
Comparison of the subcellular distribution of the wild-type and CB1 receptors with mutations in helix 8. Localization of GFP-tagged wild-type, K402Q/R405Q/R409Q, and L404A/F408A/F412A receptors (green, left), the late endosome/lysosome marker, LAMP-1 (red, middle), in HEK293 cells, and an overlay of the fluorescence images to show the extent of colocalization (yellow, right). HEK293 cells transiently expressingCB1 receptors were fixed, permeabilized, and stained with antibodies against LAMP-1, as described in Experimental Procedures. Images were chosen as representatives from at least three independent transfections. The scale bar is 15 μm.
Overall Hydrophobicity Rather Than Specific Residues Is a Major Determinant of Receptor Localization and Functional Receptor Density
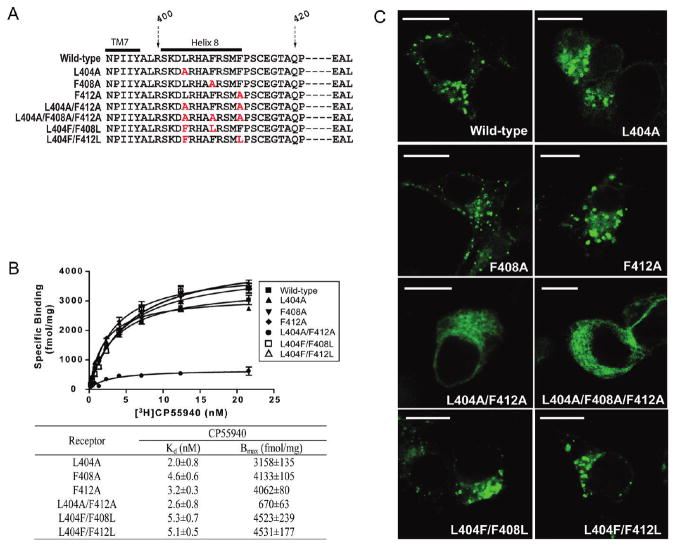
Since the L404A/F408A/F412A mutant receptor containing three amino acid substitutions exhibited altered receptor properties, it was essential to investigate whether one or more of these hydrophobic residues are specifically required or if the overall level of hydrophobicity is the critical feature for helix 8 function. To address this question, a series of mutant receptors was generated involving one, two, and three substitutions (Figure 5A) and characterized for functional expression and localization using ligand binding and confocal microscopy as described previously. Saturation binding analysis with [3H]CP55940 indicated that none of the individual substitutions (L404A, F408A, or F412A) with alanine affected the properties of binding of the receptor to CP55940. Kd values of 2.0 ± 0.8, 4.6 ± 0.6, and 3.2 ± 0.3 nM and Bmax values of 3158 ± 135, 4133 ± 105, and 4062 ± 80 fmol/mg for the L404A, F408A, and F412A receptors, respectively, were observed (Figure 5B). Interestingly, along with the triply substituted L404A/F408A/F412A mutant receptor described above, the simultaneous substitution of two of the hydrophobic residues with alanine displayed a remarkably reduced Bmax value (670 ± 63 fmol/mg) while retaining the wild-type affinity for CP55940 (Kd=2.6 ± 0.8 nM). These observations suggest that the overall hydrophobicity, rather than any specific residue side chain, mediates the functional expression of the receptor. To further support this conclusion, two mutant receptors, L404F/F408L and L404F/F412L, involving an exchange of residue position were generated and characterized for ligand binding. These mutant receptors retain the same overall composition and hydrophobicity but no longer retain the sequence of the wild-type receptor. Both L404F/F408L and L404F/F412L receptors retain the wild-type affinity for CP55940 and Bmax values (Kd values of 5.3 ± 0.7 and 5.1 ± 0.5 nM and Bmax values of 4523 ± 239 and 4531 ± 177 fmol/mg, respectively). Consistent with the ligand binding data, confocal microscopy revealed the importance of the overall hydrophobicity of helix 8 in receptor localization (Figure 5C). Cells expressing receptors with individual substitutions of hydrophobic residues (L404A, F408A, and F412A receptors) and the sequence-scrambled mutant receptors (L404F/F408L and L404F/F412L receptors) displayed receptors in intracellular vesicles and a cellular distribution comparable to that of cells expressing the wild-type receptor. However, cells expressing receptors carrying multiple alanine mutations showed significant changes in receptor localization, and no colocalization with LAMP-1 was observed (data not shown).
Figure 5.
Ligand binding and localization of receptors with different combinations of amino acid substitutions in the hydrophobic face of helix 8. (A)Amino acid sequences of the relevant portion of the C terminus of the wild-type or singly or multiply substituted mutant receptors. The amino acid numbers corresponding to the sequence of the humanCB1 receptor are shown, and the C terminus of TM7 and helix 8 are indicated by black bars. The residues substituted with alanine are highlighted in red. Several of the most C-terminal residues are represented by the dashed lines and not shown due to space limitations. (B) Saturation binding with [3H]CP55940 to a membrane prepared from HEK293 cells expressing the wild-type (■) and mutant recptors, L404A(▲), F408A(▼), F412A(◆), L404A/F412A(●), L404F/F408L(□), andL404F/F412L(△). The Kd and Bmax values were calculated by nonlinear regression (fitted to a one-site binding model) as described in Experimental Procedures and are reported as the means ± the standard error of the mean. The Kd and Bmax values for the wild-type receptor are listed in Table 1. (C) Comparison of the cellular distribution of the wild-type or helix 8mutant receptors. GFP-tagged wild-type and mutant receptors were expressed inHEK293 cells. A day after transfection, cells were fixed and mounted for confocal microscopy. Images were chosen as representatives from at least three independent transfections. The scale bar is 15 μm.
The Proximity of Helix 8 to TM7 Is Crucial
To investigate the significance of the location of the helix 8 domain relative to the C terminus of TM7, we developed a series of mutant receptors that varied in the length of a glutamine linker (two, four, or six residues) inserted between these two domains (Figure 6A). Each of these mutant receptors exhibited a Kd value similar to that of the wild-type receptor [2.9 ± 0.2, 2.4 ± 0.7, and 2.6 ± 0.5 nM for the 401(Gln)2, 401(Gln)4, and 401(Gln)6 receptors, respectively], suggesting that at least some of the receptors could fold appropriately for ligand recognition. However, we observed a progressive decrease in the Bmax values of these receptors concomitant with an increase in the number of glutamines in the linker (Figure 6B). Insertions of two, four, and six glutamine residues resulted in 18, 56, and 80% reductions in the Bmax value, respectively, relative to that of the wild-type receptor (Figure 6B), suggesting an impact on the size of the receptor population accessible to ligand. The 401(Gln)6 mutant receptor exhibited binding parameters similar to those observed for the receptors carrying multiple alanine substitutions within the hydrophobic face of helix 8. Confocal microscopy further substantiates the impact on localization. As shown in Figure 6C, cells expressing the 401(Gln)2-GFP receptor show a punctuate pattern and mostly endosomal receptor localization comparable to that of the wild type while cells expressing the 401(Gln)4, and 401(Gln)6 receptors display the same diffuse pattern observed with cells expressing the trafficking defective L404A/F412A and L404A/F408A/F412A receptors. These observations imply that the distance betweenTM7 and helix 8 (or the membrane surface) is as crucial for localization and pharmacological properties as formation of the helical conformation itself.
Figure 6.
Effect of glutamine insertions in the membrane-proximal region of the CB1 C terminus on ligand binding and localization. (A) Amino acid sequences of the wild-type and mutant receptors. The amino acid numbers corresponding to the sequence of the CB1 receptor are shown, and the C terminus of TM7 and helix 8 are indicated by black bars. The glutamine residues inserted between TM7 and helix 8 are highlighted in red, and helix 8 is shaded in gray. Several of the most C-terminal residues are represented by the dashed lines and not shown due to space limitations. (B) Saturation binding with [3H]CP55940 to the membrane prepared fromtheHEK293 cells expressing the wild-type (■) or insertion mutant receptors, 401(Gln)2 (●), 401(Gln)4 (▲), and 401(Gln)6 (▽). Kd and Bmax values were calculated by nonlinear regression (fitted to a one-site binding model) as described in Experimental Procedures and are reported as the means ± the standard error of the mean. Binding data for the wild-type receptor from Table 1 and Figure 5 were included here for the sake of comparison. (C) Comparison of the cellular distribution of the wild-type and mutant CB1 receptors carrying the glutamine insertions. GFP-tagged wild-type and mutant receptors were expressed in HEK293 cells as described in the legend of Figure 5. The scale bar is 15 μm.
DISCUSSION
In addition to the heptahelical transmembrane domains shared by all GPCRs, the X-ray crystal structures of the rhodopsins (18, 31), the adenosine receptor (32), and the β-adrenergic receptors (33–36) all identify an additional helical domain, termed helix 8, in the membrane-proximal region of the C terminus. This helical domain is amphipathic, with leucines and/or phenylalanines and basic residues common in the hydrophobic and hydrophilic faces, respectively. Previous studies have suggested these residues might participate in intramolecular interactions with other regions of the receptor and/or intermolecular interactions with other proteins. For example, Marin and colleagues demonstrated an interaction between the N terminus of helix 8 of rhodopsin and Gαt (37). de los Santos et al. also suggested that the β1-adrenergic receptor is tethered to the G protein through the hydrophilic interface of helix 8 (19). It has been reported that an intramolecular interaction including helix 8 and other domains of the receptor, such as intracellular loop 1 and the NPxxY motif of TM7, is critical for receptor signaling in some receptors (22). In addition, there is emerging evidence that the hydrophobic residues in this helix play a role as a determinant of receptor folding and/or proper trafficking. Several motifs within the C tail have been implicated as an ER export signal: F(X)6LL in the AT1R and α2B-AR (24), F(X)3F(X)3F in the dopamine D1 receptor (38), FN(X)2LL(X)3L in the vasopressin V1b/V3 receptor (39), and F(X)3V(X)3L in the vasopressin V2 receptor (25). However, the molecular mechanism underlying the impact of these hydrophobic residues on receptor expression and function remains elusive.
In this study, we probed the role of several key residues in each face of helix 8 in CB1. To differentiate between a role for specific residues and global features, we made two key mutants: one involving replacement of the hydrophobic groups, Leu404, Phe408, and Phe412, all with alanine to reduce hydrophobicity and the second involving substitution of the basic residues, Lys402, Arg405, and Arg409, all with glutamine to remove the positive charge. The former showed a very low Bmax based on binding isotherms, a minimal Emax based on G protein coupling analysis using GTPγS assays, and defective localization relative to that of the wild-type CB1 receptor. The latter mutant (K402Q/R405Q/R409Q), in marked contrast, was virtually indistinguishable from the wild-type receptor using these criteria. Using CD spectroscopy to establish the secondary structure of the corresponding sequences within the context of a full-length, otherwise wild-type CB1 C terminus (residues Arg400–Leu472), we found that the presence of highly hydrophobic residues is critical for the maintenance of the helix, whereas the positively charged residues were readily replaced with neutral polar residues with no structural consequences. These data argue that the formation of the helical domain itself is important for receptor localization and consequently ligand binding and activation as opposed to any particular set of residues in helix 8. This conclusion is underscored by additional data involving characterization of mutant receptors in which each of the hydrophobic residues is replaced individually (L404A, F408A, and F412A receptors) and found to localize and bind ligand in a manner comparable to that of the wild-type receptor. However, substitution of two of these same residues at one time (L404A/F412A receptor) led to pharmacological properties comparable to those observed when the third highly hydrophobic residue is also replaced (L404A/F408A/F412A receptor). Moreover, the sequence-scrambled mutant receptors, in which overall hydrophobicity is retained, also functioned like the wild-type receptor, further supporting the notion that sequence-specific interactions are not involved. This indicates that formation of helix 8 and localization of CB1 require a threshold level of hydrophobicity not achieved by two or three new alanines in the domain.
For the CB1 receptor, recent NMR and CD structural analyses of short synthetic peptides corresponding to wild-type helix 8 suggested the secondary structure of this region was surfactant-dependent (40–43). Although short peptides (amino acids 17–22) were used in previous studies (40–43), the findings are consistent with the data presented here using the entire C terminus in multiple detergents (16). Previously, we conducted molecular dynamics simulations of the CB1 C terminus attached to TM7 in the presence of a POPC bilayer. The data suggested that helix 8 lies parallel to the plane of the membrane with its hydrophobic face associated with the fatty acyl region of the bilayer and the hydrophilic face exposed to the surrounding solvent (16). The amphiphilic environment provided by the membrane–cytosol interface is well suited to the induction of an amphiphilic helix 8, a process that requires at least two highly hydrophobic residues. This is consistent with the requirement of helix 8 for proper folding in the ER membrane for subsequent trafficking to the cell surface and/or for endocytosis (25). A recent examination of the C5a receptor suggests that helix 8 is involved in both trafficking to the cell surface from the ER and internalization (44). Our data further indicate that the residues we examined do not contribute to a side chain-specific export signal as suggested previously (44, 45) but induce and stabilize the helical motif that is critical. Nonetheless, this does not preclude the possibility that the secondary structural unit is bound by other regions of the receptor or other proteins.
Interestingly, our results showed three positive charges in helix 8 could be removed without substantially altering structure and function, including agonist-induced activation of G protein for signaling. Other studies using co-immunoprecipitation strategies have suggested that the helix 8 sequence is critical for Gi3 interaction (46, 47). However, we cannot rule out the possibility that the wild-type level of maximal activation shown by this mutant receptor is contributed by only Gi1 and Gi2, but notGi3. This differential selective coupling may affect the binding kinetics of the signal molecule, but not the maximal activation and binding affinity indicated by Emax and EC50 values, respectively. Alternatively, additional charged residues, such as Arg400 and Asp403, may be involved in Gi-mediated signaling and contribute to the retention of the G protein coupling activity observed here. Interestingly, Mukhopadhyay and colleagues demonstrated that the peptide fragment corresponding to CB1 helix 8 with additional flanking residues directly activates G protein in N18TG2 and CHO cell membrane preparations (40). Their data suggested that Arg401 in rat CB1 (corresponding to R400 in hCB1) is the important residue for high-affinity G protein binding, whereas the truncation of the downstream positively charged residues showed an only minimal effect on G protein binding. Our mutation of the charged residues does not include this corresponding residue; rather, R400 is retained upstream of helix 8. It is important to point out that this residue is not defined within helix 8 by any NMR studies (16, 41–43).
Intriguingly, our data indicate that there is little flexibility in the location of helix 8 within the C terminus. By varying the number of glutamine residues inserted between the NPxxY motif of TM7 and helix 8 as a spatial linker, and examining cellular localization and functional properties, we found a graded drop-off in Bmax values that parallels increases in the length of the linker inserted. This suggests that the spatial relationship of helix 8 with the membrane and/or other regions of the receptor is important. In each mutant receptor [401(Gln)2, 401(Gln)4, or 401(Gln)6], the Pro residue (at position 413 in the wild-type receptor) that is conserved in GPCRs at the end of helix 8 to inhibit further helix propagation and the Cys (at position 415 in the wild-type receptor) that is potentially palmitoylated and tethered to the membrane are both retained. However, their position relative to the NPxxY motif of TM7 is shifted by the size of the linker inserted, and it is possible that this also leads to the observed defects. For other receptors, the palmitoylated Cys is thought to help anchor helix 8 to the membrane surface. Indeed, the crystal structure of squid rhodopsin suggests helix 8 sits as a cap over parts of the cytoplasmic surface of the receptor, and movement of helix 8 could accommodate activated G protein and/or proteins that assist in trafficking or endocytosis. Given the intimate association of this domain with the cytoplasmic portion of the receptor and the membrane surface, it is not surprising that a positionally constrained, amphiphilic helical motif is ideally suited for this role.
Footnotes
This work was supported in part by National Institutes of Health Grants DA020763 (to D.A.K.) and DA018428 and GM082054 (to D.F.M.).
Abbreviations: GPCR, G protein-coupled receptor; CB1, cannabinoid receptor 1; TM, transmembrane helix; CP55940, (1R,3R,4R)-3-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-4-(3-hydroxypropyl)cyclohexan-1-ol; CD, circular dichroism; HEK293, human embryonic kidney; PBS, phosphate-buffered saline; BSA, bovine serum albumin; TME, Tris/Mg2+/EDTA; GTPγS, guanosine 5′-3-O-(thio)triphosphate; ER, endoplasmic reticulum; GFP, green fluorescent protein; LAMP-1, lysosomal-associated membrane protein 1.
References
- 1.Howlett AC, Barth F, Bonner TI, Cabral G, Casellas P, Devane WA, Felder CC, Herkenham M, Mackie K, Martin BR, Mechoulam R, Pertwee RG. International Union of Pharmacology. XXVII Classification of cannabinoid receptors. Pharmacol Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- 2.Glass M, Felder CC. Concurrent stimulation of cannabinoid CB1 and dopamine D2 receptors augments cAMP accumulation in striatal neurons: Evidence for a Gs linkage to the CB1 receptor. J Neurosci. 1997;17:5327–5333. doi: 10.1523/JNEUROSCI.17-14-05327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lauckner JE, Hille B, Mackie K. The cannabinoid agonist WIN55,212-2 increases intracellular calcium via CB1 receptor coupling to Gq/11 G proteins. Proc Natl Acad Sci USA. 2005;102:19144–19149. doi: 10.1073/pnas.0509588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter AC, Felder CC. The endocannabinoid nervous system: Unique opportunities for therapeutic intervention. Pharmacol Ther. 2001;90:45–60. doi: 10.1016/s0163-7258(01)00130-9. [DOI] [PubMed] [Google Scholar]
- 5.Gardner EL, Vorel SR. Cannabinoid transmission and reward-related events. Neurobiol Dis. 1998;5:502–533. doi: 10.1006/nbdi.1998.0219. [DOI] [PubMed] [Google Scholar]
- 6.Harkany T, Guzmán M, Galve-Roperh I, Berghuis P, Devi LA, Mackie K. The emerging functions of endocannabinoid signaling during CNS development. Trends Pharmacol Sci. 2007;28:83–92. doi: 10.1016/j.tips.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Kreitzer AC, Regehr WG. Retrograde signaling by endocannabinoids. Curr Opin Neurobiol. 2002;12:324–330. doi: 10.1016/s0959-4388(02)00328-8. [DOI] [PubMed] [Google Scholar]
- 8.Pertwee RG. Ligands that target cannabinoid receptors in the brain: From THC to anandamide and beyond. Addict Biol. 2008;13:147–159. doi: 10.1111/j.1369-1600.2008.00108.x. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson SS. Evolving concepts in G protein-coupled receptor endocytosis: The role in receptor desensitization and signaling. Pharmacol Rev. 2001;53:1–24. [PubMed] [Google Scholar]
- 10.Jin W, Brown S, Roche JP, Hsieh C, Celver JP, Kovoor A, Chavkin C, Mackie K. Distinct domains of the CB1 cannabinoid receptor mediate desensitization and internalization. J Neurosci. 1999;19:3773–3780. doi: 10.1523/JNEUROSCI.19-10-03773.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu DF, Yang LQ, Goschke A, Stumm R, Brandenburg LO, Liang YJ, Höllt V, Koch T. Role of receptor internalization in the agonist-induced desensitization of cannabinoid type 1 receptors. J Neurochem. 2008;104:1132–1143. doi: 10.1111/j.1471-4159.2007.05063.x. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh C, Brown S, Derleth C, Mackie K. Internalization and recycling of the CB1 cannabinoid receptor. J Neurochem. 1999;73:493–501. doi: 10.1046/j.1471-4159.1999.0730493.x. [DOI] [PubMed] [Google Scholar]
- 13.Leterrier C, Bonnard D, Carrel D, Rossier J, Lenkei Z. Constitutive endocytic cycle of the CB1 cannabinoid receptor. J Biol Chem. 2004;279:36013–36021. doi: 10.1074/jbc.M403990200. [DOI] [PubMed] [Google Scholar]
- 14.Martini L, Waldhoer M, Pusch M, Kharazia V, Fong J, Lee JH, Freissmuth C, Whistler JL. Ligand-induced down-regulation of the cannabinoid 1 receptor is mediated by the G-protein-coupled receptor-associated sorting protein GASP1. FASEB J. 2007;21:802–811. doi: 10.1096/fj.06-7132com. [DOI] [PubMed] [Google Scholar]
- 15.Daigle TL, Kwok ML, Mackie K. Regulation of CB1 cannabinoid receptor internalization by a promiscuous phosphorylation-dependent mechanism. J Neurochem. 2008;106:70–82. doi: 10.1111/j.1471-4159.2008.05336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn KH, Pellegrini M, Tsomaia N, Yatawara AK, Kendall DA, Mierke DF. Structural analysis of the human cannabinoid receptor one carboxyl-terminus identifies two amphipathic helices. Biopolymers. 2009;91:565–573. doi: 10.1002/bip.21179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conner M, Hicks MR, Dafforn T, Knowles TJ, Ludwig C, Staddon S, Overduin M, Günther UL, Thome J, Wheatley M, Poyner DR, Conner AC. Functional and biophysical analysis of the C-terminus of the CGRP-receptor: A family B GPCR. Biochemistry. 2008;47:8434–844. doi: 10.1021/bi8004126. [DOI] [PubMed] [Google Scholar]
- 18.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 19.Delos Santos NM, Gardner LA, White SW, Bahouth SW. Characterization of the residues in helix 8 of the human β1-adrenergic receptor that are involved in coupling the receptor to G proteins. J Biol Chem. 2006;281:12896–12907. doi: 10.1074/jbc.M508500200. [DOI] [PubMed] [Google Scholar]
- 20.Zhong M, Navratil AM, Clay C, Sanborn BM. Residues in the hydrophilic face of putative helix 8 of oxytocin receptor are important for receptor function. Biochemistry. 2004;43:3490–3498. doi: 10.1021/bi035899m. [DOI] [PubMed] [Google Scholar]
- 21.Tetsuka M, Saito Y, Imai K, Doi H, Maruyama K. The basic residues in the membrane-proximal C-terminal tail of the rat melanin-concentrating hormone receptor 1 are required for receptor function. Endocrinology. 2004;145:3712–3723. doi: 10.1210/en.2003-1638. [DOI] [PubMed] [Google Scholar]
- 22.Swift S, Leger AJ, Talavera J, Zhang L, Bohm A, Kuliopulos A. Role of the PAR1 receptor 8th helix in signaling: The 7-8-1 receptor activation mechanism. J Biol Chem. 2006;281:4109–4116. doi: 10.1074/jbc.M509525200. [DOI] [PubMed] [Google Scholar]
- 23.Oksche A, Dehe M, Schülein R, Wiesner B, Rosenthal W. Folding and cell surface expression of the vasopressin V2 receptor: Requirement of the intracellular C-terminus. FEBS Lett. 1998;424:57–62. doi: 10.1016/s0014-5793(98)00140-9. [DOI] [PubMed] [Google Scholar]
- 24.Duvernay MT, Zhou F, Wu G. A conserved motif for the transport of G protein-coupled receptors from the endoplasmic reticulum to the cell surface. J Biol Chem. 2004;279:30741–30750. doi: 10.1074/jbc.M313881200. [DOI] [PubMed] [Google Scholar]
- 25.Thielen A, Oueslati M, Hermosilla R, Krause G, Oksche A, Rosenthal W, Schülein R. The hydrophobic amino acid residues in the membrane-proximal C tail of the G protein-coupled vasopressin V2 receptor are necessary for transport-competent receptor folding. FEBS Lett. 2005;579:5227–5235. doi: 10.1016/j.febslet.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abadji V, Lucas-Lenard JM, Chin C, Kendall DA. Involvement of the carboxyl terminus of the third intracellular loop of the cannabinoid CB1 receptor in constitutive activation of Gs. J Neurochem. 1999;72:2032–2038. doi: 10.1046/j.1471-4159.1999.0722032.x. [DOI] [PubMed] [Google Scholar]
- 28.Murphy JW, Kendall DA. Integrity of extracellular loop 1 of the human cannabinoid receptor 1 is critical for high-affinity binding of the ligand CP 55,940 but not SR 141716A. Biochem Pharmacol. 2003;65:1623–1631. doi: 10.1016/s0006-2952(03)00155-2. [DOI] [PubMed] [Google Scholar]
- 29.Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 30.Rozenfeld R, Devi LA. Regulation of CB1 cannabinoid receptor trafficking by the adaptor protein AP-3. FASEB J. 2008;22:2311–2322. doi: 10.1096/fj.07-102731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le TI, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 32.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 35.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 36.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marin EP, Krishna AG, Zvyaga TA, Isele J, Siebert F, Sakmar TP. The amino terminus of the fourth cytoplasmic loop of rhodopsin modulates rhodopsin-transducin interaction. J Biol Chem. 2000;275:1930–1936. doi: 10.1074/jbc.275.3.1930. [DOI] [PubMed] [Google Scholar]
- 38.Bermak JC, Li M, Bullock C, Zhou QY. Regulation of transport of the dopamine D1 receptor by a new membrane-associated ER protein. Nat Cell Biol. 2001;3:492–498. doi: 10.1038/35074561. [DOI] [PubMed] [Google Scholar]
- 39.Robert J, Clauser E, Petit PX, Ventura MA. Anovel C-terminal motif is necessary for the export of the vasopressin V1b/V3 receptor to the plasma membrane. J Biol Chem. 2005;280:2300–2308. doi: 10.1074/jbc.M410655200. [DOI] [PubMed] [Google Scholar]
- 40.Mukhopadhyay S, Cowsik SM, Lynn AM, Welsh WJ, Howlett AC. Regulation of Gi by the CB1 cannabinoid receptor C-terminal juxtamembrane region: Structural requirements determined by peptide analysis. Biochemistry. 1999;38:3447–3455. doi: 10.1021/bi981767v. [DOI] [PubMed] [Google Scholar]
- 41.Xie XQ, Chen JZ. NMR structural comparison of the cytoplasmic juxtamembrane domains of G-protein-coupled CB1 and CB2 receptors in membrane mimetic dodecylphosphocholine micelles. J Biol Chem. 2005;280:3605–3612. doi: 10.1074/jbc.M410294200. [DOI] [PubMed] [Google Scholar]
- 42.Choi G, Guo J, Makriyannis A. The conformation of the cytoplasmic helix 8 of the CB1 cannabinoid receptor using NMR and circular dichroism. Biochim Biophys Acta. 2005;1668:1–9. doi: 10.1016/j.bbamem.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 43.Grace CR, Cowsik SM, Shim JY, Welsh WJ, Howlett AC. Unique helical conformation of the fourth cytoplasmic loop of the CB1 cannabinoid receptor in a negatively charged environment. J Struct Biol. 2007;159:359–368. doi: 10.1016/j.jsb.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suvorova ES, Gripentrog JM, Oppermann M, Miettinen HM. Role of the carboxyl terminal di-leucine in phosphorylation and internalization of C5a receptor. Biochim Biophys Acta. 2008;1783:1261–1270. doi: 10.1016/j.bbamcr.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duvernay MT, Dong C, Zhang X, Zhou F, Nichols CD, Wu G. Anterograde trafficking of G protein-coupled receptors: Function of the C-terminal F(X)6LL motif in export from the endoplasmic reticulum. Mol Pharmacol. 2009;75:751–761. doi: 10.1124/mol.108.051623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukhopadhyay S, Howlett AC. CB1 receptor-G protein association. Subtype selectivity is determined by distinct intracellular domains. Eur J Biochem. 2001;268:499–505. doi: 10.1046/j.1432-1327.2001.01810.x. [DOI] [PubMed] [Google Scholar]
- 47.Anavi-Goffer S, Fleischer D, Hurst DP, Lynch DL, Barnett-Norris J, Shi S, Lewis DL, Mukhopadhyay S, Howlett AC, Reggio PH, Abood ME. Helix 8 Leu in the CB1 cannabinoid receptor contributes to selective signal transduction mechanisms. J Biol Chem. 2007;282:25100–25113. doi: 10.1074/jbc.M703388200. [DOI] [PubMed] [Google Scholar]