Abstract
Malignant primary glial and secondary metastatic brain tumors represent distinct pathological entities. Nevertheless, both tumor types induce profound angiogenic responses in the host brain microvasculature that promote tumor growth. We hypothesized that primary and metastatic tumors induce similar microvascular changes that could function as conserved angiogenesis based therapeutic targets. We previously isolated glioma endothelial marker genes (GEMs) that were selectively upregulated in the microvasculature of proliferating glioblastomas. We sought to determine whether these genes were similarly induced in the microvasculature of metastatic brain tumors. RT-PCR and quantitative RT-PCR were used to screen expression levels of 20 candidate GEMs in primary and metastatic clinical brain tumor specimens. Differentially regulated GEMs were further evaluated by immunohisto-chemistry or in situ hybridization to localize gene expression using clinical tissue microarrays. Thirteen GEMs were upregulated to a similar degree in both primary and metastatic brain tumors. Most of these genes localize to the cell surface (CXCR7, PV1) or extracellular matrix (COL1A1, COL3A1, COL4A1, COL6A2, MMP14, PXDN) and were selectively expressed by the microvasculature. The shared expression profile between primary and metastatic brain tumors suggests that the molecular pathways driving the angiogenic response are conserved, despite differences in the tumor cells themselves. Anti-angiogenic therapies currently in development for primary brain tumors may prove beneficial for brain metastases and vice versa.
Keywords: Glioblastoma, Metastatic brain tumor, Angiogenesis, Endothelial genes, Vascular genes
Introduction
Malignant gliomas, the most common type of primary brain tumor, represent a uniformly fatal form of cancer. Despite advances in neurosurgical techniques, chemotherapeutic regimens and radiotherapy protocols, little improvement has been made in 5-year relative survival rates of brain tumor patients during the past several decades. One recent study showed that overall survival of patients with newly diagnosed glioblastoma (GBM), the most common and highest grade of malignant glioma, was 42.4% at 6 months, 17.7% at 1 year and 3.3% at 2 years [1].
The management of patients with malignant brain metastases also represents a formidable challenge. Brain metastasis occurs in 20–40% of cancer patients, leading to 200,000–400,000 new cases a year in the United States or nearly 10 times the number of primary brain tumors (approximately 40,000 cases). The frequency of clinical brain metastasis is also rising as a result of advances in the systemic management for extracranial tumors. The brain often acts as a “safe harbor” for cancer cells to evade commonly used chemotherapy regimens. Few patients who develop brain metastasis from any primary tumor survive >2 years from diagnosis of metastasis and their median survival time is only 2 months [2].
One positive development in the battle against malignant primary brain tumors has been the recent clinical success of anti-angiogenesis based therapies such as bevacizumab, a monoclonal antibody targeted against vascular endothelial growth factor-A (VEGF-A). Angiogenesis is a fundamental clinical characteristic of glioma, which occurs in response to the secretion of pro-angiogenic growth factors by tumor cells, stromal cells and inflammatory cells [3]. Neovascularization in brain tumors correlates directly with their biological aggressiveness, degree of malignancy and clinical recurrence and inversely with the post-operative survival [4]. The new vasculature is comprised of endothelial cells in tight association with vascular associated cells, including smooth muscle cells and pericytes, which serve to stabilize the neo-endothelium, bone marrow-derived endothelial progenitor cells, which contribute to vascular remodeling, and astrocytic “endfeet” which induce tight junctions of the blood–brain barrier [3, 5]. Unlike normal brain blood vessels, however, tumor vessels never fully mature and the resultant abnormal angiogenesis disrupts the blood–brain barrier, producing vasogenic cerebral edema and elevating incracranial pressure. New therapeutic agents that block VEGF mediated signal transduction have been able to inhibit these effects, induce radiographic responses, prolong disease free survival, and may improve overall survival as well. Given these results we believe that angiogenesis based therapeutic targets represent an attractive approach to treating malignant brain tumors.
One approach to block angiogenesis is to target tumor induced changes in the microvascular endothelial cells themselves [6]. Endothelial cells derived from normal brain and from primary malignant brain tumors have significant phenotypic and functional distinctions [7, 8]. By using serial analysis of gene expression (SAGE), we and others have been able to determine unique gene expression targets in microvascular endothelial cells purified from clinical specimens of malignant glioma [9, 10]. We identified 122 tags representing putative glioma endothelial markers which, using a combination of beta-distribution analyses and comparisons to independent SAGE libraries from normal and cancerous endothelium, we narrowed to 20 genes that were upregulated >4-fold in glioma endothelial cells versus normal brain endothelium and met all additional statistical parameters. In this study, we have focused on those 20 promising glioma endothelial markers (GEMs) and validated their differential expression in an independent cohort of clinical samples by using quantitative RT-PCR, immunohistochemistry and in situ hybridization methods.
We were particularly interested in GEMs that were differentially regulated in the microvasculature of both primary and metastatic brain tumors. Most of the available data on microvascular angiogenesis in brain tumors has been derived from primary malignant glioma and less commonly from brain metastases [11]. We wished to determine whether these GEMs were GBM specific of were conserved in secondary metastatic tumors as well. Furthermore, we were also curious as to whether these GEMs were obligate markers of malignancy or whether they were expressed in highly angiogenic, but lower grade brain tumors as well. Toward this end we examined the expression of our candidate GEMs in clinical specimens of pilocytic astrocytoma, a histologically benign brain tumor that can be angiogenic.
Our results demonstrate that the majority of these GEMs are upregulated across tumor types in both primary and metastatic brain tumors, indicating that brain tumor angiogenesis is likely occurring along common biological pathways despite differences in the biology of the tumor cells themselves. Anti-angiogenic therapies proposed against one group of tumors, therefore, would seem to have a molecular rationale for therapeutic efficacy against other tumors in the brain.
Materials and methods
Clinical specimens
All clinical specimens were provided by the Brain Tumor Bank of the University of Pittsburgh as approved under University of Pittsburgh Institutional Review Board (IRB) protocol 021164 and the Neurosurgical Brain Tissue Bank of the University of Rochester as approved under University of Rochester Research Subjects Review Board (RSRB) protocol 00021141. All samples were de-identified according to Health Insurance Portability and Accountability regulations by a university-approved honest broker before being provided for this research. This research falls under NIH guidelines for exempt research, not requiring informed consent, and was approved by the IRB of both institutions for this exemption.
Reverse transcription PCR (RT-PCR)
Clinical samples included five GBMs, five metastatic brain tumor tissues (lung adenocarcinoma primary) and two non-neoplastic control tissues from epileptic cortex. RNA isolation, reverse transcription, PCR preparation was performed as previously described [12]. Primers are summarized in Table 1. For APOD, AKAP13, COL6A2, CXCR7, ITGA5, LAMC3, MMP14, PLXNA2, PV1, PXDN, SOX4, TEM1, THY1 and VWA1 annealing temperature was 57°C, for COL1A1, COL4A1, COL3A1, EDNRB, INSR and HSPG2 annealing temperature was 54°C.
Table 1.
Genes and primers in the present study
| Symbol | Gene | Accession no. | T/N | Forward | Reverse |
|---|---|---|---|---|---|
| AKAP13 | A kinase (PRKA) anchor protein 13 | NM_006738 | 6 | gaggttgacgtggttcctgt | aggagtctggttctccagca |
| APOD | Apolipoprotein D | NM_001647 | 7 | cctgtacctgcatcatccaa | gcagttcacctggtctgtga |
| COL1A1 | Collagen, type I, alpha 1 | NM_000088 | 5 | cctggatgccatcaaagtct | aatccatcggtcatgctctc |
| COL3A1 | Collagen, type III, alpha 1 | NM_000090 | 6 | ttgaccctaaccaaggatgc | ggaagttcaggattgccgta |
| COL4A1 | Collagen, type IV, alpha 1 | NM_001845 | 13 | ctggtccaagaggatttcca | tcattgccttgcacgtagag |
| COL6A2 | Collagen, type VI, alpha 2 | NM_001849 | 5 | gagatcgaccaggacaccat | ggtctccctgtcttcccttc |
| CXCR7 | Chemokine (C-X-C motif) receptor 7 | NM_020311 | 9 | ggctatgacacgcactgcta | ctcatgcacgtgaggaagaa |
| EDNRB | Endothelin receptor type B | NM_000115 | 14 | gcaaaagattggtggctgtt | cagagggcaaagacaaggac |
| HSPG2 | Heparan sulfate proteoglycan 2 | NM_005529 | 13 | ctgccgtaatctccaccaat | cttttggctgtgcagatgaa |
| INSR | Insulin receptor | NM_000208 | 9 | tgccagtgatgtgtttccat | tgaggaactcaatccgctct |
| ITGA5 | Integrin, alpha 5 | NM_002205 | 10 | agcctcagaaggaggaggac | ttaatggggtgattggtggt |
| LAMC3 | Laminin, gamma 3 | NM_006059 | 4 | agagaatgtggaaggcaacc | acaccttggagtggccatag |
| MMP14 | Matrix metallopeptidase 14 | NM_004995 | 8 | gagctcagggcagtggatag | ggtagcccggttctaccttc |
| PLXNA2 | Plexin A2 | NM_025179 | 6 | ttaagaacccccagttcgtg | ctccacccagctcttgtagc |
| PV1 | Plasmalemmal vesicle associated protein 1 | NM_031310 | 7 | gagctggccatcagaaactc | gggactccaggatcttcctc |
| PXDN | Peroxidasin homolog | NM_012293 | 17 | ttgcgactggactcaaacac | ggcctttcacagttcagctc |
| SOX4 | SRY (sex determining region Y)-box 4 | NM_003107 | 6 | gctggaagctgctcaaagac | accgaccttgtctcccttct |
| TEM1 | Endosialin (CD248) | NM_020404 | 7 | ctacgttggtggcttcgagt | caggcctcgtcttcatcttc |
| THY1 | thy-1 cell surface antigen | NM_006288 | 12 | gacccgtgagacaaagaagc | tggagtgcacacgtgtaggt |
| VWA1 | von Willebrand factor A domain containing 1 | NM_022834 | 4 | ggtctatgccaaggaacagc | gctgacagctccaggaagtt |
T/N Fold-induction of genes in tumor-derived endothelial cells (T) versus nonneoplastic brain endothelium (N) [10]
Quantitative RT-PCR (qPCR)
Quantitative RT-PCR was performed as previously described [13]. Each reaction was performed in triplicate and three individual values were averaged and normalized to von Willebrand factor (vWF), an endothelial control gene, to account for differences in relative vascularity. Primer sequences are listed in Table 1.
Antibodies
Antibodies used for immunohistochemical analysis are listed in Table 2. Every effort was made to ensure that commercial antibodies selected for use in the study were highly specific to the protein being tested and did not cross react with protein family members. This included choosing antibodies that had been confirmed in the literature, were raised against specific protein epitope signature tags (PrESTs) as dissimilar as possible to other proteins, or were raised against epitopes did not cross react with other proteins by BLAST search. The affinity purified TEM1 antibody was custom generated against a gene specific epitope and its specificity was confirmed by comparison with in situ mRNA hybridization staining patterns (Carson-Walter, data not shown). Biotinylated goat-anti-rabbit and goat-anti-mouse secondary antibodies were from Vector Laboratories (Burlingame, CA).
Table 2.
Summary of antibodies and working conditions for immunohistochemistry
| Antibody | Dilution | Incubation condition |
Company | Cat. number |
|---|---|---|---|---|
| COL1A1 | 1:50 | 4°C, overnight | Orbigen | PAB-10433 |
| COL3A1 | 1:400 | RT, 45 min | Lifespan | LS-B693 |
| COL4A1 | 1:400 | RT, 45 min | Lifespan | LS-B340 |
| COL6A2 | 1:15 | 4°C, overnight | Sigma | HPA007029 |
| CXCR7 | 1:100 | RT, overnight | Abcam | ab12870 |
| HSPG2 | 1:100 | RT, 30 min | Zymed | 13-4400 |
| Integrin α5 | 1:500 | 4°C, overnight | Santa Cruz | SC-10729 |
| MMP14 | 1:50 | 4°C, overnight | Abcam | ab3644 |
| SOX4 | Original | RT, 30 min | Abcam | ab52107 |
| TEM1 | 1:50 | 4°C, overnight | QCB | a |
A custom affinity-purified rabbit polyclonal antibody to TEM1 was generated by Quality Controlled Biochemicals (QCB)
Immunohistochemistry
Tissue microarray was generated by the Department of Neuropathology, University of Rochester. All specimens were reviewed by a board-certified neuro-pathologist (M.D.J.) prior to selection and inclusion in the array. The microarray contained seven non-neoplastic brain tissue specimens, four pilocytic astrocytomas, seven metastatic adenocarcinomas removed from brain (3 lung adenocarcinomas, 1 lung adenosquamous carcinoma, 1 esophageal adenocarcinoma, 1 non-small cell lung carcinoma and 1 colon adenocarcinoma) and nine GBMs. Slides were deparaffined in xylenes, dehydrated in EtOH, followed by washing in 1× PBS or TBST. Sections were blocked for 30 min with peroxidase blocking reagent (Dako, Carpinteria, CA), then blocked with 10% normal goat serum/1× PBS for 1 h. For COL1A1, COL4A1, COL3A1, COL6A2, CXCR7, ITGA5, MMP14, SOX4, citrate buffer (Zymed, San Francisco, CA), and for HSPG2, retrieval buffer (Dako), were used for antigen retrieval according to manufacturer’s protocol. Sections were incubated with primary antibodies as summarized in Table 2. Sections were washed in 1× PBS or TBST then incubated for 1 h with 1:1,000 diluted biotinylated goat-anti-rabbit/mouse antibodies (Vector). Sections were washed in 1× PBS or TBST then exposed to Vectastain ABC (Vector) for 30 min. Sections were washed in 1× PBS or TBST and incubated with NovaRed substrate (Vector) for 5 min. Slides were rinsed with distilled water and counterstained with Mayer’s Hematoxylin (Dako). Vector Mount (Vector) was applied to slides and allowed to dry overnight. Images were captured with an Olympus AX70 microscope using Spot Insight Color model 3.2.0 camera and Spot Advanced software (Diagnostic Instruments, Sterling Heights, MI). A rabbit polyclonal anti-vWF antibody (Dako), was used to confirm vascular and protein content of specimens according to manufacturer’s protocol.
In situ hybridization
In situ hybridizations were performed essentially as previously reported [12]. Briefly, probe templates blanketing the target cDNA were generated by PCR amplification with incorporation of a T7 promoter into the antisense primer. In vitro transcription was performed using the DIG RNA Labeling Kit (SP6/T7) (Roche, Indianapolis, IN) according to manufacturer’s protocol. Sections were deparaffinized in xylenes, rehydrated in graded ethanol and permeabilized with proteinase K (Invitrogen). DIG-labeled RNA probe cocktails were diluted to 200 ng/ml in RNA hybridization solution (EMD Biosciences, Madison, WI) and hybridized to slides overnight at 55°C. Slides were treated with RNase A/T1 (Ambion, Austin, TX) and stringently washed. Slides were blocked with normal rabbit IgG (Dako) diluted 1:20 in 100 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% casein (Roche). For detection, sections were incubated with rabbit HRP-anti-DIG primary (Dako) diluted 1:150 in blocking buffer. Signal was amplified by incubating each slide with one drop of biotinyl-tyramide (Dako). Slides were then incubated in rabbit HRP-anti-biotin secondary (1:150, Dako) in blocking buffer. Amplification with biotinyltyramide was repeated. Sections were incubated with rabbit AP-anti-biotin tertiary (1:75, Dako) in blocking buffer and washed. Signal was detected with Fast Red (Sigma-Aldrich, St. Louis, MO) followed by counterstaining with probe/hematoxylin counterstain (Biomeda, Foster City, CA). Slides were mounted with Crystal/Mount (Biomeda). Images were captured as described above.
Immunohistochemistry and in situ hybridization scoring
The immunohistochemical and in situ analysis were scored as follows: −−, 0 pts, no staining; +/−, 1 pt, staining in an occasional cell; +, 2 pts, staining in 10% of cells; ++,3 pts staining in 50% of the endothelial cells; +++, 4 pts, staining in 100% of endothelial cells. Numeric values were utilized for statistical analysis.
Statistical analysis
The Chi square test was used to compare the expression of each GEM as measured by immunohistochemistry and in situ hybridization on the microarray to look for significant differences in expression between normal tissue and metastatic tumor, glioblastoma, and pilocytic astrocytoma. Additional comparisons were made for each GEM to examine the ability of each to discriminate between metastatic tumor and glioblastoma. A P value of <0.05 was considered statistically significant for all tests. Analyses were performed using Statistical Analysis Software SAS v9.1 (SAS Institute, Inc., Cary, NC).
Results
mRNA expression levels
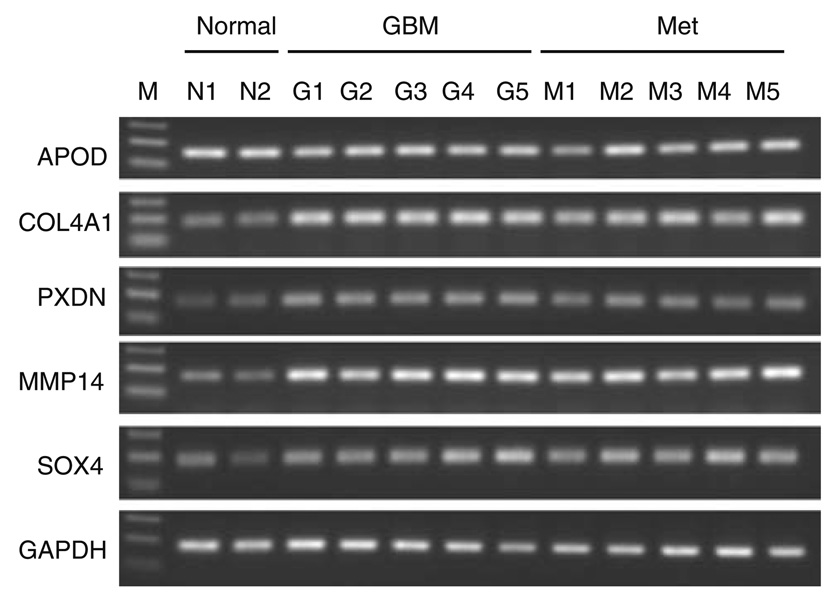
We previously described 20 gene products that were upregulated more than 4-fold and met rigid statistical filters in a comparative gene expression study of microvascular endothelial cells harvested from clinical specimens of glioblastoma and non-neoplastic brain tissue [10]. We coined the term GEMs (glioma endothelial markers) for these gene products. For the present study, we first updated the gene identities using more recent sequence and gene characterization data extracted from public databases. This resulted in the identification of new genes, such as AKAP13, that had previously existed as incomplete clonal inserts, as well as the renaming of several of our GEMs, including PXDN (previously MG50) and CXCR7 (previously RDC-1) (for full list, see Table 1). Next, to validate our SAGE data, we ran RT-PCR for each GEM on mRNA isolated from an independent cohort of fresh frozen surgical specimens of glioblastoma and non-neoplastic epileptic cortex. Epileptic cortical tissue was used as control because it represents the only non-neoplastic, non-injury disease state for which brain tissue is surgically removed. In addition, we wondered whether the induced genes were glioma specific or were similarly induced in malignant metastatic brain tumors so samples of lung adenocarcinoma metastatic to the brain were analyzed as well. We found that among our 20 candidate GEMs, 14 showed semi-quantitative upregulation in glioblastoma while showing little or no expression in non-neoplastic specimens. The remaining six GEMs, APOD, ENDRB, INSR, PLXNA2, THY1, and VWA1, were highly expressed at baseline in bulk normal tissue, making differential expression by semi-quantitative RT-PCR difficult to detect. Because high baseline expression in normal tissue would likely make therapies targeted against these molecules ineffective or highly toxic, we opted not to pursue characterization of these targets. Representative electrophoresis pictures are shown as Fig. 1 and all data are summarized in Table 3. Overall gene expression patterns were similar for GEMs in GBM and metastatic brain tumors. For example, COL4A1 showed little expression in non-neoplastic brain tissues versus strong expression in 100% (5/5) of GBM and 100% (5/5) of metastatic brain tumors (Fig. 1).
Fig. 1.
Representative gel pictures for RT-PCR. N non-neoplastic brain tissue, G GBM, M metastatic brain tumor
Table 3.
RT-PCR for 20 genes in normal, GBM and metastatic brain tumors
| Normal |
GBM |
Met |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | G1 | G2 | G3 | G4 | G5 | M1 | M2 | M3 | M4 | M5 | |
| AKAP13 | + | + | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| APOD | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| COL1A1 | + | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| COL3A1 | +/− | + | +++ | +++ | +++ | −− | +++ | +++ | +++ | +++ | +++ | +++ |
| COL4A1 | + | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| COL6A2 | + | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| CXCR7 | + | + | ++ | ++ | ++ | ++ | ++ | + | ++ | ++ | + | ++ |
| EDNRB | +++ | +++ | +++ | ++ | +++ | −− | ++ | + | ++ | ++ | ++ | ++ |
| HSPG2 | + | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| INSR | +/− | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| ITGA5 | + | +/− | ++ | ++ | ++ | −− | ++ | ++ | ++ | ++ | ++ | ++ |
| LAMC3 | +/− | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| PXDN | +/− | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| MMP14 | + | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| PLXNA2 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | −− | ++ | ++ |
| PV1 | −− | +/− | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| SOX4 | ++ | + | ++ | ++ | ++ | +++ | +++ | ++ | +++ | +++ | +++ | +++ |
| TEM1 | + | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| THY1 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| VWA1 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
Met metastatic brain tumor
We next used quantitative RT-PCR (qPCR) to better quantify the up-regulation of the 14 candidate GEMs from our semiquantitative RT-PCR results. The qPCR data are summarized in Table 4 and additional data are provided in Supplemental Material 1. Thirteen gene products, AKAP13, COL1A1, COL3A1, COL4A1, COL6A2, CXCR7, HSPG2, ITGA5, MMP14, PV1, PXDN, SOX4 and TEM1 demonstrated varying degrees of higher expression in brain tumor specimens than normal tissues. Among these, extracellular component matrix-related genes, most notably collagens, were robustly induced in brain tumors specimens compared to non-neoplastic brain specimens. Again, when we compared gene expression profiles between primary versus metastatic brain tumor groups, we found most of these up-regulated genes showed similar expression patterns. For example, MMP14 mRNA level was an average of 8.3 fold (range 3.6–16.0 fold) elevated in GBMs, and an average of 10.3 fold (range 5.3–19.0 fold) elevated in metastatic brain tumor specimens when compared to non-neoplastic control tissues. Similarly, when compared to normal (non-neoplastic) tissues, PXDN mRNA level was induced an average 13.7 fold (range 4.5–25.2 fold) in GBM and 14.6 fold (range 3.6–30.0 fold) in metastatic brain tumor tissues. CXCR7 mRNA levels were not elevated over normal in the metastatic tumors, although its expression in GBMs was up to 2-fold higher (Table 4; Supplemental Material 1). We discarded one GEM, LAMC3, from the subsequent analysis, since additional analysis of LAMC3 levels proved inconsistent in normals and tumors. As a control, qPCR on another GEM, ENDRB, confirmed our semiquantitative data that it was not differentially regulated in the sample tumors.
Table 4.
qPCR summary for 15 genes in brain tumor tissues
| Gene | Normal | GBM | Met |
|---|---|---|---|
| AKAP13 | 2.0 | 6.4 | 5.1 |
| COL1A1 | 1.0 | 19.7 | 215.6 |
| COL3A1 | 3.3 | 609 | 3025 |
| COL4A1 | 1.0 | 6.0 | 5.6 |
| COL6A2 | 3.8 | 62.9 | 147.5 |
| CXCR7 | 1.5 | 1.6 | 1.0 |
| ENDRB | 2.3 | 1.9 | 0 |
| HSPG2 | 1.7 | 16.4 | 24.6 |
| ITGA5 | 1.3 | 5.9 | 11.5 |
| LAMC3a | 10.5 | 23.1 | 22.4 |
| MMP14 | 1.0 | 8.3 | 10.3 |
| PV1 | 11.5 | 91.0 | 142.2 |
| PXDN | 1.5 | 13.7 | 14.6 |
| SOX4 | 1.0 | 1.5 | 1.6 |
| TEM1 | 1.0 | 1.4 | 1.4 |
Scoring: Average expression levels for each gene in two normal brain (normal), five GBM and five metastatic brain tumors (met). Normal levels are indicated by italic values; elevated over normal as bold values; lower than normal as underlined values. For complete values, see Supplemental Material 1
Not pursued further due to sample variation
Protein expression and localization
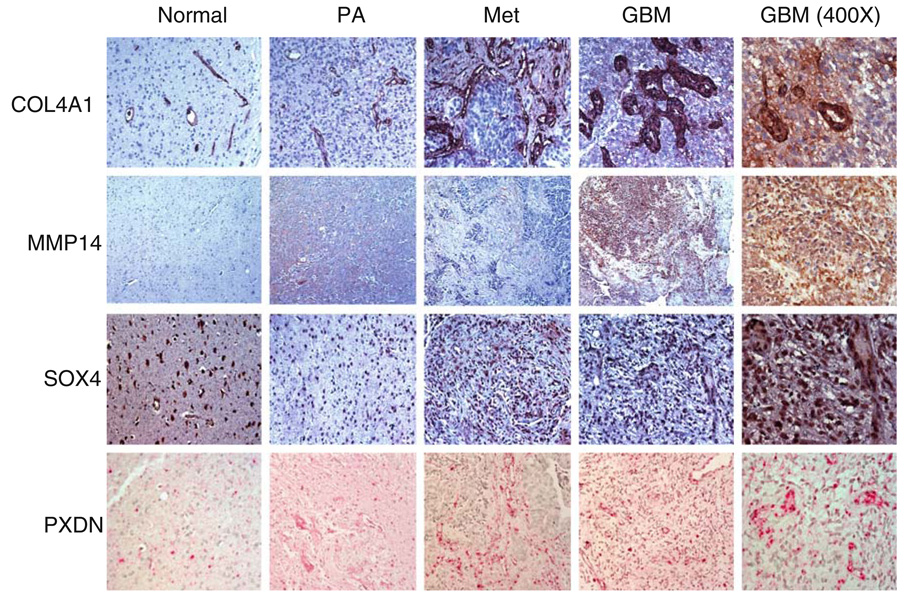
Among the GEMs confirmed by whole tissue PCR methods were several genes that regulate tumor endothelial extracellular matrix architecture or cellular signaling. To confirm that our gene expression data corresponded to alterations in protein levels and to localize these expression changes to the tumor microvasculature, we performed immunohistochemistry on an independent brain tumor tissue microarray consisting of non-neoplastic control tissue, pilocytic astrocytoma (PA), GBM and metastatic brain tumor tissue. The PA samples were included as additional controls since they are generally benign, but can be highly angiogenic, and therefore represented a clinically relevant intermediate phenotype between normal specimens and the malignant tumors. Ten of the 13 GEMs were analyzed by IHC due to the availability of compatible antibodies. Results are summarized in Table 5. We found that most of the highly expressed genes detected by SAGE and qPCR also demonstrated robust induction at the protein level. For example, 3/9 or 33% of GBMs and 7/7 or 100% of metastatic brain tumors showed moderate to robust COL4A1 staining. COL4A1 was localized to both endothelial cells and stromal cells and this was found both in metastatic brain tumors and GBMs. Only baseline vascular staining was detected in normal (non-neoplastic) brain tissue for these proteins (Fig. 2). Similar expression patterns were also observed for MMP14 and SOX4 in metastatic brain tumors and GBM (Fig. 2).
Table 5.
Immunohistochemistry and in situ hybridization results
| Case | Diagnosis | AKAP13 | COL1A1 | COL3A1 | COL4A1 | COL6A2 | CXCR7 | HSPG2 | ITGA5 | MMP14 | PV1 | PXDN | SOX4 | TEM1 | vWF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Normal | +/− | +/− | +/− | + | −− | −− | +/− | +/− | +/− | −− | +/− | −− | − | + |
| 2 | Normal | −− | +/− | +/− | + | −− | −− | +/− | +/− | −− | −− | −− | −− | − | + |
| 3 | Normal | −− | −− | −− | + | −− | −− | −− | −− | −− | −− | −− | −− | − | N/D |
| 4 | Normal | −− | + | + | + | +/− | + | + | + | + | +/− | −− | +/− | − | + |
| 5 | Normal | −− | + | +/− | + | +/− | +/− | +/− | +/− | +/− | −− | +/− | + | +/− | + |
| 6 | Normal | −− | −− | +/− | + | −− | −− | −− | −− | −− | −− | +/− | +/− | + | + |
| 7 | Normal | +/− | −− | −− | + | −− | +/− | −− | −− | −− | +/− | +/− | −− | +/− | + |
| 8 | PA | +/− | ++ | +++ | +++ | ++ | ++ | ++ | ++ | + | +/− | + | ++ | +++ | +++ |
| 9 | PA | + | + | + | + | +/− | −− | + | −− | +/− | + | + | + | + | ++ |
| 10 | PA | +/− | +/− | ++ | ++ | ++ | + | + | + | +/− | + | +/− | + | + | ++ |
| 11 | PA | +/− | −− | +/− | + | −− | −− | −− | +/− | −− | +/− | +/− | +/− | − | + |
| 12 | Met | + | ++ | +++ | +++ | +++ | +++ | ++ | +++ | +/− | +++ | ++ | ++ | +++ | +++ |
| 13 | Met | −− | ++ | +++ | +++ | +++ | +++ | + | +++ | −− | ++ | −− | + | + | +++ |
| 14 | Met | +/− | ++ | ++ | ++ | + | + | + | + | + | + | +/− | + | ++ | ++ |
| 15 | Met | +/− | + | ++ | ++ | +/− | +/− | ++ | + | +/− | +/− | +/− | + | + | ++ |
| 16 | Met | +++ | + | +++ | +++ | +++ | +/− | ++ | ++ | + | +++ | +++ | + | +++ | ++ |
| 17 | Met | ++ | +/− | ++ | ++ | ++ | −− | + | + | + | + | ++ | ++ | ++ | ++ |
| 18 | Met | +/− | ++ | ++ | +++ | ++ | +/− | −− | +/− | ++ | +/− | −− | + | + | ++ |
| 19 | GBM | +/− | + | + | + | + | −− | −− | + | +/− | +/− | + | + | + | + |
| 20 | GBM | −− | −− | +/− | + | −− | +/− | −− | +/− | −− | −− | + | +/− | ++ | ++ |
| 21 | GBM | −− | +/− | + | ++ | + | ++ | +/− | + | +/− | +/− | ++ | + | +/++ | +++ |
| 22 | GBM | +/− | +/− | + | + | +/− | −− | +/− | +/− | +/− | +/− | +/− | +/− | + | +/− |
| 23 | GBM | +/− | −− | ++ | +++ | +/− | +/− | + | ++ | +/− | + | ++ | ++ | +++ | + |
| 24 | GBM | +/− | −− | +/− | + | −− | +/− | +/− | +/− | −− | +/− | +/− | +/− | ++ | +++ |
| 25 | GBM | N/D | −− | + | + | +/− | −− | −− | +/− | ++ | N/D | +/− | +/− | + | + |
| 26 | GBM | + | +/− | ++ | ++ | + | + | +/− | + | +/− | +/− | +++ | + | +++ | ++ |
| 27 | GBM | +/− | +/− | ++ | + | +/− | −− | −− | +/− | +/− | −− | + | +/− | +++ | +++ |
| Sig. | − | M | M | M | M | − | M,G | M | M | − | G | M,G | − |
Scores:− −, 0% of vessels; +/−, occasional vessel; +, 10% of vessels; ++, 50% of vessels; +++, 100% of vessels. PXDN, PV1, and AKAP13 were measured by in situ hybridization and all others were measured by immunohistochemistry. CD31 was used as a control to ensure that viable protein and mRNA was present in each microarray spot. Vessel density was measured by vWF staining. Significance was determined using the Chi square test for each GEM to compare normal versus metastatic tumor (M) and normal versus glioblastoma (G) at a level <0.05. No markers were able to discriminate normal versus pilocytic tumor or metastatic tumor versus glioblastoma
Fig. 2.
Expression of four conserved vascular related genes in primary and metastatic brain tumors. Immunohistochemical staining for COL4A1, MMP14 and SOX4 on representative tissue microarray specimens. Localization by in situ hybridization for PXDN on representative tissue microarray specimens. PA pilocytic astrocytoma, Met metastatic brain tumor, GBM glioblastoma. Original magnification of far right column, ×400; all other images, ×200
Two of the 13 genes, COL3A1 and CXCR7, did not show completely concordant results between the degree of induction by qRT-PCR and immunohistochemistry. For example, COL3A1 displayed 90- to 1,400-fold elevated transcript levels in five of five GBMs examined (Table 4), but robust protein induction in only three of nine GBMs (33%) (Table 5). CXCR7 did not show significantly elevated expression in metastatic tumors by bulk tissue qPCR, but did demonstrate robust protein expression in three of seven independent metastatic tumors (43%). Such inconsistency is likely due to the differential detection sensitivity of the assays or sampling bias. Gene-specific expression studies using larger sample sizes will be used to clarify such discrepancies.
Interestingly, three out of four of the PA samples displayed moderately elevated gene expression profiles, more similar to the malignant tumors than the normal controls. Although PA is generally a benign tumor, it can be highly angiogenic and often demonstrates enhancement upon MRI imaging. Although the sample size was small, these data suggest that the cerebrovascular response to tumorigenesis may be phenotypically conserved between different types and grades of brain tumors, implying the presence of shared therapeutic targets.
mRNA localization
We used in situ hybridization to screen the tissue microarray for three genes for which FFPE-IH compatible antibodies were unavailable: PXDN, AKAP13 and PV1. As shown in Fig. 2, PXDN mRNA localized to endothelial cells. Moreover, PXDN was expressed in most of the brain tumor specimens, including pilocytic astrocytomas, metastatic brain tumors and GBMs, while its expression was low to absent in the non-neoplastic temporal lobe specimens. This conserved expression profile between metastatic brain tumors and GBM was also found for AKAP13 and PV1 (Table 5).
Discussion
To understand the critical gene expression changes induced in microvascular endothelial cells by brain tumors we sought to identify whether endothelial marker genes were similarly expressed in the vasculature of both primary and metastatic brain tumors and whether these genes were present in both high grade and low grade (PA) glial tumors. Our results demonstrate that a subset of 13 of our 20 putative GEMs do appear to be differentially regulated in bulk tumor and that this upregulation is preserved in both primary and metastatic brain tumors. These genes include AKAP13, COLA1, COL3A1, COL4A1, COL6A2, CXCR7, HSPG2, ITGA5, MMP14, PV1, PXDN, SOX4 and TEM1. While the majority of these genes have been previously identified by ourselves or others as being expressed by normal or tumor vascular structures, we believe this is one of the first reports of AKAP13 expression in normal brain and brain tumor blood vessels. Furthermore, we believe this study is the first confirmation of PXDN expression in brain tumor vessels. While none of the individual GEMs was able to perfectly discriminate between normal brain tissue and malignant brain tumor in all specimens, the panel of GEMs as a whole was significantly upregulated in tumor specimens compared to normals. The degree of upregulation was actually greater in our panel of metastatic brain tumors compared to primary glioblastoma. This may be due to the greater tumor heterogeneity of glioblastoma. The location of the tissue microarray specimen relative to the entire tumor as it existed in situ is unknown for these de-identified specimens. As such it is possible that the tissue microarray spot came from a more quiescent portion of the tumor that may not be as actively angiogenic. Because our mRNA samples for qPCR were generated from larger bulk tumor specimens, such microheterogeneity is less likely to influence our qPCR results that should be more reflective of average tumor gene expression levels. Interestingly, the degree of tumor GEMs upregulation was similar in our specimens of pilocytic astrocytoma, a benign but often highly angiogenic primary brain tumor, when compared to malignant brain tumors. This would seem to indicate that the gene expression changes are influenced by the level of the angiogenic process itself and not by the histologic grade of the tumor cells.
It is important to note that we did not perform transcriptional or translational studies to confirm whether the increased abundance of a particular microvascular GEM was due to elevated gene product per individual cell or due to an increased number of vessels within the bulk tumor. However, we did normalize all qPCR values to vWF, a known endothelial marker gene, in order to account for the increased vascularity of the tumor samples and make it more likely that higher expression levels were due to altered gene expression profiles and not simply an higher vascular index. Similarly, comparison of the IHC and ISH staining data to the staining score for vWF confirms that in many cases, GEMs expression profiles were regulated independently of simple tumor vessel numbers, suggesting a selective angiogenic response to CNS tumorigenesis.
We were unable to verify differential regulation of seven of our putative GEMs identified in our SAGE study. In most cases this was due to high baseline expression of the gene in question in normal brain tissue. We believe that while these GEMs may still be differentially regulated at the microvascular level as indicated in our SAGE study of purified endothelial cells, the basal expression of these genes in normal brain and tumor cells themselves, makes them difficult to assess in bulk tissue specimens or pathologic sections. Techniques such as microdissection of tumor specimens and antibody coupled magnetic bead isolation of tumor endothelial cells from gross specimens could clarify expression differences for these genes, but their high background expression levels likely indicates that they would not be selective enough targets for therapeutic manipulation.
Based on a combination of their biology and frequency of upregulation, several of our GEMs represent particularly attractive targets for both primary and metastatic brain tumors. For example, the SOX4 gene expression level in GBM and metastatic tumors was up to 3-fold higher than those in non-neoplastic tissue (Table 4; Supplemental Material 1). Furthermore, in several samples, SOX4 appeared to show expression in tumor cells themselves. The SOX proteins compose a family of more than 20 transcription factors characterized by the presence of a high-mobility-group DNA binding domain [14]. SOX4 belongs to the C group of the SOX gene family and plays a role in normal embryonic development of many tissues, including the CNS. It is over-expressed in medulloblastoma [15–17] and may be predictive of outcome in medulloblastoma, although this remains controversial [17, 18]. One interesting recent study showed that micro-RNA 335 suppressed migration and metastasis of breast cancer cells through targeting of SOX4 and extracellular matrix component [19]. SOX4 is also being evaluated as a potential vaccine target in lung cancer, further indicating its potential clinical application [20]. However, relatively little is currently known concerning the role of SOX4 in tumor angiogenesis. SOX4 induction in human bladder carcinoma HU609 cells is associated with over-expression of vascular or angiogenesis related genes including IL-8, CTGF, JAG1 and NRP2 [21]. In a mouse knockout model, lack of Sox4 led to disruption of the vascular and cardiac systems [22]. The combined data suggest that SOX4 may contribute to brain tumor angiogenesis but the mechanism remains unclear.
Our present study also confirmed upregulation of multiple cell surface proteins in the microvasculature of primary glial and metastatic brain tumors. One was PXDN, also known as MG50, one of the few melanoma-associated antigens that is not a differentiation antigen or a mutated protein. Recent genomic evidence revealed that MG50 was identical to PXDN, the human homologue of the Drosophila gene peroxidasin, an extracellular matrix-associated peroxidase. Its expression is relatively restricted to tumors such as melanoma, breast cancer, ovarian cancer as well as the glioblastoma cell line U138MG, and is absent from many normal tissues [23]. However, its function has yet to be fully defined. Mitchell et al. [24] reported that PXDN encodes an interleukin 1 receptor antagonist containing six epitopes recognized by human cytolytic T lymphocytes. Another study identified PXDN as being induced during p53-dependent apoptosis, suggesting a link between PXDN expression and reactive oxygen production in apoptosis [25]. Previously we showed that PXDN (MG50) expression was 17-fold higher in glioma endothelial cells than non-neoplastic brain endothelium [10]. In the present study, we confirmed that PXDN mRNA was up-regulated in an independent set of brain tumor samples (Table 4) and expression localizes to microvascular endothelial cells (Fig. 2). Similarly, Castronovo et al. [26] reported that PXDN localizes in vasculature structures in renal cell carcinoma, thus PXDN may function in tumor angiogenesis in multiple cancers. Since PXDN is a surface protein accessible to extracellular pharmaceutical compounds, it may prove to be a novel target for anti-angiogenesis therapy.
Extracellular matrix (ECM) remodeling is critical for the endothelial cell activation and migration that contribute to angiogenesis [27]. By using laser capture microdissection microscopy and microarray techniques, Pen et al. [28] found IGFBP7 and SPARC, both of which encode proteins that are secreted into the extracellular space, were upregulated in GBM vessels. In the present study, we found several ECM components were upregulated in brain tumor samples including four distinct members of the collagen superfamily. One of these, COL4A1, was upregulated in brain tumor samples in a manner consistent with previous reports [29]. COL4 is believed to play an important role in angiogenesis and tumor progression [30]. Both COL4 and COL5 are expressed along tumor vessels in glioma [31]. COL4 colocalizes with TEM1, another GEM, and MMP2 during fetal brain angiogenesis [32]. Small-molecule inhibitors of COL4 biosynthesis can prevent endothelial tube formation and tumor growth [33]. Structural abnormalities or cryptic sites in COL4 have been detected in the tumor microenvironment and may be therapeutically exploitable [34, 35]. Similarly, when treated with brivanib alaninate, an anti-angiogenic agent targeting vascular endothelial growth factor receptor 2 (VEGFR-2) and fibroblast growth factor receptor 1, athymic mice bearing L2987 human tumor xenografts showed significant reduction of COL4A1 at both mRNA and protein levels [36]. Thus, COL4A1 may be an important biomarker of angiogenesis in cancer as well as a viable therapeutic target.
We similarly confirmed MMP14 (membrane type 1 metalloprotease, MT1-MMP) induction in primary and metastatic brain tumor samples by both real time PCR and immunohistochemistry (Table 4; Fig. 2). This upregulation is supported by additional reports [37–39]. MMP14 is a transmembrane metalloprotease that plays a key role in the angiogenesis response through multiple steps including degradation of ECM, endothelial invasion/migration formation of capillary tubes and recruitment of accessory cells [40]. Multiple studies demonstrate the important role of MMP14 in tumor angiogenesis. In human melanoma cells, MMP14 overexpression is associated with increased in vivo tumor growth and vascularization [41]. MMP14 overexpression results in up-regulation of VEGF and promotes angiogenesis in human GBM and breast cancer xenograft models [42, 43]. In human GBM, VEGF and MMP14 often show co-expression by the same tumor cells [38]. The combined data suggest that the conserved expression pattern of MMP14 in primary and metastatic brain tumors make this gene an attractive target for CNS neoplasms.
In conclusion, the principle goal of the present investigation was to compare gene expression patterns of microvascular marker genes in primary brain tumors versus metastatic brain tumors. Primary brain tumors grow invasively as single cells along blood vessel walls, although they do not invade the vessel wall itself. Conversely, systemic metastatic tumors tend to invade the brain as small groups of cells, which show little tendency to grow along blood vessel tracks, but do invade the vessel walls [44]. Certain investigators have reported significantly different gene expression profiles in primary lung cancer specimens compared with matched metastatic brain tumors from the same patient [45, 46]. It is certainly possible that further SAGE or gene array based studies of human metastatic brain tumor endothelium or animal models of “classic” versus “cooptional” tumor angiogenesis might reveal new, metastasis-specific angiogenic genes. However, we hypothesized that brain angiogenesis would be more likely to progress along a common preserved pathway regardless of the identity and growth characteristics of the inciting tumor and thereby provide molecular targets shared by a majority of brain tumors. We demonstrate here that despite their original sites, primary and metastatic brain tumors induce similar gene expression changes in several novel microvascular protein markers. Development of new drugs targeting these genes may benefit both primary and metastatic brain tumor patients and the treatment of metastatic brain tumors using anti-angiogenic drugs currently being tested for malignant glioma is likely to have a molecular justification as well.
Supplementary Material
Acknowledgments
We thank Dr. Sumana Datta of Texas A&M University for advice regarding HSPG2 immunohistochemical staining. This work was supported by the National Institutes of Health (NINDS K08 NS046461 to K.A.W.), the American Brain Tumor Association (to K.A.W.), the Childhood Brain Tumor Foundation (to K.A.W.) and the Ronald Bittner Brain Tumor Research Fund (to K.A.W.).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11060-009-0105-0) contains supplementary material, which is available to authorized users.
Contributor Information
Yang Liu, Department of Neurosurgery, University of Rochester, 601 Elmwood Avenue, Box 670, Rochester, NY 14642, USA.
Eleanor B. Carson-Walter, Department of Neurosurgery, University of Rochester, 601 Elmwood Avenue, Box 670, Rochester, NY 14642, USA
Anna Cooper, Department of Neurosurgery, University of Rochester, 601 Elmwood Avenue, Box 670, Rochester, NY 14642, USA.
Bethany N. Winans, Department of Neurosurgery, University of Rochester, 601 Elmwood Avenue, Box 670, Rochester, NY 14642, USA
Mahlon D. Johnson, Department of Neuropathology, University of Rochester, Rochester, NY, USA
Kevin A. Walter, Email: kevin_walter@urmc.rochester.edu, Department of Neurosurgery, University of Rochester, 601 Elmwood Avenue, Box 670, Rochester, NY 14642, USA; James P. Wilmot Cancer Center, University of Rochester, Rochester, NY, USA.
References
- 1.Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, Baeza N, Pisani P, Yonekawa Y, Yasargil MG, Lutolf UM, Kleihues P. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64(19):6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 2.Hall WA, Djalilian HR, Nussbaum ES, Cho KH. Longterm survival with metastatic cancer to the brain. Med Oncol. 2000;17(4):279–286. doi: 10.1007/BF02782192. [DOI] [PubMed] [Google Scholar]
- 3.Kim WY, Lee HY. Brain angiogenesis in developmental and pathological processes: mechanism and therapeutic intervention in brain tumors. FEBS J. 2009;276(17):4653–4664. doi: 10.1111/j.1742-4658.2009.07177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer I, Gagner JP, Law M, Newcomb EW, Zagzag D. Angiogenesis in gliomas: biology and molecular pathophysiology. Brain Pathol. 2005;15(4):297–310. doi: 10.1111/j.1750-3639.2005.tb00115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong ML, Prawira A, Kaye AH, Hovens CM. Tumour angiogenesis: its mechanism and therapeutic implications in malignant gliomas. J Clin Neurosci. 2009;16(9):1119–1130. doi: 10.1016/j.jocn.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Jansen M, de Witt Hamer PC, Witmer AN, Troost D, van Noorden CJ. Current perspectives on antiangiogenesis strategies in the treatment of malignant gliomas. Brain Res Brain Res Rev. 2004;45(3):143–163. doi: 10.1016/j.brainresrev.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Charalambous C, Chen TC, Hofman FM. Characteristics of tumor-associated endothelial cells derived from glioblastoma multiforme. Neurosurg Focus. 2006;20(4):E22. doi: 10.3171/foc.2006.20.4.e22. [DOI] [PubMed] [Google Scholar]
- 8.Charalambous C, Hofman FM, Chen TC. Functional and phenotypic differences between glioblastoma multiforme-derived and normal human brain endothelial cells. J Neurosurg. 2005;102(4):699–705. doi: 10.3171/jns.2005.102.4.0699. [DOI] [PubMed] [Google Scholar]
- 9.Beaty RM, Edwards JB, Boon K, Siu IM, Conway JE, Riggins GJ. PLXDC1 (TEM7) is identified in a genome-wide expression screen of glioblastoma endothelium. J Neurooncol. 2007;81(3):241–248. doi: 10.1007/s11060-006-9227-9. [DOI] [PubMed] [Google Scholar]
- 10.Madden SL, Cook BP, Nacht M, Weber WD, Callahan MR, Jiang Y, Dufault MR, Zhang X, Zhang W, Walter-Yohrling J, Rouleau C, Akmaev VR, Wang CJ, Cao X, St Martin TB, Roberts BL, Teicher BA, Klinger KW, Stan RV, Lucey B, Carson-Walter EB, Laterra J, Walter KA. Vascular gene expression in nonneoplastic and malignant brain. Am J Pathol. 2004;165(2):601–608. doi: 10.1016/s0002-9440(10)63324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fidler IJ, Yano S, Zhang RD, Fujimaki T, Bucana CD. The seed and soil hypothesis: vascularisation and brain metastases. Lancet Oncol. 2002;3(1):53–57. doi: 10.1016/s1470-2045(01)00622-2. [DOI] [PubMed] [Google Scholar]
- 12.Carson-Walter EB, Hampton J, Shue E, Geynisman DM, Pillai PK, Sathanoori R, Madden SL, Hamilton RL, Walter KA. Plasmalemmal vesicle associated protein-1 is a novel marker implicated in brain tumor angiogenesis. Clin Cancer Res. 2005;11(21):7643–7650. doi: 10.1158/1078-0432.CCR-05-1099. [DOI] [PubMed] [Google Scholar]
- 13.Shue EH, Carson-Walter EB, Liu Y, Winans BN, Ali ZS, Chen J, Walter KA. Plasmalemmal vesicle associated protein-1 (PV-1) is a marker of blood-brain barrier disruption in rodent models. BMC Neurosci. 2008;9:29. doi: 10.1186/1471-2202-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27(6):1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokota N, Mainprize TG, Taylor MD, Kohata T, Loreto M, Ueda S, Dura W, Grajkowska W, Kuo JS, Rutka JT. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene. 2004;23(19):3444–3453. doi: 10.1038/sj.onc.1207475. [DOI] [PubMed] [Google Scholar]
- 16.Lee CJ, Appleby VJ, Orme AT, Chan WI, Scotting PJ. Differential expression of SOX4 and SOX11 in medulloblastoma. J Neurooncol. 2002;57(3):201–214. doi: 10.1023/a:1015773818302. [DOI] [PubMed] [Google Scholar]
- 17.de Bont JM, Kros JM, Passier MM, Reddingius RE, Sillevis Smitt PA, Luider TM, den Boer ML, Pieters R. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro Oncol. 2008;10(5):648–660. doi: 10.1215/15228517-2008-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neben K, Korshunov A, Benner A, Wrobel G, Hahn M, Kokocinski F, Golanov A, Joos S, Lichter P. Microarray-based screening for molecular markers in medulloblastoma revealed STK15 as independent predictor for survival. Cancer Res. 2004;64(9):3103–3111. doi: 10.1158/0008-5472.can-03-3968. [DOI] [PubMed] [Google Scholar]
- 19.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman RS, Bangur CS, Zasloff EJ, Fan L, Wang T, Watanabe Y, Kalos M. Molecular and immunological evaluation of the transcription factor SOX-4 as a lung tumor vaccine antigen. J Immunol. 2004;172(5):3319–3327. doi: 10.4049/jimmunol.172.5.3319. [DOI] [PubMed] [Google Scholar]
- 21.Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjot L, Orntoft T. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66(7):3434–3442. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- 22.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380(6576):711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 23.Hutchins JT, Deans RJ, Mitchell MS, Uchiyama C, Kan-Mitchell J. Novel gene sequences expressed by human melanoma cells identified by molecular subtraction. Cancer Res. 1991;51(5):1418–1425. [PubMed] [Google Scholar]
- 24.Mitchell MS, Kan-Mitchell J, Minev B, Edman C, Deans RJ. A novel melanoma gene (MG50) encoding the interleukin 1 receptor antagonist and six epitopes recognized by human cytolytic T lymphocytes. Cancer Res. 2000;60(22):6448–6456. [PubMed] [Google Scholar]
- 25.Horikoshi N, Cong J, Kley N, Shenk T. Isolation of differentially expressed cDNAs from p53-dependent apoptotic cells: activation of the human homologue of the Drosophila peroxidasin gene. Biochem Biophys Res Commun. 1999;261(3):864–869. doi: 10.1006/bbrc.1999.1123. [DOI] [PubMed] [Google Scholar]
- 26.Castronovo V, Waltregny D, Kischel P, Roesli C, Elia G, Rybak JN, Neri D. A chemical proteomics approach for the identification of accessible antigens expressed in human kidney cancer. Mol Cell Proteomics. 2006;5(11):2083–2091. doi: 10.1074/mcp.M600164-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Wang D, Anderson JC, Gladson CL. The role of the extracellular matrix in angiogenesis in malignant glioma tumors. Brain Pathol. 2005;15(4):318–326. doi: 10.1111/j.1750-3639.2005.tb00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pen A, Moreno MJ, Martin J, Stanimirovic DB. Molecular markers of extracellular matrix remodeling in glioblastoma vessels: microarray study of laser-captured glioblastoma vessels. Glia. 2007;55(6):559–572. doi: 10.1002/glia.20481. [DOI] [PubMed] [Google Scholar]
- 29.Ljubimova JY, Lakhter AJ, Loksh A, Yong WH, Riedinger MS, Miner JH, Sorokin LM, Ljubimov AV, Black KL. Over-expression of alpha4 chain-containing laminins in human glial tumors identified by gene microarray analysis. Cancer Res. 2001;61(14):5601–5610. [PubMed] [Google Scholar]
- 30.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3(6):422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 31.Bellon G, Caulet T, Cam Y, Pluot M, Poulin G, Pytlinska M, Bernard MH. Immunohistochemical localisation of macromolecules of the basement membrane and extracellular matrix of human gliomas and meningiomas. Acta Neuropathol. 1985;66(3):245–252. doi: 10.1007/BF00688590. [DOI] [PubMed] [Google Scholar]
- 32.Virgintino D, Girolamo F, Errede M, Capobianco C, Robertson D, Stallcup WB, Perris R, Roncali L. An intimate interplay between precocious, migrating pericytes and endothelial cells governs human fetal brain angiogenesis. Angiogenesis. 2007;10(1):35–45. doi: 10.1007/s10456-006-9061-x. [DOI] [PubMed] [Google Scholar]
- 33.Haralabopoulos GC, Grant DS, Kleinman HK, Lelkes PI, Papaioannou SP, Maragoudakis ME. Inhibitors of basement membrane collagen synthesis prevent endothelial cell alignment in matrigel in vitro and angiogenesis in vivo. Lab Invest. 1994;71(4):575–582. [PubMed] [Google Scholar]
- 34.Baluk P, Morikawa S, Haskell A, Mancuso M, McDonald DM. Abnormalities of basement membrane on blood vessels and endothelial sprouts in tumors. Am J Pathol. 2003;163(5):1801–1815. doi: 10.1016/S0002-9440(10)63540-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mancuso MR, Davis R, Norberg SM, O’Brien S, Sennino B, Nakahara T, Yao VJ, Inai T, Brooks P, Freimark B, Shalinsky DR, Hu-Lowe DD, McDonald DM. Rapid vascular regrowth in tumors after reversal of VEGF inhibition. J Clin Invest. 2006;116(10):2610–2621. doi: 10.1172/JCI24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayers M, Fargnoli J, Lewin A, Wu Q, Platero JS. Discovery and validation of biomarkers that respond to treatment with brivanib alaninate, a small-molecule VEGFR-2/FGFR-1 antagonist. Cancer Res. 2007;67(14):6899–6906. doi: 10.1158/0008-5472.CAN-06-4555. [DOI] [PubMed] [Google Scholar]
- 37.Lampert K, Machein U, Machein MR, Conca W, Peter HH, Volk B. Expression of matrix metalloproteinases and their tissue inhibitors in human brain tumors. Am J Pathol. 1998;153(2):429–437. doi: 10.1016/S0002-9440(10)65586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munaut C, Noel A, Hougrand O, Foidart JM, Boniver J, Deprez M. Vascular endothelial growth factor expression correlates with matrix metalloproteinases MT1-MMP, MMP-2 and MMP-9 in human glioblastomas. Int J Cancer. 2003;106(6):848–855. doi: 10.1002/ijc.11313. [DOI] [PubMed] [Google Scholar]
- 39.Guo P, Imanishi Y, Cackowski FC, Jarzynka MJ, Tao HQ, Nishikawa R, Hirose T, Hu B, Cheng SY. Up-regulation of angiopoietin-2, matrix metalloprotease-2, membrane type 1 metalloprotease, and laminin 5 gamma 2 correlates with the invasiveness of human glioma. Am J Pathol. 2005;166(3):877–890. doi: 10.1016/s0002-9440(10)62308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genis L, Galvez BG, Gonzalo P, Arroyo AG. MT1-MMP: universal or particular player in angiogenesis? Cancer Metastasis Rev. 2006;25(1):77–86. doi: 10.1007/s10555-006-7891-z. [DOI] [PubMed] [Google Scholar]
- 41.Sounni NE, Baramova EN, Munaut C, Maquoi E, Frankenne F, Foidart JM, Noel A. Expression of membrane type 1 matrix metalloproteinase (MT1-MMP) in A2058 melanoma cells is associated with MMP-2 activation and increased tumor growth and vascularization. Int J Cancer. 2002;98(1):23–28. doi: 10.1002/ijc.10134. [DOI] [PubMed] [Google Scholar]
- 42.Sounni NE, Devy L, Hajitou A, Frankenne F, Munaut C, Gilles C, Deroanne C, Thompson EW, Foidart JM, Noel A. MT1-MMP expression promotes tumor growth and angiogenesis through an up-regulation of vascular endothelial growth factor expression. FASEB J. 2002;16(6):555–564. doi: 10.1096/fj.01-0790com. [DOI] [PubMed] [Google Scholar]
- 43.Deryugina EI, Soroceanu L, Strongin AY. Up-regulation of vascular endothelial growth factor by membrane-type 1 matrix metalloproteinase stimulates human glioma xenograft growth and angiogenesis. Cancer Res. 2002;62(2):580–588. [PubMed] [Google Scholar]
- 44.Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36(6):1046–1069. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 45.Kikuchi T, Daigo Y, Ishikawa N, Katagiri T, Tsunoda T, Yoshida S, Nakamura Y. Expression profiles of metastatic brain tumor from lung adenocarcinomas on cDNA microarray. Int J Oncol. 2006;28(4):799–805. [PubMed] [Google Scholar]
- 46.Zohrabian VM, Nandu H, Gulati N, Khitrov G, Zhao C, Mohan A, Demattia J, Braun A, Das K, Murali R, Jhanwar-Uniyal M. Gene expression profiling of metastatic brain cancer. Oncol Rep. 2007;18(2):321–328. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.