Abstract
Based on studies in yeast and mammalian cells the Elongator complex has been implicated in functions as diverse as histone acetylation, polarized protein trafficking and tRNA modification. Here we show that Arabidopsis mutants lacking the Elongator subunit AtELP3/ELO3 have a defect in tRNA wobble uridine modification. Moreover, we demonstrate that yeast elp3 and elp1 mutants expressing the respective Arabidopsis Elongator homologues AtELP3/ELO3 and AtELP1/ELO2 assemble integer Elongator complexes indicating a high degree of structural conservation. Surprisingly, in vivo complementation studies based on Elongator-dependent tRNA nonsense suppression and zymocin tRNase toxin assays indicated that while AtELP1 rescued defects of a yeast elp1 mutant, the most conserved Elongator gene AtELP3, failed to complement an elp3 mutant. This lack of complementation is due to incompatibility with yeast ELP1 as coexpression of both plant genes in an elp1 elp3 yeast mutant restored Elongator's tRNA modification function in vivo. Similarly, AtELP1, not ScELP1 also supported partial complementation by yeast–plant Elp3 hybrids suggesting that AtElp1 has less stringent sequence requirements for Elp3 than ScElp1. We conclude that yeast and plant Elongator share tRNA modification roles and propose that this function might be conserved in Elongator from all eukaryotic kingdoms of life.
Introduction
Studies in fields as disparate as leaf development (Nelissen et al., 2003; 2005;) and drought resistance (Chen et al., 2006) in plants, neurodegeneration in humans (Anderson et al., 2001; Slaugenhaupt et al., 2001), cytotoxicity of a fungal toxin (Frohloff et al., 2001) and transcription elongation (Otero et al., 1999) have surprisingly converged on a conserved protein complex termed Elongator. The protein complex is composed of two subcomplexes with subunits Elp1, Elp2, and Elp3 as well as Elp4, Elp5 and Elp6 forming the hexameric holo-Elongator (Petrakis et al., 2004). Elp3 has a GNAT-type histone acetyltransferase (HAT) domain and has also been named Kat9 K-acetyltransferase to conform with recent acetyltransferase nomenclature (Allis et al., 2007). The Elongator complex was initially identified in yeast to interact with hyperphosphorylated RNA polymerase II and implicated in chromatin modification during transcription elongation (Otero et al., 1999; Wittschieben et al., 1999; Kristjuhan et al., 2002; Winkler et al., 2002; Gilbert et al., 2004; Kristjuhan and Svejstrup, 2004).
Surprisingly, it turned out that yeast Elongator mutants defective in any of the Elongator subunit genes (ELP1 to ELP6) are lacking tRNA modifications at wobble uridines or thiouridines in position 34 of the anticodon (Huang et al., 2005). One of these tRNA modifications, 5-methoxycarbonyl-methyl-2-thiouridine (mcm5s2U), renders Saccharomyces cerevisiae sensitive to a toxin (zymocin) secreted by Kluyveromyces lactis (reviewed in Schaffrath and Breunig, 2000) and leads to the identification of ELP/TOT genes in a screen for zymocin-resistant target of toxin (tot) mutants (Frohloff et al., 2001). The γ-subunit (γ-toxin) of the zymocin is a tRNA endonuclease that cleaves a subset of tRNAs carrying the modified wobble uridine (mcm5s2U) (Huang et al., 2005; Lu et al., 2005; 2008; Jablonowski et al., 2006).
Genomic sequence analysis suggests that an Elongator complex exists in all eukaryotes. In plants, Elongator mutants have been isolated in a screen for mutants with aberrant leaf morphology. The Arabidopsis thaliana elongata (elo) class of leaf mutants consisting of four loci, elo1 to elo4, is characterized by narrow and elongated shape of leaves and deficiencies in organ growth. ELO1 identified the Arabidopsis homologue of yeast ELP4, ELO2 is homologous to ELP1, and ELO3 to ELP3 (Nelissen et al., 2005). Moreover, the DEFORMED ROOTS AND LEAVES (DRL1) gene is allelic to elo4 and shows sequence similarity to yeast KTI12, which encodes an Elongator partner protein (Fichtner et al., 2002a; Nelissen et al., 2003). The similar phenotypes of these mutants support the view that the gene products function in a complex and suggest that plant Elongator is similar to the one in yeast.
So far the molecular function of the complex is not well defined. Apparently, its acetyltransferase activity is essential but the substrate spectrum may be wider than initially anticipated (Svejstrup, 2007). Phenotypes associated with Elongator deficiency range from impaired zygotic paternal genome demethylation (Okada et al., 2010) to altered microtubule dynamics in human and Caenorhabditis elegans (Creppe et al., 2009; Solinger et al., 2010). Recently, mammalian Elongator was linked to acetylation of α-tubulin and shown to affect the migration and differentiation of cortical neurons (Creppe et al., 2009). In C. elegans Elongator deficiency was also associated with neurological and developmental defects and tRNA modification defects (Chen et al., 2009). C. elegans mutants still contain acetylated α-tubulin although the level may be reduced (Chen et al., 2009; Solinger et al., 2010). It has also been questioned whether histones are primary substrates for the acetyltransfer reaction because transcription-related phenotypes in yeast Elongator mutants could be suppressed by tRNA overexpression (Esberg et al., 2006).
Whether protein acetylation is the primary and only biochemical activity of Elongator in eukaryotes is unknown. The pleiotropic phenotypes of yeast Elongator mutants in processes as diverse as translation, exocytosis, filamentous growth and transcriptional silencing (Rahl et al., 2005; Johansson et al., 2008; Abdullah and Cullen, 2009; Li et al., 2009) might be explained by consequential effects of improper tRNA modification or by multiple acetylation substrates for Elongator, but there is also evidence for additional biochemical activities (Huang et al., 2005; Li et al., 2009; Lipardi and Paterson, 2009).
Here we have addressed the question whether the tRNA modification function is conserved between yeast and plants. We show that in an A. thaliana elp3 mutant, tRNA wobble uridine modifications including mcm5s2U are compromised. By complementing yeast elp mutants with A. thaliana ELO/AtELP genes we demonstrate that the yeast subunits can assemble with plant polypeptides to form hybrid Elongator complexes indicating high structural similarity between yeast and plant Elongator. Strikingly, despite the fact that AtElp3/ELO3, the most conserved subunit, could structurally replace yeast Elp3 functional complementation with AtELP3/ELO3 was not observed unless ELP1 was simultaneously replaced by AtELP1/ELO2. Taken together the data strongly support the view that the tRNA modification function of Elongator is conserved between yeast and plants and most likely among all eukaryotes.
Results
Formation of a yeast Elongator complex containing AtELP3
The A. thaliana genome contains one and only one homologue for each of the yeast ELP genes and evidence for a similar hexameric complex, composed of two subcomplexes, was recently obtained by tandem affinity purification (Nelissen et al., 2010). We tried to complement yeast elp mutant strains with the corresponding plant cDNAs fused to a yeast promoter. Sensitivity to γ-toxin, the active component of the K. lactis killer toxin zymocin provided a sensitive assay for Elongator function. If the heterologous protein integrated into the yeast Elongator complex and functioned in restoring tRNA modification, we expected reversion of the toxin resistance phenotype of the Elongator mutant. Because the AtELP3 subunit is most similar to its yeast homologue, we first tried to complement the elp3 mutant. A c-myc-tagged version of the AtELP3 protein could be produced at levels comparable to those of yeast Elp3-c-myc (see below), but the toxin resistance of the yeast elp3Δ mutant was unaffected by the AtELP3-c-myc gene (not shown) or an untagged AtELP3 allele (Fig. 1A). Likewise, thermosensitivity and hypersensitivity to caffeine, additional phenotypes of Elongator mutants, were not altered by the plant gene (Fig. 1B). Reintroduction of the yeast ELP3 gene into the elp3Δ reporter, however, fully complemented all three phenotypes (Fig. 1A and B).
Fig. 1.
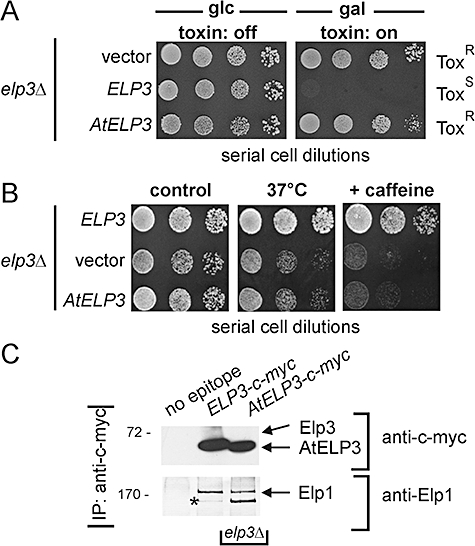
Failure of AtELP3 to complement the elp3Δ mutant despite interaction with yeast Elp1 protein. A. To test for functional complementation the elp3Δ mutant (CMY135) was transformed with plasmids containing ELP3 (pFF9), AtELP3 (YEpA4) and vector control (YEplac195) and subsequently with the GAL1 promoter driven γ-toxin expression plasmid pHMS14. Transformants were spotted in replica onto glucose-repressing (glc) or galactose-inducing (gal) media and grown for 4 days at 30°C. Growth on galactose indicates γ-toxin resistance (ToxR) and no growth corresponds to γ-toxin sensitivity (ToxS). B. To test for thermosensitivity and hypersensitivity to caffeine strains were serially diluted and replica spotted on YPD plates lacking (control) or containing 7.5 mM caffeine (right) and incubated for 4 days at 30°C and 37°C (middle). C. Anti-c-myc immunoprecipitates (IP) of strains containing chromosomally tagged ELP3-c-myc (FFY3t), AtELP3-c-myc on a plasmid in an elp3Δ background (CMY307 + yatELP3M) and wild-type parent without epitope (FY1679-8A) were analysed on Western blots probed with anti-c-myc or anti-Elp1 antibodies. Positions of the respective proteins are indicated by arrows. Protein extracts from cells without epitope-tag served as negative control. The asterisk denotes an N-terminally truncated Elp1 form (Fichtner et al., 2003). The relative abundance of this truncated form varies between experiments but is probably not dependent on which Elp3 peptide is present.
The failure of plant AtELP3 to substitute for yeast Elp3 function was not due to instability of the protein as AtELP3-c-myc was precipitated from total yeast protein extracts at levels comparable to Elp3-c-myc (Fig. 1C, top panel). c-myc-tagged Elp3 and AtElp3 both could co-precipitate the largest Elongator subunit Elp1 indicating interaction between AtELP3 and Elp1 (Fig. 1C, bottom panel).
To analyse whether AtELP3-Elp1 interaction occurred in the context of the Elongator complex we made use of the fact that interaction between the subunits Elp5 and Elp2 depends on the structural integrity of the complex and the presence of Elp3 (Frohloff et al., 2003; Petrakis et al., 2004). We constructed elp3Δ reporter strains expressing a c-myc-tagged version of Elp2 and an HA-tagged version of Elp5. As expected, co-immunoprecipitation of Elp2-c-myc with Elp5-HA was not observed when Elp3 was lacking (Fig. 2A, lane 4) but was found when the elp3Δ mutant was complemented with the yeast ELP3 gene on a plasmid (Fig. 2A, lane 3). When AtELP3 or AtELP3-c-myc alleles were introduced instead of ELP3, pull-down of Elp2-c-myc by Elp5-HA was less efficient (Fig. 2A, lanes 5 and 6), but significantly higher than in the empty vector control (Fig. 2A, lane 4). Thus, plant AtELP3 promotes Elp5-Elp2 interaction in a yeast elp3Δ mutant background.
Fig. 2.
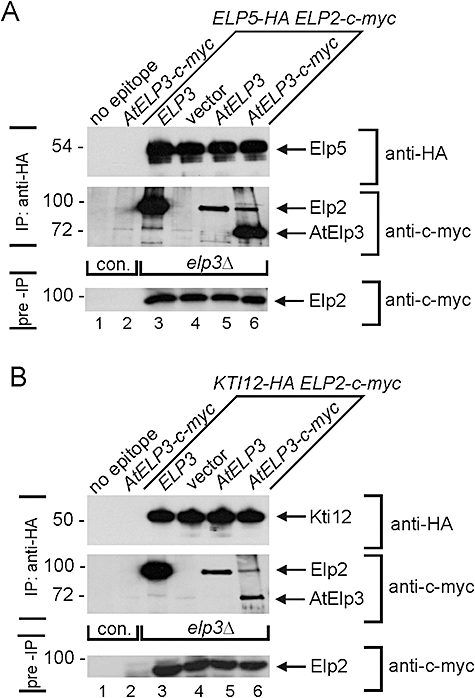
Reconstitution of Elongator subunit interactions by AtELP3 demonstrating AtELP3 integration in the yeast Elongator complex. A. Protein extracts from cells expressing HA-tagged Elp5 together with c-myc-tagged Elp2 in an elp3Δ background (strains CMY304 and CMY301, respectively) and transformed with plasmids containing ELP3 (pFF9), AtELP3 (YEpA4), AtELP3-c-myc (yatELP3M) or vector control (YEplac195) were immunoprecipitated using anti-HA antibody and analysed by Western blotting. B. Same as (A) but with strain CMY301, which expresses HA-tagged Kti12 instead of Elp5-HA. The anti-HA antibody was used to detect Elp5-HA or Kti12-HA and anti-c-myc antibodies recognized Elp2 and AtELP3. Protein extracts from cells without epitope tag and cells expressing only AtELP3-c-myc served as negative controls. The pre-IPs served as loading control (bottom panels).
Using the same approach we also tested whether the chimeric complex was able to interact with Kti12, a protein that associates with the Elongator complex (Fichtner et al., 2002a; Petrakis et al., 2005). As with Elp5-HA, Kti12-HA was able to pull-down Elp2-c-myc in the presence but not in the absence of yeast Elp3 (Fig. 2B, lanes 3 and 4) and again AtELP3 could substitute for yeast Elp3 in this interaction assay (Fig. 2B, lanes 5 and 6). The latter finding is particularly intriguing because Elp2–Kti12 interaction relies on the association between the two Elongator subcomplexes Elp1–Elp2–Elp3 and Elp4–Elp5–Elp6 (Fichtner et al., 2002b; Frohloff et al., 2003). We conclude that AtELP3 can structurally replace Elp3 in the yeast Elongator complex but apparently the complex is not functional.
Plant AtELP1 restores Elongator subunit interactions in an elp1Δ yeast mutant
Incorporation of AtElp3 into yeast Elongator requires interaction with much less conserved components. Elp1 and AtELP1 display only 19% amino acid identity compared with 67% between Elp3 and AtELP3. To analyse whether AtELP1 could also structurally replace yeast Elp1 in the complex, an elp1Δ strain that contained the epitope-tagged Elongator subunits Elp3-HA and Elp2-c-myc was transformed with an AtELP1 cDNA clone. Consistent with previous reports (Frohloff et al., 2003; Petrakis et al., 2004), we found that the Elp1 subunit is not only required for Elp3-Elp2 interaction (Fig. 3A, lane 2) but also for stability of yeast Elp3. Hence, an HA-tagged version of Elp3 was not detectable in total protein extracts of the elp1Δ mutant (Fig. 3A, bottom panel, lane 2). Reintroduction of the yeast ELP1 gene restored Elp3-HA stability and interaction between Elp2 and Elp3 (Fig. 3A, lane 4). Remarkably, the same held true when AtELP1 was introduced (Fig. 3A, lane 3). Expression of the plant gene from the inducible GAL1 promoter in the elp1Δ background allowed Elp2-c-myc to precipitate Elp3-HA. AtELP1 expression also restored the interaction between Elp2 and Kti12 (Fig. 3B).
Fig. 3.
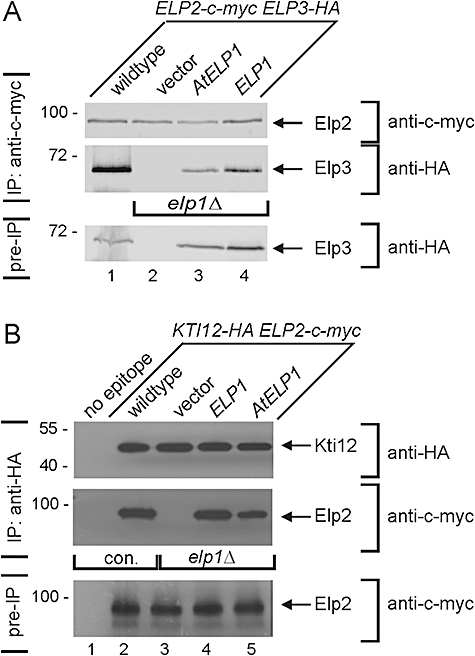
Restoration of Elp3 stability and Elongator subunit interactions in an elp1Δ strain expressing AtElp1. A. AtElp1 is able to replace yeast Elp1 and to support Elp2–Elp3 interaction and Elp3 protein stability. Protein extracts from cells expressing chromosomally c-myc-tagged Elp2 together with HA-tagged Elp3 in an elp1Δ background (FFY2/3-dt-1d) and transformed with plasmids containing ELP1 (pFF13), AtELP1 (pDJ98) or vector control (YEplac181) were immunoprecipitated using anti-c-myc antibodies. Western blots of the precipitates were probed with anti-c-myc or anti-HA antibodies to detect Elp2 and Elp3 respectively (arrows). Instability of Elp3 in the elp1Δ strain is revealed in pre-IPs (bottom panel, lane 3). B. AtElp1 is able to restore Kti12-Elp2 interaction. Protein extracts from cells expressing c-myc-tagged Elp2 together with HA-tagged Kti12 in an elp1Δ background (CMY300), and transformed with plasmids containing ELP1 (pFF13), AtELP1 (pDJ98) or vector control (YEplac181) were immunoprecipitated and analysed as in (A).
Again, as in the AtELP3-expressing elp3 mutant, the efficiency of subunit interactions was somewhat reduced compared with that of the ELP1 transformants. Nonetheless, our data show that AtELP1 like AtELP3 are assembled into complexes where they are able to structurally replace the respective yeast Elongator subunits.
Together, plant AtELP1 and AtELP3 support tRNA modification in yeast
Because Elp3 requires Elp1 for stability, its function may depend on specific contacts between these two proteins explaining the failure of AtELP3 to functionally complement the elp3Δ mutant in combination with yeast Elp1. To address this possibility we tested whether AtELP3 might function in yeast when supplied with AtELP1. FLAG-AtELP1 was expressed together with the AtELP3-c-myc gene in a yeast elp1Δelp3Δ double mutant. FLAG-AtELP1 could be detected in the anti-c-myc precipitates together with AtELP3-c-myc (Fig. 4A). Like in the single mutants, interaction between the epitope-tagged subunits Elp5-HA and Elp2-c-myc (Fig. 4B) as well as Kti12-HA and Elp2-c-myc (Fig. 4C) could be restored in the double mutant by the simultaneous expression of untagged (lanes 5) or tagged (lanes 6) versions of AtELP1 and AtELP3. Elp2-c-myc (lanes 3, 5 and 6) and AtElp3-c-myc (lane 6) could be detected in the anti-HA precipitates of strains containing yeast (lane 3) or A. thaliana Elp1 and Elp3. We conclude that a chimeric yeast–plant Elongator complex can form under these conditions. Intriguingly, AtELP1 and AtELP3 together were able to restore γ-toxin sensitivity in the yeast elp1 elp3 double mutant indicating that the chimeric Elongator complex was functional (Fig. 5A). Because toxin sensitivity requires the Elongator-dependent tRNA wobble uridine modification mcm5s2U (Lu et al., 2005), the A. thaliana subunits can reasonably be assumed to carry out a function in tRNA modification that is equivalent to that of the yeast homologues.
Fig. 4.
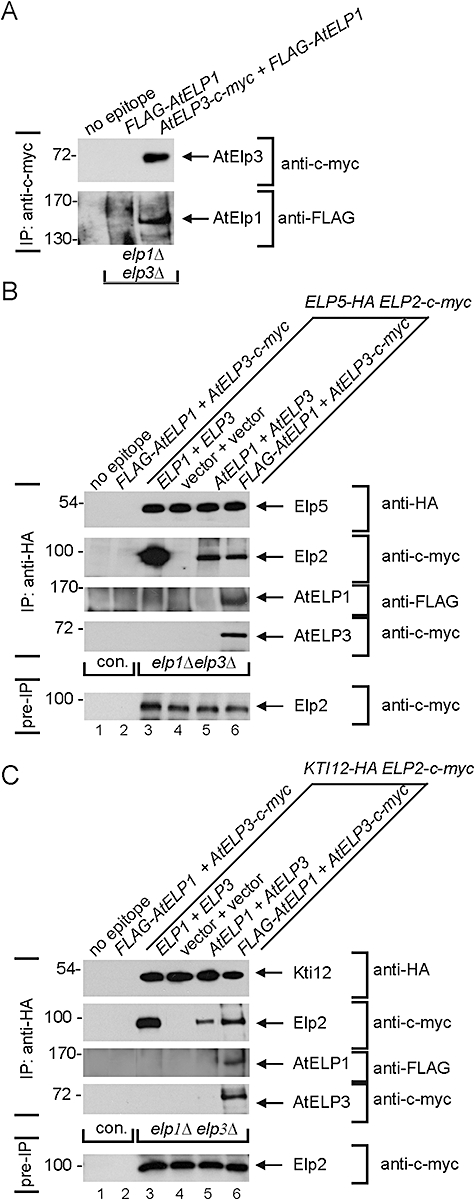
Formation of a chimeric Elongator complexes composed of yeast and plant subunits. A. AtELP1 and AtELP3 interact with each other in yeast. Anti-c-myc immunoprecipitates of elp1Δ elp3Δ strains (CMY134) containing FLAG-AtELP1 (pJET13) and AtELP3-c-myc were probed with anti-c-myc antibodies to detect AtELP3-c-myc and with anti-FLAG-antibodies to detect FLAG-AtELP1. Immunoprecipitates with anti-c-myc antibodies from elp1Δelp3Δ cells containing pFLAG-AtELP1 (lane 2) or no epitope-tag (lane1) served as negative controls. B. AtELP1 and AtELP3 restore Elp2–Elp5 interaction in the elp1Δ elp3Δ mutant. Protein extracts from cells expressing HA-tagged Elp5 together with c-myc-tagged Elp2 in an elp1Δ elp3Δ double mutant background (CMY305) and transformed with plasmids containing ELP1 (pFF13), ELP3 (pFF9), AtELP1 (pDJ98), AtELP3 (YEpA4), FLAG-AtELP1 (pJET13), AtELP3-c-myc (pELO3-myc) and vector controls (YEplac181, YEplac195) were immunoprecipitated using anti-HA antibodies. The antibodies used to detect the indicated proteins by Western blotting are marked on the right. Protein extracts from cells without epitope-tag and cells expressing only AtELP3-c-myc served as negative controls. Pre-IPs served as loading control (bottom panel). C. AtELP1 and AtELP3 restore Elp2–Kti12 interaction. Protein extracts from strain CMY302 expressing HA-tagged Kti12 together with c-myc-tagged Elp2 in an elp1Δ elp3Δ background and transformed (see B) were analysed as described in (B).
Fig. 5.
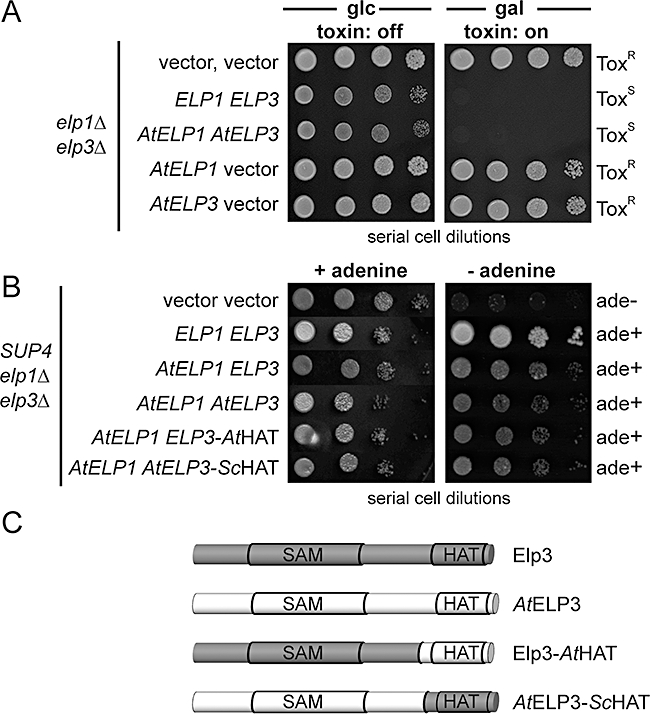
Restoration of the tRNA modification function of Elongator by complementation of a yeast elp1Δelp3Δ double mutant with AtELP1 and AtELP3. A. Restoration of toxin sensitivity. The elp1Δ elp3Δ (CMY134) strain was transformed with plasmids containing ELP1 (pFF13), ELP3 (pFF9), AtELP1 (pCM26), AtELP3 (YEpA4) and vector controls (YEplac181 and YEplac195) in the indicated combinations. The γ-toxin was expressed from pHMS14 (Frohloff et al., 2001) on galactose medium. Growth on galactose after 4 days at 30°C indicates γ-toxin resistance (ToxR) and no growth equals γ-toxin sensitivity (ToxS). B. Restoration of SUP4 suppressor tRNA function. SUP4 elp1Δ elp3Δ (CMY160) cells were transformed with plasmids containing wild-type ELP1 and ELP3 (pFF13 and p424TDH-ScElp3myc), the respective empty vectors (YEplac181 and p424TDH), the plant homologues AtELP1 and AtELP3 (pCM26 and p424TDH-AtElp3myc) or AtELP1 and the swapped HAT domain variants pElp3-AtHAT and pAtElp3-ScHAT. To check for suppression of the ade2-1 (UAA) ochre mutation (Huang et al., 2005) serial dilutions of the transformants were spotted on SC and SC-Ade plates and cultivated for 4 days at 30°C. SUP4 suppressor tRNA function results in growth on SC-Ade plates (Ade+ phenotype). C. Schematic representation of the Elp3 domain structure and domain swapped hybrids generated between AtELP3 and ScElp3 used as in (B).
To support this conclusion, we used a second assay monitoring Elongator-dependent tRNA modification that is based on the SUP4 suppressor tRNA gene (Huang et al., 2005). The SUP4 allele codes for a UAA ochre suppressor tRNATyr with a G34-to-U34 exchange in the anticodon. Suppression of ochre nonsense codons by SUP4 requires Elongator-dependent wobble uridine modification (Huang et al., 2005). In an elp1Δelp3Δ double deletion strain (CMY160) carrying the ochre mutation ade2-1 the SUP4 tRNA suppressor was inactive resulting in an Ade- phenotype (Fig. 5B, vector control). Transformation with two plasmids encoding either yeast Elp1 and Elp3 or AtElp1 and AtElp3 restored SUP4 tRNA suppression, allowing for ade2-1 readthrough and growth on adenine-lacking medium. Although growth in the latter case was weak we conclude that the AtElp1-AtElp3 containing chimeric Elongator can functionally replace yeast Elongator and promote ochre suppression by SUP4. Yeast Elp1 with AtElp3 was inactive in the adenine growth assay (data not shown) as in the γ-toxin assay (Fig. 5A).
Elp3 contains two protein motifs that are found in other functional contexts, the C-terminal GNAT-like HAT domain related to Gcn5 histone acteyltransferase (Wittschieben et al., 1999) and a central domain found in radical S-adenosyl-L-methionine enzymes (radical SAM domain) (Chinenov, 2002; Paraskevopoulou et al., 2006). To test which part of AtElp3 is not compatible with ScElp1 we constructed two hybrid variants with the HAT domain of yeast Elp3 replaced by that of AtELP3 and vice versa (Fig. 5C). Like AtELP3, both chimeric Elp3 variants were unable to complement the elp1 elp3 double mutant in the presence of yeast Elp1 (data not shown). However, in combination with AtELP1, AtELP3 and both hybrid proteins allowed for weak growth on adenine-deficient medium indicative of SUP4-dependent ochre suppression of the ade2-1 mutation (Fig. 5B). Apparently, AtELP1 has a less stringent requirement with respect to Elp3 sequence than ScElp1. The weak Ade+ phenotype in combination with AtELP1 but not with yeast ELP3 was highly reproducible; however, we could not confirm reconstitution of tRNA modification by the hybrid Elp3 variants using the γ-toxin assay (data not shown). Probably partial complementation results in hypomodification of tRNAs and gives zymocin resistance
tRNA modification is affected in Elongator-deficient plant cells
Restoration of ochre suppression and γ-toxin sensitivity of the yeast elp1Δelp3Δ double mutant by AtELP1 and AtELP3 suggested that plant Elongator may also affect tRNA modification in plant cells. For all but one tRNA modification genes of yeast (TRM genes) homologues can be found in plant genomes suggesting that plant tRNAs are modified in a similar way. To test whether Elongator deficiency in A. thaliana had any impact on tRNA modification RNA preparations enriched for small stable RNA were compared between wild-type and homozygous elo3 mutants. RNA was isolated from leaves of several independent plants, degraded to nucleosides and analysed by HPLC. The elution profiles were very similar for both samples except for the parts shown in Fig. 6, where 5-carbamoyl-methyluridine (ncm5U) (Fig. 6A and B) and mcm5s2U (Fig. 6C and D) eluted. Both modified uridine nucleosides (compare Fig. 6E) were lacking in the sample from the elo3 mutant. These data clearly show that the elo3 mutation prevented the ncm5 and mcm5s2-uridine modifications from being generated supporting the view that plant Elongator is involved in tRNA modification in a way similar to its yeast homologue. This indicated that the role of Elongator in tRNA modification might be conserved among lower and higher eukaryotes.
Fig. 6.
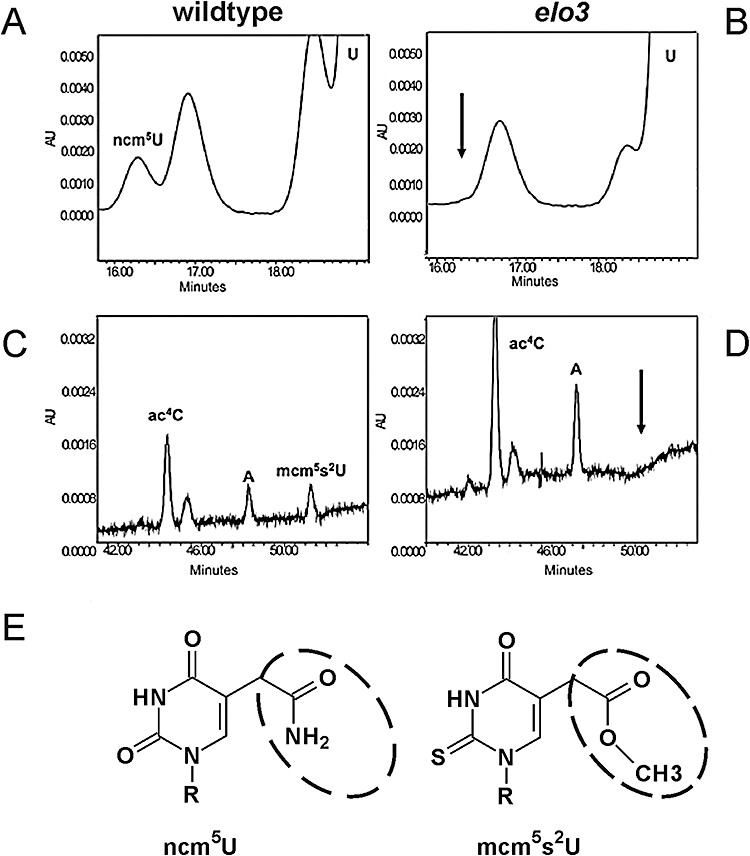
AtELP3/ELO3 is required for formation of mcm5s2U and ncm5U modified nucleosides in tRNA. A.–D. Total tRNA isolated from wild-type and elo3 plants was analysed by HPLC (see Experimental procedures). Wild-type profiles are shown on the left, elo3 profiles in the right panels. A. and B. The parts of the chromatograms between retention times 15.5 and 19.6 min are displayed. The arrow in (B) indicates the expected retention time of ncm5U. Chromatograms were monitored at 254 nm. C. and D. The parts of the chromatograms between retention times 40.0 and 52.5 min are displayed. The arrow in (D) indicates the expected retention time of mcm5s2U. Chromatograms were monitored at 314 nm. E. Uridine modifications found at the wobble position of eukaryotic tRNAs. Elongator-dependent side groups are circled (Huang et al., 2005).
Discussion
Biochemical characterizations of yeast and human Elongator and genomic sequences have indicated that the subunit composition of this complex is conserved among eukaryotes (reviewed in (Svejstrup, 2007). The data presented in this paper reinforce this view and provide evidence for a remarkable structural conservation between plant and yeast Elongator complexes. We further demonstrate that an A. thaliana Elongator mutant is lacking the very same tRNA modifications previously shown in yeast to be Elongator-dependent (Huang et al., 2005). Together with similar deficiencies reported for C. elegans Elongator mutants (Chen et al., 2009), these data strongly support the view that tRNA modification might be a function of Elongator that is conserved in animals, fungi and plants.
A crucial question is how Elongator affects tRNA modification. The acetyltransferase activity of the Elp3 subunit has been shown to be required (Huang et al., 2005). One possibility is that biosynthesis or activity of the enzymes generating the ncm5 or mcm5 side chains on the tRNA wobble uridines depend on acetylation. Alternatively, Elongator may directly participate in the tRNA modification reaction. The Elongator subunits Elp1 and Elp3 could be specifically cross-linked to tRNAGlu but not tRNAMet (Huang et al., 2005) supporting the latter model. A radical mechanism based on the putative radical SAM domain flanking the HAT motif in Elp3 (Chinenov, 2002; Paraskevopoulou et al., 2006) might be involved.
We have used yeast as a tool to analyse whether AtELP3 was able to function in the context of a heterologous Elongator complex. Two sensitive assays that allow to monitor the Elongator-dependent tRNA modification were used. The first one detects mcm5s2U-tRNAGlu and, to a lesser extent, mcm5s2U-tRNALys and mcm5s2U-tRNAGln. These tRNAs are the primary targets of the tRNA endonuclease activity associated with the K. lactis zymocin. Defects in formation of the modified uridine and typical of Elongator mutants cause zymocin resistance (Lu et al., 2005; 2008; Jablonowski et al., 2006; Studte et al., 2008). Restoration of zymocin sensitivity in resistant mutants indicates efficient formation of the modified tRNAs. The second assay makes use of the SUP4 ochre suppressor tRNA, which also depends on the mcm5 modification function of Elongator (Huang et al., 2005). Both assays are sensitive to a point mutation in the HAT domain of yeast Elp3 (Frohloff et al., 2001; Huang et al., 2005). Under conditions of partial complementation the two assays are expected to differ in sensitivity. Hypomodified tRNAs would confer (some degree of) resistance in the zymocin assay (i.e. the mutant phenotype) by escaping endonucleolytic cleavage but might support some growth on selective plates (the wild-type phenotype) in the nonsense suppression assay.
We have shown here that a double mutant lacking Elp3 and Elp1 can be complemented by the Arabidopsis homologues AtELP1 and AtELP3/ELO3, which together restore zymocin sensitivity and SUP4-mediated nonsense suppression. Thus, these two subunits cooperate to functionally replace yeast Elp1 and Elp3 indicating that they have a similar role in the plant Elongator complex.
Surprisingly, despite 67% sequence identity between ScElp3 and AtELP3, the latter was unable to complement the yeast elp3 mutation. Only in combination with AtELP1 could complementation of the Elongator mutant phenotypes by the AtELP3 genes be achieved. AtELP3-c-myc alone was incorporated into the yeast complex as shown by its ability to restore association between the large and small subcomplexes and between Elongator and its partner protein Kti12. Both types of interactions require Elp3 and the structural integrity of the complex (Frohloff et al., 2001; Fichtner et al., 2002a; Fichtner et al., 2003; Petrakis et al., 2005). We conclude that, although an Elongator complex in which AtELP3 replaces Elp3 appears structurally intact, the AtELP3 containing chimeric complex is unable to provide the activity required for tRNA modification.
The lack of functional complementation by AtELP3 is probably not due to the reduced efficiency of Elp2–Elp5 or Elp2–Kti12 interactions as the same reduction was observed in the AtELP3 and AtELP1 containing chimeric complex, which is functional. AtELP3 appears stable in yeast in the presence of ScElp1 excluding the possibility that lack of function is due to protein degradation, which is observed for ScElp3 in the absence of ScElp1 (Fig. 3).
To determine whether sequence divergence in the HAT domain was responsible for the lack of complementation, domain swapped variants with ScHAT replaced by AtHAT and vice versa were tested. Like AtELP3, both variants were inactive in the presence of ScELP1. This failure of the AtHAT to replace the corresponding yeast domain contrasts with the ability of the HsELP3-HAT to do so (Li et al., 2005). However surprisingly, in the SUP4 tRNA suppression assay (but not in the zymocin assay), both hybrid ELP3 genes were able to partially complement the yeast elp3 mutation in combination with AtELP1. This indicates weak U34-tRNA modification mediated by AtELP1 in combination with multiple structural variants of Elp3 including ScElp3 (Chen et al., 2006). AtELP1 appears more tolerant towards sequence variations in the Elp3 protein. The crucial role of ScElp1 phosphorylation/dephosphorylation (Mehlgarten et al., 2009) might be important in this context. Alternatively but less likely, AtELP1 may contribute to complementation in yeast via an additional function not found in ScElp1, like the RNA-dependent RNA polymerase activity recently reported for Drosophila Elp1 (Lipardi and Paterson, 2009).
A current challenge is to understand the biological function(s) of the Elongator complexes in the various organisms and their possible divergence during evolution. If, as proposed here and supported by studies in C. elegans (Chen et al., 2009), tRNA modification defects are a general consequence of Elongator deficiency in eukaryotes it will be essential to determine whether the mutant phenotypes are causally related to this defect. Studies in yeast support such a view. Pleiotropic phenotypes of yeast Elongator mutants including hypoacetylation of histones, delayed adaptation of transcription to changing environmental conditions as well as delocalization of the secretory protein Sec2 could all be suppressed by overexpression of two tRNAs, tRNALys and tRNAGln, both of which in wild-type cells undergo Elongator-dependent anticodon modifications (Esberg et al., 2006). Based on these data it has been proposed that the effects of Elongator mutants on transcription and secretion are indirect consequences of influences on translation caused by inappropriate tRNA modification. Apparently, U34 modifications in the anticodon increase the decoding efficiency of A- and G-ending codons (Yokoyama et al., 1985; Lim, 1994; Durant et al., 2005; Esberg et al., 2006; Begley et al., 2007; Johansson et al., 2008). Thus, the translation of individual proteins might be differentially affected depending on their amino acid composition and codon usage. Moreover, yeast elp mutants are synthetic lethal with mutations affecting thiolation of U34 (Esberg et al., 2006; Johansson et al., 2008) emphasizing the importance of the wobble uridine modifications.
A causal relationship between tRNA hypomodification and Elongator phenotypes does not exclude that Elongator has additional activities or multiple targets for the acetyltransferase reaction. In plants, yeast and human cells the complex is also found in the nucleus and its depletion affects transcription (Winkler et al., 2002; Close et al., 2006; Svejstrup, 2007; Nelissen et al., 2010). Transcriptome analysis in those organisms revealed that only a small set of genes is affected by Elongator depletion with little apparent overlap of functional categories (Krogan and Greenblatt, 2001; Close et al., 2006; Nelissen et al., 2010). This argues against a general role of Elongator in transcription elongation.
In A. thaliana several auxin-related genes were found to be differentially expressed in wild-type and elo mutants, and at least two of these genes, SHY2/IAA3 and LAX2, were also affected in histone H3K14 acetylation, which is a predominant target of the ScELP3-HAT (Nelissen et al., 2010). The data were taken as evidence for a role of Elongator in transcription elongation of specific genes, which may explain the observed influences on cell proliferation and development in elo mutants. How specificity for these genes might be achieved is currently unknown. Because auxin signalling is specific to plants whereas neuron development is specific to animals studies on the role of Elongator in these diverse processes might reveal how a protein complex with (a) conserved molecular function(s) has evolved to fulfil these kingdom specific functions. We believe that the answer to these questions can be found at the cellular level.
Taken together, our data indicate that the Elongator complexes are structurally and functionally highly conserved between yeast and plants, such that yeast subunits can be replaced by their plant orthologues. This offers the opportunity to take advantage of the yeast model system in structure–function studies of Elongator complexes from other kingdoms. It is likely that eukaryotic Elongator complexes have a conserved biochemical activity. Based on reconstitution of U34 tRNA modification in yeast by plant Elongator subunits and the requirement of AtElp3/ELO3 for this modification in plants, we favour the view that this conserved activity is directly related to tRNA modification.
Experimental procedures
Yeast strains, media and general methods
Yeast strains used are listed in Table 1. Yeast was grown in rich media containing yeast extract, peptone and 2% dextrose (YPD) or 2% galactose (YPG) or on synthetic complete medium (SC) (Sherman, 1991).
Table 1.
Yeast strains.
| Strain | Description | Source/reference |
|---|---|---|
| K. lactis | ||
| AWJ137 | αleu2 trp1[k1+ k2+] killer and zymocin producer | Karin D. Breunig |
| S. cerevisiae | ||
| FY1679-08A | MATa ura3-52 leu2Δ1 trp1-Δ63 his3-Δ200 GAL | Euroscarf |
| CMY307 | as FY1679-08A, but elp3Δ::natNT2 | This study |
| FFY3t | as FY1679-08A, but ELP3-(cmyc)3::SpHIS3 | Frohloff et al. (2001) |
| CMY306 | as FY1679-08A, but elp1Δ::TRP1, elp3Δ::natNT2 | This study |
| FFY2/3-dt | as FY1679-08A, but ELP2-(cmyc)3::SpHIS3, ELP3-(HA)6::KlTRP1 | Fichtner et al. (2002a) |
| FFY2/3-dt-1d | as FFY2/3-dt, but elp1Δ::KlURA3 | Fichtner et al. (2002b) |
| FFY2/4dt | as FY1679-08A, but ELP2-(cmyc)3::SpHIS3, KTI12-(HA)6::KlTRP1 | Fichtner et al. (2002a) |
| CMY300 | as FFY2/4dt, but elp1Δ::hphNT1 | This study |
| CMY301 | as FFY2/4dt, but elp3Δ::natNT2 | This study |
| CMY302 | As FFY2/4dt, but elp1Δ::hphNT1, elp3Δ::natNT2 | This study |
| FFY2/5dt | as FY1679-08A, but ELP2-(cmyc)3::SpHIS3, ELP5-(HA)6::KlTRP1 | Frohloff et al. (2003) |
| CMY304 | as FFY2/5dt, but elp3Δ::natNT2 | This study |
| CMY305 | as FFY2/5dt, but elp1Δ::hphNT1, elp3Δ::natNT2 | This study |
| W303-1a | MATa ade2-1 his3-11, 15 leu2-3, -112 trp1-1 ura3-1 can1-100 | Laboratory stock |
| CMY135 | as W303, but elp3Δ::natNT2 | This study |
| CMY134 | as W303, but elp1Δ::hphNT1, elp3Δ::natNT2 | This study |
| UMY2893 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | Huang et al. (2005) |
| UMY2916 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 elp3Δ::KanMX4 | Huang et al. (2005) |
| CMY160 | as UMY2916, but elp1Δ::HIS3 | This study |
Thermosensitivity was assayed on YPD medium at 30 or 38°C for 2–3 days. Drug sensitivity was assayed at 30°C with 5 mM caffeine. ade2-1 ochre stop codon suppression by the SUP4 gene was tested as described (Huang et al., 2005). Yeast was transformed with plasmid DNA or polymerase chain reaction (PCR) products according to a previous protocol (Schiestl and Gietz, 1989). Primers are listed in Table S1.
Arabidopsis thaliana strains and growth conditions
An A. thaliana homozygous T-DNA insertion mutant in At5g50230 (AtELP1) derived from GABI-Kat Line 636H02 and the corresponding wild type (Columbia ecotype) were analysed for tRNA modification. Total RNA was prepared from leaves of several independent plants grown under short day conditions.
Plasmid constructions
pFF9, a YEplac195-based plasmid carrying ELP3 has been described (Frohloff et al., 2001). A. thaliana AtELP3/ELO3 was amplified from cDNA clone RAFL0811J12 (http://www.brc.riken.jp) with primers cDNAfw and cDNA2re inserting flanking SalI restriction sites. The SalI fragment was fused to the yeast ADH1 promoter and integrated into YEplac195 to give YEpA4. The AtELP3 open reading frame was verified by DNA sequencing. In yatELP3 the ADH1 promoter and 5′ end of the AtELP3 gene from YEpA4 was replaced after cleavage with HindIII and AgeI by the yeast ELP3 promoter and 5′ end using primers E3′-fw and E3′-re and pFF9 as template. yatELP3M is a triple c-myc-tagged variant of yatELP3. pFF13 is a YEplac181-based plasmid, carrying the yeast ELP1 gene (Fichtner et al., 2002a). pDJ98 is a multicopy plasmid for galactose-regulated expression of the AtELP1 gene carrying the selection markers URA3 and leu2d (Chen et al., 2006). pJET13, carrying FLAG-AtELP1 under control of the GAL1 promoter was obtained by PCR amplification of the coding sequence from pFLAG-ELO2 (Z. Gong, unpublished) and cloned into SalI-cleaved pCM22. In the latter, the GAL1 promoter was introduced on a EcoRI-BamHI fragment from plasmid pRB1438 (a kind gift from Mike Stark, University of Dundee, UK) into YEplac181 (LEU2). pCM22 served also as destination vector to clone untagged AtELP1 from pDJ98 via SalI restriction sites to obtain pCM26 for the SUP4 suppression assay. The centromeric plasmid p424TDH (Mumberg et al., 1995) served as the destination vector for ELP3 variants in double complementation studies. p424TDH-ScElp3myc encodes a C-terminal triple c-myc-tagged variant of ScELP3 (kindly provided by O. Onuma, Univ. Halle), p424TDH-AtElp3myc a C-terminal triple c-myc-tagged variant of AtELP3. PCR-mediated domain swap of the AtELP3 HAT domain (amino acids 436–565) into p424TDH-ScElp3myc (1–426) and of ScElp3-HAT (382–557) into p424TDH-AtElp3myc (1–390) resulted in plasmids pELP3-AtHAT and pAtELP3-ScHAT respectively.
Yeast genetic manipulations
Defined elp1Δ and elp3Δ null alleles and genetic variants encoding hemagglutinin (HA6) or c-myc3 epitope-tagged proteins were obtained after transformation of PCR fragments generated with template plasmids containing the marker genes YDp-KlU (URA3), YDp-SpH (HIS3), pFA6a-hphNT1, pFA6a-natNT2 (for deletions) or pYM3 and pYM5 (for HA and c-myc epitope-tagging) (Knop et al., 1999; Frohloff et al., 2001; Jablonowski et al., 2001; Janke et al., 2004). Manipulations were verified by PCR and by killer eclipse assays (Kishida et al., 1996) to test for biological functionality.
Elongator complementation studies in yeast
To analyse the function of Arabidopsis ELO2 and ELO3 encoded gene products, the γ-toxin sensitivity was assayed (Frohloff et al., 2001). In detail, strains CMY135 (elp3Δ) and CMY134 (elp1Δelp3Δ) were transformed with pDJ98 (ELO2/AtELP1), YEpA4 (ELO3/AtELP3), pFF13 (ELP1) and pFF9 (ELP3) (Frohloff et al., 2001; Fichtner et al., 2002a; Chen et al., 2006) or the respective empty vector controls. Subsequently, plasmid pHMS14 (Frohloff et al., 2001) expressing the γ-toxin subunit of K. lactis zymocin under the GAL1 promoter was introduced into the transformed strains. Strains were grown on 2% (w/v) glucose SC medium under selective conditions and 10-fold serial dilutions were spotted on glucose and galactose medium. The response to γ-toxin induction was monitored on galactose plates after 3 to 4 days at 30°C.
Immunological techniques
Detection of tagged proteins used anti-c-myc (A-14) and anti-HA (F-7) (Santa Cruz) antibodies. Anti-Elp1 antibodies (Otero et al., 1999; Wittschieben et al., 1999) were kindly provided by Dr J. Svejstrup (London Research Institute, Cancer Research, UK). An anti-FLAG antibody (Sigma–Aldrich) was used for detection of FLAG-ELO2. Protein concentrations were determined by the method of Bradford (Bradford, 1976). Antibody cross-linking to protein A-Sepharose, preparation of protein extract and co-immunoprecipitations were performed as described previously (Zachariae et al., 1996; Frohloff et al., 2001).
tRNA isolation and HPLC analysis
tRNA was prepared from total RNA preparations as described (Bjork et al., 2001). Purified tRNA was digested with Nuclease P1 for 16 h at 37°C and then treated with bacterial alkaline phosphatase for 2 h at 37°C. The hydrolysate was analysed by high-pressure liquid chromatography with a Develosil C-30 reverse-phase column as described (Gehrke et al., 1982; Gehrke and Kuo, 1990).
Acknowledgments
The authors would like to thank Osita Onuma for providing plasmid p424TDH-ScELP3myc, Jesper Q. Svejstrup for anti-Elp1 antibodies, Jens-Eike-Täubert and Sabrina Zink for help and advice and Mieke van Lijsebettens for discussion and communication of results prior to publication. We thank Sabine Rosahl (Institute of Plant Biochemistry, Halle) for providing the A. thaliana T-DNA insertion line. This work was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) in SFB 648 ‘Molekulare Mechanismen der Informationsverarbeitung in Pflanzen’ to K.D.B and R.S. and by the Sachsen-Anhalt federal graduate programme ‘Structures and Mechanisms of Biological Information Processing’. In addition, R.S. acknowledges support by the BBSRC (BB/F019106/1). A.B. is supported by grants from the Swedish Cancer Foundation (07 0637), Swedish Research Council (621-2006-4269) and Bernhard and Signe Bäckström Foundation (223-438-07).
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abdullah U, Cullen PJ. The tRNA modification complex elongator regulates the Cdc42-dependent mitogen-activated protein kinase pathway that controls filamentous growth in yeast. Eukaryot Cell. 2009;8:1362–1372. doi: 10.1128/EC.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, et al. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet. 2001;68:753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, et al. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen C, Tuck S, Bystrom AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009;5:e1000561. doi: 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang H, Jablonowski D, Zhou X, Ren X, Hong X, et al. Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol Cell Biol. 2006;26:6902–6912. doi: 10.1128/MCB.00433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y. A second catalytic domain in the Elp3 histone acetyltransferases: a candidate for histone demethylase activity? Trends Biochem Sci. 2002;27:115–117. doi: 10.1016/s0968-0004(02)02058-3. [DOI] [PubMed] [Google Scholar]
- Close P, Hawkes N, Cornez I, Creppe C, Lambert CA, Rogister B, et al. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol Cell. 2006;22:521–531. doi: 10.1016/j.molcel.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry. 2005;44:8078–8089. doi: 10.1021/bi050343f. [DOI] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJ, Bystrom AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Fichtner L, Frohloff F, Bürkner K, Larsen M, Breunig KD, Schaffrath R. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol Microbiol. 2002a;43:783–791. doi: 10.1046/j.1365-2958.2002.02794.x. [DOI] [PubMed] [Google Scholar]
- Fichtner L, Frohloff F, Jablonowski D, Stark MJ, Schaffrath R. Protein interactions within Saccharomyces cerevisiae Elongator, a complex essential for Kluyveromyces lactis zymocicity. Mol Microbiol. 2002b;45:817–826. doi: 10.1046/j.1365-2958.2002.03055.x. [DOI] [PubMed] [Google Scholar]
- Fichtner L, Jablonowski D, Schierhorn A, Kitamoto HK, Stark MJ, Schaffrath R. Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol. 2003;49:1297–1307. doi: 10.1046/j.1365-2958.2003.03632.x. [DOI] [PubMed] [Google Scholar]
- Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001;20:1993–2003. doi: 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohloff F, Jablonowski D, Fichtner L, Schaffrath R. Subunit communications crucial for the functional integrity of the yeast RNA Polymerase II Elongator (gamma-Toxin Target (TOT)) complex. J Biol Chem. 2003;278:956–961. doi: 10.1074/jbc.M210060200. [DOI] [PubMed] [Google Scholar]
- Gehrke CW, Kuo KCT. Chromatography and Modification of Nucleosides. Amsterdam: Elsevier; 1990. [Google Scholar]
- Gehrke CW, Kuo KC, McCune RA, Gerhardt KO, Agris PF. Quantitative enzymatic hydrolysis of tRNAs: reversed-phase high-performance liquid chromatography of tRNA nucleosides. J Chromatogr. 1982;230:297–308. [PubMed] [Google Scholar]
- Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- Huang B, Johansson MJ, Bystrom AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski D, Fichtner L, Martin VJ, Klassen R, Meinhardt F, Stark MJ, Schaffrath R. Saccharomyces cerevisiae cell wall chitin, the Kluyveromyces lactis zymocin receptor. Yeast. 2001;18:1285–1299. doi: 10.1002/yea.776. [DOI] [PubMed] [Google Scholar]
- Jablonowski D, Zink S, Mehlgarten C, Daum G, Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol Microbiol. 2006;59:677–688. doi: 10.1111/j.1365-2958.2005.04972.x. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida M, Tokunaga M, Katayose Y, Yajima H, Kawamura-Watabe A, Hishinuma F. Isolation and genetic characterization of pGKL killer-insensitive mutants (iki) from Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 1996;60:798–801. doi: 10.1271/bbb.60.798. [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristjuhan A, Walker J, Suka N, Grunstein M, Roberts D, Cairns BR, Svejstrup JQ. Transcriptional inhibition of genes with severe histone H3 hypoacetylation in the coding region. Mol Cell. 2002;10:925–933. doi: 10.1016/s1097-2765(02)00647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Greenblatt JF. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:8203–8212. doi: 10.1128/MCB.21.23.8203-8212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Lu J, Han Q, Zhang G, Huang B. The Elp3 subunit of human Elongator complex is functionally similar to its counterpart in yeast. Mol Genet Genomics. 2005;273:264–272. doi: 10.1007/s00438-005-1120-2. [DOI] [PubMed] [Google Scholar]
- Li Q, Fazly AM, Zhou H, Huang S, Zhang Z, Stillman B. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 2009;5:e1000684. doi: 10.1371/journal.pgen.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim VI. Analysis of action of wobble nucleoside modifications on codon-anticodon pairing within the ribosome. J Mol Biol. 1994;240:8–19. doi: 10.1006/jmbi.1994.1413. [DOI] [PubMed] [Google Scholar]
- Lipardi C, Paterson BM. Identification of an RNA-dependent RNA polymerase in Drosophila involved in RNAi and transposon suppression. Proc Natl Acad Sci USA. 2009;106:15645–15650. doi: 10.1073/pnas.0904984106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lu J, Huang B, Esberg A, Johansson MJ, Bystrom AS. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Esberg A, Huang B, Bystrom AS. Kluyveromyces lactis gamma-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res. 2008;36:1072–1080. doi: 10.1093/nar/gkm1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlgarten C, Jablonowski D, Breunig KD, Stark MJ, Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol Microbiol. 2009;73:869–881. doi: 10.1111/j.1365-2958.2009.06811.x. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nelissen H, Clarke JH, De BM, De BS, Vanderhaeghen R, Zielinski RE, et al. DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell. 2003;15:639–654. doi: 10.1105/tpc.007062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Fleury D, Bruno L, Robles P, De VL, Traas J, et al. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci USA. 2005;102:7754–7759. doi: 10.1073/pnas.0502600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, De Groeve S, Fleury D, Neyt P, Bruno L, Bitonti MB, et al. Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc Natl Acad Sci USA. 2010;107:1678–1683. doi: 10.1073/pnas.0913559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, et al. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Paraskevopoulou C, Fairhurst SA, Lowe DJ, Brick P, Onesti S. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol. 2006;59:795–806. doi: 10.1111/j.1365-2958.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- Petrakis TG, Wittschieben BO, Svejstrup JQ. Molecular architecture, structure-function relationship, and importance of the Elp3 subunit for the RNA binding of holo-elongator. J Biol Chem. 2004;279:32087–32092. doi: 10.1074/jbc.M403361200. [DOI] [PubMed] [Google Scholar]
- Petrakis TG, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Physical and functional interaction between Elongator and the chromatin-associated Kti12 protein. J Biol Chem. 2005;280:19454–19460. doi: 10.1074/jbc.M413373200. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Chen CZ, Collins RN. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell. 2005;17:841–853. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Schaffrath R, Breunig KD. Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet Biol. 2000;30:173–190. doi: 10.1006/fgbi.2000.1221. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Sherman F. Guide to yeast genetics and molecular biology. Getting started with yeast. Methods Enzymol. 1991;194:3–20. [PubMed] [Google Scholar]
- Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Paolinelli R, Kloss H, Scorza FB, Marchesi S, Sauder U, et al. The Caenorhabditis elegans Elongator complex regulates neuronal alpha-tubulin acetylation. PLoS Genet. 2010;6:e1000820. doi: 10.1371/journal.pgen.1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studte P, Zink S, Jablonowski D, Bar C, von der HT, Tuite MF, Schaffrath R. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol Microbiol. 2008;69:1266–1277. doi: 10.1111/j.1365-2958.2008.06358.x. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ. Elongator complex: how many roles does it play? Curr Opin Cell Biol. 2007;19:331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci USA. 2002;99:3517–3522. doi: 10.1073/pnas.022042899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben BO, Otero G, de BT, Fellows J, Erdjument-Bromage H, Ohba R, et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Watanabe T, Murao K, Ishikura H, Yamaizumi Z, Nishimura S, Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc Natl Acad Sci USA. 1985;82:4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 1996;274:1201–1204. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


