Abstract
We report the first high-resolution structural data for the β/γ-peptide 13-helix (i,i+3 C=O···H-N H-bonds), a secondary structure that is formed by oligomers with a 1:1 alternation of β- and γ-amino acid residues. Our characterization includes both crystallophaphic and 2D NMR data. Previous studies suggested that β/γ-peptides constructed from conformationally flexible residues adopt a different helical secondary structure in solution. Our design features preorganized β- and γ-residues, which strongly promote 13-helical folding by the 1:1β:γ backbone.
Identification of new types of foldamers with strong and discrete secondary structural propensities is a subject of ongoing research.1 These studies enhance our understanding of the relationship between local conformational preferences and molecular shape. In addition, new folding patterns can be valuable for specific applications.2,3 Foldamers that contain more than one type of subunit, i.e., oligomers that have heterogeneous backbones, have been a subject of extensive recent interest.1e Most examples involve combination of α-amino acid residues with other types of subunits, including those derived from β-4 or γ-amino acids5 or other building blocks.6 Heterogeneous backbones that do not include α-amino acid residues have received relatively limited attention,5e,7 perhaps because α-amino acids are far more available than are other building blocks. Backbones with alternating β- and γ-amino acid residues (β/γ-peptides) are of particular interest because a β/γ-dipeptide has the same number of atoms between the N- and C-termini as an α-tripeptide.5b An extended β/γ-peptide can in principle form a helix containing 13-membered ring backbone H-bonds (C=O(i)--H-N(i+3)) that are analogous to the 13-membered ring backbone H-bonds characteristic of the α-helix (C=O(i)--H-N(i+4)). However, Sharma, Kunwar et al.5e have recently reported that flexible β/γ-peptides adopt a different type of helical conformation in solution. Here we show that β/γ-peptides containing appropriately preorganized subunits do indeed adopt the 13-helix in solution and the solid state.
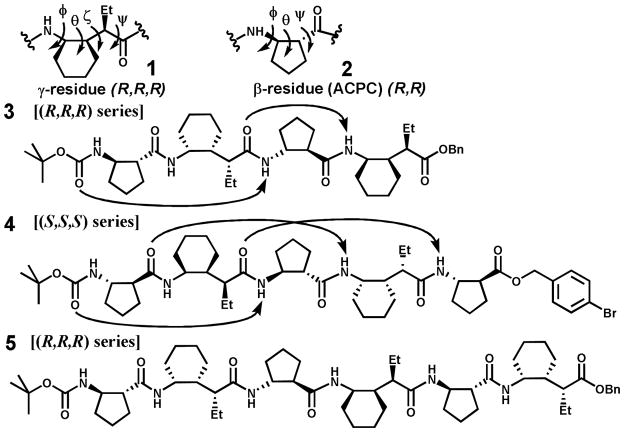
The β/γ-peptide 13-helix is predicted by Hofmann et al.5d to have g+,g+ or g−,g− local conformations about the Cα−Cβ (ζ) and Cβ−Cγ (θ) bonds in the γ-residues and a Cα−Cβ torsion angle of ~90° in the β-residues. Based on these predictions and available data for the conformational propensities of constrained β-and γ-residues in other contexts, we concluded that combining (R,R,R) γ-residue 1 (Figure 1), which has recently become available,5i,8 with (R,R)-trans-2-aminocyclopentanecarboxylic acid (ACPC, 2) should favor formation of the left-handed β/γ-peptide 13-helix (the right-handed helix should be favored by residues with S configurations). This hypothesis was tested by preparation and analysis of tetramer 3, pentamer 4 and hexamer 5 (Figure 1).
Figure 1.
Structures of β/γ peptides 3, 4, 5 (arrows indicate H-bonds in the crystal structures of 3 and 4)
The crystal structure of β/γ-peptide 3 contains two molecules in the asymmetric unit; the two conformations are very similar (Figure 2). Each independent molecule forms one 13-atom H-bonded ring, involving the NH group of the second ACPC residue and the carbonyl of the N-terminal Boc group. The other possible 13-atom ring H-bond does not form in either case [N--O distance~ 4.9Å]; instead, each molecule contains an 8-atom ring H-bond involving the carbonyl of the first γ-residue and the NH group of second γ-residue. Despite this deviation from the 13-helical H-bonding pattern, the backbone torsion angles for the β- and γ-residues in 3 generally fall in ranges predicted by Hofmann et al.5d for the β/γ-peptide 13-helix.9
Figure 2.
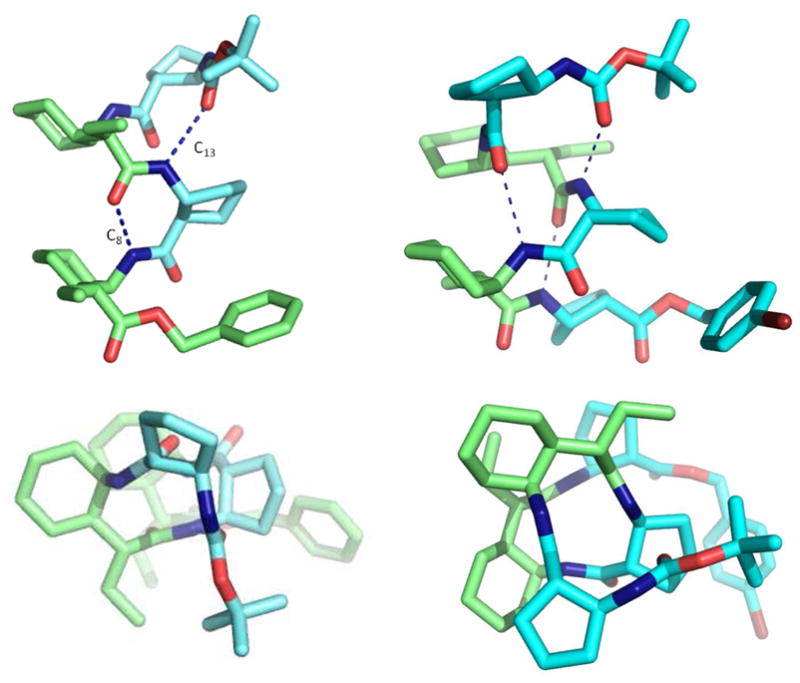
Crystal structures of 3 (left) and 4 (right): (top) views perpendicular to helical axis; (bottom) views along the helical axis.
Pentamer 4, containing β- and γ-residues with S configurations, adopts the right-handed 13-helix in the crystalline state. All three of the possible C=O(i)--H-N(i+3) H-bonds are formed (Figure 2). Table 1 compares backbone torsion angles for the β- and γ-residues in pentamer 4 with analogous values from the computational work of Hofmann et al.5d and from the NMR analysis of flexible β/γ-peptides in organic solvent by Sharma, Kunwar et al.5e The preorganized γ-residues in 4 display g+,g+ local conformations about the Cα−Cβ (ζ) and Cβ−Cγ (θ) bonds, and ψ and φ near −120°, with a somewhat wider distribution for the latter torsion angle. These values are consistent with the predictions for the 13-helical conformation from Hofmann et al.5d In contrast, the helical conformations deduced via NMR for flexible β/γ-peptides feature opposite signs for the ζ and θ torsion angles (g−,g+), and opposite signs for the ψ and φ torsion angles. The helical conformation deduced for these flexible β/γ-peptides has a distinctive H-bonding pattern with two types of interaction: C=Oγ (i)--H-Nγ (i−1) and C=Oγ (i)--H-Nγ (i+3).
Table 1.
Backbone Torsion Angles (deg) † from β/γ Peptides
| Peptides | residues | φ | θ | ζ | ψ |
|---|---|---|---|---|---|
| β/γ pentamer 4 | β | −107.7 | 93.3 | −128.3 | |
| γ | −134.7 | 60.1 | 59.8 | −121.0 | |
| β | −133.6 | 113.5 | −85.7 | ||
| γ | −147.3 | 57.9 | 46.5 | −129.8 | |
| β | −167.9 | 141.4 | −155.0 | ||
|
| |||||
| computational Study ‡,5d | β | 89.1 | −94.1 | 121.9 | |
| γ | 124.9 | −60.4 | −62.2 | 132.0 | |
|
| |||||
| flexible β/γ (NMR)5e | β | 120 | 60 | 0 | |
| γ | 120 | −60 | 60 | −120 | |
Nomenclature for the backbone torsion angles in β,γ-residues is as described in Figure 1.
Average backbone torsion angles
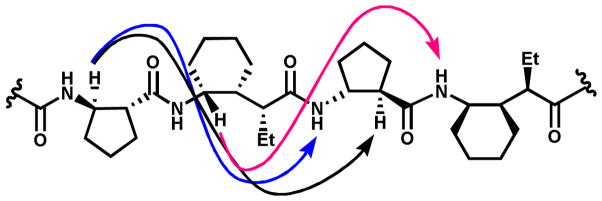
Hexamer 5 did not produce high-quality crystals, but 2D 1H NMR analysis in pyridine-d5 solution indicated that the 13-helix is significantly populated under these conditions. Among the unambiguous NOEs involving backbone protons, six strong NOEs were observed between protons from residues that are not adjacent in the sequence: CβH(1)--NH(3), CβH(1)--CαH(3), CγH(2)--NH(4), CβH(3)--NH(5), CβH(3)--CαH(5), and CγH(4)--NH(6) (Figure 3). These NOEs are consistent with intramolecular proton-proton distances in the crystal structure of pentamer 4: CβH(1)--NH(3) = 3.5 Å, CβH(1)--CαH(3) = 2.7 Å, CγH(2)--NH(4) = 2.8 Å, CβH(3)--NH(5) = 2.3 Å and CβH(3)--CαH(5) = 2.2 Å. Thus, the three NOE patterns observed for 5, CβH(i)--NH(i+2) and CβH(i)--CαH(i+2) for β-residues and CγH(i)--NH(i+2) for γ-residues, appear to be general indicators of β/γ-peptide 13-helical secondary structure.
Figure 3.
Characteristic NOEs patterns observed for the 1:1 β/γ-peptide hexamer 5 in pyridine-d5.
The β/γ-peptide helix we have documented is interesting because of its relationship to the α-helix formed by pure α-residue backbones. Both helices contain 13-atom ring H-bonds. Detailed comparison of the two helices reveals further similarities: both have a rise-per-turn of 5.4 Å, and the radii are similar (2.5 vs. 2.3 Å).9 These parameters suggest that the β/γ-peptide 13-helix may be a promising scaffold for functional mimicry of natural α-helices.2b,3
Our results show that appropriately preorganized residues promote the formation of the 13-helical conformation in short β/γ-peptides. This secondary structure was anticipated (along with alternative helices) in computational studies,5c,d and hints of 13-helical propensity can be found in the local conformations observed in crystal structures for isolated β-γ segments,5b,g but the only previous analysis of β/γ-peptide oligomer folding indicated the formation of a different helical conformation, containing both 11- and 13-membered ring H-bonds.5e Conformationally constrained β-amino acid residues have been shown to induce novel secondary structures,1a,e,10 and the present studies highlight the prospect that constrained γ-amino acid residues will be similarly useful in controlling molecular shape.
Supplementary Material
Acknowledgments
This research was supported by NSF (CHE-0848847). NMR spectrometers were purchased with partial support from NIH and NSF.
Footnotes
Supporting Information Available: Experimental procedures and compound characterizations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Gellman SH. Acc Chem Res. 1998;31:173. [Google Scholar]; (b) Seebach D, Beck AK, Bierbaum DJ. Chem Biodivers. 2004;1:1111. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]; (c) Hecht S, Huc I, editors. Foldamers: Structure, Properties and Applications. Wiley-VCH Weinheim; Germany: 2007. [Google Scholar]; (d) Goodman CM, Choi S, Shandler S, DeGrado WF. Nat Chem Biol. 2007;3:252. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Horne WS, Gellman SH. Acc Chem Res. 2009;41:1399. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Recent examples of biologically active foldamers: Claudon P, Violette A, Lamour K, Decossas M, Fournel S, Heurtault B, Godet J, Mely Y, Jamart-Gregoire B, Averlant-Petit M-C, Briand J-P, Duportail G, Monteil H, Guichard G. Angew Chem, Int Ed. 2010;49:333. doi: 10.1002/anie.200905591.Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Proc Natl Acad Sci USA. 2009;106:14751. doi: 10.1073/pnas.0902663106.Jochim AL, Miller SE, Angelo NG, Arora PS. Bioorg Med Chem Lett. 2009;19:6023. doi: 10.1016/j.bmcl.2009.09.049.Choi S, Isaacs A, Clements D, Liu DH, Kim H, Scott RW, Winkler JD, DeGrado WF. Proc Natl Acad Sci USA. 2009;106:6968. doi: 10.1073/pnas.0811818106.Bautista AD, Stephens OM, Wang LG, Domaoal RA, Anderson KS, Schepartz A. Bioorg Med Chem Lett. 2009;19:3736. doi: 10.1016/j.bmcl.2009.05.032.Brown NJ, Wu CW, Seurynck-Servoss SL, Barron AE. Biochemistry. 2008;47:1808. doi: 10.1021/bi7021975.(g) For earlier examples, see ref. 1d.
- 3.Sadowsky JD, Fairlie WD, Hadley EB, Lee HS, Umezawa N, Nikolovska-Coleska Z, Wang SM, Huang DCS, Tomita Y, Gellman SH. J Am Chem Soc. 2007;129:139. doi: 10.1021/ja0662523. [DOI] [PubMed] [Google Scholar]
- 4.De Pol S, Zorn C, Klein CD, Zerbe O, Reiser O. Angew Chem, Int Ed. 2004;43:511. doi: 10.1002/anie.200352267.Hayen A, Schmitt MA, Ngassa FN, Thomasson KA, Gellman SH. Angew Chem, Int Ed. 2004;43:505. doi: 10.1002/anie.200352125.Sharma GVM, Nagendar P, Jayaprakash P, Krishna PR, Ramakrishna KVS, Kunwar AC. Angew Chem, Int Ed. 2005;44:5878. doi: 10.1002/anie.200501247.Mandity IM, Weber E, Martinek TA, Olajos G, Toth GK, Vass E, Fulop F. Angew Chem, Int Ed. 2009;48:2171. doi: 10.1002/anie.200805095.(e) For a heterogeneous backbone review, see ref. 1e.
- 5.(a) Hagihara M, Anthony NJ, Stout TJ, Clardy J, Schreiber SL. J Am Chem Soc. 1992;114:6568. [Google Scholar]; (b) Karle IL, Pramanik A, Banerjee A, Bhattacharjya S, Balaram P. J Am Chem Soc. 1997;119:9087. [Google Scholar]; (c) Ananda K, Vasudev PG, Sengupta A, Raja KMP, Shamala N, Balaram P. J Am Chem Soc. 2005;127:16668. doi: 10.1021/ja055799z. [DOI] [PubMed] [Google Scholar]; (d) Baldauf C, Gunther R, Hofmann HJ. J Org Chem. 2006;71:1200. doi: 10.1021/jo052340e. [DOI] [PubMed] [Google Scholar]; (e) Sharma GVM, Jadhav VB, Ramakrishna KVS, Narsimulu K, Subash V, Kunwar AC. J Am Chem Soc. 2006;128:14657. doi: 10.1021/ja064875a. [DOI] [PubMed] [Google Scholar]; (f) Baruah PK, Sreedevi NK, Gonnade R, Ravindranathan S, Damodaran K, Hofmann HJ, Sanjayan GJ. J Org Chem. 2007;72:636. doi: 10.1021/jo062032w. [DOI] [PubMed] [Google Scholar]; (g) Vasudev PG, Ananda K, Chatterjee S, Aravinda S, Shamala N, Balaram P. J Am Chem Soc. 2007;129:4039. doi: 10.1021/ja068910p. [DOI] [PubMed] [Google Scholar]; (h) Chatterjee S, Vasudev PG, Raghothama S, Ramakrishnan C, Shamala N, Balaram P. J Am Chem Soc. 2009;131:5956. doi: 10.1021/ja900618h. [DOI] [PubMed] [Google Scholar]; (i) Guo L, Chi Y, Almeida AM, Guzei IA, Parker BK, Gellman SH. J Am Chem Soc. 2009;131:16018. doi: 10.1021/ja907233q. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Chakraborty TK, Rao KS, Kiran MU, Jagadeesh B. Tetrahedron Lett. 2009;50:4350. [Google Scholar]; (k) Araghi RR, Jackel C, Cofen H, Salwiczek M, Vokel A, Wagner SC, Wieczorek S, Baldauf C, Koksch B. ChemBioChem. 2010;11:335. doi: 10.1002/cbic.200900700. [DOI] [PubMed] [Google Scholar]
- 6.(a) Yang D, Li W, Qu J, Luo SW, Wu YD. J Am Chem Soc. 2003;125:13018. doi: 10.1021/ja036136p. [DOI] [PubMed] [Google Scholar]; (b) Chowdhury S, Schatte G, Kraatz HB. Angew Chem, Int Ed. 2006;45:6882. doi: 10.1002/anie.200602248. [DOI] [PubMed] [Google Scholar]; (c) Olsen CA, Bonke G, Vedel L, Adsersen A, Witt M, Franzhk H, Jaroszewski JW. Org Lett. 2007;9:1549. doi: 10.1021/ol070316c. [DOI] [PubMed] [Google Scholar]; (d) Zhao Y, Zhong ZQ, Ryu EH. J Am Chem Soc. 2007;129:218. doi: 10.1021/ja0671159. [DOI] [PubMed] [Google Scholar]; (e) Angelici G, Luppi G, Kaptein B, Broxterman QB, Hofmann HJ, Tomasini C. Eur J Org Chem. 2007:2713. [Google Scholar]; (f) Sakai N, Mareda J, Matile S. Acc Chem Res. 2008;41:1354. doi: 10.1021/ar700229r. [DOI] [PubMed] [Google Scholar]; (g) Sharma GVM, Babu BS, Ramakrishna KV, Nagendar P, Kunwar AC, Schramm P, Baldauf C, Hofmann HJ. Chem Eur J. 2009;15:5552. doi: 10.1002/chem.200802078. [DOI] [PubMed] [Google Scholar]; (h) Sharma GVM, Babu BS, Chatterjee D, Ramakrishna KVS, Kunwar AC, Schramm P, Hofmann HJ. J Org Chem. 2009;74:6703. doi: 10.1021/jo901277a. [DOI] [PubMed] [Google Scholar]; (i) Hetenyi A, Toth GK, Somlai C, Vass E, Martinek TA, Fulop F. Chem Eur J. 2009;15:10736. doi: 10.1002/chem.200900724. [DOI] [PubMed] [Google Scholar]
- 7.(a) Gong B, Zeng H, Zhu J, Yuan L, Han Y, Cheng S, Furukawa M, Parra RD, Kovalevsky AY, Mills JL, Skrzypczak-Jankun E, Martinovic S, Smith RD, Zheng C, Szyperski T, Zeng XC. Proc Natl Acad Sci USA. 2002;99:11583. doi: 10.1073/pnas.162277099. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Delsuc N, Godde F, Kauffmann B, Leger JM, Huc I. J Am Chem Soc. 2007;129:11348. doi: 10.1021/ja074285s. [DOI] [PubMed] [Google Scholar]
- 8.A complementary example: Nodes WJ, Nutt DR, Chippindale AM, Cobb AJA. J Am Chem Soc. 2009;131:16016. doi: 10.1021/ja9070915.
- 9.See the Supporting Information.
- 10.Schmitt MA, Choi SH, Guzei IA, Gellman SH. J Am Chem Soc. 2005;127:13130. doi: 10.1021/ja0536163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.