Abstract
For most of the single-molecule fluorescence studies to date, biomolecules of interest are labeled with small organic dyes which suffer from their limited photostability evidenced by blinking and photobleaching. An enzymatic oxygen scavenging system of glucose oxidase and catalase is widely used to improve the dye photostability but with the unfavorable side effect of producing gluconic acid. It is known that accumulation of this byproduct in solution can lead to considerable acidification, but the uncertainty in its severity under experimentally relevant conditions has been a long-standing area of concern due to the lack of a suitable assay. In this paper we report a fluorescence-based analytical assay for quantitatively assessing the acidification of oxygen scavenging systems in situ. By using a ratiometric, dual-emission dye, SNARF-1, we observed the presence and, for the first time, measured the severity of solution acidification due to the oxygen scavenging system for a number of conditions relevant to single-molecule studies. On the basis of the quantitative analysis of the acidification profile under these conditions, practical guidelines for optimizing the oxygen scavenging system are provided. This in situ assay should be applicable to a large variety of future single-molecule fluorescence studies.
In the past decade, single-molecule fluorescence techniques have attracted widespread interest and provided numerous insights into many important biological problems, including DNA replication,1,2 transcription,(3) viral infection,4–6 translation,7,8 etc. From the original optical detection of individual biomolecules, such as DNA and protein, to real-time observation of their interactions by FRET and from conventional confocal and total internal reflection microscopies to the recently developed super-resolution imaging9,10 as well as the fluorescence-tweezers hybrid,(11) the potentials of these techniques are rising to new levels. In most of these studies, biomolecules of interest are labeled with small fluorescent dyes to report their localizations and dynamics. However, one fundamental problem with these organic dye reporters is their limited photostability as evidenced by the blinking and photobleaching found in single-molecule trajectories. Employing an enzymatic oxygen scavenging system effectively reduces photobleaching. In addition, using Trolox,(12) a vitamin E analogue, or a coupled reducing−oxidizing system of ascorbic acid and methyl viologen(13) has been shown to eliminate blinking and further reduce photobleaching. The most popular oxygen scavenging system in use today employs glucose oxidase and catalase to remove molecular O2 by oxidizing glucose (Scheme 1)(14) but with the unfavorable side effect of producing gluconic acid.(15) It is well-known that the acidity of this byproduct (pKa = 3.86)(16) can lead to considerable acidification if the solution is well exposed to air, but the conditions in real experiments are often different. The uncertainty in the severity of acidification under experimental conditions has been a long-standing area of concern mostly due to the lack of a suitable assay. Since considerable acidification can introduce into single-molecule studies undesirable heterogeneities in the protein side chain protonation state, enzymatic activity, reporter photophysics, etc., this problem requires a quantitative analysis.
Scheme 1. Enzymatic Reactions Involved in the Oxygen Scavenging System of Glucose, Glucose Oxidase, and Catalase.
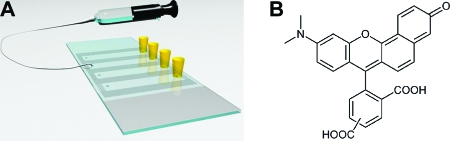
Single-molecule studies are often performed in sample chambers that are designed to accommodate certain practical needs. The example shown in Figure 1A illustrates the flow chamber used in many of our single-molecule FRET experiments with which total internal reflection microscopy(14) can be carried out conveniently. Like the chambers used by other researchers, our design provides a fairly confined environment while allowing for stepwise additions of different sample solutions. However, monitoring the real-time acidification profile of the oxygen scavenging system in the chamber is inaccessible to the conventional methods of using a pH paper or common laboratory electrode. First, both methods provide only a one-time value, while a continuous readout over a long duration is often required depending on the length of single-molecule experiments. In addition, both pH paper and the common electrode would be used with the solution exposed to air, which can accelerate the acidification process, while using a noninvasive probe in situ is much more desirable. Moreover, the amount of sample available from the chamber, typically tens of microliters, may pose a further challenge to the usage of a common pH electrode given its volume requirement, while pH paper may suffer from its limited accuracy. Note that continuous measurement in the chamber is possible with a specialized pH microelectrode, although this may depend on the specific chamber design involved. The measurement will require certain special instruments that may not be available in most single-molecule laboratories, and it may prove to be more difficult for more complicated chamber designs.
Figure 1.
(A) Flow chamber used in our single-molecule experiments with which total internal reflection microscopy can be carried out conveniently. A small sample reservoir shown in yellow and a piece of tubing connected to a syringe are attached to the inlet and outlet of the channel, respectively, to facilitate sample flow. The tubing and syringe are shown for one channel only for clarity. (B) Structure of the ratiometric, dual-emission probe 5-(and-6)-carboxy SNARF-1.
Here, we report a fluorescence-based analytical assay for quantitatively assessing the acidification of oxygen scavenging systems used in single-molecule fluorescence studies. This assay utilizes a commercially available, pH-sensitive fluorescent dye, 5-(and-6)-carboxy SNARF-1 (Figure 1B), that exhibits ratiometric, dual emission with a near-neutral pKa of 7.5,(17) relevant to the buffer condition in many single-molecule studies. A dual-emission probe provides two emission signals simultaneously as the readout,18−20 with each serving as an internal standard for the other when the ratio of these two intensities is taken. Thus, any intensity drift caused by photobleaching of the probe or fluctuations in excitation power is canceled out, offering this type of probe a clear advantage over single-emission probes, such as fluorescein. Furthermore, fluorescence-based sensing provides a continuous and noninvasive way to monitor the acidification process of oxygen scavenging systems in situ and, therefore, overcomes all the challenges mentioned above. In the literature, there have been a number of reports discussing the application of SNARF-1 in pH sensing, even at the single-molecule level.21,22 Nevertheless, applying this method to assessing the acidification of oxygen scavenging systems in situ has not to our knowledge been demonstrated previously. Note that, in this study, the term in situ sensing means to examine the acidification process exactly in place where it occurs, i.e., inside the actual experimental chamber, versus taking the analyte out to somewhere else as mentioned above. It does not necessarily imply that the acidification measurements would be part of a simultaneous single-molecule experiment. With this assay, we observed the presence and, for the first time, measured the severity of solution acidification due to the oxygen scavenging system under a number of different conditions that are relevant to single-molecule studies today. On the basis of the quantitative analysis of the acidification profile under these conditions, practical guidelines for optimizing the oxygen scavenging system are provided. The in situ assay established here should be applicable to a large variety of future single-molecule fluorescence studies.
Experimental Section
Materials
5-(and-6)-carboxy SNARF-1 was purchased from Invitrogen (Carlsbad, CA), and 10 μM SNARF-1 was used in all pH-sensing experiments. Glucose oxidase from Aspergillus niger and catalase from bovine liver were both purchased from Sigma (St. Louis, MO), and 1.0 mg/mL6 (168.8 U/mL) glucose oxidase and 0.04 mg/mL6 catalase (1,404 U/mL) were used in the oxygen scavenging system unless otherwise indicated. 6-Hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox) was purchased from Aldrich (St. Louis, MO) and dissolved slowly at 2 mM concentration in the desired buffer with gentle shaking at room temperature overnight.(23) Protocatechuic acid (PCA) and protocatechuate 3,4-dioxygenase (PCD) were generously provided by Paul Selvin’s laboratory (University of Illinois, Urbana, IL), and 2.5 mM PCA and 50 nM PCD were used in the oxygen scavenging system.(24) Cy3-Top18-biotin and Cy5-D27-biotin DNA oligos were purchased from Integrated DNA Technologies (Coralville, IA), and 100 pM DNA was used in all single-molecule experiments: (Cy3-Top18-biotin) 5′-Cy3 GCC TCG CTG CCG TCG CCA biotin-3′, (Cy5-D27-biotin) 5′-biotin ACA AGT ATA GGA TCC CCG AGA ACC GAG Cy5-3′.
Flow Chamber and Fluorescence Measurement
Four-channel chambers were assembled using a quartz slide and a cover glass, with strips of double-sided tape and fast-curing epoxy applied to the long and short edges of the channel, respectively (Figure 1A). Except for single-molecule experiments, the slide and cover glass were not subjected to a prior PEGylation procedure for surface passivation purposes.(14) Unless otherwise stated, a small sample reservoir and a piece of tubing connected to a syringe were attached to the inlet and outlet of the channel, respectively, to facilitate sample flow.
For all fluorometer measurements, a small transparent window was created for the channel in use by masking the rest of the chamber with black electric tape. This chamber was then mounted on a 1933 solid-sample holder (Horiba Jobin Yvon, Edison, NJ) at a 60° angle relative to excitation to reduce reflected and scattered light. The fluorescence emission spectra of SNARF-1 upon 488 nm excitation were collected using a Fluoromax-4 spectrofluorometer (Horiba Jobin Yvon). The excitation and emission monochromator slits were set to 2 and 10 nm, respectively. A blank spectrum collected with the buffer alone was subtracted from those with 10 μM SNARF-1 in the same buffer. The fluorescence intensities at 568 and 616 nm were determined for the two emission bands of SNARF-1, A and B, respectively. To calibrate the ratiometric dual emission of SNARF-1 as a function of pH, we measured the spectra of the probe in a series of standard solutions with pH ranging from 8.00 to 5.99 at 0.2 pH intervals. All these solutions contain 25 mM Tris, 5 mM MgCl2, 0.8% (w/v) dextrose, and 2 mM Trolox, with no oxygen scavenging system included. The ratio of the two intensities, IB/IA, was then calculated for each spectrum, plotted against the corresponding pH, and fit to17,25
 |
where c is the slope, RA/B is the intensity ratio IA/IB, i.e., the inverse of the ratio IB/IA above, with the limiting values of Rmin and Rmax, and Iacid and Ibase are the fluorescence intensities at 616 nm in acid and base, respectively. Thus, a continuous IB/IA−pH mapping curve was obtained. Note that a correction of the spectra for the fluorometer’s detection efficiency as a function of the wavelength would produce more accurate line shapes and peak positions, but would have no effect on the quantitative analysis in this work and thus was not carried out. This is because the same correction factor for the ratiometric readout from the calibration experiment would apply to the one from the sensing experiment below as well.
Sensing the Acidification of the Oxygen Scavenging System
Immediately before the experiment, glucose oxidase and catalase were added to a buffer with a starting pH of 8.0 that contains 10 μM SNARF-1, 5 mM MgCl2, 0.8% (w/v) dextrose, 2 mM Trolox, and the buffering agent at the concentration specified in the text. This mixture was then transferred into the flow chamber rapidly. Alternatively, PCA and PCD were added to a buffer with the same starting pH, containing 10 μM SNARF-1, 5 mM MgCl2, 2 mM Trolox, and 25 mM Tris. Fluorescence emission spectra of SNARF-1 in these solutions inside the flow chamber were taken at 5 min intervals for up to 1 h. Note that a short time delay was always present for the spectrum taken at time zero, due to the time required to mix the oxygen scavenging system and transfer the sample into the chamber; nonetheless, this delay does not significantly affect the kinetics of acidification observed. For each time point monitored, the ratiometric readout IB/IA was determined from the spectrum and converted into the corresponding pH value using the IB/IA−pH calibration curve described above. This 1 h measurement was repeated at least four times for each condition to ensure the reproducibility and provide the basis for statistical analysis. The characteristic acidification time, t0.5pH, is defined as the time needed, under a specific experimental condition, for the solution pH to drop by half a unit. The average value and standard deviation of this parameter was obtained from the multiple independent measurements performed under the same experimental conditions.
Analysis of the Single-Molecule Photobleaching Time
Cy3-Top18-biotin or Cy5-D27-biotin DNA was immobilized on a PEG-coated surface through biotin-neutravidin binding using 100 pM concentration of DNA.(14) Prior to imaging, the sample was first thoroughly rinsed with a pH 8.0 T50 buffer, containing 10 mM Tris and 50 mM NaCl, and then incubated in a pH 8.0 imaging buffer containing 25 mM Tris, 5 mM MgCl2, 0.8% (w/v) dextrose, 2 mM Trolox, 0.04 mg/mL catalase, and glucose oxidase at the concentrations specified in the text. This two-step buffer exchange procedure was carried out for each of the three glucose oxidase concentrations examined, ranging from 1.0 to 0.25 mg/mL.
Single-molecule experiments were performed with prism-type total internal reflection microscopy.(14) In brief, a 532 nm DPSS laser or 633 nm HeNe laser was used to excite the fluorophores on immobilized DNA molecules through total internal reflection. The fluorescence was collected using a 60×, 1.2 NA, water immersion objective lens from Olympus America (Center Valley, PA) and detected up to 60 s at 30 ms time resolution with an EMCCD camera from Andor (Belfast, Northern Ireland). Single molecules were identified and their intensity traces were extracted from the raw data using a custom program written in IDL. The total intensity of all the molecules identified from within the imaging area was then obtained using a program written in MATLAB. The decay in the average intensity of all molecules as a function of time was fit to a single exponential, with the decay lifetime representing the single-molecule photobleaching time, τphotobleaching. This single-molecule measurement was repeated five times in different imaging areas for both fluorophores and all glucose oxidase concentrations to provide the basis for statistical analysis.
Results and Discussion
SNARF-1’s pH-Dependent Dual Emission
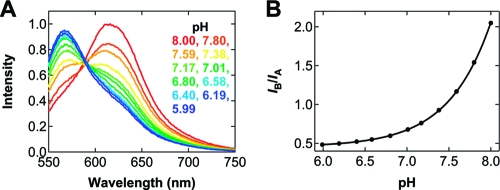
To use SNARF-1’s ratiometric, dual emission for sensing pH, we first measured the emission spectra of this probe in a series of standard solutions with the pH ranging from 8.00 to 5.99 (Figure 2A). At pH 8.00, one band at 616 nm, denoted B, dominates the spectrum. As the pH is lowered, another band at 568 nm, denoted A, becomes visible; the intensity of this band increases while that of band B decreases at the same time. At about pH 7.5, the intensities of both bands are equal, consistent with the probe’s pKa. Below this pH, band A starts to dominate, and this trend continues to the lowest pH we characterized, 5.99, where the ratio of the two bands barely changes due to the diminished sensitivity of SNARF-1 here as expected from its pKa. Note that, if other pH ranges are in question, researchers can choose a probe sensitive in that range such as SNARF-4F (pKa 6.4, Invitrogen) or LysoSensor Yellow/Blue DND-160 (pKa 4.2, Invitrogen).
Figure 2.
(A) Fluorescence emission spectra of SNARF-1 upon 488 nm excitation in a series of standard solutions with the pH ranging from 8.00 (red) to 5.99 (blue). These solutions contain 25 mM Tris, 5 mM MgCl2, 0.8% (w/v) dextrose, and 2 mM Trolox, with no oxygen scavenging system included. The bands at 568 and 616 nm are denoted A and B, respectively. (B) Ratio of the two intensities, IB/IA, as a function of pH (circles) and a continuous IB/IA−pH mapping curve obtained by fitting the data to eq 1.
The ratio of the two intensities, IB/IA, was calculated from these spectra for SNARF-1 at each pH and is plotted in Figure 2B. Clearly, this ratio decreases monotonically as the pH is lowered. We fit these data to eq 1, determined that pKa′ = 7.36, c = −1.15, Rmin = 0.01, and Rmax = 2.20, and then obtained a continuous curve that associates the ratiometric readout IB/IA directly with the corresponding pH. This IB/IA−pH mapping curve allows us to extract the pH value of an imaging buffer containing an oxygen system after a certain amount of incubation time as shown below.
Sensing the Acidification of the Oxygen Scavenging System in Situ
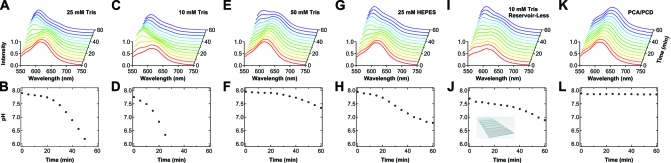
Here we first characterize the acidification behavior of a pH 8.0 imaging buffer used in our single-molecule FRET experiments, which contains 25 mM Tris, 5 mM MgCl2, 0.8% (w/v) dextrose, 2 mM Trolox, 1.0 mg/mL glucose oxidase, and 0.04 mg/mL catalase. After mixing, this solution was added to the flow chamber immediately, and the fluorescence emission spectra of SNARF-1 were obtained at 5 min intervals over the course of 1 h (Figure 3A). Right after time zero, SNARF-1’s spectrum mimics the one taken in the pH 8.0 standard solution shown in Figure 2A, indicating still a minimal change in pH by this time. As the oxygen scavenging system continues to remove O2, the amount of gluconic acid produced accumulates and SNARF-1’s dual emission becomes more appreciable with an increase in band A and a concomitant decrease in band B. After about 30 min of incubation in the chamber, the probe’s spectrum starts to be dominated by band A, and this trend continues through the end of the hour, suggesting that a significant amount of acidification has occurred during this time window. Note that toward the end of the hour the probe’s spectrum barely changes any further due to the diminished sensitivity of SNARF-1 in this low-pH region, far from its pKa; nevertheless, the acidification of the oxygen scavenging system certainly continues beyond this point.
Figure 3.
(Top) Fluorescence emission spectra of SNARF-1 taken from time zero (red) to 1 h (purple) at 5 min intervals in an imaging buffer with a starting pH of 8.0 inside a flow chamber. These buffers contain (A) 25 mM Tris, (C, I) 10 mM Tris, (E) 50 mM Tris, and (G) 25 mM HEPES as well as 5 mM MgCl2, 0.8% (w/v) dextrose, 2 mM Trolox, 1.0 mg/mL glucose oxidase, and 0.04 mg/mL catalase. For panel K, the buffer contains 25 mM Tris, 5 mM MgCl2, 2 mM Trolox, 2.5 mM PCA, and 50 nM PCD. The flow chamber shown in Figure 1A was used in all cases except panel I, where the one illustrated in the inset to panel J was used instead. (Bottom) Corresponding pH of the imaging buffer solution as a function of the incubation time in the flow chamber obtained by using the IB/IA−pH mapping curve shown in Figure 2B.
To quantify these results, we calculated the intensity ratio IB/IA at each time point and converted it into the corresponding pH using the IB/IA−pH mapping curve in Figure 2B. As shown in Figure 3B, only a modest drop in pH is observed at the beginning of the incubation, when the pH of the imaging buffer still lies close to the pKa of Tris (8.1), where the buffer capacity of Tris is maximal. After this initial stage of about 30 min, the drop in buffer pH speeds up substantially, mostly due to the decreasing buffer capacity of Tris in this region (Figure 3B). At the end of the hour, the buffer pH even falls below 6.0 and outside the range detectable by SNARF-1. Repeated experiments under the same condition show good reproducibility of our assay (Figure S-1, Supporting Information) and provide the basis for statistical analysis. Clearly, if a single-molecule experiment requires incubation over tens of minutes under the conditions described here, a serious pH change will occur and can significantly affect the experimental result.
Single-molecule experiments have been carried out in our laboratory using imaging buffers containing glucose oxidase and catalase at the concentrations specified above,6,26 while the buffering agent, buffer strength, initial pH, flow chamber design, etc. are often varied to satisfy the needs of a specific experiment. Each of these parameters can have an impact on the rate of acidification and thus are surveyed below. To provide a common platform for comparison, we define the characteristic acidification time, t0.5pH, as the time it takes the solution pH to drop by half a unit due to the oxygen scavenging system under a specific condition. From a practical point of view, this characteristic time also provides a measure of the useful experimental time window, within which the pH of the imaging buffer is considered to be stable for certain biological systems. For the imaging buffer described here, t0.5pH = 27 ± 3 min as determined from multiple independent experiments performed under the same conditions.
Survey of Common Experimental Variables
Using our in situ assay, we then examined the acidification of the oxygen scavenging system under several other experimental conditions that are relevant to single-molecule studies being performed by us and other researchers.
As one of the parameters most commonly varied in single-molecule imaging buffers, the buffer strength has a large impact on the stability of the buffer pH and, thus, the acidification kinetics. We first characterized the performance of a 10 mM Tris buffer which has been employed in many single-molecule studies previously.11,27 The buffer we studied here contains the same components as the 25 mM Tris buffer above. As can be seen in Figure 3C, the acidification of the 10 mM Tris buffer is much faster than that of the 25 mM Tris buffer. Even 15 min of incubation in the flow chamber leads to a drastic change in the probe’s spectrum, and by 30 min, the solution pH has dropped to below 6.0 (Figure 3D). Consistent with the faster acidification seen here, the characteristic time t0.5pH shortens to 12 ± 3 min, giving researchers a fairly narrow time window with a stable pH. Conversely, increasing the buffer strength to 50 mM Tris while keeping the other components the same slows the probe’s spectral progression dramatically (Figure 3E), and t0.5pH lengthens to 51 ± 2 min accordingly. These results clearly demonstrate the effect of the buffer strength on the imaging buffer’s acidification behavior. This effect needs serious attention especially for those single-molecule studies where a low-strength imaging buffer is in use.
In addition to the buffer strength, the type of buffering agent, in particular its pKa, and the desired solution pH are often varied in practice and may also play roles in modulating the acidification kinetics. As an example, we studied the acidification behavior of a 25 mM HEPES buffer with the same components and starting pH as the 25 mM Tris buffer used in Figure 3A. As shown in Figure 3G, in the beginning the probe’s spectrum evolves at a rate comparable to that of the Tris buffer, but during the second 30 min the slope of the pH-vs-time curve appears to be less steep for HEPES than Tris (Figure 3H). This interesting difference can be explained by the different pKa of HEPES (7.5) and, thus, its better buffer capacity in the pH range just below its pKa, while it is further away from Tris’s pKa (8.1). In general, it is expected that a buffering agent with a pKa close to or even slightly lower than the desired pH is more advantageous in maintaining the buffer pH than the one with a pKa greater than the desired pH, for example, a pH 7.5 Tris buffer.(6) It is worth noting that in practice single-molecule studies are sometimes carried out at temperatures other than room temperature and the pKa of some buffering agents, such as Tris,(28) changes as a function of temperature. Still, matching the desired buffer pH for the experiment at a specific temperature to the buffering agent’s pKa at that temperature will be beneficial in maintaining the buffer pH stability. In addition, a quantitative analysis can be made with the assay described in this study to characterize the acidification process at any temperature of interest.
In single-molecule experiments, many different chamber designs have been employed to satisfy specific practical needs such as optics positioning, sample flow, solution mixing, etc., and this difference can potentially affect the acidification behavior of the oxygen scavenging system therein. As an example, we subjected a simplified version of the flow chamber used in our laboratory to our in situ assay. As illustrated in the inset to Figure 3J, this type of chamber omits the attachment of the sample reservoir and tubing shown in Figure 1A. With the same 10 mM Tris buffer as used in Figure 3C,D, the probe’s spectral progression and pH change (Figure 3I,J) are much slower in this chamber than in the other one mentioned above, reflecting a difference in the oxygen exchange rate that is allowed by the corresponding port dimensions. Consistent with these observations, the characteristic acidification time t0.5pH is now lengthened to 18 ± 6 min (cf. 12 ± 3 min for the case shown in Figure 3C,D), providing a slightly wider time window with this reservoirless chamber. The chamber designs used by other researchers can in principle exhibit even larger differences in acidification kinetics from the ones examined here; thus, a case-by-case evaluation is necessary.
Lastly, it is noteworthy that besides the oxygen scavenging system we have characterized thus far, an alternative system containing PCD and PCA(24) has been adapted by some researchers recently. Using our assay, we examined the performance of a pH 8.0 buffer that contains 2.5 mM PCA, 50 nM PCD, and the same other components as in the buffer for Figure 3A minus dextrose, glucose oxidase, and catalase. As seen in Figure 3K, the probe’s spectrum remains intact throughout the entire time course monitored, and because of the lack of an appreciable pH change in this case (Figure 3L), the characteristic time t0.5pH is estimated to be much longer than 60 min if there is any pH change. Note that this excellent pH stability is likely to be intrinsic to this enzymatic system.(24) For our current system, we also lowered the concentration of glucose oxidase to see if a similar pH stability can be achieved without compromising the dye photostability (see the section below on photobleaching time analysis). With the amount of glucose oxidase reduced from 1.0 to 0.25 mg/mL while the other components were kept the same as in Figure 3A,B, a noticeable improvement in the buffer pH stability was observed (Figure S-2A,B, Supporting Information), although it still does not reproduce the same exceptional pH stability seen for the PCA/PCD system, which is essentially non-acidifying. On the other hand, it is noteworthy that, according to a recent study by Aitken et al,(24) in comparison to the current system, the PCA/PCD system provides a comparable to even modestly improved photostability for common fluorophores including Cy3, Cy5, and Alexa488. Given the excellent pH stability provided by the PCA/PCD system at the least, it is worth considering this alternative system in future single-molecule studies; however, its compatibility with a specific biological system is not warranted and needs to be assessed.
Single-Molecule Photobleaching Time
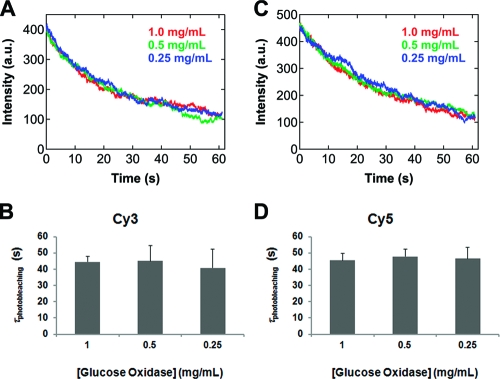
To test the effect of lowering the glucose oxidase concentration on the dye photostability in single-molecule experiments, we immobilized Cy3- or Cy5-labeled DNA molecules on a PEG-coated surface and measured the intensity traces of hundreds of isolated molecules in imaging buffers that contain different amounts of glucose oxidase. As expected, the average intensity of these molecules decays with time (Figure 4A,C), and these data can be fit to a single exponential with the lifetime named as the single-molecule photobleaching time, τphotobleaching. Interestingly, for both Cy3 and Cy5 τphotobleaching is found to be independent of the glucose oxidase concentration used in the imaging buffer in the range from 1.0 to 0.25 mg/mL (Figure 4B,D). Thus, the apparent dye photostability will not be compromised by using less glucose oxidase in this range at least for the two cyanine fluorophores examined here. Likewise, for the PCA/PCD enzymatic system mentioned above, Aitken et al. reported that lowering the enzyme concentration by a factor of 5 produces a similar photostability for Cy3, Cy5, and Alexa488, even though the initial rate of oxygen scavenging is 5 times slower.(24) Therefore, these results suggest that the rate of oxygen scavenging in both systems under the currently employed conditions exceeds the minimum requirements for dye photostability in single-molecule fluorescence experiments.(24) As a control, we also measured the intensity traces of Cy3 and Cy5 in the imaging buffer containing no glucose oxidase. For Cy3 in the absence of glucose oxidase, τphotobleaching is only 8% of that in the presence of 1.0 mg/mL glucose oxidase (data not shown), while for Cy5 the photobleaching time with no glucose oxidase was too short to be measured accurately. Note that the concentration of 1.0 mg/mL for glucose oxidase has been adapted by our laboratory for some time mostly for practical reasons. The data presented here suggest that, when the absence of a batch-to-batch variation in enzymatic activity can be assumed for the glucose oxidase obtained from a commercial source, a low concentration of glucose oxidase such as 0.25 mg/mL may be used in the imaging buffer together with a photostability-enhancing agent, such as Trolox,12,23 for future single-molecule fluorescence experiments.
Figure 4.
Average intensity of isolated (A) Cy3- and (C) Cy5-labeled DNA molecules as a function of time in imaging buffers containing 1.0 (red), 0.5 (green), and 0.25 (blue) mg/mL glucose oxidase. For each glucose oxidase concentration, only the data from one imaging area are shown for clarity. Single-molecule photobleaching time, τphotobleaching, for (B) Cy3 and (D) Cy5 at different glucose oxidase concentrations obtained by fitting the decay in average intensity to a single exponential. The error bars are the standard deviation of the fitted lifetime determined from five data sets.
Potential Single-Molecule Acidification Assays
In addition to the assay established in this study that works at the bulk level, analytical assays at the single-molecule level can also be designed for probing the acidification of the oxygen scavenging system in situ. For example, the pH-dependent dual emission of individual SNARF-1 molecules(25) may be used to report on the acidification process even locally. It should be possible to attach SNARF-1 functionalized with certain groups, such as a succinimidyl ester, to DNA molecules containing the corresponding modification, i.e., an amine. Such SNARF-1-labeled DNA molecules could then be anchored on the surface and subjected to single-molecule imaging. On the basis of the study by Brasselet and Moerner(25) on the pH-dependent fluorescence characteristics of individual SNARF-1 molecules, we believe that the brightness and photostability of this probe should be sufficient for a single-molecule acidification assay in situ. Besides sensing the pH with a ratiometric dual-emission probe, it is possible to utilize the pH-dependent properties of certain single-molecule probes as well. Vogelsang et al.(29) recently reported that the pH affects the blinking kinetics, in particular the dwell time of the “off” state, of an oxazine dye, ATTO655. Thus, this property can be used to sense the acidification of the oxygen scavenging system in situ, although such an assay may suffer from its limited sensitivity, which is on the order of 0.2 pH unit.(29)
Conclusions
In this paper, we established a fluorescence-based assay for quantitatively assessing the acidification of oxygen scavenging systems in single-molecule fluorescence studies. By using a ratiometric, dual-emission probe, we measured the acidification profile in situ in real time under a variety of experimental conditions relevant to single-molecule studies today that were inaccessible to conventional methods. Although performed on a spectrofluorometer here, our assay can be conveniently adapted to other instrument platforms, for example, fluorescence microscopes commonly found in single-molecule fluorescence laboratories. With this assay we surveyed the effects of several common experimental variables on the acidification kinetics and found that increasing the buffer strength, choosing a buffering agent with a pKa close to the desired pH, optimizing the chamber design, lowering the concentration of glucose oxidase to a certain extent, or employing the PCA/PCD system can improve the buffer pH stability and extend the effective observation time window in single-molecule experiments. In practice, the acidification profile in other researchers’ experiments can be collectively affected by the combination of all these critical parameters plus those not surveyed in this work, such as the ionic strength, glucose concentration, catalase concentration, temperature, β-mercaptoethanol concentration,(12) etc. A quantitative analysis can be made with our assay to precisely pinpoint the acidification process therein, making it valuable in the design of many future single-molecule studies of important biological systems. Finally, in addition to enzymatic oxygen scavenging, physical means of deoxygenation have been shown to effectively remove molecular oxygen as well, e.g., via buffer sonication and purging the sample reservoir with argon gas(30) or via ventilating a microfluidic device with nitrogen.(31) These methods can be promising alternatives to the current enzymatic systems, and their physical nature clearly makes them free of potential concerns that may be associated with the enzymatic system, including acidification and compatibility with the biological system under investigation. Therefore, they are worth considering in future single-molecule studies as well.
Acknowledgments
This work was supported by the NIH (Grant GM65367). We are grateful to Hyunjoon Kong’s laboratory for generously providing us access to their fluorometer and Paul Selvin’s laboratory for giving us PCA/PCD samples as a gift.
Supporting Information Available
Figures to illustrate the good reproducibility of our in situ assay and the improvement in buffer pH stability by lowering the glucose oxidase concentration. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Eid J.; Fehr A.; Gray J.; Luong K.; Lyle J.; Otto G.; Peluso P.; Rank D.; Baybayan P.; Bettman B.; Bibillo A.; Bjornson K.; Chaudhuri B.; Christians F.; Cicero R.; Clark S.; Dalal R.; Dewinter A.; Dixon J.; Foquet M.; Gaertner A.; Hardenbol P.; Heiner C.; Hester K.; Holden D.; Kearns G.; Kong X.; Kuse R.; Lacroix Y.; Lin S.; Lundquist P.; Ma C.; Marks P.; Maxham M.; Murphy D.; Park I.; Pham T.; Phillips M.; Roy J.; Sebra R.; Shen G.; Sorenson J.; Tomaney A.; Travers K.; Trulson M.; Vieceli J.; Wegener J.; Wu D.; Yang A.; Zaccarin D.; Zhao P.; Zhong F.; Korlach J.; Turner S. Science 2009, 323, 133–138. [DOI] [PubMed] [Google Scholar]
- Schwartz J. J.; Quake S. R. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 20294–20299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis A. N.; Margeat E.; Ho S. O.; Kortkhonjia E.; Weiss S.; Ebright R. H. Science 2006, 314, 1144–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbondanzieri E. A.; Bokinsky G.; Rausch J. W.; Zhang J. X.; Le Grice S. F.; Zhuang X. Nature 2008, 453, 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Abbondanzieri E. A.; Rausch J. W.; Le Grice S. F.; Zhuang X. Science 2008, 322, 1092–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S.; Cui S.; Cornish P. V.; Kirchhofer A.; Gack M. U.; Jung J. U.; Hopfner K. P.; Ha T. Science 2009, 323, 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard S. C.; Kim H. D.; Gonzalez R. L. Jr.; Puglisi J. D.; Chu S. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 12893–12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard S. C.; Gonzalez R. L.; Kim H. D.; Chu S.; Puglisi J. D. Nat. Struct. Mol. Biol. 2004, 11, 1008–1014. [DOI] [PubMed] [Google Scholar]
- Betzig E.; Patterson G. H.; Sougrat R.; Lindwasser O. W.; Olenych S.; Bonifacino J. S.; Davidson M. W.; Lippincott-Schwartz J.; Hess H. F. Science 2006, 313, 1642–1645. [DOI] [PubMed] [Google Scholar]
- Rust M. J.; Bates M.; Zhuang X. Nat. Methods 2006, 3, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohng S.; Zhou R.; Nahas M. K.; Yu J.; Schulten K.; Lilley D. M.; Ha T. Science 2007, 318, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnik I.; McKinney S. A.; Ha T. Nat. Methods 2006, 3, 891–893. [DOI] [PubMed] [Google Scholar]
- Vogelsang J.; Kasper R.; Steinhauer C.; Person B.; Heilemann M.; Sauer M.; Tinnefeld P. Angew. Chem., Int. Ed. 2008, 47, 5465–5469. [DOI] [PubMed] [Google Scholar]
- Roy R.; Hohng S.; Ha T. Nat. Methods 2008, 5, 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesch R. E.; Benesch R. Science 1953, 118, 447–448. [DOI] [PubMed] [Google Scholar]
- Meyer A.; Hoffler S.; Fischer K. J. Chromatogr., A 2007, 1170, 62–72. [DOI] [PubMed] [Google Scholar]
- Whitaker J. E.; Haugland R. P.; Prendergast F. G. Anal. Biochem. 1991, 194, 330–344. [DOI] [PubMed] [Google Scholar]
- Hanson G. T.; McAnaney T. B.; Park E. S.; Rendell M. E.; Yarbrough D. K.; Chu S.; Xi L.; Boxer S. G.; Montrose M. H.; Remington S. J. Biochemistry 2002, 41, 15477–15488. [DOI] [PubMed] [Google Scholar]
- McAnaney T. B.; Park E. S.; Hanson G. T.; Remington S. J.; Boxer S. G. Biochemistry 2002, 41, 15489–15494. [DOI] [PubMed] [Google Scholar]
- McAnaney T. B.; Shi X.; Abbyad P.; Jung H.; Remington S. J.; Boxer S. G. Biochemistry 2005, 44, 8701–8711. [DOI] [PubMed] [Google Scholar]
- Boldt F. M.; Heinze J.; Diez M.; Petersen J.; Borsch M. Anal. Chem. 2004, 76, 3473–3481. [DOI] [PubMed] [Google Scholar]
- Fu Y.; Collinson M. M.; Higgins D. A. J. Am. Chem. Soc. 2004, 126, 13838–13844. [DOI] [PubMed] [Google Scholar]
- Cordes T.; Vogelsang J.; Tinnefeld P. J. Am. Chem. Soc. 2009, 131, 5018–5019. [DOI] [PubMed] [Google Scholar]
- Aitken C. E.; Marshall R. A.; Puglisi J. D. Biophys. J. 2008, 94, 1826–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasselet S.; Moerner W. E. Single Mol. 2000, 1, 17–23. [Google Scholar]
- Myong S.; Bruno M. M.; Pyle A. M.; Ha T. Science 2007, 317, 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S.; Rasnik I.; Joo C.; Lohman T. M.; Ha T. Nature 2005, 437, 1321–1325. [DOI] [PubMed] [Google Scholar]
- Bates R. G.; Hetzer H. B. J. Phys. Chem. 1961, 65, 667–671. [Google Scholar]
- Vogelsang J.; Cordes T.; Tinnefeld P. Photochem. Photobiol. Sci. 2009, 8, 486–496. [DOI] [PubMed] [Google Scholar]
- Sabanayagam C. R.; Eid J. S.; Meller A. J. Chem. Phys. 2005, 123, 224708. [DOI] [PubMed] [Google Scholar]
- Lemke E. A.; Gambin Y.; Vandelinder V.; Brustad E. M.; Liu H. W.; Schultz P. G.; Groisman A.; Deniz A. A. J. Am. Chem. Soc. 2009, 131, 13610–13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.