Abstract
The Escherichia coli d-galactose and d-glucose receptor is an aqueous sugar-binding protein and the first component in the distinct chemosensory and transport pathways for these sugars. Activation of the receptor occurs when the sugar binds and induces a conformational change, which in turn enables docking to specific membrane proteins. Only the structure of the activated receptor containing bound d-glucose is known. To investigate the sugar-induced structural change, we have used 19F NMR to probe 12 sites widely distributed in the receptor molecule. Five sites are tryptophan positions probed by incorporation of 5-fluorotryptophan; the resulting 19F NMR resonances were assigned by site-directed mutagenesis. The other seven sites are phenylalanine positions probed by incorporation of 3-fluorophenylalanine. Sugar binding to the substrate binding cleft was observed to trigger a global structural change detected via 19F NMR frequency shifts at 10 of the 12 labeled sites. Two of the altered sites lie in the substrate binding cleft in van der Waals contact with the bound sugar molecule. The other eight altered sites, specifically two tryptophans and six phenylalanines distributed equally between the two receptor domains, are distant from the cleft and therefore experience allosteric structural changes upon sugar binding. The results are consistent with a model in which multiple secondary structural elements, known to extend between the substrate cleft and the protein surface, undergo shifts in their average positions upon sugar binding to the cleft. Such structural coupling provides a mechanism by which sugar binding to the substrate cleft can cause structural changes at one or more docking sites on the receptor surface.
While important structural features of sensory and signaling proteins have now been elucidated in a number of systems, much less is known about the structural changes these proteins undergo when they are activated. Such structural changes play a key functional role, since they in turn trigger changes in the interaction between an activated protein and its target proteins or nucleic acids. The present study illustrates a 19F NMR approach that can be used to probe structural changes within macromolecules in solution, especially those too large (>30 kDa) for structure determination by multidimensional NMR. This approach is designed to identify the structural regions within a macromolecule that are altered by a conformational change.
The focus of this study is the Escherichia coli aqueous receptor for d-galactose and d-glucose, a component of the bacterial chemosensory pathway for these sugars (Hazelbauer & Adler, 1971; Scholle et al., 1987). This pathway has been shown to illustrate many of the fundamental questions in biological signaling. In addition, due to the ease of molecular cloning and mutagenesis in E. coli, much has been learned about the structures of the protein components of the pathway, including primary structures, domain structures, transmembrane structures, and in certain cases crystal structures [reviewed by Stock et al. (1990), Bourret et al. (1989), Meister et al. (1989), Parkinson (1988), Koshland (1988), Eisenbach and Matsumura (1988), Taylor et al. (1988), Stewart and Dahlquist (1987), Macnab (1987), Quiocho et al. (1987), and Ames (1986)]. As a result the system is ideally suited for molecular studies of biological signaling mechanisms.
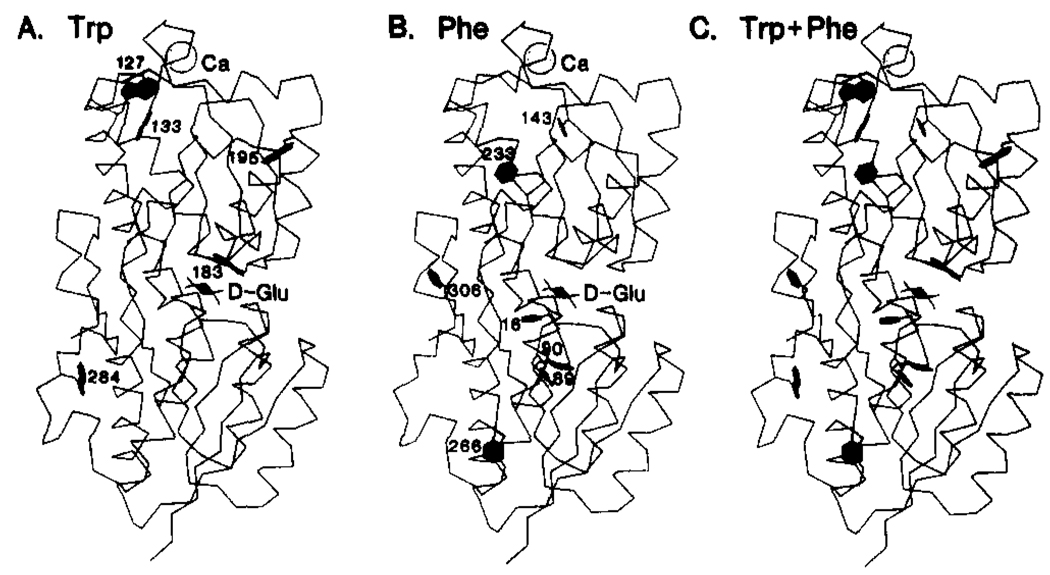
The E. coli aqueous receptor for d-galactose and d-glucose illustrates a common situation in which the activating structural change is not understood because structural information is available for only one conformer. Quiocho and co-workers have determined the crystal structure of the receptor containing bound d-glucose to 1.9-Å resolution (Vyas et al., 1987, 1988) and have previously solved the crystal structures of several structurally similar bacterial receptors (Quiocho et al., 1987). In no case, however, have the active (substrate site occupied) and inactive (substrate site empty) structures of the same receptor been solved; thus important elements of the structural change triggered by substrate binding remain uncharacterized. Also needed are solution studies of these structural changes. The d-galactose and d-glucose receptor is localized in the aqueous periplasmic compartment between the inner and outer bacterial membranes, where it is activated by the binding of a sugar molecule and then docks to one of two integral proteins of the inner membrane. Docking to the chemotaxis transducer Trg initiates the chemosensory pathway controlling the cellular swimming response to these sugars (Macnab, 1987); alternatively, docking to the Mgl transport complex initiates transport of the bound sugar into the cycloplasm (Ames, 1986). The two-domain structure of the receptor, illustrated in Figure 1 as the α-carbon backbone trace, is typical of the periplasmic aqueous receptors (or binding proteins) (Quiocho et al., 1987). The sugar binding cleft lies between the domains, which are connected by three strands of polypeptide. The domains are structurally homologous, each being constructed in a three-layered pattern consisting of two layers of α-helices separated by a layer of (β-sheet. In general the helices and βS-strands are oriented nearly perpendicular to the sugar binding cleft, which is composed largely of side chains from turns. The receptor also contains a Ca(II)-binding site similar in structure to the EF-hand class of eukaryotic Ca(II) sites (Vyas et al., 1987). The present paper focuses on the structural changes triggered by sugar binding to the cleft.
FIGURE 1.
Structure of the E. coli d-galactose and d-glucose receptor. Shown is the α-carbon backbone structure of the receptor (Vyas et al., 1987). The structure contains a d-glucose molecule bound in the substrate cleft between the two domains and Ca(II) ion in the metal binding site. Also shown are the five Trp and seven Phe residues that served as fluorine labeling sites in the present study.
A simple model has been presented for the structural change triggered by substrate binding to the periplasmic aqueous receptors (Mao et al., 1982; Miller et al., 1983). This model is similar to that described for a broad class of two-domain proteins that bind substrate in a cleft between domains (Bennett & Huber, 1984). The model proposes that the hinge connecting the two domains is flexible while the domains themselves are essentially rigid. The empty cleft is proposed to exist in a stable open conformation; then when substrate binds, the cleft closes and traps the substrate inside. Such a model requires that the structural changes triggered by substrate binding will be localized to the substrate cleft and the hinge region. Here we also consider an alternative model in which the domains are not assumed to be rigid, such that structural changes are transmitted from the hinge and cleft to other regions of the protein. For example, the α-helices and β-strands that terminate near the cleft could be shifted to new relative positions upon sugar binding, thereby generating structural changes that span the entire length of these secondary structure units. Similar structural changes have been observed in other cleft proteins (Lesk & Chothia, 1984). The distinction between these models is an important one for an understanding of the docking of the receptor to the membrane proteins it activates, since if the rigid-domain model is correct, then the surface structure changes that regulate docking must involve the hinge and cleft regions only. To distinguish these models, we have applied 19F NMR to the d-galactose and d-glucose receptor in order to ascertain the regions of the receptor structure altered by sugar binding.
19F NMR holds great promise as a probe of the structure and mechanism of large biomolecules, as illustrated by recent studies of large proteins by Ho and co-workers (Rule et al., 1987; Peersen et al., 1990). Fluorine is small and chemically inert, can replace hydrogen, can be introduced by biosynthetic incorporation of fluorinated amino acids, and its NMR frequency is highly sensitive to the local environment. F-Phe-, F-Trp-, and F-Tyr-labeled proteins have been successfully used in pioneering 19F NMR studies by several groups (Sykes et al., 1974; Pratt & Ho, 1975; Lu et al., 1976; Post et al., 1984; Wilson & Dahlquist, 1985; Wacks & Schachman, 1985; Peersen, et al., 1990). However, the potential of this approach to characterize functionally significant conformational changes in proteins has yet to be fully explored. The isomer of fluorotryptophan possessing a fluorine at position five of the indole ring yields a greater chemical shift dispersion than the other fluorinated amino acids we have tested in the d-galactose and d-glucose receptor (4F-Trp, 5F-Trp, 6F-Trp, and 3F-Phe); thus the present study employs primarily 5F-Trp. Each of the five Trp residues in the receptor are found in interesting regions of the structure (Figure 1A): one in the sugar binding cleft interacting directly with bound sugar, one near the hinge connecting the two domains, two within 10 Å of bound Ca(II), and one on an α-helix extending from the sugar binding cleft. Also utilized is the 3F-Phe probe, which enables investigation of the seven Phe positions located at positions generally complementary to the Trp locations (Figure 1B). By combining the information from the 5F-Trp and 3F-Phe probes, it is possible to examine nearly all regions of the receptor structure (Figure 1C).
The results of this approach indicate that sugar binding in the substrate cleft triggers widespread structural changes, altering even regions of the receptor distant from the substrate cleft and hinge. These structural changes are slow on the NMR time scale, indicating the presence of a substantial barrier between the sugar-occupied and sugar-empty conformers. Quite different results are observed for the binding of metals to the Ca(II) site, which, as described in the following paper, generates a highly localized structural change. Together these studies illustrate the unique advantages of 19F NMR as a qualitative probe of both allosteric and nonallosteric structural changes in macromolecules.
EXPERIMENTAL PROCEDURES
Cloning
The wild-type protein was expressed from plasmid pVB2, generously provided by Dr. W. Boos, Universität Konstanz (Scholle et al., 1987). For the construction and expression of mutant proteins, plasmid pSF5 was generated by standard techniques (Maniatis et al., 1982). This plasmid contained the E. coli natural promoter and the gene for the d-galactose and d-galactose receptor in the phagemid plasmid pTZ18U (Bio-Rad Laboratories). Briefly, a fragment containing the promotor and the gene was constructed as previously described (Snyder et al., 1990) and then cloned into the PstI/SacI sites of the pTZ18U polylinker. The resulting pSF5 plasmid was used to generate single-stranded template DNA for oligonucleotide-directed mutagenesis.
Mutagenesis
The 19F NMR resonances of the 5F-Trp-labeled receptor were assigned to specific Trp residues in the primary structure by site-directed mutagenesis. Mutagenic oligonucleotides 19 bases in length, designed to change each Trp codon to Tyr or Phe, were synthesized on an Applied Biosystems DNA synthesizer. These oligo’s were used to construct six mutant genes by using a single-stranded pSF5 template DNA and the uracil selection method of Kunkel (1988). The mutagenesis reagents and protocol were from Bio-Rad Laboratories. Mutant plasmids were identified by dideoxy DNA sequencing of the plasmid DNA with a primer oligo end labeled with 32P (Sanger et al., 1977). Sequenase sequencing reagents and protocol were from the United States Biochemical Corp. The mutant genes obtained were W127Y, W133Y, W183Y, W195Y, and W284Y; W127F was also generated when it was discovered that the W127Y gene did not express a sufficient amount of protein for NMR, perhaps due to altered folding.
Isolation of the Fluorine-Labeled Receptor
Large quantities of 5F-Trp-labeled wild-type receptor were obtained by expression of plasmid pVB2 in the strain E. coli W3110 trp A33, a tryptophan auxotroph [Drapeau et al. (1968), provided by Dr. C. Yanofsky, Stanford University]. Media for the growth of the 5F-Trp-labeled wild-type receptor contained 65 µg/mL 5F-tryptophan (Sigma), 16 µg/mL tryptophan (Sigma), M9 salts (Miller, 1972) with twice the standard phosphate salts, 2% (w/v) casamino acids (Difco Laboratories), 1% (v/v) glycerol as a carbon source, 20 µg/mL methionine, 10 µg/mL thiamine, 0.5 mM d-(+)-fucose [an inducer of the receptor promoter (Scholle et al., 1987)], and 50 µg/mL ampicillin. (In one case the media contained 32 µg/mL 5F-Trp instead of 65 µg/mL in order to vary the extent of fluorine incorporation; see Figure 2C). Six 400-mL cultures were grown with vigorous aeration at 37 °C for 24 h. After harvesting the cells, standard procedures were used to lyse the E. coli outer membrane to release the contents of the periplasm, including the receptor (Kellerman & Ferenci, 1982). PMSF was added to 0.5 mM, and the resulting supernatant was concentrated to 5 mL by ultrafiltration (Amicon apparatus and YM10 membranes) and then dialyzed against a series of buffers, each for 6–12 h at 4 °C in a volume at least 50-fold that of the sample. Unfolding buffer (two changes) denatured the protein and released bound substrate and contained (Miller et al., 1980) 3.0 M guanidine HCl, 100 mM KCl, 20 mM EDTA, 10 mM Tris, pH 7.1 with HCl, 0.5 mM phenyl-methanesulfonylfluoride. Refolding buffer (four changes) renatured the protein in the presence of Ca(II) and contained 100 mM KCl, 10 mM Tris, pH 7.1 with HCl, and 0.5 mM CaCl2. Dialyzed samples were further concentrated to yield a receptor concentration of 200–500 µ M for NMR.
FIGURE 2.
19F NMR spectra of 3F-Phe- and 5F-Trp-labeled receptors. Indicated for each spectrum are the extent of incorporation of the fluorinated amino acid and all the known assignments (from Figures 3 and 4). No additional resonances were observed when the spectral window was shifted 10 ppm upfield or downfield. All samples contained 100 mM KCl, 10 mM Tris, pH 7.1, 10% D2O, 0.5 mM CaCl2, and 1.0 mM d-glucose; spectra were recorded at 470 MHz and 25 °C. (A) The receptor was labeled with 20 ± 10% 3F-Phe at the phenylalanine positions. Included was 75 µM free 5F-Trp as an internal standard. A total of 5500 scans were acquired. (B) The receptor was labeled with 65 ± 10% 5F-Trp at the tryptophan positions. The internal standard was 75 µM free 3F-Phe. A total of 4000 scans were acquired. (C) Same as (B) except the receptor was labeled with 40 ± 10% 5F-Trp. A total of 12 000 scans were acquired.
5F-Trp-labeled mutant receptors were expressed from the appropriate pSF5-derived plasmid in the same strain. Production of large quantities of fluorine-labeled mutant receptors required a richer media than that used for the wild-type and contained 200 µg/mL 5F-Trp, 1.5% casamino acids, 0.25% bactotryptone (Difco Laboratories; provides ~87 µg/mL tryptophan), 140 mM NaCl, 75 µg/mL sodium ampicillin, and 5 mM each of MgCl2, CaCl2, and SrCl2. The latter three metals were included to maximize occupancy of the Ca(II) site, which was found to be required for the correct folding of certain mutant receptors. Cells were grown and mutant receptors were isolated, dialyzed, and concentrated as described above. Interestingly, the media used for 5F-Trp labeling of the wild-type receptor (preceding paragraph) and the mutant receptors (this paragraph) yielded different populations for the two structural states observed for the 5F-Trp127 and 5F-Trp133 residues (compare Figures 2 and 5 obtained with the former media and Figures 3 and 4 obtained with the latter media). The difference is not related to different extents of 5F-Trp incorporation, because varying the 5F-Trp to Trp ratio in either media has no effect on the relative populations (Figure 2B,C; also data not shown). We have identified the bactotryptone in the mutant media as the source of this difference but do not yet know the molecular basis of this effect.
FIGURE 5.
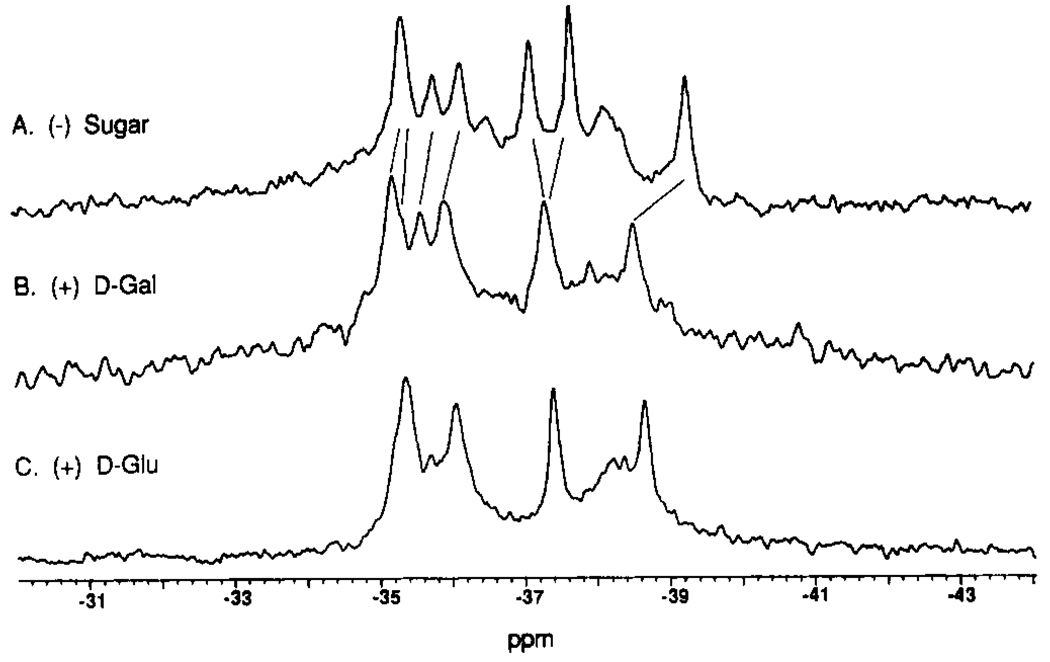
Effect of sugar binding on the 19F NMR resonances of the 5F-Trp-labeled receptor. Shown are spectra for the 5F-Trp-labeled receptor in the presence of different sugar substrates. Indicated by lines between spectra are the frequency shifts caused by sugar binding. All samples contained 100 mM KCl, 10 mM Tris, pH 7.1, 10% D2O, 0.5 mM CaCl2, and 75 µM 3F-Phe as an internal frequency reference; spectra were recorded at 470 MHz and 25 °C. The samples also contained (A) no sugar, (B) 1.0 mM d-galactose, (C) 1.0 mM d-glucose, and (D) a substoichiometric mixture of d-galactose and d-glucose, each 0.25 mole equivalents per receptor.
FIGURE 3.
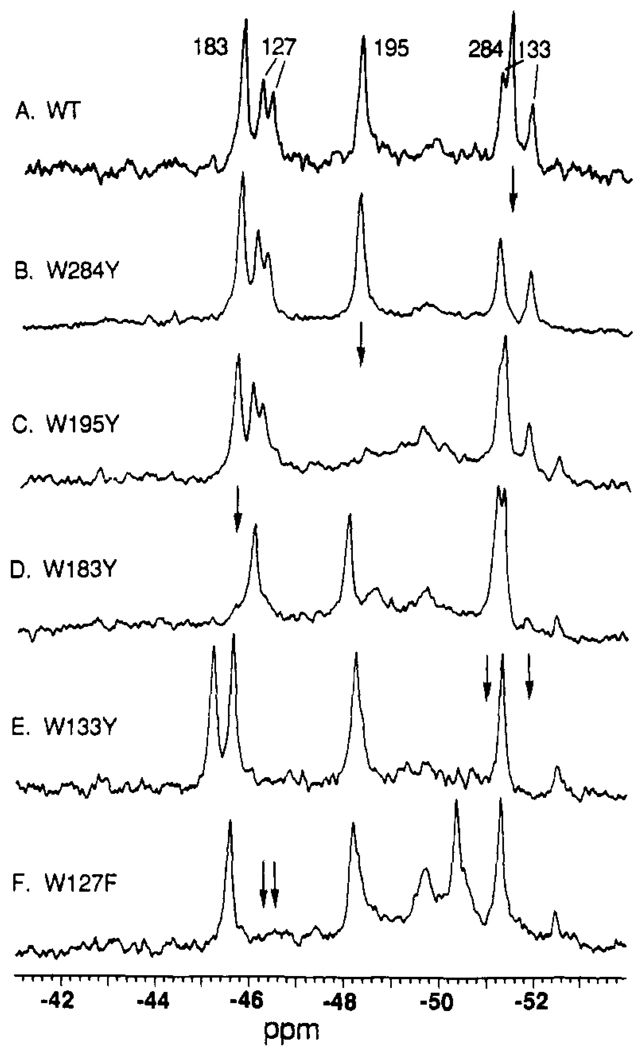
Assignment of 5F-Trp 19F NMR resonances by site-directed mutagenesis: the empty sugar cleft conformer. Shown are spectra for the wild-type and indicated mutant receptors, each labeled with 5F-Trp. The positions of resonances deleted by mutation are indicated by arrows. All samples contained 100 mM KCl, 10 mM Tris, pH 7.1, 10% D2O, 0.5 mM CaC12, and 75 µM 3F-Phe as an internal frequency reference; spectra were recorded at 470 MHz and 25 °C.
FIGURE 4.
Assignment of 5F-Trp 19F NMR resonances by site-directed mutagenesis: the bound d-glucose conformer. Conditions are as in Figure 3, with the addition of 1.0 mM d-glucose to each receptor.
3F-Phe-labeled wild-type receptors were expressed from plasmid pVB2 in the strain E. coli KA197, a phenylalanine auxotroph [Yale E. coli Genetic Stock Center, strain no. 5243; Hoekstra et al. (1974)]. The expression media contained 190 µg/mL 3F-Phe (Sigma), 1.2% (v/v) glycerol, 0.1% bactotryptone (provides ~35 µg/mL phenylalanine), 0.05% yeast extract (provides an undetermined amount of phenylalanine), 86 mM NaCl, 10 mM CaCl2, 3 µg/mL methionine, 1.5 µg/mL thiamine, and 100 µg/mL ampicillin. Growth of cells, as well as isolation, dialysis, and concentration of the receptor were performed as described above.
The purity of each isolated receptor preparation was quantitated by SDS–polyacrylamide gel electrophoresis on 12% Laemmli gels (Laemmli, 1980) stained with Coomassie Blue R-250 (Bio-Rad Laboratories). Typical purities were >90%, as verified by the lack of contaminating resonances in 19F NMR spectra.
Measurement of Sugar Binding, Metal Dissociation, and Structural Stability
d-Galactose binding to the sugar site was monitored by the resulting increase in intrinsic tryptophan fluorescence according to a published procedure (Boos et al., 1972). Sugar was titrated into each sample, and the resulting fluorescence changes were normalized to an identical reference sample titrated with sugar-free buffer. Fluorescence measurements used excitation at 285 nm and emission at 338 nm with 5-nm bandwidths and a 3-s response time on a Perkin-Elmer MPF 43A fluorometer. Sample components are given in Table I.
Table I.
Effects of Fluorine Labeling on Sugar and Metal Binding Sitesa
| Model I | Model II | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| label | incorp. | no. perturbing fluorines |
fract. pop. | KDd-Gal (µM) | τoff Tb(III) (min) |
no. perturbing fluorines |
fract. pop. | KDd-Gal (µM) | τoff Tb(III) (min) |
| 5F-Trp | L = 0.65 | 1 | 0.65 | 1.2 ± 0.4 | 2 ± 1 | 1–5 | 0.995 | 0.9 ± 0.2 | 3 ± 1 |
| 0 | 0.35 | 0.4 ± 0.1 | 5 ± 1 | 0 | 0.005 | 0.4 ± 0.1 | 5 ± 1 | ||
| 3F-Phe | L = 0.20 | 1 | 0.20 | 1.6 ± 0.2 | 7 ± 1 | 1–7 | 0.79 | 0.6 ± 0.1 | 6 ± 1 |
| 0 | 0.80 | 0.4 ± 0.1 | 5 ± 1 | 0 | 0.21 | 0.4 ± 0.1 | 5 ± 1 | ||
In order to estimate the perturbation due to fluorine in a heterogeneously fluorinated population, the following two models were best fit to d-galactose binding data and Tb(IIl) dissociation kinetics by nonlinear least-squares analysis. Each model assumes two subpopulations of binding sites: one perturbed by fluorine labeling and the other unperturbed, equivalent to the native site. The characteristics of the unperturbed site were determined by direct measurement with the unlabeled receptor. Model I. The perturbation is localized to a single labeling position in the molecule; the other labeling positions yield a negligible perturbation. In this case the perturbed fractional population is F′ = L or simply the extent of fluorine incorporation at the key labeling position, while the unperturbed fractional population is F =1−L. This model gives the maximum perturbation due to labeling at a single position. Model II. All molecules containing one or more fluorines are perturbed to the same extent. In this case the perturbed fractional population is F′ = 1 − (1 − L)n, where n is number of labeling sites, while the unperturbed fractional population is F = (1 − L)n d-Galactose binding. The equation y = F′[x(x + KD)−1] + F[x(x + 0.4)−1] was best fit to the binding data, where x is the free sugar concentration, KD is the dissociation constant of the perturbed site, and the dissociation constant of the unperturbed (native) site is 0.4 µM. The sample contained 150 nM receptor, 150 mM NaCl, 10 mM Tris, pH 7.1, and 100 µM CaCl2, at 25 °C. Tb(III) dissociation kinetics. The equation y = F′[exp(−t/τoff)] + F[exp(−t/5.0)] was best fit to the dissociation time course, where t is the time after initiating dissociation, τoff is the residence time of Tb(III) in the perturbed site, and the residence time of the unperturbed site is 5.0 min. The sample contained 10 µM receptor, 100 mM KCl, 10 mM PIPES, pH 6.0, and 1 mM d-glucose, at 25 °C. At t = 0 EDTA was added to obtain a 1.0 mM concentration.
The Tb(III) off rate from the metal site was measured by monitoring the decrease in bound Tb(III) fluorescence following the addition of excess (1 mM) EDTA. When an excitation wavelength of 295 nm was used, bound Tb(III) was selectively excited by fluorescence energy transfer from the nearby trp127 and −133 residues as previously described (Snyder et al., 1990); bound Tb(III) emission was measured at 543 nm with a 0.3-s response time and 5-nm bandwidths. Sample components are given in Table I.
The denaturation free energy of the receptor was measured by preparing a series of identical receptor samples containing 0–8 M urea; the samles were incubated at 25 °C for 48 h, when equilibrium was reached. Subsequently, the decrease in intrinsic tryptophan fluorescence due to unfolding was measured with an excitation wavelength of 278 nm and an emission wavelength of 335 nm with 5-nm bandwidths and a 0.3-s response time. The resulting fluorescence data were corrected for an increase in the fluorescence quantum yield caused by addition of urea; the maximum correction at 8 M urea was 30%. Free energies of unfolding were calculated by using a two-state folding model for unfolding at urea concentrations near the midpoint of the titration, and they were used to extrapolate the free energy of unfolding at zero urea concentration as previously described (Pace, 1986). Samples were prepared in medium containing 2 µM receptor in 100 mM KCl, 10 mM Tris pH 7.1, 0.5 mM CaCl2, and 1 mM d-glucose, at 25 °C.
Each of the described substrate-binding and stability assays utilized tryptophan fluorescence; thus it was important to consider potential differences between Trp and 5F-Trp. The excitation and emission parameters of these chromophores were found to be nearly identical (see Results). Heterogeneity due to energy transfer from Tyr to Trp or 5F-Trp was judged to be negligible by the following analysis. The known structure of the receptor indicates that five of the seven Tyr residues lie ≥ 15 Å from the nearest Trp and are thus expected to have transfer efficiencies of <5% (assuming a typical R0 of ~9 Å). Furthermore, Tyr excitation is 4.2-fold weaker than Trp or 5F-Trp excitation (Cantor & Schimmel, 1980). It follows that >90% of the Trp or 5F-Trp fluorescence stems from direct excitation.
NMR Measurements
19F NMR spectra were obtained at 470 MHz on a Varian VXR 500 spectrometer with a 5-mm 1H/19F probe or at 282 MHz on a Varian VXR 300 spectrometer with a 10-mm 1H/19F dual-coil probe (Z-Spec Probes, Inc.). Samples containing 100–500 µM receptor were prepared by adding 10% D2O (v/v) as the lock solvent to the receptor in the final dialysis buffer. Also added was an internal frequency and integration standard (75 µM 3F-Phe for 5F-Trp-labeled receptor or 75 µM 5F-Trp for 3F-Phe-labeled receptor), which was referenced to its known chemical shift relative to TFA at 0 ppm (−38 ppm or −49.5 ppm, respectively), thereby enabling direct comparison of chemical shifts in different spectra. Standard undecoupled spectral parameters were as follows: 12000-Hz spectral width, 16K data points, 60° pulse width, 0.68-s acquisition time, 1.0-s relaxation delay, 25-Hz line broadening, and temperature control at 25 °C. Receptor 19F NMR resonances were observed to have longitudinal relaxation times T1 of ~0.7 s, while the line widths at half-height, corrected for line broadening due to the FID weighting, were 35–65 Hz (Luck and Falke, unpublished results). No significant saturation of resonances was detected by the following test. Spectra were obtained with the pulse width decreased to 45° and the relaxation delay increased to 5.0 s. The resulting resonance intensities did not differ significantly from those of standard-parameter spectra within the error of integration (discussed below).
Determination of the Extent of Fluorine Incorporation
To quantitate the extent of fluorine incorporation, the 19F NMR spectrum of a given sample was obtained and then the concentration of incorporated 19F nuclei was quantitated by comparing the integral of a well-resolved 19F protein resonance (or the entire 3F-Phe protein spectrum) to the integral of an internal standard of known concentration. The concentration of the receptor was determined by routine assays including [3H] d-galactose binding measurements, A280, and total protein analysis (Falke group, unpublished results). Together, the 19F and receptor concentrations enabled the calculation of the extent of incorporation as the mole ratio of incorporated 19F per labeling site.
Protein Graphics and Distance Measurements
Receptor coordinates graciously supplied by Quiocho and co-workers (Vyas et al., 1987) were displayed by Biosym Technologies Insight graphics software on an Evans and Sutherland PS-300 color graphics system driven by a DEC VAXstation 3500.
Error Estimates
Integrations of individual 19F NMR resonances exhibited a relative error of ±20%, determined by measuring the ratio of two typical integrals from the same spectrum and then comparing this ratio to analogous ones from other spectra (n ≥ 4). The standard deviation inherent in measurements of 19F frequency shifts is ±0.1 ppm, determined by comparing identical measurements from multiple spectra (n ≥ 3). Parameters determined by curve fits are given as ±1 standard error, determined by nonlinear least-squares error analysis (n ≥ 6).
RESULTS
Features of 19F NMR
The usefulness of 19F NMR as a probe of structural changes stems from the unique properties of the 19F nucleus. This spin 1/2 nucleus exhibits high NMR sensitivity due to its large natural abundance (~ 100%) and large magnetic dipole (94% that of 1H). Furthermore 19F resonances are generally easily resolved and assigned because this nucleus possesses a chemical shift range up to two orders of magnitude larger than that of 1H and is generally absent in biomolecules unless introduced by design. The 19 NMR frequency exhibits a strong environmental dependence controlled largely by the fluorine lone-pair electrons, which provide a large paramagnetic term in the shielding formula (Carrington & McLachlan, 1979). These lone pairs directly sense their nonbonded environment via hydroen bonding, electrostatics, and van der Waals interactions. Aromatic fluorines are further sensitive to changes in the electron density of the adjacent π system (Brownlee & Taft, 1970). In short, multiple factors make the fluorine resonance frequency highly sensitive to the environment, so that it can be used as a correspondingly sensitive detector of local structural changes.
Also utilized in the present study were general relationships that provide information about the time scales of structural changes detected in NMR spectra (Wagner & Wuthrich, 1986). If two structures exhibit resonance frequencies υa and υb and are allowed to interconvert at the rate υic, the resulting lineshapes will reveal which of the following limits applies:
| (1a) |
| (1b) |
| (1c) |
In the slow interconversion limit, distinct resonances are observed at υa and υb; in the intermediate interconversion limit, the resonances are significantly broadened and may disappear; in the rapid interconversion limit, a single resonance is observed at a frequency that is a weighted average of υa and υb. The NMR time scale relevant to all three limits is simply τ = |υa − υb|−1.
Incorporation of 5F-Trp and 3F-Phe and Their Effects on Receptor Structure
Fluorinated amino acids were biosynthetically incorporated into the receptor by expression of the cloned receptor gene in a tryptophan or phenylalanine auxotroph strain of E. coli. The extent of incorporation was less than 100% because the unlabeled amino acid was required during biosynthesis for efficient protein production. Except where noted otherwise, a 4:1 ratio of 5F-Trp to Trp was used for biosynthesis, yielding 65 ± 10% incorporation of 5F-Trp per labeling site (see Experimental Procedures). A ~5:1 ratio of 3F-Phe to Phe yielded 20 ± 10% incorporation of 3F-Phe, less than that of 5F-Trp due to the ability of the biosynthetic machinery to selectively incorporate natural Phe in preference to the fluorinated analogue. To determine whether fluorine labeling caused important structural perturbations, we compared the binding of sugar and metal substrates to the native and fluorine-labeled proteins. In addition, the effects of fluorine labeling on overall receptor stability were determined.
Substrate binding is perhaps the most sensitive assay of perturbation. Incorporation of 5F-Trp or 3F-Phe is expected to have a relatively large effect on sugar binding, since the bound sugar backbone is sandwiched directly between Trp183 and Phe16, in van der Waals contact with both aromatic rings. An intrinsic Trp fluorescence increase that occurs upon binding of d-galactose (Boos et al., 1972) can be used to monitor sugar binding to both the unlabeled and the fluorine-labeled receptor because the fluorescence excitation and emission spectra of Trp and 5F-Trp are nearly identical [the 5F-Trp excitation and emission maxima are red-shifted 3 nm and 5 nm, respectively, and the quantum-yield change is <10%; (Luck and Falke, unpublished results)]. It was anticipated that the substrate affinities of the binding site population would be heterogeneous due to substoichiometric fluorine incorporation: given that the extent of labeling is L and the number of labeling sites is n, the n + 1 terms in the binomial distribution [(1−L) + L]n yield the fraction of receptors containing respectively 0, 1, 2,…,n fluorines. Additional heterogeneity is provided by the multiple ways that fluorines can be distributed among n distinguishable sites. It is not possible to fully account for this heterogeneity when analyzing sugar binding data; however, two extreme models enable estimation of the range of perturbations caused by fluorine labeling.
Each model assumes two subpopulations of binding sites: one perturbed by fluorine labeling and the other unperturbed, equivalent to the native site. In model I the perturbation is assumed to stem from a single labeling position; all other labeling positions yield no perturbation. In this case the fractional populations of the perturbed and unperturbed sites are L and 1 − L, respectively. This model lumps all the perturbation into a single site and thus places an upper limit on the perturbation due to a single fluorine. Model II assumes that all molecules containing one or more fluorines are perturbed to the same extent. In this case the fractional population of the fluorinated perturbed sites is 1 − (1 − L)n while the fractional population lacking fluorine is (1 − L)n. This model distributes the perturbation equally across the entire labeled population and thus yields the minimum perturbation capable of fitting the heterogeneous binding data. When models I and II are used for a best fit of the d-galactose binding data by nonlinear least-squares analysis, the following ranges of fluorine perturbation are obtained (details in Table I): (a) the 5F-Trp-labeled receptor has a 2.2- to 3.0-fold lower affinity for the sugar than does the unlabeled receptor and (b) the 3F-Phe-labeled receptor has a 1.5- to 4-fold lower affinity than does the unlabeled receptor. These results suggest that the majority of fluorine-labeled receptors are similar to the unlabeled receptor in sugar binding affinity. Not ruled out is the possibility of a minor labeled population that is significantly more perturbed than the indicated range, but 19F NMR confirms that the entire fluorine-labeled receptor population binds d-glucose and d-galactose (below, Figures 5 and 6).
FIGURE 6.
Effect of sugar binding on the 19F NMR resonances of the 3F-Phe-labeled receptor. Shown are spectra for the 3F-Phe-labeled receptor in the presence of different sugar substrates. The resonances have not been assigned, but the simplest model for the frequency shifts caused by sugar binding is indicated by the lines connecting resonances. All samples contained 100 mM KCl, 10 mM Tris, pH 7.1, 10% D2O, 0.5 mM CaCl2, and 75 µM 5F-Trp as an internal frequency reference; spectra were recorded at 470 MHz and 25 °C. The samples also contained (A) no sugar, (B) 1.0 mM d-galactose, and (C) 1.0 mM d-glucose.
The Ca(II) site contains no Trp or Phe ligands, but the Trp133, Trp127, and Phe143 residues each lie near the base of the Ca(II)-binding loop within 10 Å of the bound metal, thus metal binding was also examined for perturbations due to fluorine labeling. The off rate of Tb(III) bound in the site, which provides information regarding the kinetic stability of the substrate-occupied structure, was measured as the decay of the bound Tb(III) fluorescence after addition of EDTA. The resulting dissociation time courses were best fit by using the same two models described for sugar binding. The following ranges of fluorine perturbation were obtained: (a) Tb(III) dissociation from the 5F-Trp-labeled receptor was 1.6-to 2.5-fold faster than that from the native site, while (b) the 3F-Phe label had no significant effect on Tb(III) dissociation. These results suggest that fluorine labeling does not greatly perturb the Ca(II) site. Additional evidence is provided by 19F NMR experiments in which all detected fluorine-labeled receptors bind Ca(II) (accompanying article, Figures 1 and 3).
The effect of fluorine labeling on the overall structural stability of the receptor was investigated by using intrinsic tryptophan fluorescence to monitor urea denaturation. Assuming a simple equilibrium between one folded and one unfolded state, the free energy of unfolding was determined for different urea concentrations and then extrapolated to zero urea (Pace, 1986). The resulting free energy of unfolding of the native receptor is ΔG°UH2O = 5.9 ± 0.4 kcal/mol. Fluorine incorporation is observed to reduce the macroscopic stability of the receptor population by 1.2 ± 0.8 kcal/mol for the 5F-Trp label and by 0.8 ± 1.0 kcal/mol for the 3F-Phe label. At least part of this destabilization is likely to stem from stabilization of the unfolded state, due to the increased solubility of the fluorinated side chains in water: the free amino acids 5F-Trp and 3F-Phe are ~1.5-fold more soluble than Trp and Phe, respectively, in 25 mM NaPi, pH 7.0 (Luck and Falke, unpublished results). In addition, destabilization contains contributions from multiple fluorines; simply dividing the total destabilization by the number of 5 F-Trp-labeling positions, for example, yields a 0.24 kcal/mol destabilization per fluorine. This approximation suggests that the hydrogen-to-fluorine substitution has a significantly smaller impact on protein stability than more drastic side-chain alterations including mutagenesis and covalent modification (Bowie et al., 1990; Sandberg & Terwilliger, 1989). Together the substrate binding, substrate dissociation and denaturation results indicate that fluorine incorporation does not greatly disrupt the structure or the dynamics of the receptor.
19F NMR Spectra of the 5F-Trp- and 3F-Phe-Labeled Receptors
Figure 2 presents the 19F NMR spectrum of 5F-Trp- and 3F-Phe-labeled receptors containing bound d-glucose and Ca(II). All five of the 5F-Trp residues exhibit well-resolved resonances. Integration of the individual resonances indicates that 5 F-Trp is incorporated equally well at the five different Trp positions (within the relative error of ±20% for integration). Comparison of receptors containing 65 ± 10% and 40 ± 10% 5F-Trp shows that different levels of incorporation produce indistinguishable spectra (Figure 2), suggesting that the structure of the folded receptor is not a sensitive function of fluorine incorporation.
The 19F NMR spectrum of the 3F-Phe-labeled receptor also shows distinct resonances (Figure 2), although they are not as well resolved as those of the 5F-Trp receptor. In part, the greater complexity of this spectrum may arise from the tendency of some Phe positions to undergo slow 180° ring-flip motions (Wagner & Wüthrich, 1986), which would expose the fluorine at the 3 position to two environments, yielding a twin resonance or an exchange-broadened single resonance (eq 1).
Assignment of the 19F NMR Resonances to Specific 5 F-Trp Residues
The 5F-Trp-labeled receptor was chosen for resonance assignments due to its superior 19F NMR resolution and fluorine incorporation (Figure 2). Oligonucleotide-directed mutagenesis was used to replace each Trp residue with a Phe or Tyr residue, and the resulting mutants were labeled with 5F-Trp. All of the labeled mutant receptors retain functional sugar- and metal-binding sites (Figures 3 and 4 and data not shown), and their 19F NMR spectra are generally similar to that of the native 5F-Trp receptor, except that a single resonance or pair of resonances associated with the missing Trp position is deleted. The resulting resonance assignments are shown in Figure 3 for the sugar-empty, Ca(II)-bound conformer and in Figure 4 for the d-glucose-bound, Ca(II)-bound conformer.
All of the major resonances have been assigned by Trp substitution; four minor resonances with integrated intensities <20% of the assigned resonances were also detected. Two of these minor resonances were reproducible: one is seen 0.5 ppm upfield from the 5F-Trp183 resonance in the sugar-occupied cleft (Figures 2B,C and 5B,C), suggesting that this resonance stems from a minor conformer of Trp183, while the other occurs at −49.6 ppm, which is the same frequency observed for solvent-exposed 5F-Trp (Figures 2 and 5A,D), suggesting that this resonance is associated with unfolded or degraded protein [see the companion paper, Luck and Falke (1991)]. Two less reproducible resonances observed at −46.8 and −50.8 ppm (Figure 2 C and B, respectively) represent minor contaminating proteins. The subsequent discussion will focus only on the major resonances.
Structural Effects of Tryptophan Substitution
In addition to deleting major resonances, Trp substitution in some cases yielded (a) changes in the relative populations of multiple conformers, and (b) structural perturbations at distant 5F-Trp positions. Both effects provide evidence for direct or allosteric coupling between two structural locations. The former effect is illustrated by the region of the Ca(II) site, which yields double resonances for the 5F-Trp127 and 5F-Trp133 residues at the base of the Ca(II)-binding loop. These double resonances indicate the existence of two conformers in this region of the receptor that interconvert slowly on the NMR time scale of 7 ms (eq 1); the population ratio for the two conformers is ~60:40 (Figures 3 and 4). The distant W183Y substitution in the sugar binding cleft shifts this ratio to ~90:10. Even more dramatic are the effects caused by substitution of either 5F-Trp127 or 5F-Trp133, which collapses the two conformers to a unique or rapidly averaged structure giving rise to a single resonance. These observations suggest that the Trp127 and Trp133 residues interact strongly; in fact they are in van der Waals contact in the crystal structure (Vyas et al., 1987). A plausible explanation for the two conformers is proline cistrans isomerization, since Pro231 is in van der Waals contact with Trp133.
An example of a frequency shift generated by substitution of a distant 5F-Trp residue is provided by the W183Y mutant. This substitution in the sugar binding cleft significantly shifts the frequency of the 5F-Trp195 resonance by 0.3 ± 0.1 ppm, providing evidence that the relative position of the α-helix containing 5F-Trp195 is sensitive to structural changes in the cleft. Additional examples of frequency shifts are observed in Figures 3 and 4 for the W183Y, W133Y, and W127Y substitutions.
Structural Effects of Sugar Binding
The above assignments enable the five 5F-Trp resonances to be used as probes of structural changes at five known locations in the native receptor molecule. In order to ascertain the locations perturbed by sugar binding, Figure 5 compares the 19F NMR spectra of 5F-Trp-labeled receptor containing Ca(II) in the metal site and the indicated sugar (or no sugar) in the sugar cleft. Widespread structural effects are observed at the 5F-Trp positions upon sugar binding. The resonance of 5F-Trp183 in the sugar binding cleft shows the largest frequency change upon sugar binding (+3.8 ± 0.1 ppm for d-galactose; +2.8 ± 0.1 ppm for d-glucose) and possesses a different local environment when d-galactose is replaced by d-glucose. The structural difference between these two sugars is simply a switch of the -H and -OH substituents above and below the ring plane at the C4 position of the sugar backbone, and the frequency difference observed between the sugars indicates both the sensitivity of the technique to subtle structural changes and the direct structural interaction between the 5F-Trp183 residue and the bound sugar. In contrast 5F-Trp284 near the hinge exhibits the same large resonance frequency shift upon the binding of either sugar (−1.2 ± 0.1 ppm), suggesting that the conformational change in the hinge region is the same for different sugars. The resonance of 5F-Trp195 from an α-helix abutting the sugar cleft also undergoes a reproducible frequency shift that is essentially the same for d-galactose and d-glucose (−0.3 ± 0.1 ppm) as does 5F-Trp133 at the base of the Ca(II) site, which yields the smallest detectable shift (−0.1 ± 0.1 ppm). Together these observations indicate that structural changes occur not only in the sugar binding cleft and the hinge but in distant regions of the protein as well, where the structural changes triggered by the binding of different sugars are similar or identical.
The structural effects observed upon sugar binding to the 3F-Phe-labeled receptor yield an analogous picture. 19F NMR frequency changes and in some cases lineshape changes are observed for the 3F-Phe resonances when d-galactose binds to the receptor, as shown in Figure 6. Also indicated in Figure 6 is the simplest pattern of frequency shifts consistent with the observed spectra. This pattern suggests that all seven 3F-Phe residues detect structural changes upon d-galactose or d-glucose binding.
Kinetics of the Structural Changes Induced by Sugar Binding
Information regarding the kinetics of interconversion of the observed receptor conformers is revealed by the addition of a substoichiometric mixture of d-galactose and d-glucose to the 5F-Trp-labeled receptor. The resulting spectrum yields distinct 5F-Trp183 resonances for the d-galactose-occupied, the d-glucose-occupied, and the empty-site conformers, all coexisting in the same sample (Figure 5D). It follows that the rate of interconversion of the three conformers is slow relative to the differences in their 5F-Trp183 NMR frequencies (eq 1). The application of eq 1a to the d-galactose/empty site exchange process yields an interconversion time of τic = υic−1 ≫ 0.6 ms and to the d-glucose/empty site exchange process yields a time of τic ≫ 0.8 ms. These results are consistent with a simple interpretation of earlier rapid kinetics data that suggested bound sugar lifetimes of 220 ms for d-galactose and 710 ms for d-glucose (Miller et al., 1980).
DISCUSSION
The results illustrate the usefulness of 19F NMR as a probe of ligand-induced structural changes at multiple sites in a 36-kDa receptor protein. The environmental dependence of the 19F spin system is observed to make the 19F NMR frequency a sensitive detector of changes in local structure triggered by substrate binding. The substitution of fluorine for hydrogen is observed to yield relatively small effects on substrate binding equilibria, substrate dissociation, and the free energy of receptor unfolding, particularly when compared to mutagenesis and extrinsic labeling. It has previously been observed in other well-characterized fluorine-labeled proteins that perturbations due to fluorine are minor (Lu et al., 1976; Wacks & Schachman, 1985; Rule et al., 1987), suggesting that small and localized effects on structure are a general feature of fluorine incorporation.
The 19F NMR resonances of the fluorine-labeled d-galactose and d-glucose receptor reveal that the binding of d-galactose or d-glucose triggers a dramatic global change in the average solution structure of the receptor. This structural change extends from the 5F-Trp183 residue in van der Waals contact with the bound sugar to distant residues: the 5F-Trp284 residue is 13 Å from the bound sugar but lies in a surface loop nearby the interdomain hinge, while the 5F-Trp195 residue is located 15 Å away from the bound sugar, near the opposite end of a helix also containing 5F-Trp183. Similarly all seven 3F-Phe residues experience structural perturbations despite the fact that these residues are located at diverse sites, ranging from one in contact with the bound sugar to several at distant sites: 0, 6, 10, 12, 13, 15, and 18 Å from bound d-glucose, respectively. Together the three 5F-Trp and seven 3F-Phe positions that are perturbed by sugar binding represent a large fraction of the receptor structure and are equally distributed between the two domains (Figure 1). The smallest perturbations are observed for 5F-Trp127 and −133 at the base of the Ca(II) site. This region of the protein thus appears to be locked in a relatively rigid structure by Ca(II) binding.
The widespread allosteric effects of sugar binding to the substrate cleft provide strong suport for the involvement of secondary structural elements. Most of these elements originate or terminate in turns at the substrate cleft and extend outward to the receptor surface. Other elements involved in the interdomain hinge effectively provide a wall at the back of the cleft (Figure 1). In total, a group of 11 β-strands and 9 α-helices are intimately associated with the cleft. This architecture guarantees the transmission of structural changes from the substrate cleft to distant regions through spatial shifts of the secondary structure elements. Such structural coupling is capable of generating changes in surface structure on any region of the receptor surface, so that in principle sugar binding could regulate one or more surface docking sites. Consistent with this idea is the proposal that different docking surfaces are used in receptor docking to the transmembrane transducer and transport proteins (Kossman et al., 1988). Allosteric shifts of secondary structures have also been proposed in other protein systems containing substrate clefts (Lesk & Chothia, 1984) and may be a generally important feature in biological signaling systems.
The long residence time of bound sugar in the substrate cleft (0.2–0.7 s) (Miller et al., 1983), confirmed by the observation that the sugar-occupied and sugar-empty conformers are in slow exchange on the NMR time scale, is likely to play two important functional roles. First, the long lifetime of bound sugar enables the receptor to diffuse long distances in the activated conformation during its search for the appropriate membrane docking site. An interesting question is whether other types of docking proteins regulated by substrate binding will generally exhibit long bound-substrate lifetimes. Second, the long lifetime of the bound sugar together with the significant in vivo receptor concentration in the periplasmic compartment will maintain a high equilibrium sugar concentration in the periplasm and will slow the kinetics of diffusional sugar loss in the event that the cell encounters a decrease in the surrounding sugar concentration.
In contrast to the global structural change triggered by sugar binding to the substrate cleft, the 19F NMR study in the following article indicates that metal binding to the Ca(II) site triggers a highly localized structural change in the region containing the Ca(II) site. Thus the 19F NMR approach can be used to classify structural changes as global or local, as well as to characterize each class. A future goal is the use of 19F NMR to measure distances, thereby enabling quantitation of structural changes in sensory and signaling proteins. These techniques will be applied to other proteins in the bacterial chemosensory pathway, as well as cloned eukaryotic signaling proteins that can be expressed in bacteria or yeast, in order to further investigate the molecular mechanisms underlying biological signaling.
ACKNOWLEDGMENTS
We are indebted to the following co-workers: Claire Careaga, Eric Snyder, and Kit Swaggert for molecular biology and mutagenesis, Kay Thatcher and Stefan Voertler for Tb-(III) emission measurements, and Claire Careaga for helpful discussions. Also instrumental were Dr. Florante A. Quiocho and his co-workers for providing crystallographic coordinates. Thanks to Drs. Daniel Herschlag and Daniel Celander for comments on the manuscript.
Registry No. d-Galactose, 59-23-4; d-glucose, 50-99-7.
Footnotes
This investigation was supported by the National Institutes of Health Grant R01-GM40731 and by a University of Colorado CRCW Junior Faculty Development Award (J.J.F.).
REFERENCES
- Ames GF-L. Annu. Rev. Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- Bennett WS, Huber R. Crit. Rev. Biochem. 1984;15:291–384. doi: 10.3109/10409238409117796. [DOI] [PubMed] [Google Scholar]
- Boos W, Gordon AS, Hall RE, Price DH. J. Biol. Chem. 1972;247:917–924. [PubMed] [Google Scholar]
- Bourret R, Hess J, Borkovitch K, Simon MI. J. Biol. Chem. 1989;264:7085–7088. [PubMed] [Google Scholar]
- Bowie JU, Reidharr-Olson JF, Lim WA, Sauer RT. Science. 1990;247:1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- Brownlee RTC, Taft RW. J. Am. Chem. Soc. 1970;92:7007–7019. [Google Scholar]
- Cantor CR, Schimmel PR. Biophysical Chemistry. San Francisco: W. H. Freeman and Co.; 1980. pp. 443–454. [Google Scholar]
- Carrington A, McLachlan AD. Magnetic Resonance. New York: John Wiley and Sons, Inc.; 1979. pp. 54–63. [Google Scholar]
- Dettman HD, Wiener JH, Sykes BD. Biophys. J. 1982;37:243–251. doi: 10.1016/S0006-3495(82)84673-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau GR, Brammer WJ, Yanofsky C. J. Mol. Biol. 1968;35:357–367. doi: 10.1016/s0022-2836(68)80030-0. [DOI] [PubMed] [Google Scholar]
- Eisenbach M, Matsumura P. Bot. Acta. 1988;101:105–110. [Google Scholar]
- Hazelbauer GL, Adler J. Nat. New Biol. 1971;230:101–104. doi: 10.1038/newbio230101a0. [DOI] [PubMed] [Google Scholar]
- Kellerman OK, Ferenci T. Methods Enzymol. 1982;90:459–463. doi: 10.1016/s0076-6879(82)90171-9. [DOI] [PubMed] [Google Scholar]
- Koshland DE., Jr. Biochemistry. 1988;27:5829–5834. doi: 10.1021/bi00416a001. [DOI] [PubMed] [Google Scholar]
- Kossman M, Wolff C, Manson MD. J. Bacteriol. 1988;170:4516–4521. doi: 10.1128/jb.170.10.4516-4521.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lesk AM, Chothia C. J. Mol. Biol. 1984;174:175–191. doi: 10.1016/0022-2836(84)90371-1. [DOI] [PubMed] [Google Scholar]
- Lu P, Jarema MC, Mosar K, Daniel WE. Proc. Natl. Acad. Sci. U.S.A. 1976;73:3471–3475. doi: 10.1073/pnas.73.10.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck LA, Falke JJ. Biochemistry. 1991 (following paper in this issue). [Google Scholar]
- Macnab RM. In: E. coli and S. typhimurium. Neidhardt FL, editor. Vol. 1. Washington, D.C.: American Society for Microbiology; 1987. pp. 732–759. [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1982. [Google Scholar]
- Mao B, Pear MR, McCammon JA, Quiocho FA. J. Biol. Chem. 1982;257:1131–1133. [PubMed] [Google Scholar]
- Meister M, Caplan SR, Berg HC. Biophys. J. 1989;55:905–914. doi: 10.1016/S0006-3495(89)82889-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Olson JS, Quiocho FA. J. Biol. Chem. 1980;255:2465–2471. [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1972. [Google Scholar]
- Pace CN. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- Parkinson JS. Cell. 1988;53:1–2. doi: 10.1016/0092-8674(88)90478-3. [DOI] [PubMed] [Google Scholar]
- Peersen OB, Pratt EA, Truong HTN, Ho C, Rule GS. Biochemistry. 1990;29:3256–3262. doi: 10.1021/bi00465a017. [DOI] [PubMed] [Google Scholar]
- Post JFM, Cottam PF, Simplaceau V, Ho C. J. Mol. Biol. 1984;179:729–743. doi: 10.1016/0022-2836(84)90164-5. [DOI] [PubMed] [Google Scholar]
- Pratt EA, Ho C. Biochemistry. 1975;14:3035–3040. doi: 10.1021/bi00684a037. [DOI] [PubMed] [Google Scholar]
- Quiocho FA, Vyas NK, Sack JS, Vyas MN. Cold Spring Harbor Symp. Quant. Biol. 1987;52:453–463. doi: 10.1101/sqb.1987.052.01.052. [DOI] [PubMed] [Google Scholar]
- Robertson DE, Kroon PA, Ho C. Biochemistry. 1977;16:1443–1451. doi: 10.1021/bi00626a032. [DOI] [PubMed] [Google Scholar]
- Rule GS, Pratt EA, Simplaceau V, Ho C. Biochemistry. 1987;26:549–556. doi: 10.1021/bi00376a029. [DOI] [PubMed] [Google Scholar]
- Sandberg WS, Terwilliger TC. Science. 1989;245:54–57. doi: 10.1126/science.2787053. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle A, Vreemann J, Blank V, Nold A, Boos W, Manson MD. Mol. Gen. Genet. 1987;208:247–253. doi: 10.1007/BF00330450. [DOI] [PubMed] [Google Scholar]
- Snyder EE, Buoscio BW, Falke JJ. Biochemistry. 1990;29:3937–3943. doi: 10.1021/bi00468a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RC, Dahlquist FW. Chem. Rev. 1987;87:997–1025. [Google Scholar]
- Stock JB, Stock AM, Mottonen JM. Nature. 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- Sykes BD, Wingarten HJ, Schlesinger M. Proc. Natl. Acad. Sci. U.S.A. 1974;71:469–473. doi: 10.1073/pnas.71.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Johnson MS, Smith JM. Bot. Acta. 1988;101:101–104. [Google Scholar]
- Vyas NK, Vyas MN, Quiocho FA. Nature. 1987;327:635–638. doi: 10.1038/327635a0. [DOI] [PubMed] [Google Scholar]
- Vyas NK, Vyas MN, Quiocho FA. Science. 1988;242:1290–1295. doi: 10.1126/science.3057628. [DOI] [PubMed] [Google Scholar]
- Wagner G, Wüthrich K. Methods Enzymol. 1986;131:307–326. doi: 10.1016/0076-6879(86)31047-4. [DOI] [PubMed] [Google Scholar]
- Wilson ML, Dahlquist FW. Biochemistry. 1985;24:1920–1928. doi: 10.1021/bi00329a018. [DOI] [PubMed] [Google Scholar]