Abstract
The transmissible spongiform encephalopathies (TSEs) arise from conversion of the membrane-bound prion protein from PrPC to PrPSc. Examples of the TSEs include mad cow disease, chronic wasting disease in deer and elk, scrapie in goats and sheep, and kuru and Creutzfeldt-Jakob disease in humans. Although the precise function of PrPC in healthy tissues is not known, recent research demonstrates that it binds Cu(II) in an unusual and highly conserved region of the protein termed the octarepeat domain. This review describes recent connections between copper and PrPC, with an emphasis on the electron paramagnetic resonance elucidation of the specific copper-binding sites, insights into PrPC function, and emerging connections between copper and prion disease.
Keywords: transmissible spongiform encephalopathies, electron paramagnetic resonance, octarepeat domain, neuroprotection, apoptosis
Introduction
Prion diseases result from the accumulation of a misfolded form of the endogenous prion protein (PrP) and present an ongoing threat to human health, and agricultural and wildlife animals (1–5). Referred to as the transmissible spongiform encephalopathies (TSEs), these fatal, neurodegenerative diseases share pathologies with Alzheimer's and Parkinson's disease (6). What sets the TSEs apart, however, is that the misfolded prion protein is an infectious agent. As such, single instances of mad cow disease (bovine spongiform encephalopathy) create immediate concern with regard to the safety of our food supply and greatly affect the cattle industry (7). Chronic wasting disease has infected upward of 15% of deer and elk in the U.S. and Canadian Rocky Mountain regions (8). Although prion diseases in humans are rare, those cases in which transmission has taken place certainly motivate extreme caution. Approximately 140 young individuals in the United Kingdom have died from variant Creutzfeldt-Jakob disease (vCJD) contracted from bovine spongiform encephalopathy–contaminated food (9, 10). Surgical procedures involving corneal transplants and dura mater grafts have transmitted disease to healthy individuals, as have contaminated surgical instruments and cadaver-derived hormones (5). At this time, it is unknown whether blood transfusions are able to transmit prion diseases (11).
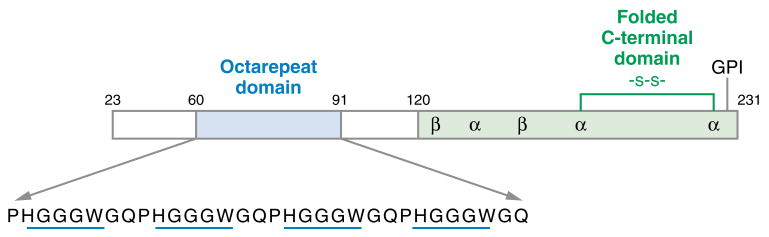
PrP is a membrane-bound glycoprotein found in the central nervous system (CNS) of all mammals and avian species (3, 5, 12). Post-translational modification removes the first 22 residues to give mature PrP(23–231). The protein is tethered to the outside surface of cellular membranes by a glycosylphosphatidylinositol anchor at its C terminus (Figure 1). The normal cellular form is designated PrPC. The pathogenic form (the prion), responsible for the TSEs, is designated PrPSc (scrapie form) and differs from PrPC only in conformation (2, 3). Nuclear magnetic resonance (NMR) studies on recombinant human PrPC demonstrate that the C-terminal region, starting at approximately residue 125, adopts a globular fold that is largely helical, but with a small two-strand β-sheet (13). Similar structures are found for hamster and mouse PrPC (14–16). There are currently no high-resolution structures for PrPSc, but recent electron-crystallography experiments suggest that residues 89–175 refold into a β-helix (17, 18).
Figure 1.
Diagram indicating secondary structure and other features of the prion protein (PrP). The C-terminal domain contains three α-helical segments, two short β-strands, a disulfide bond, and a glycosylphosphatidylinositol (GPI) anchor that tethers PrP to the membrane surface. The N-terminal segment, up to approximately residue 120, is largely unstructured and contains the octarepeat domain that binds Cu2+.
PrP's N-terminal region, up to approximately residue 110,is unstructured and flexible in solution (19). A hallmark of this region is the so-called octarepeat domain composed of tandem repeats of the fundamental eight-residue sequence PHGGGWGQ (Figure 1). Most species, including humans, have four or five repeat segments. Interestingly, the octarepeat domain is among the most highly conserved regions of the prion protein (20).
Prion diseases, the TSEs, may be sporadic, inherited, or infectious (21). Sporadic disease, in which PrPC spontaneously misfolds to PrPSc, is by far the most common, accounting for roughly 85% of cases in humans (21). Inherited diseases typically arise from mutations in the C-terminal domain (2); by contrast, polymorphisms or mutations in the octarepeat domain are rare (20). Sporadic and inherited prion diseases fall into the same class as Alzheimer's and Parkinson's disease insofar that all are associated with the accumulation of endogenous protein aggregates (21). As noted above, what sets prion diseases apart is that they are infectious. The passage of brain material from diseased animals (mice, hamsters, primates) into healthy animals causes the symptoms of the TSEs, along with the associated buildup of infectious PrPSc (2, 5). Treatment of brain-derived inoculants with radiation or other means of destroying nucleic acids fails to reduce the prion titer (3). Conversely, treatment with certain protein denaturants fully inactivates PrPSc (22). These unusual findings motivate the protein-only prion hypothesis, which posits that PrP alone, in the form of PrPSc, constitutes the infectious prion particle (5, 21). Researchers have now demonstrated that under partially denaturing conditions, amyloid fibrils generated from recombinant-mouse PrP indeed produce infectious material, the so-called synthetic prions (23).
The specific function of PrPC in healthy tissues is unknown—determining this function represents one of the great challenges in prion biology. Interestingly, the functions of the proteins responsible for Parkinson's disease and the extracellular deposits in Alzheimer's disease are also unknown. However, within the past several years, it has become clear that the prion protein binds copper in vivo, and the interaction between PrPC and copper requires the highly conserved, N-terminal octarepeat domain (24–29). Brown et al. (24) report that brain extracts from PrP-knockout mice have lower copper content than wild type, and they also exhibit reduced copper-metalloprotein activity. PrPC is constitutively cycled from the cell surface to the interior through endocytosis. The addition of Cu2+ rapidly stimulates this process, resulting in significant PrPC internalization (30). Neurons in cell culture are able to sequester Cu2+ at the plasma membrane in concentrations dictated by the level of PrPC expression (31). The PrP octarepeats are highly selective for Cu2+, with affinity for Cu+ or other metal-ion species either weak or nonexistent (27, 29, 32).
Within the CNS, PrPC is concentrated at presynaptic membranes (33), where neurotransmitter release drives communication between neurons. Interestingly, the presynaptic membrane is also a region of high copper localization and flux (34– 36). Copper moves from the cell interior to the synaptic space through both exocytosis and neuronal depolarization. Copper efflux may be an obligatory event associated with vesicle fusion, leading to neurotransmitter release (37). There are ongoing experiments to determine exact copper concentrations at the synapse. The best measurements place the resting concentration at approximately 1.0 μM and peak concentrations between 3.0 μM and 250 μM (35, 36).
Several lines of investigation have firmly established that PrPC helps maintain the integrity of neurons (i.e., the protein acts as a neuroprotectant). Compared with PrP knockouts, wild-type nerve cells in culture are much more resistant to deleterious oxidative chemistry mediated by copper or reactive oxygen species; this resistance requires the octarepeat domain (38). Transgenic mice lacking PrP, as they age, present widespread tissue damage, mainly through protein and lipid oxidation (39).
The findings discussed above and numerous other studies show that PrPC selectively binds copper, and this interaction is required for neuroprotective function. So what is the precise function of PrPC? There are a number of intriguing possibilities and a good bit of controversy. One prominent idea advanced by Brown and colleagues (40) is that PrPC is a neuronal superoxide dismutase (SOD) that converts O2− to peroxide and oxygen. Despite the attractiveness of this proposal, other labs have failed to demonstrate SOD activity (41, 42). Another possibility is that PrPC endocytosis transports copper from the extracellular space to the cell interior (29–31, 43, 44). However, measurements of copper transport do not show any dependence on PrP expression levels. PrPC is also implicated in copper buffering (32), copper sensing (37), signal transduction (45), the suppression of apoptosis (46), copper-reductase activity (47), and neuron development (48).
Elucidating the structural features of the copper centers in PrP is clearly essential for fleshing out function and for testing hypotheses related to neuroprotection. Although X-ray crystallography and NMR are the canonical approaches for determining protein structures, each method encounters challenges when applied to the copper-occupied, full-length prion protein. PrP has proven difficult to crystallize and, to date, only two PrP X-ray structures have been reported, both of the ovine C-terminal domains (49, 50). High-resolution NMR, conversely, is confounded by the presence of paramagnetic Cu2+, which leads to significant line broadening. To investigate structural features of the PrP copper sites, our laboratory applies a wide range of electron paramagnetic resonance (EPR) techniques, in conjunction with recombinant PrP and designed peptides. In the sections below, I review the basic EPR methodologies, newly identified PrP structural features, the affinity and cooperativity of copper binding, potential functions of the cellular prion protein, and possible connections between copper and the TSEs.
Electron Paramagnetic Resonance Methodologies
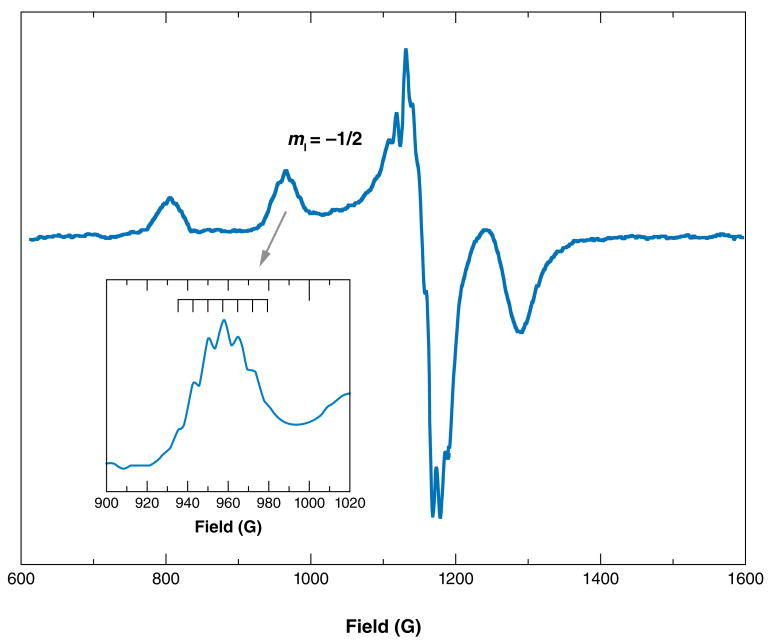
PrP takes up Cu2+, the dominant form of exchangeable copper in the extracellular space. Cu2+ is a d9 transition-metal center with a single unpaired electron. The two naturally occurring isotopes of copper, 63Cu and 65Cu, have nuclear spins, so each isotopic species gives rise to nearly equivalent four hyperfine line EPR spectra. As demonstrated many years ago by Peisach & Blumberg (51), both the g-tensor and hyperfine tensor (A) are sensitive to the equatorial-coordination environment. Analysis of the g‖ and A‖ features, usually derived from low-temperature spectra (77 K), provides an estimate of the number of coordinating nitrogen and oxygen atoms and the overall charge of the copper center. Moreover, examination of the parallel region often reveals whether there is more than one type of coordination environment and if pH or other factors influence the various coordination modes.
For example, Figure 2 shows the EPR spectra of a peptide corresponding to the PrP octarepeat domain, PrP(57–91), complexed with copper, as a function of pH. As indicated by the hyperfine splittings, both g‖ and A‖ shift with increasing pH. This is attributable to a progressive transition from 4O coordination at low pH to 4N coordination at high pH (52, 53). There are several archetypal copper centers readily identified by their EPR spectra. Type 1 centers involve sulfur coordination, as found in blue copper proteins, and are characterized by low A‖ values, typically around 50 G. Type 2 centers give spectra similar to those in Figure 1, and they arise from nitrogen and oxygen coordination and have larger A‖ values in excess of 150 G (54).
Figure 2.
Electron paramagnetic resonance spectra PrP(57–91) complexed with Cu2+, at 77 K, as a function of pH. Note the systematic changes in g‖ and A‖.
14N has a nuclear spin of I = 1 and, when coordinated to copper, often reveals additional superhyperfine couplings. Such couplings are small and typically on the order of 10–15 G. In some cases, these couplings are observable in the intense perpendicular region of the EPR spectrum between 3200 and 3400 G. Under ideal circumstances, the superhyperfine multiplets reflect the number of coordinated nitrogens, with a single nitrogen giving a three-line multiplet, two nitrogens giving a five-line multiplet, and so on. However, if Axx ≠ Ayy, the perpendicular region exhibits overlapping superhyperfine features, thus obscuring detailed couplings.
Superhyperfine interactions are best obtained from the parallel region of the Cu2+ spectrum. Unfortunately, inhomogeneous broadening owing to random variations in the coordination environments gives rise to distributions of g‖ and A‖ values, which are termed g strain and A strain, respectively, in turn masking subtle spectral features. Froncisz and Hyde (55) recognized, however, that for certain 63Cu hyperfine lines, g‖ and A‖ shift in opposite directions from systematic changes in the ligand field. Moreover, they reasoned that because g‖ depends on magnetic field, and A‖ does not, proper selection of magnetic field leads to a mutual cancellation of the g and A strain, thus revealing the desired superhyperfine interactions. This is achieved for the hyperfine line at low S-band spectrometer frequencies (56). Figure 3 shows an example of an S-band EPR spectrum obtained at 3.4 GHz. Expansion of the hyperfine line reveals subtle splittings indicative of a multiplet structure arising from coordinated 14N. The seven-line pattern, with a separation of approximately 10 G, suggests that three, or perhaps four, nitrogens are directly coordinated to the copper center. As shown below, selective 15N labeling in conjunction with S-band EPR directly identifies nitrogens coordinated to the Cu2+ center.
Figure 3.
Low-frequency S-band electron paramagnetic resonance spectrum of a copper center. The line is particularly sensitive to nitrogen superhyperfine couplings. The inset shows the expansion of the line, revealing seven lines consistent with three 14N atoms in equatorial coordination to the Cu2+.
Electron spin echo–envelope modulation (ESEEM), a pulsed EPR technique, is particularly useful for copper centers. If a copper center is magnetically coupled to a 14N nucleus with a hyperfine energy approximately equal to the nuclear Zeeman energy, the height of a spin echo from a two- or three-pulse sequence exhibits modulations as a function of timing between the pulses (57–59). Near this so-called exact cancellation condition, the frequencies of these modulations, as revealed by Fourier transform, correspond to the energies of the pure 14N quadrupolar transitions. In such cases, ESEEM actually reveals 14N NMR-like transitions, but they are detected by EPR (57–59).
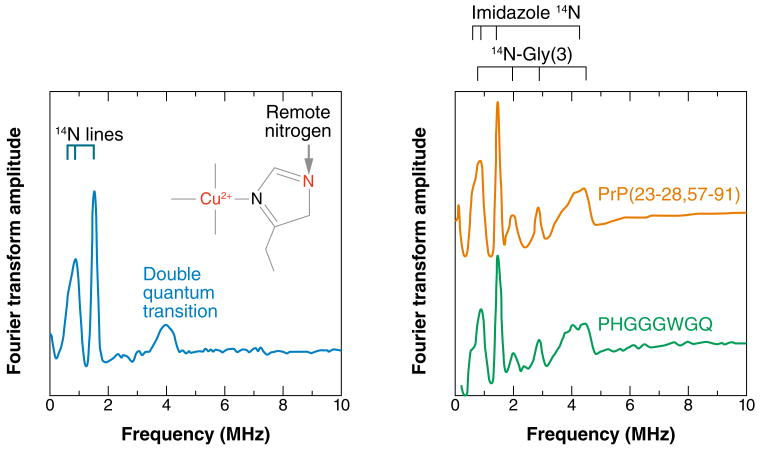
ESEEM is marvelously diagnostic for identifying imidazole coordination to copper arising from histidine (His) side chains. The remote, noncoordinated nitrogen is just the right distance from the copper center to satisfy the exact cancellation condition and maximize the ESEEM signal; Figure 4 shows a characteristic spectrum. The three sharp lines below 2.0 MHz result from transitions among the three nuclear-spin states of the 14N. The broad feature at approximately 4.0 MHz is called the double quantum, or Δm = 2, transition, the intensity of which often reflects the number of coordinated imidazoles (60).
Figure 4.
(Left panel) Characteristic electron spin echo–envelope modulation (ESEEM) spectrum of a copper center containing an equatorial imidazole. The observed frequencies arise from the remote nitrogen, as indicated. The lines below 2 MHz arise from transitions among the three 14N nuclear-spin states. The double quantum transition is often enhanced if there is more than one imidazole coordinated to the copper center. (Right panel) ESEEM spectra of the octarepeat domain and an individual octarepeat. Equivalence of these spectra shows that the HGGGW coordination mode is preserved in the full domain. The second set of lines arises from the noncoordinated nitrogen from the third glycine (Gly).
Structural Features and Affinities of the PrP Cu2+ Sites
Previous investigations suggest that copper sites are localized to the octarepeat domain (24–26, 28).To identify the specific residues, we examined the octarepeat peptide PrP(23–28, 57–91), where residues 57–91 encompass the octarepeat domain, and the segment 23–28 is included to improve solubility (52). Titration experiments demonstrated that the octarepeat domain takes up four equivalents of copper, in which each exists in a coordination environment comprising approximately three nitrogen atoms and one oxygen atom. By scanning a broad series of octarepeat peptides with EPR, we further showed that the residues HGGGW in each repeat constitute the fundamental Cu2+-binding unit. Imidazole coordination was unambiguously identified through ESEEM.
To further localize the coordinating atoms, we prepared a library of site specifically 15N-labeled octarepeat peptides, enabling us to identify nitrogens through superhyperfine couplings (43). With S-band EPR, a change from 14N to 15N alters the superhyperfine multiplets in the Cu2+ hyperfine line only for directly coordinated nitrogen atoms. Examination of the spectra in Figure 5 shows that the two glycine (Gly) residues following the His, in each octarepeat segment, coordinate through their amide nitrogens. Using Raman spectroscopy, Miura et al. (61) also identified amide-nitrogen coordination, although their modeling studies suggested the involvement of the second and third Gly residues following the His.
Figure 5.
The hyperfine line from the S-band electron paramagnetic resonance spectrum of PHGGGWGQ, with singly 15N-labeled octarepeat peptides (labeled at the underlined residue). The change in multiplet structure arising from labeling at the first and second glycine residues demonstrates that these amino acids coordinate Cu2+ through their amide nitrogens.
Although the ESEEM spectra of the octarepeat domain clearly identify coordination from the His side chains, there are additional features beyond those expected from a single imidazole group (Figure 4). Using the library of 15N-labeled octarepeat peptides, we showed that the additional features are from the 14N of the third Gly following each His (43). This particular nitrogen is not coordinated to the Cu2+ center and, alternatively, is approximately 4.0 Å away, appropriate for a strong ESEEM signal.
Figure 6 shows the Cu2+-coordination environment, based on EPR experiments, along with a crystal structure of the Cu2+-HGGGW complex (43). There is complete agreement between the EPR-defined contacts and the crystal structure. Computational (62) and NMR studies (63) provide further support for this type of coordination environment. Moreover, experiments on selectively labeled PrP(23–28, 57–91) proved that the coordination environment in Figure 6 is preserved in the full octarepeat domain (43). Using full-length recombinant PrP, we also found a nonoctarepeat site involving His-96 (64). The location of this particular site has been controversial, with Jones et al. (65) arguing that Cu2+ coordinates at His-111 with higher affinity than at His-96. Figure 7 shows our model of full-length PrP, with all copper sites occupied.
Figure 6.
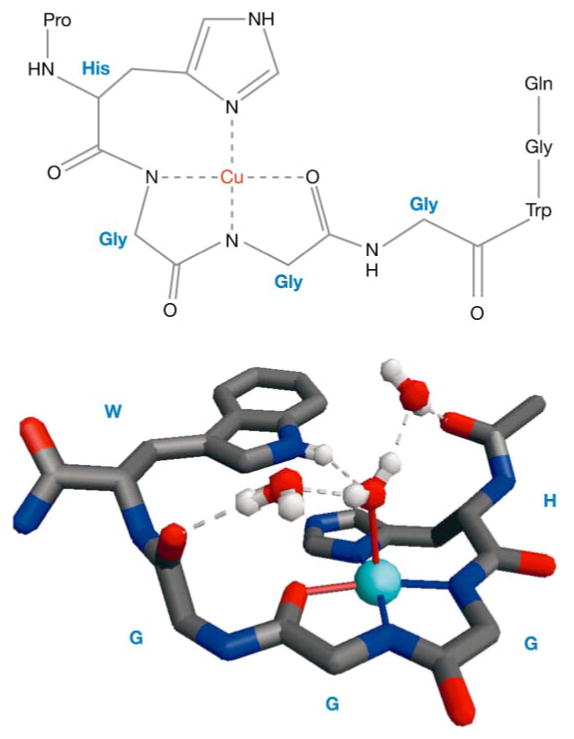
Chemical details of the Cu2+-octarepeat interaction (top panel), determined from electron paramagnetic resonance (EPR) constraints, and the X-ray crystal structure (bottom panel) of the Cu2+-HGGGW complex. The molecular features of the crystal structure are fully consistent with the structural details elucidated by EPR. Gly, glycine; His, histidine; Pro, proline; Trp, tryptophan.
Figure 7.

A structural model of the prion protein with its full complement of copper. Each copper in the octarepeat domain interacts with the HGGGW residues as shown in Figure 6. Electron paramagnetic resonance studies on full-length recombinant protein identified an additional nonoctarepeat binding site involving His-96.
The experiments above were performed on fully Cu2+ -occupied octarepeat complexes and thus do not address the details of how PrP responds to intermediate copper concentrations. If each HGGGW copper-binding module sequentially takes up a single equivalent, EPR spectra should be invariant as a function of Cu2+ occupancy. Instead, our EPR experiments showed that spectral features vary significantly as a function of copper load, demonstrating an unambiguous rearrangement of the octarepeat domain. Using a series of designed peptides and the EPR methodologies above, we identified three distinct binding components (Figure 8) (66). At low-Cu2+ occupancy, which gives rise to the component 3 spectrum, a single Cu2+ coordinates through multiple His side chains; at high occupancy, the component 1 spectrum dominates and reflects the interaction with a single His and deprotonated amide side chains, as described in the sections above. Component 2 is an intermediate state in which each Cu2+ is coordinated by two His residues, thus forming large intervening loops. Consistent with our findings, potentiometric titrations identify distinct deprotonated binding states depending on the ratio of copper to octarepeat (67). X-ray absorption spectra (68), molecular-modeling studies (69), and NMR-lineshape-broadening experiments (70) also find evidence of multiple His coordination at intermediate Cu2+ concentrations.
Figure 8.
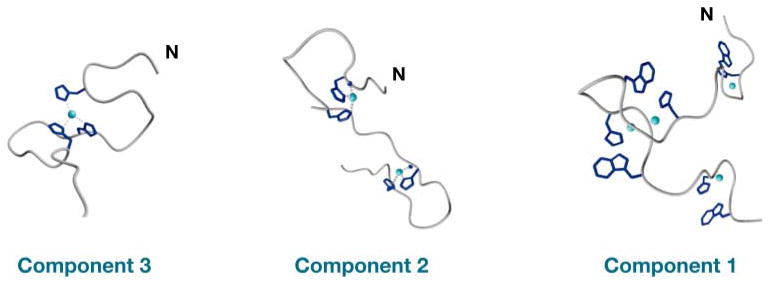
Distinct octarepeat Cu2+ binding modes with three, two, or one coordinated His residues, respectively. Low-Cu2+ occupancy favors component 3; high occupancy favors component 1.
Assessing PrP's copper-binding affinity and cooperativity has proven to be a thorny issue. Clearly, an accurate assessment of these properties is essential for corroborating potential function. For an enzyme, one expects high affinity (low-dissociation constant), whereas buffering or sensing functions require a lower affinity matched to the concentration of extracellular copper. Current estimates place the dissociation constant between 10−6 M (29, 71) and 10−14 M (69), a lack of agreement spanning eight orders of magnitude! There are also reports of highly positive binding cooperativity, in which the affinity ramps up with the addition of each Cu2+ (24, 29, 72). However, the molecular mechanism by which this might take place has not been identified.
Previous measurements of PrP's affinity for Cu2+ did not take into account the distinct binding modes, no doubt contributing to the disagreement among the published values. EPR methodologies allow us to perform binding measurements on each of the well-defined coordination modes. Using spectral decomposition, we determined the concentrations of all species as a function of Cu2+ concentration (Figure 9) (72a). Next, using peptide constructs designed to favor specific binding modes, we determined the precise affinity for each. At low-copper occupancy, which favors multiple His coordination, the octarepeat domain binds Cu2+ with a dissociation constant (Kd) of 0.12 nM.
Figure 9.
Relative populations of components 1, 2, and 3 (see Figure 8) as a function of total bound Cu2+. The dipolar-coupled spectrum occurs at full copper occupancy of PrP(23–28, 57–97) and arises from copper-copper couplings between component 1 centers. The significant population of intermediate species, such as component 3, is consistent with negative cooperativity.
In contrast, high-copper occupancy, involving coordination through deprotonated amide nitrogens, exhibits a weaker affinity characterized by a Kd in the range 7.0 μM to 12.0 μM. The decrease in affinity (increase in Kd) is consistent with negative cooperativity and a Hill coefficient (at half maximum binding) of approximately 0.7 (72a). The concentration distribution in Figure 9, showing significant populations of intermediates, provides further evidence of negative cooperativity because positive cooperativity suppresses intermediates and favors an equilibrium dominated by the fully unbound and fully bound species. Taken together, these findings demonstrate that PrP responds to an exceptionally wide range of copper concentrations, covering approximately five orders of magnitude.
Insights into PrP Function
PrP's nanomolar Cu2+ affinity and localization in synaptic regions have lead to the suggestion that the protein is a neuronal metalloenzyme with SOD activity (40, 73, 74). However, direct assays of SOD activity have been inconsistent with some measurements identifying weak dismutase function (73) and others finding no activity whatsoever (41, 42). Known SODs usually coordinate Cu2+ through His side chains in a tetrahedral environment, thus facilitating redox activity between the Cu+ and Cu2+ states. There are several aspects of the copper-coordination features identified by our experiments that are inconsistent with known SODs. First, the variable structures within the octarepeat domain (Figure 8), which depend on copper concentration, are certainly not characteristic of a well-defined metal-ion-coordination site typically required for catalysis. Moreover, even the highest affinity determined from our measurements is significantly less than that found for copper-dependent SODs, which bind with Kd values on the order of 10−14 M. Finally, the strong ligand field provided by the negatively charged amide nitrogens, in the dominant component 1–coordination mode, preferentially stabilizes the Cu2+-oxidation state, and direct electrochemical measurements have confirmed this (75).
PrP protects neurons against oxidative stress from reactive oxygen species or redox active metal ions. For example, it is well established that weakly complexed copper in the micromolar range is toxic to neurons, and cells expressing PrP are much more resistant to this oxidative stress than PrP knockouts (38). Yet, as noted above, there is significant copper efflux at synapses as a consequence of neuronal activity, and the peak synaptic concentrations are likely to exceed 10 μM. We believe that our results point toward an antioxidant function, as opposed to enzymatic function, in which PrP binds extracellular Cu2+, thus quenching copper's intrinsic redox activity (66). Our structural studies show that component 1 is dominant at high occupancy and thus maximizes PrP's antioxidant character at elevated Cu2+ concentrations. With these chemical and structural considerations, PrP's localization at synapses may be part of a protective mechanism in which copper sequestration ameliorates deleterious redox activity during synaptic depolarization. PrP may also play a role in trafficking copper back to the cell interior.
The protein composition of the cerebrospinal fluid in the CNS also points toward a copper-sequestering role for PrP. In the blood and blood plasma, exchangeable copper is bound to a number of molecular species, including amino acids, but the primary copper-buffering component is serum albumin, which takes up one equivalent of Cu2+ at its N terminus (76, 77). The concentration of albumin is approximately 600 μM, and its copper-coordination mode stabilizes Cu2+, much like the component 1 complex in PrP. Interestingly, cerebrospinal fluid also contains amino acids but lacks high concentrations of albumin and other copper-binding proteins normally found in blood. Consequently, membrane-bound PrP may, in part, play an albumin-like role, thus helping to maintain copper homeostasis.
Consideration of PrP's octarepeat domain sequence provides further insight into relevant Cu2+-binding modes. Within the octarepeat domain, component 1 coordination requires the residues HGGGW. His anchors Cu2+, and the torsional flexibility of the Gly residues allows for the planar equatorial configuration observed in Figure 6. Garnett & Viles (72) have shown that the replacement of Gly with chiral amino acids such as alanine (Ala) results in an alteration of the copper-coordination environment. Interestingly, in the HGGGW sequence, the His, Gly, and tryptophan (Trp) residues that make direct contact with Cu2+ are absolutely conserved in all mammalian sequences (20). The only variability is at the noncoordinating third Gly, where some species, such as mouse, have a Ser in two of the four repeats.
Component 3 coordination, conversely, depends not on the precise details of each octarepeat, but on the number of octarepeats, because each provides a single His; octapeptide sequences with less than three repeats fail to exhibit the component 3–binding mode. Here again, there is absolute sequence conservation among the mammalian sequences with all species possessing four or five repeats (20). These sequence-based observations suggest that the capacity for both component 1– and component 3–coordination modes have been preserved through evolution and are likely involved in PrP function (with component 2 serving as an intermediate). As the discussion above suggests, if component 1 is critical for suppressing Cu2+-redox activity, the role of component 3 may be to confer negative cooperativity, thereby spreading out PrP's response to varying copper concentrations.
Alternatively, the transition among binding modes may serve as a copper-dependent switch that facilitates specific cellular processes. For example, copper concentrations in excess of 100 μM stimulate PrP endocytosis (30); the transition from component 3 to component 1 coordination may be the trigger for this process. Although there is doubt as to whether copper bound to PrP is trafficked by endocytosis, Takeuchi and colleagues (47) recently suggested that, once in the endosome, component 3 coordination facilitates reduction to Cu+ as part of copper transport to the intracellular space.
Programmed cell death, termed apoptosis, is an essential process for regulating the number of cells in an organ. Several recent studies show that PrP is antiapoptotic (46, 78–80); it protects cells from signals that would otherwise trigger apoptosis. For example, PrP protects against the cellular death brought on by the expression of the Doppel protein (a homolog of C-terminal PrP) (78) or from serum deprivation (80). Mutagenesis experiments show that the PrP octarepeat domain is required for protection against Doppel-protein toxicity (78).
Investigations into the X-linked inhibitor of apoptosis (XIAP), an intracellular protein that negatively regulates apoptotic enzymes, recently identified an intimate link between copper homeostasis and the regulation of cell death (81). Specifically, intracellular copper binds directly to XIAP, causing a conformational change that accelerates its degradation, thus promoting apoptosis. The authors of this study argued that the build up of copper in Wilson's disease, for example, might upregulate apoptosis, resulting in tissue and organ damage. Thus, copper toxicity may occur through two primary mechanisms: its inherent redox activity and a lowering of the apoptosis threshold. In turn, PrP's antiapoptotic function may arise from its ability to sense copper at the extracellular-membrane surface or to regulate the transmembrane movement of copper.
Copper and PrPSc
Experiments into the connection between copper and PrPSc formation reveal contradictory results. It is simply not known whether copper promotes or inhibits prion disease. For example, the copper-binding octarepeats are not necessary for propagating prion infection (82), yet humans with octarepeat expansions are prone to inherited CJD (83). Copper treatment of extracted PrPC converts the protein to a protease-resistant form, perhaps an intermediate en route to PrPSc (84). Conversely, the addition of copper to protocols used for creating synthetic prions greatly slows amyloid formation, suggesting that copper inhibits PrPSc (85). In scrapie-infected cellular preparations, the addition of copper reduces the accumulation of PrPSc (86), yet copper is found in PrPSc deposits (87), and the removal of copper by chelation seems to delay the onset of prion disease (88).
Several aspects of copper binding likely help explain these divergent results. First, the copper site involving His-96 is in the region of PrP that typically remains after proteolytic cleavage, liberating amyloidogenic, C-terminal PrPSc. Thus, copper may modulate PrPSc formation even in the absence of the octarepeats, as Cox et al. (89) recently suggested. Another critical issue, discussed above, is that PrPC binds copper with several unique coordination modes (66). It is certainly possible that each mode exhibits distinct characteristics with regard to prion conversion.
The TSEs are often considered amyloid diseases in which ordered fibrils are a required component of the prion lesions. However, this is not strictly correct. There are many instances of prion diseases, such as certain types of CJD, in which accumulated PrPSc is devoid of the fibrillar structure of amyloid (21). Moreover, transgenic mice engineered to produce PrPC without the glycosylphosphatidylinositol-membrane anchor, exhibit widespread amyloid accumulation when infected with PrPSc, but without the usual, profound spongiform degeneration (90).
Prion specialists have long suspected that, in addition to amyloid, small PrPSc oligomers likely contribute to neuron deterioration. Interestingly, new avenues into the mechanism of neurodegeneration in Alzheimer's disease also focus on prefibrillar aggregates of the Aβ peptide, as opposed to high-molecular-weight fibrils (91, 92). Another aspect of these diseases under current consideration is that loss of function of the normal endogenous peptide or protein creates cell stress that contributes to neuronal death (93). In support of this, PrPSc added to cellular preparations inhibits copper binding to endogenous PrPC, thus rendering cells susceptible to copper toxicity (94). Moreover, cross-linking PrPC with monoclonal antibodies triggers rapid neuronal apoptosis (79). If PrPC protects neuronal integrity, perhaps by suppressing apoptosis, loss of this function may leave cells susceptible to stress, thus contributing to neurodegeneration. As such, the link between PrP and copper is certain to not only elucidate PrPC function, but also to provide important new concepts in the development of neurodegenerative disease.
Summary Points
The prion protein (PrP), responsible for the TSEs, is a copper-binding protein of unknown function. In its normal cellular form, PrPC, the protein acts as a neuroprotectant, thereby maintaining the integrity of cells in the CNS. The protein may function as a SOD, a copper transporter, a copper reductase, or a copper scavenger.
Most of the copper ions bind in a domain composed of tandem PHGGGWGQ repeats. At low-Cu2+ occupancy, equatorial coordination is provided by three or four His imidazoles; at high occupancy, equatorial coordination is from the His imidazole and deprotonated amide-nitrogen atoms from the two Gly residues that immediately follow His. The high-occupancy coordination mode stabilizes Cu2+ over Cu+ and thus suppresses copper-redox activity.
The affinity for Cu2+ varies significantly with Kd values of 0.12 nM at low occupancy and 7–10 μM at high occupancy. Such affinities are well matched to the known Cu2+ concentrations in the synapse where PrP is localized.
Although there is good evidence that PrP may function as a copper scavenger, much like albumin in the blood, we believe that the protein may also be involved in modulating signals for apoptosis by regulating membrane-surface-bound Cu2+.
Copper may also play a role in disease, although it is currently not clear whether the interaction between PrP and Cu2+ facilitates or inhibits prion formation. Recent evidence suggests that a loss of normal PrPC function may contribute to cell stress, thus enhancing neurodegeneration.
Acknowledgments
This work was supported by NIH grant GM065790 and NSF instrumentation grant DBI-0217922. I also thank Dr. Madhuri Chattopadhyay, Dr. Eric Walter, and Ms. Mira Patel for discussion and editorial comments on the manuscript.
- PrP
prion protein
- TSE
transmissible spongiform encephalopathy
- CJD
Creutzfeldt-Jakob disease
- CNS
central nervous system
- PrPC
cellular isoform of PrP
- PrPSc
scrapie isoform of PrP
- SOD
superoxide dismutase
- EPR
electron paramagnetic resonance
- PrP(57–91)
residues 57 through 91 of PrP
- Kd
dissociation constant
Literature Cited
- 1.Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–22. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. Prion diseases and the BSE crisis. Science. 1997;278:245–51. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weissmann C. The state of the prion. Nat Rev Microbiol. 2004;2:861–71. doi: 10.1038/nrmicro1025. [DOI] [PubMed] [Google Scholar]
- 5.Prusiner SB. Prion Biology and Diseases. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2003. p. 800. [Google Scholar]
- 6.Lansbury PTJ. Structural neurology: Are seeds at the root of neuronal degeneration? Neuron. 1997;19:1151–54. doi: 10.1016/s0896-6273(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 7.Pauli G. Tissue safety in view of CJD and variant CJD. Cell Tissue Bank. 2005;6:191–200. doi: 10.1007/s10561-005-0336-z. [DOI] [PubMed] [Google Scholar]
- 8.Miller MW, Williams ES, McCarty CW, Spraker TR, Kreeger TJ, et al. Epizootiology of chronic wasting disease in free-ranging cervids in Colorado and Wyoming. J Wildl Dis. 2000;36:676–90. doi: 10.7589/0090-3558-36.4.676. [DOI] [PubMed] [Google Scholar]
- 9.Horby P. Variant Creutzfeldt-Jakob disease: an unfolding epidemic of misfolded proteins. J Paediatr Child Health. 2002;38:539–42. doi: 10.1046/j.1440-1754.2002.00055.x. [DOI] [PubMed] [Google Scholar]
- 10.Ironside JW. The spectrum of safety: variant Creutzfeldt-Jakob disease in the United Kingdom. Semin Hematol. 2003;40:16–22. doi: 10.1016/s0037-1963(03)80748-5. [DOI] [PubMed] [Google Scholar]
- 11.Ironside JW, Head MW. Variant Creutzfeldt-Jakob disease: risk of transmission by blood and blood products. Haemophilia. 2004;10(Suppl. 4):64–69. doi: 10.1111/j.1365-2516.2004.00982.x. [DOI] [PubMed] [Google Scholar]
- 12.Flechsig E, Weissmann C. The role of PrP in health and disease. Curr Mol Med. 2004;4:337–53. doi: 10.2174/1566524043360645. [DOI] [PubMed] [Google Scholar]
- 13.Zahn R, Liu A, Luhrs T, Riek R, von Schroetter C, et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci USA. 2000;97:145–50. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James TL, Liu H, Nikolai BU, Farr-Jones S, Zhang H, et al. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci USA. 1997;94:10086–91. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP(121–321) Nature. 1996;382:180–82. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 16.Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231) FEBS Lett. 1997;413:282–88. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 17.Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed β-helices into trimers. Proc Natl Acad Sci USA. 2004;101:8342–47. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wille H, Michelitsch MD, Guenebaut V, Supattapone S, Serban A, et al. Structural studies of the scrapie prion protein by electron crystallography. Proc Natl Acad Sci USA. 2002;99:3563–68. doi: 10.1073/pnas.052703499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, et al. Structure of the recombinant full-length hamster prion protein PrP(29–231): The N-terminus is highly flexible. Proc Natl Acad Sci USA. 1997;94:13452–56. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wopfner F, Weidenhofer G, Schneider R, von Brunn A, Gilch S, et al. Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J Mol Biol. 1999;289:1163–78. doi: 10.1006/jmbi.1999.2831. [DOI] [PubMed] [Google Scholar]
- 21.Prusiner SB. Shattuck lecture: neurodegenerative diseases and prions. N Engl J Med. 2001;344:1516–26. doi: 10.1056/NEJM200105173442006. [DOI] [PubMed] [Google Scholar]
- 22.Prusiner SB, Groth D, Serban A, Stahl N, Gabizon R. Attempts to restore scrapie prion infectivity after exposure to protein denaturants. Proc Natl Acad Sci USA. 1993;90:2793–97. doi: 10.1073/pnas.90.7.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, et al. Synthetic mammalian prions. Science. 2004;305:673–76. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 24.Brown DR, Qin K, Herms JW, Madlung A, Manson J, et al. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–87. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]; This seminal report was the first to draw a convincing connection between PrP and in vivo copper.
- 25.Hornshaw MP, McDermott JR, Candy JM. Copper binding to the n-terminal tandem repeat regions of mammalian and avian prion protein. Biochem Biophys Res Comm. 1995;207:621–29. doi: 10.1006/bbrc.1995.1233. [DOI] [PubMed] [Google Scholar]
- 26.Hornshaw MP, McDermott JR, Candy JM, Lakey JH. Copper binding to the N-terminal tandem repeat region of mammalian and avian prion protein: structural studies using synthetic peptides. Biochem Biophys Res Commun. 1995;214:993–99. doi: 10.1006/bbrc.1995.2384. [DOI] [PubMed] [Google Scholar]
- 27.Stöckel J, Safar J, Wallace AC, Cohen FE, Prusiner SB. Prion protein selectively binds copper(II) ions. Biochemistry. 1998;37:7185–93. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 28.Viles JH, Cohen FE, Prusiner SB, Goodin DB, Wright PE, Dyson HJ. Copper binding to the prion protein: structural implications of four identical cooperative binding sites. Proc Natl Acad Sci USA. 1999;96:2042–47. doi: 10.1073/pnas.96.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whittal RM, Ball HL, Cohen FE, Burlingame AL, Prusiner SB, Baldwin MA. Copper binding to octarepeat peptides of the prion protein monitored by mass spectrometry. Protein Sci. 2000;9:332–43. doi: 10.1110/ps.9.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pauly PC, Harris DA. Copper stimulates endocytosis of the prion protein. J Biol Chem. 1998;273:33107–19. doi: 10.1074/jbc.273.50.33107. [DOI] [PubMed] [Google Scholar]
- 31.Rachidi W, Vilette D, Guiraud P, Arlotto M, Riondel J, et al. Expression of prion protein increases cellular copper binding and antioxidant enzyme activities but not copper delivery. J Biol Chem. 2003;278:9064–72. doi: 10.1074/jbc.M211830200. [DOI] [PubMed] [Google Scholar]
- 32.Millhauser GL. Copper binding in the prion protein. Acc Chem Res. 2004;37:79–85. doi: 10.1021/ar0301678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herms J, Tings T, Gall S, Madlung A, Giese A, et al. Evidence of presynaptic location and function of the prion protein. J Neurosci. 1999;19:8866–75. doi: 10.1523/JNEUROSCI.19-20-08866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartter DE, Barnea A. Evidence for release of copper in the brain: depolarization-induced release of newly taken-up 67copper. Synapse. 1988;2:412–15. doi: 10.1002/syn.890020408. [DOI] [PubMed] [Google Scholar]
- 35.Hopt A, Korte S, Fink H, Panne U, Niessner R, et al. Methods for studying synaptosomal copper release. J Neurosci Methods. 2003;128:159–72. doi: 10.1016/s0165-0270(03)00173-0. [DOI] [PubMed] [Google Scholar]
- 36.Kardos J, Kovacs I, Hajos F, Kalman M, Simonyi M. Nerve endings from rat brain tissue release copper upon depolarization: a possible role in regulating neuronal excitability. Neurosci Lett. 1989;103:139–44. doi: 10.1016/0304-3940(89)90565-x. [DOI] [PubMed] [Google Scholar]
- 37.Vassallo N, Herms J. Cellular prion protein function in copper homeostasis and redox signaling at the synapse. J Neurochem. 2003;86:538–44. doi: 10.1046/j.1471-4159.2003.01882.x. [DOI] [PubMed] [Google Scholar]
- 38.Brown DR, Schmidt B, Kretzschmar HA. Effects of copper on survival of prion protein knockout neurons and glia. J Neurochem. 1998;70:1686–93. doi: 10.1046/j.1471-4159.1998.70041686.x. [DOI] [PubMed] [Google Scholar]
- 39.Klamt F, Dal-Pizzol F, Conte DA, Frota JML, Walz R, et al. Imbalance of antioxidant defense in mice lacking cellular prion protein. Free Radic Biol Med. 2001;30:1137–44. doi: 10.1016/s0891-5849(01)00512-3. [DOI] [PubMed] [Google Scholar]
- 40.Daniels M, Brown DR. Purification and preparation of prion protein: synaptic superoxide dismutase. Methods Enzymol. 2002;349:258–67. doi: 10.1016/s0076-6879(02)49340-8. [DOI] [PubMed] [Google Scholar]
- 41.Hutter G, Heppner FL, Aguzzi A. No superoxide dismutase activity of cellular prion protein in vivo. Biol Chem. 2003;384:1279–85. doi: 10.1515/BC.2003.142. [DOI] [PubMed] [Google Scholar]
- 42.Jones S, Batchelor M, Bhelt D, Clarke AR, Collinge J, Jackson GS. Recombinant prion protein does not possess SOD-1 activity. Biochem J. 2005;392:309–12. doi: 10.1042/BJ20051236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burns CS, Aronoff-Spencer E, Dunham CM, Lario P, Avdievich NI, et al. Molecular features of the copper binding sites in the octarepeat domain of the prion protein. Biochemistry. 2002;41:3991–4001. doi: 10.1021/bi011922x. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first detailed elucidation of the Cu2+-binding features in an octarepeat segment.
- 44.Waggoner DJ, Drisaldi B, Bartnikas TB, Casareno RLB, Prohaska JR, et al. Brain copper content and cuproenzyme activity do not vary with prion protein expression level. J Biol Chem. 2000;275:7455–58. doi: 10.1074/jbc.275.11.7455. [DOI] [PubMed] [Google Scholar]
- 45.Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche JL, Lehmann S, et al. Signal transduction through prion protein. Science. 2000;289:1925–28. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- 46.Roucou X, Gains M, LeBlanc AC. Neuroprotective functions of prion protein. J Neurosci Res. 2004;75:153–61. doi: 10.1002/jnr.10864. [DOI] [PubMed] [Google Scholar]
- 47.Miura T, Sasaki S, Toyama A, Takeuchi H. Copper reduction by the octapeptide repeat region of prion protein: pH dependence and implications in cellular copper uptake. Biochemistry. 2005;44:8712–20. doi: 10.1021/bi0501784. [DOI] [PubMed] [Google Scholar]
- 48.Kanaani J, Prusiner SB, Diacovo J, Baekkeskov S, Legname G. Recombinant prion protein induces rapid polarization and development of synapses in embryonic rat hippocampal neurons in vitro. J Neurochem. 2005;95:1373–86. doi: 10.1111/j.1471-4159.2005.03469.x. [DOI] [PubMed] [Google Scholar]
- 49.Eghiaian F, Grosclaude J, Lesceu S, Debey P, Doublet B, et al. Insight into the PrPC–>PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants. Proc Natl Acad Sci USA. 2004;101:10254–59. doi: 10.1073/pnas.0400014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haire LF, Whyte SM, Vasisht N, Gill AC, Verma C, et al. The crystal structure of the globular domain of sheep prion protein. J Mol Biol. 2004;336:1175–83. doi: 10.1016/j.jmb.2003.12.059. [DOI] [PubMed] [Google Scholar]
- 51.Peisach J, Blumberg WE. Structural implications derived from the analysis of electron paramagnetic resonance spectra of natural and artificial copper proteins. Arch Biochem Biophys. 1974;165:691–708. doi: 10.1016/0003-9861(74)90298-7. [DOI] [PubMed] [Google Scholar]
- 52.Aronoff-Spencer E, Burns CS, Avdievich NI, Gerfen GJ, Peisach J, et al. Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy. Biochemistry. 2000;39:13760–71. doi: 10.1021/bi001472t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Doorslaer S, Cereghetti GM, Glockshuber R, Schweiger A. Unraveling the Cu2+ binding sites in the C-terminal domain of the murine prion protein: a pulse EPR and ENDOR study. J Phys Chem. 2001;105:1631–39. [Google Scholar]
- 54.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. Mill Valley, CA: Univ. Sci. Books; 1994. p. 411. [Google Scholar]
- 55.Froncisz W, Hyde JS. Broadening by strains of lines in the g-parallel region of Cu2+ ion EPR spectra. J Phys Chem. 1980;73:3123–31. [Google Scholar]
- 56.Yuan H, Antholine WE, Kroneck PMH. Complexation of type 2 copper by cytochrome c oxidase: probing of metal-specific binding sites by electron paramagnetic resonance. J Inorg Biochem. 1998;71:99–107. doi: 10.1016/s0162-0134(98)10038-7. [DOI] [PubMed] [Google Scholar]
- 57.Flanagan HL, Singel DJ. Analysis of 14N ESEEM patterns of randomly oriented solids. J Chem Phys. 1987;87:5606–16. [Google Scholar]
- 58.Mims WB, Peisach J. The nuclear modulation effect in electron spin echoes for complexes of Cu2+ and imidazole with 14N and 15N. J Chem Phys. 1978;69:4921–30. [Google Scholar]
- 59.Mims WB, Peisach J. Electron spin echo spectroscopy and the study of metalloproteins. In: Berliner LJ, Reuben J, editors. Biological Magnetic Resonance. New York: Plenum; 1981. [Google Scholar]
- 60.McCracken J, Pember S, Benkovic SJ, Villafranca JJ, Miller RJ, Peisach J. Electron-spin echo studies of the copper-binding site in phenylalanine-hydroxylase from chromobacterium-violaceum. J Am Chem Soc. 1988;110:1069–74. [Google Scholar]
- 61.Miura T, Hori-i A, Mototani H, Takeuchi H. Raman spectroscopic study on the copper(II) binding mode of prion octapeptide and its pH dependence. Biochemistry. 1999;38:11560–69. doi: 10.1021/bi9909389. [DOI] [PubMed] [Google Scholar]
- 62.Pushie MJ, Rauk A. Computational studies of Cu(II)[peptide] binding motifs: Cu[HGGG] and Cu[HG] as models for Cu(II) binding to the prion protein octarepeat region. J Biol Inorg Chem. 2003;8:53–65. doi: 10.1007/s00775-002-0386-7. [DOI] [PubMed] [Google Scholar]
- 63.Zahn R. The octapeptide repeats in mammalian prion protein constitute a pH-dependent folding and aggregation site. J Mol Biol. 2003;334:477–88. doi: 10.1016/j.jmb.2003.09.048. [DOI] [PubMed] [Google Scholar]
- 64.Burns CS, Aronoff-Spencer E, Legname G, Prusiner SB, Antholine WE, et al. Copper coordination in the full-length, recombinant prion protein. Biochemistry. 2003;42:6794–803. doi: 10.1021/bi027138+. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identified copper-binding sites outside of the octarepeat domain.
- 65.Jones CE, Klewpatinond M, Abdelraheim SR, Brown DR, Viles JH. Probing Cu2+ binding to the prion protein using diamagnetic Ni2+ and 1H NMR: the unstructured N terminus facilitates the coordination of six Cu2+ ions at physiological concentrations. J Mol Biol. 2005;346:1393–407. doi: 10.1016/j.jmb.2004.12.043. [DOI] [PubMed] [Google Scholar]
- 66.Chattopadhyay M, Walter ED, Newell DJ, Jackson PJ, Aronoff-Spencer E, et al. The octarepeat domain of the prion protein binds Cu(II) with three distinct coordination modes at pH7.4. J Am Chem Soc. 2005;127:12647–56. doi: 10.1021/ja053254z. [DOI] [PMC free article] [PubMed] [Google Scholar]; EPR methodologies show that the molecular features of Cu2+ coordination in the octarepeat domain depend on the amount of bound copper.
- 67.Valensin D, Luczkowski M, Mancini FM, Legowska A, Gaggelli E, et al. The dimeric and tetrameric octarepeat fragments of prion protein behave differently to its monomeric unit. Dalton Trans. 2004;2004:284–93. doi: 10.1039/b402090a. [DOI] [PubMed] [Google Scholar]
- 68.Morante S, Gonzalez-Iglesias R, Potrich C, Meneghini C, Meyer-Klaucke W, et al. Inter- and intraoctarepeat Cu(II) site geometries in the prion protein: implications in Cu(II) binding cooperativity and Cu(II)-mediated assemblies. J Biol Chem. 2004;279:11753–59. doi: 10.1074/jbc.M312860200. [DOI] [PubMed] [Google Scholar]
- 69.Jackson GS, Murray I, Hosszu LLP, Gibbs N, Waltho JP, et al. Location and properties of metal-binding sites on the human prion protein. Proc Natl Acad Sci USA. 2001;98:8531–35. doi: 10.1073/pnas.151038498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wells MA, Jackson GS, Jones S, Hosszu LL, Craven CJ, et al. A reassessment of copper (II) binding in the full-length prion protein. Biochem J. 2006 doi: 10.1042/BJ20060458. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kramer ML, Kratzin HD, Schmidt B, Romer A, Windl O, et al. Prion protein binds copper within the physiological concentration range. J Biol Chem. 2001;276:16711–19. doi: 10.1074/jbc.M006554200. [DOI] [PubMed] [Google Scholar]
- 72.Garnett AP, Viles JH. Copper binding to the octarepeats of the prion protein. Afffinity, specificity, folding and cooperativity: insights from circular dichroism. J Biol Chem. 2003;278:6795–802. doi: 10.1074/jbc.M209280200. [DOI] [PubMed] [Google Scholar]
- 72a.Walter ED, Chattopadhyay M, Millhauser GL. The affinity of copper binding to the prion protein octarepeat domain: evidence for negative cooperativity. Biochemistry. 2006 doi: 10.1021/bi060948r. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown DR, Wong BS, Hafiz F, Clive C, Haswell SJ, Jones IM. Normal prion protein has an activity like that of superoxide dismutase. Biochem J. 1999;344:1–5. [PMC free article] [PubMed] [Google Scholar]
- 74.Brown DR. Prion and prejudice: normal protein and the synapse. Trends Neurosci. 2001;24:85–90. doi: 10.1016/s0166-2236(00)01689-1. [DOI] [PubMed] [Google Scholar]
- 75.Bonomo RP, Impellizzeri G, Pappalardo G, Rizzarelli E, Tabbi G. Copper(II) binding modes in the prion octarepeat PHGGGWGQ: a spectroscopic and voltammetric study. Chem Eur J. 2000;6:4195–202. doi: 10.1002/1521-3765(20001117)6:22<4195::aid-chem4195>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 76.May PM, Linder PW, Williams DR. Computer simulation of metal-ion equilibria in biofluids: models for the low-molecular-weight complex distribution of calcium(II), magnesium(II), manganese(II), iron(III), copper(II), zinc(II), and lead(II) ions in human blood plasma. J Chem Soc Dalton Trans. 1977;1977:588–95. [Google Scholar]
- 77.Harford C, Sarkar B. Amino terminal Cu(II)- and Ni(II)-binding (ATCUN) motif of proteins and peptides: metal binding, DNA cleavage, and other properties. Acc Chem Res. 1997;30:123–30. [Google Scholar]
- 78.Drisaldi B, Coomaraswamy J, Mastrangelo P, Strome B, Yang J, et al. Genetic mapping of activity determinants within cellular prion proteins: N-terminal modules in PrPC offset proapoptotic activity of the Doppel helix B/B′ region. J Biol Chem. 2004;279:55443–54. doi: 10.1074/jbc.M404794200. [DOI] [PubMed] [Google Scholar]; These experiments with the doppel protein show that PrP protects against apoptosis, and this activity requires the octarepeat domain.
- 79.Solforosi L, Criado JR, McGavern DB, Wirz S, Sanchez-Alavez M, et al. Cross-linking cellular prion protein triggers neuronal apoptosis in vivo. Science. 2004;303:1514–16. doi: 10.1126/science.1094273. [DOI] [PubMed] [Google Scholar]
- 80.Kim BH, Lee HG, Choi JK, Kim JI, Choi EK, et al. The cellular prion protein (PrPC) prevents apoptotic neuronal cell death and mitochondrial dysfunction induced by serum deprivation. Brain Res Mol Brain Res. 2004;124:40–50. doi: 10.1016/j.molbrainres.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 81.Mufti AR, Burstein E, Csomos RA, Graf PC, Wilkinson JC, et al. XIAP is a copper binding protein deregulated in Wilson's disease and other copper toxicosis disorders. Mol Cell. 2006;21:775–85. doi: 10.1016/j.molcel.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 82.Flechsig E, Shmerling D, Hegyi I, Raeber AJ, Fischer M, et al. Neuron. Vol. 27. 2000. Prion protein devoid of the octapeptide repeat region restores susceptibility to scrapie in PrP knockout mice; pp. 399–408. [DOI] [PubMed] [Google Scholar]; Prion diseases develop more slowly in transgenic animals that express PrP lacking the octarepeats.
- 83.Goldfarb LG, Brown P, McCombie WR, Goldgaber D, Swergold GD, et al. Transmissible familial Creutzfeldt-Jakob disease associated with five, seven, and eight extra octapeptide coding repeats in the PRNP gene. Proc Natl Acad Sci USA. 1991;88:10926–30. doi: 10.1073/pnas.88.23.10926. [DOI] [PMC free article] [PubMed] [Google Scholar]; Humans with octarepeat expansions are susceptible to CJD.
- 84.Quaglio E, Chiesa R, Harris DA. Copper converts the cellular prion protein into a protease-resistant species that is distinct from the scrapie isoform. J Biol Chem. 2001;276:11432–38. doi: 10.1074/jbc.M009666200. [DOI] [PubMed] [Google Scholar]
- 85.Bocharova OV, Breydo L, Salnikov VV, Baskakov IV. Copper(II) inhibits in vitro conversion of prion protein into amyloid fibrils. Biochemistry. 2005;44:6776–87. doi: 10.1021/bi050251q. [DOI] [PubMed] [Google Scholar]; Copper may slow the formation of infectious prions.
- 86.Hijazi N, Shaked Y, Rosenmann H, Ben-Hur T, Gabizon R. Copper binding to PrPC may inhibit prion disease propagation. Brain Res. 2003;993:192–200. doi: 10.1016/j.brainres.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 87.Wadsworth DF, Hill AF, Joiner S, Jackson GS, Clarke AR, Collinge J. Strain-specific prion-protein conformation determined by metal ions. Nat Cell Biol. 1999;1:55–59. doi: 10.1038/9030. [DOI] [PubMed] [Google Scholar]
- 88.Sigurdsson EM, Brown DR, Alim MA, Scholtzova H, Carp R, et al. Copper chelation delays the onset of prion disease. J Biol Chem. 2003;278:46199–202. doi: 10.1074/jbc.C300303200. [DOI] [PubMed] [Google Scholar]
- 89.Cox DL, Pan J, Singh RR. Amechanism for copper inhibition of infectious prion conversion. Biophys J. 2006;91:L11–13. doi: 10.1529/biophysj.106.083642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308:1435–39. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 91.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid β-protein (Aβ) assembly: Aβ40 and Aβ42 oligomerize through distinct pathways. Proc Natl Acad Sci USA. 2003;100:330–35. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–89. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 93.Bush AI. The metallobiology of Alzheimer's disease. Trends Neurosci. 2003;26:207–14. doi: 10.1016/S0166-2236(03)00067-5. [DOI] [PubMed] [Google Scholar]
- 94.Rachidi W, Mange A, Senator A, Guiraud P, Riondel J, et al. Prion infection impairs copper binding of cultured cells. J Biol Chem. 2003;278:14595–98. doi: 10.1074/jbc.C300092200. [DOI] [PubMed] [Google Scholar]