Abstract
Sexual selection is an important force driving the evolution of morphological and genetic traits. To determine the importance of male–male, postcopulatory sexual selection in natural populations of house mice, we estimated the frequency of multiple paternity, defined as the frequency with which a pregnant female carried a litter fertilized by more than one male. By genotyping eight microsatellite markers from 1095 mice, we found evidence of multiple paternity from 33 of 143. Evidence for multiple paternity was especially strong for 29 of these litters. Multiple paternity was significantly more common in higher-density vs. lower-density populations. Any estimate of multiple paternity will be an underestimate of the frequency of multiple mating, defined as the frequency with which a female mates with more than a single male during a single oestrus cycle. We used computer simulations to estimate the frequency of multiple mating, incorporating observed reductions in heterozygosity and levels of allele sharing among mother and father. These simulations indicated that multiple mating is common, occurring in at least 20% of all oestrus cycles. The exact estimate depends on the competitive skew among males, a parameter for which we currently have no data from natural populations. This study suggests that sperm competition is an important aspect of postcopulatory sexual selection in house mice.
Keywords: multiple mating, multiple paternity, Mus domesticus
Introduction
Sexual selection is an important evolutionary force and may act before or after copulation. For example, sperm competition is primarily an intrasexual postcopulatory form of sexual selection (Parker 1970; Birkhead & Pizzari 2002), although female choice may also play an important role in the outcome of sperm competition (Eberhard 1996; Clark & Begun 1998). In mammals, sperm competition occurs when ejaculates from multiple males overlap in the reproductive tract of a single female during a single oestrus cycle. Under such conditions, sperm from different ejaculates effectively compete for fertilization of ova.
The house mouse, Mus domesticus, offers a promising model system to tease apart the evolutionary patterns and processes of sperm competition. Resources include a sequenced genome (Waterston et al. 2002), a dense microsatellite map (Dietrich et al. 1992, 1996; Schalkwyk et al. 1999), single nucleotide polymorphisms (Wade et al. 2002; Ideraabdullah et al. 2004), expression information (Schultz et al. 2003; Khil et al. 2004; Su et al. 2004), ecological and physiological information (Bronson 1979, 1989; Sage 1981; Silver 1995), phylogenetic hypotheses (Boursot et al. 1993, 1996; Din et al. 1996; Prager et al. 1998), and amenability to laboratory manipulation. Comparison of mouse and rat genomes revealed abundant evidence for rapid evolution of reproductive genes (Waterston et al. 2002; Gibbs et al. 2004). Postcopulatory, intrasexual selection may explain this rapid evolution these patterns; however, the frequency of multiple mating in natural populations of house mice is unknown and has never been studied directly.
Several observations provide indirect evidence that M. domesticus females mate with multiple males during a single oestrus cycle. Ejaculates form copulatory plugs, which presumably inhibit fertilizations from a second male (Hartung & Dewsbury 1978; Dewsbury 1984). Ejaculations are usually preceded by a variable number of intromissions, whereby the penis enters the vagina but no sperm are transferred (Dewsbury 1984). In rats, such behaviour removes copulatory plugs from previous males, and morphological features such as penile spines may facilitate such removal (Mosig & Dewsbury 1970). However, it is not clear whether these mating characteristics represent adaptation to a system of multiple mating. For example, tactile stimulation is necessary to induce physiological changes that prepare the uterus for implantation and pregnancy (Bronson 1979).
A few laboratory experiments have produced direct evidence of multiple mating in females, although the relevance to natural populations is not clear. Given a choice, nine of 39 female M. domesticus mated with two tethered males (Egid & Brown 1989). Using large enclosures, females mated with more than a single male in 13 of 198 (7%), 68 of 349 (19%), and 12 of 15 litters (80%) (Oakeshott 1974; Carroll et al. 2004; Ehman & Scott 2004). Which, if any, of these estimates applies to natural populations is an open question.
Furthermore, there is unlikely to be a single estimate of multiple mating that pertains to all population types. In high-density populations, dominant males may exert their control more effectively, preventing mating by subordinates (Bronson 1979). Therefore, multiple mating may be higher in low-density populations, where turnover precludes a stable dominance hierarchy. On the other hand, there should be more opportunities for a female to encounter multiple males in a high-density population, suggesting multiple mating may be higher in high-density populations. Furthermore, high-density populations may be less stable because they experience more mortality (Southwick 1955), perhaps due to increased aggression among individual mice (Gerlach 1996).
The frequency of multiple paternity, defined as the frequency with which a pregnant female carries a litter sired by more than one male, can be estimated by inferring the minimum number of fathers per litter from genotypes of mothers and their offspring. However, this strategy will always underestimate the frequency of multiple mating, defined as the frequency with which a female mates with more than a single male during a single oestrus cycle. Even if multiple mating occurs, one father may sire the entire litter. Such a competitive skew may arise through genetic effects, mating order effects, or chance. Multiple paternity may also go undetected if mothers share alleles with potential fathers, which might be common in inbred populations.
Here, we estimated the frequency of multiple paternity by genotyping 143 mothers pregnant with at least three embryos. We found evidence of multiple paternity in 33 litters (23%). Evidence for multiple paternity was especially strong for 29 of these litters (20%). We then estimated the true frequency of multiple mating through computer simulations conditioned upon our observations. We inferred that multiple mating occurs in 20–100% of all oestrus cycles, suggesting that sperm competition is a powerful evolutionary force in mice.
Materials and methods
Sampling
Mice from Arizona were collected in Sherman traps from April 2004 through February 2006. Mice from Iowa, Maryland, New Hampshire, New Jersey, and Tennessee, and Australia were collected between 1991 and 1995 (Ardlie & Silver 1998). Embryos were dissected from pregnant females and preserved individually. Litters were only included if embryos were large enough to rule out contamination with maternal tissue. In addition to mothers and their litters, we genotyped 114 males and nonpregnant females from Arizona. All mice, excluding embryos, were used to estimate allele frequencies. Only pregnant females were available from non-Arizona localities for estimating allele frequencies.
We used trapping data to define two main groups of populations — those coming from relatively high-density and those coming from relatively low-density populations. Arizona mice were divided into these two categories based on the number of mice caught per trapnight (high density > 0.5 mice caught per trapnight; low density < 0.5 mice per trapnight). All non-Arizona mice were from Ardlie & Silver (1998), who defined low-density populations as those where 60 or fewer total mice were captured over a period of 5 days of ‘saturation trapping’. High-density populations were those where greater than 60 total mice were captured. Since the number of traps used by Ardlie & Silver (1998) was not recorded, the identification of populations as either high density or low density is not quantitative. We use it here only as a rough guide and we make no claims about the specific number of mice inhabiting an area. The density data offer a rough test of the hypothesis that multiple paternity should be more frequent in high-density populations (Bronson 1979).
Genotyping
DNA was extracted with the PureGene kit (Gentra) following the ‘DNA purification from solid tissues’ protocol with slight modifications. We genotyped eight microsatellite loci: D1Mit303 + D15Mit174, D8Mit121 + D11Mit4, D6Mit138 + D2Mit277 and D14Mit132 + D4Mit199 (Dietrich et al. 1992, 1996). Primer pairs sharing an underline were assayed together in single 10-μL reactions consisting of approximately 35 ng genomic DNA, 0.4–2.4 pmol each primer, 2 nmol each dNTP, 20 nmol MgSO4, ¼ unit Hi-Fi Platinum Taq (Invitrogen), 1.0 μL 10× buffer (Invitrogen), and water. The forward primer of the first locus in each pair above was dyed with 6-FAM, while that of the second locus was dyed with HEX. Reactions were subjected to 30 cycles of 94 °C for 5 s and 55 °C for 1 min (Yoshida & Awaji 2000). Following polymerase chain reaction (PCR), the first four primer pairs above were combined and the second four primers above were combined to create two multiplexed reactions. To avoid overlap of alleles, we redesigned the reverse primer of D6Mit138 as TTGTTTTTTAAATAATGTGGAGGG. Multiplexes were genotyped on an ABI 3100 with a 500 ROX ladder (which allows differentiation of alleles that differ by as little as a single base pair), and alleles scored with Genotyper (ABI) and inspected manually. These markers consist primarily of dinucleotide CA repeats; the exceptions are D2Mit277, which consists primarily of GA and GT repeats, and D6Mit138, which consists primarily of GA and GAAA repeats.
Through the course of our studies, 2272 PCR genotypes were determined twice, many from new DNA extractions. The primary goal of regenotyping was to confirm multiple paternity when it occurred (see below), but also included randomly resampled specimens. These reactions were used to estimate repeatability of genotyping.
Multiple paternity
There are several alternative strategies to test whether a litter was fertilized by more than a single male. Maximum-likelihood models have been developed for inferring multiple paternity (Queller & Goodnight 1989; Kichler et al. 1999; Jones & Clark 2003), but preliminary analyses suggested these were not appropriate for our data given the small average litter size of mice. Furthermore, panmixia is an important assumption in these methods, which was violated in our data (see Results). Therefore, multiple paternity was identified by subtracting the maternal allele from each embryo and inferring a paternal allele. Inference of at least three paternal alleles from at least one locus from a litter was taken as evidence of multiple paternity. In cases of ambiguity, such as when heterozygous embryos shared the genotype of their heterozygous mothers, paternal alleles were inferred in a way that minimized the total number of paternal alleles from the litter. This method is conservative and probably underestimated the true frequency of multiple paternity.
Estimating mutation rates
Estimating mutation rates was not a specific goal of this study, but it should be noted that a mutation during gametogenesis might spuriously lead to inference of multiple paternity. For example, three paternal alleles could be contributed by a single male if a mutational event occurred during spermatogenesis. If three paternal alleles are observed in multiple embryos at each of several loci, however, this is strong evidence for multiple paternity since mutations are rare events. We estimated the number of mutations in two ways. First, we identified litters that showed a third paternal allele in only one embryo at one of the eight loci, where the third allele was separated by a single mutational step from another inferred paternal allele. Here, we assumed one mutational step was 1 repeat unit in the microsatellite, although such a stepwise-mutation model may not be fully appropriate. Second, we asked how often an embryo did not carry a maternal allele, but carried an allele separated by one mutational step from a maternal allele.
Reductions in heterozygosity and allele sharing
It is important to consider biological patterns that will reduce our power to detect multiple paternity when it occurs, including reductions in observed heterozygosity and increased allele sharing among mother and father(s). Reductions in heterozygosity and allele sharing are related measures. However, in nonequilibrium populations, such as those experiencing regular migration, reductions in heterozygosity do not necessarily predict allele sharing among parents. Both patterns result in fewer alleles present in any single litter. To test for reductions in heterozygosity within each of the eight broad geographic regions (Table 1), we first removed all embryos from the data set, which represent nonindependent estimates of allele frequency, then analysed the remaining data with genepop version 3.3 (Rousset & Raymond 1995; run from http://wbiomed.curtin.edu.au/genepop/). Significant departures from expectation were judged by a Markov chain permutation test.
Table 1.
Mice sampled, levels of multiple paternity, and observed and expected heterozygosities for each locality
| Locality | #mice* | #mothers* | #embryos* | #mp litters† | Mdist (km)‡ | HE§ | HO¶ | P-value** |
|---|---|---|---|---|---|---|---|---|
| Arizona | ||||||||
| (all) | 519 (33) | 62 (7) | 343 (26) | 16 | 16 | 0.80 | 0.68 | < 0.001 |
| (nonzoo) | 212 (15) | 22 (2) | 121 (13) | 3 | 16 | 0.79 | 0.71 | < 0.001 |
| (zoo) | 307 (18) | 40 (5) | 222 (13) | 13 | 0.1 | 0.75 | 0.66 | < 0.001 |
| Iowa | 72 | 9 | 63 | 1 | 0.1 | 0.65 | 0.64 | 0.56 |
| Maryland | 143 (13) | 23 (3) | 120 (10) | 5 | unknown | 0.65 | 0.54 | < 0.001 |
| New Hampshire | 11 | 2 | 9 | 0 | 0.1 | NA†† | NA†† | NA†† |
| New Jersey | 202 (11) | 34 (3) | 168 (8) | 8 | 0.1 | 0.67 | 0.64 | < 0.006 |
| Tennessee | 64 | 9 | 55 | 1 | 0.1 | 0.58 | 0.58 | 0.72 |
| Australia | 84 | 11 | 73 | 2 | 1150 | 0.71 | 0.55 | < 0.001 |
| Total | 1095 | 143 | 831 | 33 |
Numbers in parentheses indicate litters which either showed evidence of mutational events or only had two embryos; these litters were excluded from studies of multiple paternity (see text).
The number of litters that showed evidence of multiple paternity.
The maximum distance between collecting localities within a population.
Expected frequency of heterozygotes. Average HE = 0.69.
Observed frequency of heterozygotes. Average HO = 0.62.
Significant departures from expected and observed frequency of heterozygotes judged by Markov chain permutation tests (see text).
Not applicable since expected and observed heterozygosities could not be reliably estimated from only two mothers.
To test whether mother and inferred father shared alleles significantly more than expected, we calculated the average allele sharing across the eight microsatellite loci, using a modification of Li et al. (1993), from all singly sired litters. Briefly, a score of 0 indicated no alleles in common between mother and inferred father, 2 indicated two heterozygotes with one allele in common, 3 indicated a heterozygote and homozygote with two total alleles, and 4 indicated identical genotypes. We calculated the average allele sharing across all singly sired litters. To construct a null distribution, we simulated 10 000 data sets of 143 litters. In each of these 10 000 runs, litter size was drawn without replacement from the empirical distribution of litter sizes and genotypes sampled randomly from the empirical distributions of allele frequencies. Information from the father was then discarded, and the father's multilocus genotype re-inferred from a comparison of mother to embryos.
Multiple mating
Any estimate of multiple paternity will underestimate the frequency of multiple mating. Competitive skew among males may arise by chance, or through mating order effects, which in rodents appears to give an advantage to the first male to mate (Levine 1967; Dewsbury 1984; Lacey et al. 1997). Genetic make-up of males, or interactions between male and female genotypes may also lead to competitive skew (Edwards 1955; Levine 1967; Clark et al. 1995, 1999; Clark & Begun 1998; Fiumera et al. 2005). Therefore, we incorporated our estimate of multiple paternity into Monte Carlo simulations, written in PYTHON (www.python.org), to infer the rate of multiple mating. These simulated data sets drew from observed levels of heterozygosity, allele sharing, allele frequencies, and litter sizes. Therefore, demographic or selective forces that may have given rise to genetic differences among populations (or deviations from Hardy–Weinberg equilibrium within populations) were accounted for, and panmixia was not a necessary assumption underlying these simulations. In other words, we were not interested in simulating specific genotypes and population structure per se, but in simulating a realistic power to detect multiple paternity.
Mothers were simulated by randomly drawing from the empirical distribution of heterozygosity scores and allele frequencies for each locus. Given a particular mother, two potential fathers were simulated by randomly drawing from the empirical distribution of allele sharing scores and allele frequencies for each locus. Embryos were simulated by randomly drawing one maternal and one paternal allele, with litter size sampled without replacement from the observed distribution of litter sizes. This process was repeated until 143 litters were simulated (our observed data set). For each litter, we discarded all information about potential father(s), then re-inferred the paternal genotype and calculated the frequency of multiple paternity as described previously.
We explored all combinations of multiple mating, ranging from a probability of 0 (no multiple mating) to 1 (a female mates with both males in every oestrus cycle), and competitive skew, ranging from 0.5 (when multiple mating occurs, each male has equal probability of fertilizing any one ovum) to 1 (one male always fertilizes all ova), moving along both axes in units of 0.05. For each particular combination of multiple mating and competitive skew, we simulated 1000 data sets, each data set consisting of 143 litters, to produce a null distribution. When the observed proportion of multiply sired litters (33/143 = 0.23 or 29/143 = 0.20, see below) fell in the top or bottom 5% of this distribution, that combination of multiple mating and competitive skew was considered unlikely. These simulations provided a maximum-likelihood estimate of the frequency of multiple mating as well as 95% credibility intervals that were consistent with our observed data set.
Results
Sampling
We collected 150 pregnant females, carrying an average of 5.6 pups each, with embryos large enough for confident analysis. Seven females carried only two embryos and were not included in estimates of multiple paternity since it is impossible to infer three paternal alleles in such cases. In Arizona, the Reid Park Zoo population (zoo) was the only locality inferred to be at high density. The probability of catching a mouse per trap per night was significantly higher at the zoo compared to all pooled nonzoo populations from Arizona (zoo: median = 0.69 mice per trapnight, range = 0–1.47, N = 27 collecting nights; nonzoo: median = 0.06, range = 0–0.33, N = 37; Kolmogorov–Smirnov D = 0.85, P < 0.0001). An average of 35.7 traps were placed each collecting night at all localities. Mice from the zoo were caught from enclosed aviaries that excluded predators and competitors, and food and water were essentially ad libitum. Mice from nonzoo localities were collected from feed stores, farms, horse stables, etc., where predators were probably more abundant and food/water was scarcer.
Genotyping
We genotyped a total of 1095 mice, including all mothers (N = 150), their embryos (N = 831), and other mice (N = 114). We found an average of 17.4 alleles per locus. Within each broadly defined region (Table 1), we found an average of 5.9 alleles per locus. Average observed heterozygosity, HO = 0.62, was lower than average expected heterozygosity, HE = 0.69 (Table 1). Although the presence of null alleles may account for the discrepancy between observed and expected heterozygosities, mice show strong population substructure and reductions in heterozygosity are expected.
We regenotyped 2272 PCRs. A total of 2215 reactions corroborated the initial genotype, 943 of which were from independent DNA extractions. Of the 67 reactions that did not corroborate the original score, 52 were cases where an individual was initially genotyped as a homozygote and later genotyped as a heterozygote (or vice versa). This result indicated stochastic variation in the amplification success of both alleles in heterozygous individuals. In these 52 cases, we assumed the heterozygous genotype was correct. Overall, the corroboration rate from regenotyping was over 97%.
Multiple paternity
Using the conservative approach of subtracting maternal alleles from each embryo, we found at least three paternal alleles in at least one locus in 33 of 143 litters that had more than two embryos (23%). Multiply sired litters averaged 5.9 pups, a nonsignificant difference from the average of 5.7 pups found in all other litters (Mann–Whitney U = 1912, P = 0.64). Multiple paternity was found in all populations except New Hampshire (Table 1). Of these, 17 litters showed at least three paternal alleles at one of the eight loci, eight litters at two loci, four litters at three loci, two litters at four loci, and two litters at five loci.
For all litters that were inferred to be sired by more than a single male, we regenotyped them for the eight micro-satellite loci, then re-inferred multilocus paternal genotypes. In all cases, multiple paternity was confirmed.
Spurious inference of multiple paternity may result if one of three paternal alleles arose through a mutational event. For litters where at least three paternal alleles were inferred from more than one locus, this explanation is extremely unlikely, as it requires multiple independent mutational events. However, this alternative hypothesis may apply to at least some of the 17 litters where three paternal alleles were inferred from a single locus. To test this alternative hypothesis, we genotyped an additional eight microsatellite loci (D13Mit186 + D19Mit75, D9Mit217 + D3Mit22, D16Mit139 + D10Mit42 and D7Mit69 + D18Mit149, following the conditions described in the Materials and methods) from these 17 litters. Three litters showed at least three paternal alleles at an additional locus, five litters at two loci, and two litters at three loci, confirming multiple paternity in 10 of the 17 litters. For the remaining seven litters, we asked whether one of the three inferred paternal alleles was separated by any other inferred paternal allele by more than one mutational step, here assumed to be 1 repeat unit in the microsatellite. Three of these seven litters showed three paternal alleles that were separated by more than a single repeat unit, making the mutational explanation less likely. For the remaining four litters, the third paternal allele was only observed in one embryo; furthermore, multiple paternity was inferred for only 1 of 16 loci (the original 8 plus the additional 8 loci assayed). It is possible that some of these inferences resulted from mutational events, and not from multiple paternity. Removing these litters leads to a minimum estimate of multiple paternity of 29/143 = 20%.
Multiple paternity was significantly more frequent in relatively high-density vs. low-density populations. Of 81 litters collected from high-density populations, 25 (31%) showed evidence of multiple paternity. Of these 25 litters, 13 were from Arizona, four from Maryland, and eight from New Jersey. In contrast, 6 of 51 litters (12%) collected from low-density populations showed evidence of multiple paternity. Of these six litters, three were from Arizona, one from Iowa, one from Maryland, and one from Tennessee. The number of total litters does not sum to 143, and the number of multiply sired litters does not sum to 33, because trapping data were not available for all litters. There was a significantly higher probability of observing multiple paternity in relatively high-density populations vs. low-density populations (Fisher's two-tailed exact test P ≈ 0.01), even after removing the four litters where evidence of multiple paternity was weak (of which all four were from high-density populations) (P < 0.05). This result was probably not due to differences in power to detect multiple paternity since HO = 0.66 in both high-density and low-density populations.
Estimating mutation rates
To avoid spurious inference of multiple paternity, it was important to estimate the number of mutations. If we assume the four litters mentioned above resulted from mutation, the estimated rate of paternally contributed mutations is calculated as four mutations per (817 embryos * 8 microsatellite loci genotyped per embryo), which equals 6.1 × 10−4 mutations per locus per genotype. We also estimated the frequency of maternally contributed mutations by quantifying maternal mismatch. Of the 143 mothers and their 817 embryos (excluding two-embryo litters), there were 34 cases where an embryo did not carry a maternal allele at a particular locus from 12 litters. Of the 34 maternal mismatches, 23 were cases where either the mother or the embryo was inferred to be a homozygote, and may have resulted from the presence of null alleles. Alternatively, these could have arisen through mutation; in 13 of these 23 cases, a maternal allele and an embryonic allele were separated by one mutational step. In the other 11 cases of maternal mismatch, occurring in six different litters and four different loci, the mother and embryo were both heterozygotes that shared no alleles. Of these 11 embryos, five carried an allele separated by one mutational step from one maternal allele. These five cases provided support for a maternally contributed mutation. Thus, maternally contributed mutations occurred at a rate of at least five maternal mismatches per (817 embryos * 8 microsatellite loci genotyped per embryo), or 7.6 × 10−4 mutations per locus genotype. These inferred frequencies of mutations were similar to previous estimates of dinucleotide mutation rates (Whittaker et al. 2003).
Reductions in heterozygosity and allele sharing
Significant discrepancies between observed and expected levels of heterozygosity were observed. Within all broad geographic regions except Iowa and Tennessee, there was a significant reduction in observed heterozygosity, averaging 7% (Table 1). One explanation for significant reductions in heterozygosity within populations is inbreeding. Another is unrecognized population structure. We expect population structure to be more prominent in localities where distances among precise collecting sites are greater. Consistent with this prediction, all localities where Mdist > 0.1 show significant reductions in heterozygosity. Conversely, only one of the three localities where Mdist = 0.1 (i.e. those localities where all mice were collected within 100 m of each other) showed a significant reduction in heterozygosity (Table 1), possibly due to the reduced effects of unrecognized population structure. There was significantly more allele sharing between mother and inferred fathers than expected (empirical average of allele sharing = 2.33, randomized null range = 0.89–1.26, P < 0.001).
We estimated the power to detect multiple paternity using computer simulations, incorporating reductions in heterozygosity and allele sharing between parents. For each simulated litter, we removed genotypic information about the fathers, and then re-inferred the paternal genotype. The proportion of times we observed at least three paternal alleles in at least one locus was taken as the power to detect multiple paternity. These simulations were also used to generate the number of loci expected to show evidence of multiple paternity. The power to detect at least three paternal alleles in one or more loci was dependent on the litter size (Table 2). With five embryos in a litter, there was a 96% chance that multiple paternity was detected, under the assumption of no competitive skew (competitive skew = 0.5). Table 2 shows that with increasing litter size, more loci are expected to show evidence of multiple paternity. In contrast, increasing competitive skew decreased the number of loci expected to show multiple paternity. We chose two values of competitive skew, 0.50 and 0.99, as two possible extremes, but no estimates of competitive skew exist for natural populations of house mice.
Table 2.
Power to detect multiple paternity
| Competitive skew* | Proportion of litters† |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Litter size | 1 locus | 2 loci | 3 loci | 4 loci | 5 loci | 6 loci | 7 loci | 8 loci | power‡ | |
| 0.50 | 3 | 0.39 | 0.24 | 0.08 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.73 |
| 4 | 0.27 | 0.31 | 0.21 | 0.08 | 0.02 | 0.00 | 0.00 | 0.00 | 0.90 | |
| 5 | 0.17 | 0.29 | 0.27 | 0.16 | 0.05 | 0.01 | 0.00 | 0.00 | 0.96 | |
| 6 | 0.11 | 0.24 | 0.28 | 0.21 | 0.10 | 0.03 | 0.00 | 0.00 | 0.98 | |
| 7 | 0.08 | 0.18 | 0.28 | 0.25 | 0.14 | 0.05 | 0.01 | 0.00 | 0.99 | |
| 0.99 | 3 | 0.38 | 0.24 | 0.08 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.73 |
| 4 | 0.29 | 0.33 | 0.18 | 0.07 | 0.02 | 0.00 | 0.00 | 0.00 | 0.89 | |
| 5 | 0.23 | 0.31 | 0.25 | 0.11 | 0.03 | 0.00 | 0.00 | 0.00 | 0.93 | |
| 6 | 0.18 | 0.31 | 0.26 | 0.13 | 0.05 | 0.01 | 0.00 | 0.00 | 0.95 | |
| 7 | 0.17 | 0.29 | 0.27 | 0.15 | 0.06 | 0.01 | 0.00 | 0.00 | 0.96 | |
When multiple mating occurs, the probability that one male sires each embryo in a litter (see text).
The proportion of 10 000 simulated litters for which multiple paternity was successfully inferred at the number of loci indicated.
The proportion of 10 000 simulated litters for which multiple paternity was successfully inferred at one or more loci.
The distributions in Table 2 suggest that more litters showing multiple paternity at a single locus were observed than expected (expected proportion averaged across both competitive skews and all litter sizes = 0.23, observed: 17/33 = 0.52). One explanation is that mutations inflated the cell where observed multiple paternity was detected at only one locus.
We also quantified the power to detect five or more paternal alleles (i.e. triple paternity). With eight loci, the power to detect triple paternity asymptotically approached 75% as litter size increased, and was only 5% in a litter of five, 13% in a litter of six, and 21% in a litter of seven pups, under the conservative assumption of no competitive skew. Thus, the small litter sizes of mice, reductions of observed heterozygosity, and allele sharing among parents made detection of triple paternity if it occurred very unlikely. Nevertheless, triple paternity was detected in a single litter, consisting of eight pups. This litter showed five paternal alleles at one locus, as well as three paternal alleles at two additional loci. However, one of these five paternal alleles was separated by a maternal allele by one mutational step, so this could have been a mutational event.
Multiple mating
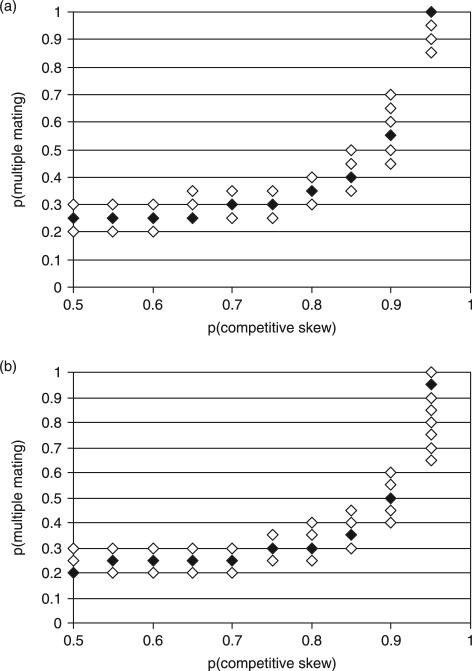
Maximum-likelihood estimates of the frequency of multiple mating, given observed levels of multiple paternity, heterozygosity, allele sharing, litter sizes, and allele frequencies, are shown in Fig. 1. The magnitude of the discrepancy between multiple mating and multiple paternity depended upon competitive skew. As competitive skew increased, increasing values of multiple mating were required to account for our observed level of multiple paternity. For example, at a competitive skew of 0.9 (in cases of multiple mating, one male had a 90% chance of fertilizing each embryo), multiple mating had to occur in 45–70% of all encounters to be consistent with our observed level of multiple paternity (23%). While no experimental data exist on competitive skew in wild mice, it may be as high as 0.95 in laboratory strains of mice (Levine 1967). At a competitive skew of 0.95, multiple mating occurs 85–100% of oestrus cycles in our simulations (Fig. 1a).
Fig. 1.
(a) The combinations of multiple mating and competitive skew that were consistent with an observed level of multiple paternity of 33/143 = 0.23. The criterion for multiple paternity was inferring three paternal alleles from at least one locus. Black diamonds indicate the maximum-likelihood estimate of multiple mating given a particular value of competitive skew; open diamonds indicate 95% credible intervals around this estimate. Regions of the figure without diamonds were inconsistent with our observed level of multiple paternity. (b) The combinations of multiple mating and competitive skew that were consistent with the more conservative estimate of multiple paternity, 29/143 = 0.20.
We also estimated the frequency of multiple mating using the more conservative estimate of multiple paternity of 29/143, excluding the six litters with presumed mutations (Fig. 1b). Similar patterns were found, and multiple mating was inferred to occur frequently, depending on the competitive skew.
Discussion
By genotyping mothers and their litters, we found clear evidence for multiple paternity in 29 of 143, or 0.20% of wild-caught litters. The true frequency of multiple mating depends on the average level of competitive skew, a parameter that has yet to be estimated from natural populations. Even with no competitive skew, multiple paternity and multiple mating are clearly common in house mice. By extension, sperm competition must be an evolutionarily important component of sexual selection in natural populations of house mice. Below, we discuss these results in light of mouse population structure. We then discuss the importance of competitive skew and the evolutionary consequences of multiple mating.
Multiple paternity and population structure
House mice are known to exhibit significant population structuring, meaning limited dispersal results in genetic differences among distant populations (Sage 1981). Even within single barns, mouse populations show structuring; mice from the west side of a barn showed significant differences from east-side mice in allozyme frequencies (Selander 1970). The finding of reduced heterozygosity in the present study is consistent with population structuring and inbreeding.
Strong local genetic differentiation is at least partially explained by social structure of mice. Local structure usually consists of a dominant male, as males will fight aggressively until a single dominant emerges (Brown 1953; Sage 1981). Once established, the boundaries of dominant males are surprisingly stable, moving less than 5 cm over 2 months in one experiment (Mackintosh 1970). There is effectively no migration of subordinate males over such boundaries. Female mice and juveniles move freely between territories of dominant males (Mackintosh 1970; Potts et al. 1991), but unfamiliar individuals tend to encounter more aggressive encounters from incumbent mice (Smith et al. 1994; Hurst & Barnard 1995). Estimates of individual movement ranges from approximately 3–19 feet (Young et al. 1950; Reimer & Petras 1968), and the average home range appears to be on the order of 20–50 square feet (Southern & Laurie 1946; Selander 1970).
Females often pair nonrandomly with dominant males, and sometimes assist dominant males in territory defence (Penn & Potts 1998, 1999; Drickamer et al. 2000; Penn 2002; Candolin 2003; Leinders-Zufall et al. 2004). In the laboratory, females mate indiscriminately with males until about 2 h prior to oestrus, at which time they mate almost exclusively with dominant males (Mossman & Drickamer 1996; Drickamer et al. 2000; Rolland et al. 2003). This time period is significant because females are most receptive to fertilizations approximately 2 h prior to oestrus. Females may also show preferences for mating with genetically unrelated males (Egid & Brown 1989; Potts et al. 1991; Penn 2002).
Given this type of social hierarchy, it has been suggested that dominant males are the only effectively breeding males in the population (Bronson 1979). However, while sexual development of subordinate males is suppressed, they regularly achieve fertilizations (DeFries & McClearn 1970; Oakeshott 1974), especially during initial establishment of dominance hierarchy (Wolff 1985). Our study clearly shows that females regularly mate with more than a single male. While we find evidence of multiple paternity, we cannot determine the social status of fathers. In fact, we cannot confidently assign specific sires to individual embryos due to extensive allele sharing.
Bronson (1979) suggested that at high densities, dominant males may exert greater control over subordinates more effectively, thus reducing their opportunity for multiple mating. This hypothesis leads to the prediction that multiple mating would be less common in high-density populations, the opposite of what we observed. Here, we find that multiple paternity was significantly more frequent in relatively high-density vs. low-density populations. This result suggests that there are more opportunities for multiple mating in high-density populations.
Competitive skew
Competitive skew is a fundamental parameter in the estimation of multiple mating. The mean and variance of competitive skew are unknown in wild mice. In laboratory mice, Levine (1967) mated single female Mus domesticus to two males in succession, and found competitive skew between 0.8 and 0.95. In Drosophila, competitive skew ranged between 0.49 and 1.00, with a median of 0.93 (Fiumera et al. 2005). A high competitive skew may be a general phenomenon among species with sperm competition.
Competitive skew may arise through many factors. One factor is mating order, which in mammals appears to provide an advantage to the first male (Levine 1967; Dewsbury 1984; Lacey et al. 1997). Others include genetic, environmental, or interaction factors that give some males an advantage over others. In Drosophila, allelic variation was significantly associated with at least some aspects of sperm competition (Clark et al. 1995, 2000; Fiumera et al. 2005), and it is possible that such associations also exist in house mice. In mammals, advantageous phenotypes in sperm competition could take the form of larger ejaculates (Delbarco-Trillo & Ferkin 2004), larger testes (Harcourt et al. 1981), shorter refractory periods between male re-mating, incapacitation of sperm from competing males, copulatory plug formation (Ramm et al. 2005), behavioural aspects such as mate guarding (Stockley & Preston 2004), or morphological features of sperm (Anderson & Dixson 2002; Moore et al. 2002). The relative frequency of such strategies in nature remains unknown. There is likely to be a mosaic of strategies employed by males depending on natural conditions and population characteristics.
Evolutionary consequences of multiple mating
Why might females mate multiply with different males in a single oestrus cycle? Possible explanations include fertilization assurance (Hunter et al. 1993; Hoogland 1998), increased genetic diversity of a litter (Madsen et al. 1992; Zeh & Zeh 2001; Tregenza & Wedell 2002; Head et al. 2005), and increased probability of finding genetically compatible gametes (Colegrave et al. 2002; Mays & Hill 2004). Females may mate multiply to conceal true paternity and avoid infanticide (Wolff & Macdonald 2004), a common behaviour in house mice, especially during times of dominance turnover (Silver 1995). Multiple mating in females could also be driven indirectly, by selection favouring multiple mating in males (Halliday & Arnold 1987).
In addition, multiple mating may be a strategy by which females select against selfish DNA elements. Males that harbour meiotic drivers, such as the well-characterized t allele in mice (Silver 1993; Lyon 2003), often include sperm with reduced fitness in their ejaculates. By mating multiply, a female may place ejaculates in competition and favour fertilization by wild-type sperm (Haig & Bergstrom 1995). The high frequency of multiple mating estimated in the present study may help explain the low frequency of the t allele in natural populations of house mice, especially in high-density populations. Consistent with this hypothesis, Ardlie & Silver (1996) found that three litters with unusually low transmission of the t allele were multiply sired.
In addition to affecting the reproductive fitness of an individual, multiple mating is expected to influence genetic variation on a population level. Multiple mating by females increases the effective population size of a species relative to monogamous and polygynous mating systems, since more alleles will be represented in the next generation (Sugg & Chesser 1994). For genes directly involved in sperm competition, selection may maintain variation through heterosis or nontransitivity (Prout & Bundgaard 1977; Clark et al. 2000).
Direct evidence for multiple mating exists for other mammal species. Using genetic and/or behavioural data, multiple mating has been inferred in marsupials (Kraaijeveld-Smit et al. 2002), shrews (Tegelström et al. 1991; Stockley et al. 1993), snowshoe hares (Burton 2002), and several rodent species (Birdsall & Nash 1973; Hanken & Sherman 1981; Boellstorff et al. 1994; Lacey et al. 1997; Baker et al. 1999; Bartmann & Gerlach 2001; Haynie et al. 2003; Shurtliff et al. 2005). Even in mammals where social pair bonding occurs, such as marmots, females may participate in extra-pair mating (Goossens et al. 1998). The present study adds house mice to this list, lending powerful resources of this model system to a better understanding of the evolutionary patterns and processes of sperm competition.
In the context of sperm competition, achieving fertilizations is clearly a desirable outcome among competing males. An especially interesting research avenue is to study competitive skew in natural populations. Of particular interest is the question of whether natural genetic variation can be associated with variation in competitive skew among males. Such studies have been conducted in Drosophila (Clark et al. 1995; Fiumera et al. 2005) and the results presented here suggest that such studies may reveal interesting evolutionary patterns in mice.
Acknowledgements
S. Barton, A. D'Urso, A. Kurosaki, J. Melcher-Post, T. Salcedo, and K. Smith assisted in trapping and/or preparation of specimens. We would like to thank the many people who gave us permission to trap mice on their property. J. Melcher-Post helped with genotyping. K. Kiesler, K. Chistopherson, and A. Baker provided technical input. R. Ree fielded many Python questions. Members of the Nachman laboratory provided critical discussion. J. Good read an early version of the manuscript. Three anonymous reviewers helped improve the manuscript. Funding was provided by an NIH postdoctoral fellowship F32GM070246-02 (M.D.D.), an NSF grant DEB0213013 (M.W.N.), and an NIH grant RO1GM074245-01 (M.W.N.).
Biography
M. D. Dean is a postdoctoral fellow in the laboratory of M. W. Nachman. He investigates evolutionary genetics of male reproductive traits, focusing on the mouse as a model system.
K. G. Ardlie studies mouse evolutionary and population genetics, with a particular interest in understanding the basis of common disease.
M. W. Nachman studies population, evolutionary and ecological genetics and genomics in mice and humans.
References
- Anderson MJ, Dixson AF. Sperm competition: motility and the midpiece in primates. Nature. 2002;416:496. doi: 10.1038/416496a. [DOI] [PubMed] [Google Scholar]
- Ardlie KG, Silver LM. Low frequency of mouse t haplotypes in wild populations is not explained by modifiers of meiotic drive. Genetics. 1996;144:1787–1797. doi: 10.1093/genetics/144.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie KG, Silver LM. Low frequency of t haplotypes in natural populations of house mice (Mus musculus domesticus). Evolution. 1998;52:1185–1196. doi: 10.1111/j.1558-5646.1998.tb01844.x. [DOI] [PubMed] [Google Scholar]
- Baker RJ, Makova KD, Chesser RK. Microsatellites indicate a high frequency of multiple paternity in Apodemus (Rodentia). Molecular Ecology. 1999;8:107–111. doi: 10.1046/j.1365-294x.1999.00541.x. [DOI] [PubMed] [Google Scholar]
- Bartmann S, Gerlach G. Multiple paternity and similar variance in reproductive success of male and female wood mice (Apodemus sylvaticus) housed in an enclosure. Ethology. 2001;107:889–899. [Google Scholar]
- Birdsall DA, Nash D. Occurrence of successful multiple insemination of females in natural population of deer mice (Peromyscus maniculatus). Evolution. 1973;27:106–110. doi: 10.1111/j.1558-5646.1973.tb05922.x. [DOI] [PubMed] [Google Scholar]
- Birkhead TR, Pizzari T. Postcopulatory sexual selection. Nature Reviews Genetics. 2002;3:262–273. doi: 10.1038/nrg774. [DOI] [PubMed] [Google Scholar]
- Boellstorff DE, Owings DH, Penedo MCT, Hersek MJ. Reproductive behaviour and multiple paternity of California ground squirrels. Animal Behaviour. 1994;47:1057–1064. [Google Scholar]
- Boursot P, Auffray J-C, Britton-Davidian J, Bonhomme F. The evolution of house mice. Annual Review of Ecology and Systematics. 1993;24:119–152. [Google Scholar]
- Boursot P, Din W, Anand R, et al. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. Journal of Evolutionary Biology. 1996;9:391–415. [Google Scholar]
- Bronson FH. The reproductive ecology of the house mouse. Quarterly Review of Biology. 1979;54:265–299. doi: 10.1086/411295. [DOI] [PubMed] [Google Scholar]
- Bronson FH. Mammalian Reproductive Biology. University of Chicago Press; Chicago, Illinois: 1989. [Google Scholar]
- Brown RZ. Social behavior, reproduction, and populations changes in the house mouse (Mus musculus L.). Ecological Monographs. 1953;23:217–240. [Google Scholar]
- Burton C. Microsatellite analysis of multiple paternity and male reproductive success in the promiscuous snowshoe hare. Canadian Journal of Zoology. 2002;80:1948–1956. [Google Scholar]
- Candolin U. The use of multiple cues in mate choice. Biological Reviews of the Cambridge Philosophical Society. 2003;78:575–595. doi: 10.1017/s1464793103006158. [DOI] [PubMed] [Google Scholar]
- Carroll LS, Meagher S, Morrison L, Penn DJ, Potts WK. Fitness effects of a selfish gene (the Mus t complex) are revealed in an ecological context. Evolution. 2004;58:1318–1328. doi: 10.1111/j.0014-3820.2004.tb01710.x. [DOI] [PubMed] [Google Scholar]
- Clark AG, Aguadé M, Prout T, Harshman LG, Langley CH. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Begun DJ. Female genotypes affect sperm displacement in Drosophila. Genetics. 1998;149:1487–1493. doi: 10.1093/genetics/149.3.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Begun DJ, Prout T. Female–male interactions in Drosophila sperm competition. Science. 1999;283:217–220. doi: 10.1126/science.283.5399.217. [DOI] [PubMed] [Google Scholar]
- Clark AG, Dermitzakis ET, Civetta A. Nontransitivity of sperm precedence in Drosophila. Evolution. 2000;54:1030–1035. doi: 10.1111/j.0014-3820.2000.tb00102.x. [DOI] [PubMed] [Google Scholar]
- Colegrave N, Kotiaho JS, Tomkins JL. Mate choice or polyandry: reconciling genetic compatibility and good genes sexual selection. Evolutionary Ecology Research. 2002;4:911–917. [Google Scholar]
- DeFries JC, McClearn GE. Social dominance and Darwinian fitness in the laboratory mouse. American Naturalist. 1970;104:408–411. [Google Scholar]
- Delbarco-Trillo J, Ferkin MH. Male mammals respond to a risk of sperm competition conveyed by odours of conspecific males. Nature. 2004;431:446–449. doi: 10.1038/nature02845. [DOI] [PubMed] [Google Scholar]
- Dewsbury DA. Sperm competition in muroid rodents. In: Smith RL, editor. Sperm Competition and the Evolution of Animal Mating Systems. Academic Press; New York: 1984. pp. 547–571. [Google Scholar]
- Dietrich W, Katz H, Lincoln SE, et al. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich WF, Miller J, Steen R, et al. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Din W, Anand R, Boursot P, et al. Origin and radiation of the house mouse: clues from nuclear genes. Journal of Evolutionary Biology. 1996;9:519–539. [Google Scholar]
- Drickamer LC, Gowaty PA, Holmes CM. Free female mate choice in house mice affects reproductive success and offspring viability and performance. Animal Behaviour. 2000;59:371–378. doi: 10.1006/anbe.1999.1316. [DOI] [PubMed] [Google Scholar]
- Eberhard WG. Female Control: Sexual Selection by Cryptic Female Choice. Princeton University Press; Princeton, New Jersey: 1996. [Google Scholar]
- Edwards RG. Selective fertilization following the use of sperm mixtures in the mouse. Nature. 1955;175:215–216. doi: 10.1038/175215b0. [DOI] [PubMed] [Google Scholar]
- Egid K, Brown JL. The major histocompatibility complex and female mating preferences in mice. Animal Behaviour. 1989;38:548–550. [Google Scholar]
- Ehman KD, Scott ME. Microsatellite analysis reveals that female mice are indiscriminate when choosing infected or dominant males in an arena setting. Parasitology. 2004;129:723–731. doi: 10.1017/s0031182004006195. [DOI] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics. 2005;169:243–257. doi: 10.1534/genetics.104.032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach G. Emigration mechanisms in feral house mice — a laboratory investigation of the influence of social structure, population density, and aggression. Behavioral Ecology and Sociobiology. 1996;39:159–170. [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, et al. Genome sequence of the brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Goossens B, Graziani L, Waits LP, et al. Extra-pair paternity in the monogamous Alpine marmot revealed by nuclear DNA microsatellite analysis. Behavioral Ecology and Sociobiology. 1998;43:281–288. [Google Scholar]
- Haig D, Bergstrom CT. Multiple mating, sperm competition, and meiotic drive. Journal of Evolutionary Biology. 1995;8:265–282. [Google Scholar]
- Halliday R, Arnold SJ. Multiple mating by females: a perspective from quantitative genetics. Animal Behaviour. 1987;35 [Google Scholar]
- Hanken J, Sherman PW. Multiple paternity in Belding's ground squirrel litters. Science. 1981;212:351–353. doi: 10.1126/science.7209536. [DOI] [PubMed] [Google Scholar]
- Harcourt AH, Harvey PH, Larson SG, Short RV. Testis weight, body weight and breeding system in primates. Nature. 1981;293:55–57. doi: 10.1038/293055a0. [DOI] [PubMed] [Google Scholar]
- Hartung TG, Dewsbury DA. A comparative analysis of copulatory plugs in muroid rodents and their relationship to copulatory behavior. Journal of Mammalogy. 1978;59:717–723. [Google Scholar]
- Haynie ML, Van Den Bussche RA, Hoogland JL, Gilbert DA. Parentage, multiple paternity, and breeding success in Gunnison's and Utah prarie dogs. Journal of Mammalogy. 2003;84:1244–1253. [Google Scholar]
- Head ML, Hunt J, Jennions MD, Brooks R. The indirect benefits of mating with attractive males outweigh the direct costs. PLoS Biology. 2005;3:e33. doi: 10.1371/journal.pbio.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogland JL. Why do female Gunnison's prairie dogs copulate with more than one male? Animal Behaviour. 1998;55:351–359. doi: 10.1006/anbe.1997.0575. [DOI] [PubMed] [Google Scholar]
- Hunter FM, Petrie M, Otronen M, Birkhead TR, Møller AP. Why do females copulate repeatedly with one male? Trends in Ecology & Evolution. 1993;8:21–26. doi: 10.1016/0169-5347(93)90126-A. [DOI] [PubMed] [Google Scholar]
- Hurst JL, Barnard CJ. Kinship and social tolerance among female and juvenile wild house mice: kin bias but not kin discrimination. Behavioral Ecology and Sociobiology. 1995;36:333–342. [Google Scholar]
- Ideraabdullah FY, de la Casa-Esperon E, Bell TA, et al. Genetic and haplotype diversity among wild-derived mouse inbred strains. Genome Research. 2004;14:1880–1887. doi: 10.1101/gr.2519704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Clark AG. Bayesian sperm competition estimates. Genetics. 2003;163:1193–1199. doi: 10.1093/genetics/163.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nature Genetics. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- Kichler K, Holder MT, Davis SK, Márquez MR, Owens DW. Detection of multiple paternity in the Kemp's ridley sea turtle with limited sampling. Molecular Ecology. 1999;8:819–830. [Google Scholar]
- Kraaijeveld-Smit FJL, Ward SJ, Temple-Smith PD. Multiple paternity in a field population of a small carnivorous marsupial, the agile antechinus, Antechinus agilis. Behavioral Ecology and Sociobiology. 2002;52:84–91. [Google Scholar]
- Lacey EA, Wieczorek JR, Tucker PK. Male mating behaviour and patterns of sperm precedence in Arctic ground squirrels. Animal Behaviour. 1997;53:767–779. [Google Scholar]
- Leinders-Zufall T, Brennan P, Widmayer P, et al. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306:1033–1037. doi: 10.1126/science.1102818. [DOI] [PubMed] [Google Scholar]
- Levine L. Sexual selection in mice. IV. Experimental demonstration of selective fertilization. American Naturalist. 1967;101:289–294. [Google Scholar]
- Li CC, Weeks DE, Chakravart A. Similarity of DNA fingerprints due to chance and relatedness. Human Heredity. 1993;43:45–52. doi: 10.1159/000154113. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Transmission ratio distortion in mice. Annual Review of Genetics. 2003;37:393–408. doi: 10.1146/annurev.genet.37.110801.143030. [DOI] [PubMed] [Google Scholar]
- Mackintosh JH. Territory formation by laboratory mice. Animal Behaviour. 1970;18:177–183. [Google Scholar]
- Madsen T, Shine R, Loman J, Håkansson T. Why do female adders copulate so frequently? Nature. 1992;355:440–441. [Google Scholar]
- Mays HLJ, Hill GE. Choosing mates: good genes versus genes that are a good fit. Trends in Ecology & Evolution. 2004;19:554–559. doi: 10.1016/j.tree.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Moore H, Dvorakova K, Jenkins N, Breed W. Exceptional sperm cooperation in the wood mouse. Nature. 2002;418:174–177. doi: 10.1038/nature00832. [DOI] [PubMed] [Google Scholar]
- Mosig DW, Dewsbury DA. Plug fate in the copulatory behavior of rats. Psychonomic Science. 1970;20:315–316. [Google Scholar]
- Mossman CA, Drickamer LC. Odor preferences of female house mice (Mus domesticus) in seminatural enclosures. Journal of Comparative Psychology. 1996;110:131–138. doi: 10.1037/0735-7036.110.2.131. [DOI] [PubMed] [Google Scholar]
- Oakeshott JG. Social dominance, aggressiveness and mating success among male house mice (Mus musculus). Oecologia. 1974;15:143–158. doi: 10.1007/BF00345742. [DOI] [PubMed] [Google Scholar]
- Parker GA. Sperm competition and its evolutionary consequences in the insects. Biological Reviews of the Cambridge Philosophical Society. 1970;45:525–567. [Google Scholar]
- Penn DJ. The scent of genetic compatibility: sexual selection and the major histocompatibility complex. Ethology. 2002;108:1–21. [Google Scholar]
- Penn D, Potts W. MHC-disassortative mating preferences reversed by cross-fostering. Proceedings of the Royal Society B: Biological Sciences. 1998;265:1299–1306. doi: 10.1098/rspb.1998.0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn DJ, Potts WK. The evolution of mating preferences and major histocompatibility complex genes. American Naturalist. 1999;153:145–164. doi: 10.1086/303166. [DOI] [PubMed] [Google Scholar]
- Potts WK, Manning CJ, Wakeland EK. Mating patterns in seminatural populations of mice influenced by MHC genotype. Nature. 1991;352:619–621. doi: 10.1038/352619a0. [DOI] [PubMed] [Google Scholar]
- Prager EM, Orrego C, Sage RD. Genetic variation and phylogeography of central Asian and other house mice, including a major new mitochondrial lineage in Yemen. Genetics. 1998;150:835–861. doi: 10.1093/genetics/150.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prout T, Bundgaard J. The population genetics of sperm displacement. Genetics. 1977;85:95–124. doi: 10.1093/genetics/85.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller DC, Goodnight KF. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Ramm SA, Parker GA, Stockley P. Sperm competition and the evolution of male reproductive anatomy in rodents. Proceedings of the Royal Society B: Biological Sciences. 2005;272:949–955. doi: 10.1098/rspb.2004.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer JD, Petras ML. Some aspects of commensal populations of Mus musculus in southwestern Ontario. Canadian Field-Naturalist. 1968;82:32–42. [Google Scholar]
- Rolland C, MacDonald DW, de Fraipont M, Berdoy M. Free female choice in house mice: leaving the best for last. Behaviour. 2003;140:1371–1388. [Google Scholar]
- Rousset F, Raymond M. Testing heterozygote excess and deficiency. Genetics. 1995;140:1413–1419. doi: 10.1093/genetics/140.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RD. Wild mice. In: Foster HL, Small JD, Fox JG, editors. The Mouse in Biomedical Research. Academic Press; New York: 1981. pp. 39–90. [Google Scholar]
- Schalkwyk LC, Jung M, Daser A, et al. Panel of microsatellite markers for whole-genome scans and radiation hybrid mapping and a mouse family tree. Genome Research. 1999;9:878–887. doi: 10.1101/gr.9.9.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proceedings of the National Academy of Sciences, USA. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander RK. Behavior and genetic variation in natural populations. American Zoologist. 1970;10:53–66. doi: 10.1093/icb/10.1.53. [DOI] [PubMed] [Google Scholar]
- Shurtliff QR, Pearse DE, Rogers DS. Parentage analysis of the canyon mouse (Peromyscus crinitus): evidence for multiple paternity. Journal of Mammalogy. 2005;86:531–540. [Google Scholar]
- Silver LM. The peculiar journey of a selfish chromosome: mouse t haplotypes and meiotic drive. Trends in Genetics. 1993;9:250–254. doi: 10.1016/0168-9525(93)90090-5. [DOI] [PubMed] [Google Scholar]
- Silver L. Mouse Genetics: Concepts and Applications. Oxford University Press; Oxford: 1995. [Google Scholar]
- Smith J, Barnard CJ, Hurst JL. Kin-biased behaviour in male wild house mice: mixed-paternity grouping a group member versus kin discrimination. Ethology. 1994;97:141–160. [Google Scholar]
- Southern HN, Laurie EMO. The house mouse in (Mus musculus) in corn ricks. Journal of Animal Ecology. 1946;7:134–149. [Google Scholar]
- Southwick CH. Regulatory mechanisms of house mouse populations: social behavior affecting litter survival. Ecology. 1955;36:627–634. [Google Scholar]
- Stockley P, Preston BT. Sperm competition and diversity in rodent copulatory behaviour. Journal of Evolutionary Biology. 2004;17:1048–1057. doi: 10.1111/j.1420-9101.2004.00742.x. [DOI] [PubMed] [Google Scholar]
- Stockley P, Searle JB, MacDonald DW, Jones CS. Female multiple mating behaviour in the common shrew as a strategy to reduce inbreeding. Proceedings of the Royal Society B: Biological Sciences. 1993;254:173–179. doi: 10.1098/rspb.1993.0143. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences, USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugg DW, Chesser RK. Effective population sizes with multiple paternity. Genetics. 1994;137:1147–1155. doi: 10.1093/genetics/137.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegelström H, Searle J, Brookfield J, Mercer S. Multiple paternity in wild common shrews (Sorex araneus) is confirmed by DNA-fingerprinting. Heredity. 1991;66(3):373–379. doi: 10.1038/hdy.1991.47. [DOI] [PubMed] [Google Scholar]
- Tregenza T, Wedell N. Polyandrous females avoid costs of inbreeding. Nature. 2002;415:71–73. doi: 10.1038/415071a. [DOI] [PubMed] [Google Scholar]
- Wade CM, Kulbokas EJ, 3rd, Kirby AW, et al. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Whittaker JC, Harbord RM, Boxall N, et al. Likelihood-based estimation of microsatellite mutation rates. Genetics. 2003;164:781–787. doi: 10.1093/genetics/164.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff RJ. Mating behaviour and female choice: their relation to social structure in wild caught house mice (Mus musculus) housed in a semi-natural environment. Journal of Zoology. 1985;207:43–51. [Google Scholar]
- Wolff JO, Macdonald DW. Promiscuous females protect their offspring. Trends in Ecology & Evolution. 2004;19:127–134. doi: 10.1016/j.tree.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Awaji M. Improvement of polymerase chain reaction condition to detect polymorphic dinucleotide repeat microsatellite DNA marker in the puffer fish Fugu rubripes. Fisheries Science. 2000;66:397–399. [Google Scholar]
- Young H, Strecker RL, Emlen JT., Jr Localization of activity in two indoor populations of house mice, Mus musculus. Journal of Mammalogy. 1950;31:403–410. [Google Scholar]
- Zeh JA, Zeh DW. Reproductive mode and the genetic benefits of polyandry. Animal Behaviour. 2001;61:1051–1063. [Google Scholar]