Abstract
Ligand binding to the periplasmic domain of the transmembrane aspartate receptor generates an intramolecular conformational change which spans the bilayer and ultimately signals the cytoplasmic CheA histidine kinase, thereby triggering chemotaxis. The receptor is a homodimer stabilized by the interface between its two identical subunits: the present study investigates the role of the periplasmic and transmembrane regions of this interface in the mechanism of transmembrane signaling. Free cysteines and disulfide bonds are engineered into selected interfacial positions, and the resulting effects on the transmembrane signal are assayed by monitoring in vitro regulation of kinase activity. Three of the 14 engineered cysteine pairs examined, as well as six of the 14 engineered disulfides, cause perturbations of the interface structure which essentially destroy transmembrane regulation of the kinase. The remaining 11 cysteine pairs, and eight engineered disulfides covalently linking the two subunits at locations spanning positions 18–75, are observed to retain significant transmembrane kinase regulation. The eight functional disulfides positively identify adjacent faces of the two N-terminal helices in the native receptor dimer and indicate that large regions of the periplasmic and transmembrane subunit interface remain effectively static during the transmembrane signal. The results are consistent with a model in which the subunit interface plays a structural role, while the second membrane-spanning helix transmits the ligand-induced signal across the bilayer to the kinase binding domain. The effects of engineered cysteines and disulfides on receptor methylation in vitro are also measured, enabling direct comparison of the in vitro methylation and phosphorylation assays.
The aspartate receptor of Escherichia coli, Salmonella typhimurium, and related bacteria serves as an initial chemosensor in a pathway which regulates taxis toward chemoattractants and away from repellents [reviewed by Stock and Surrette (1995), Parkinson (1993), Hazelbauer (1992), and Bourret et al. (1991)]. The periplasmic domain of this transmembrane receptor binds aspartate and transmits a conformational signal through the bilayer to its cytoplasmic domain, which together with the histidine kinase CheA and the coupling protein CheW exists as a kinetically stable ternary complex (Schuster et al., 1993; Gegner et al., 1992; Ninfa et al., 1991; Borkovich & Simon, 1989). The function of this receptor–CheW–CheA ternary complex is to phosphorylate the response regulator protein CheY in a reaction which is inhibited by ligand binding to the receptor. Ultimately, the steady-state level of phospho-CheY controls the switching of the flagellar motor between its two directions of rotation, thereby enabling chemotaxis. Homologous histidine kinase signaling pathways regulated by specific environmental or physiological stimuli are present in all prokaryotic organisms examined to date and appear to be widespread in eukaryotes as well (Alex & Simon, 1994; Ota & Varshavsky, 1993; Chang et al., 1993).
The present work investigates the mechanism of transmembrane signaling by the aspartate receptor. The focus is the structural interface joining the two subunits of this homodimeric protein, where the following questions are addressed. Does the signal require a change in the association state of the dimer or a rearrangement of the packing interface between subunits within the dimer? Does the signal trigger movement of specific transmembrane helices outside the subunit interface? The answers to such questions have broader relevance, since the aspartate receptor is representative of a large class of membrane-spanning receptor proteins exhibiting direct primary and transmembrane structure homologies, found in both the prokaryotic and eukaryotic versions of histidine kinase pathways [reviewed by Ecker (1995), Stock and Surrette (1995), Alex and Simon (1994), Parkinson (1993), and Hazelbauer (1992)].
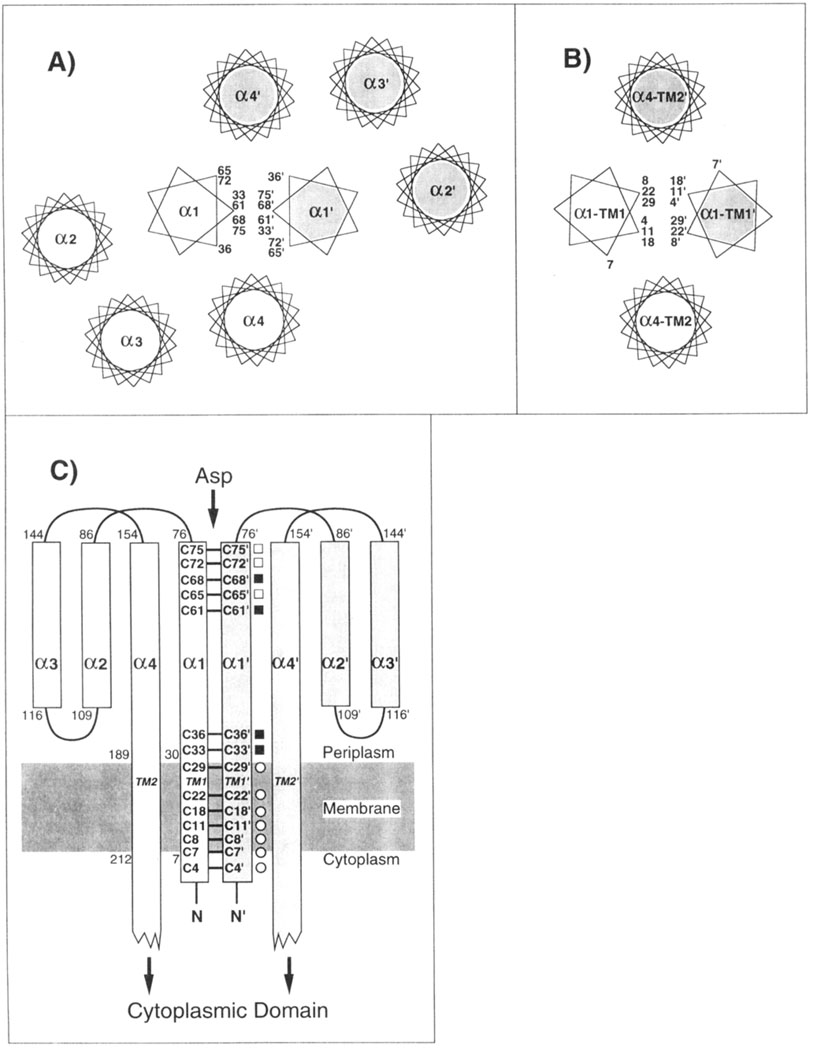
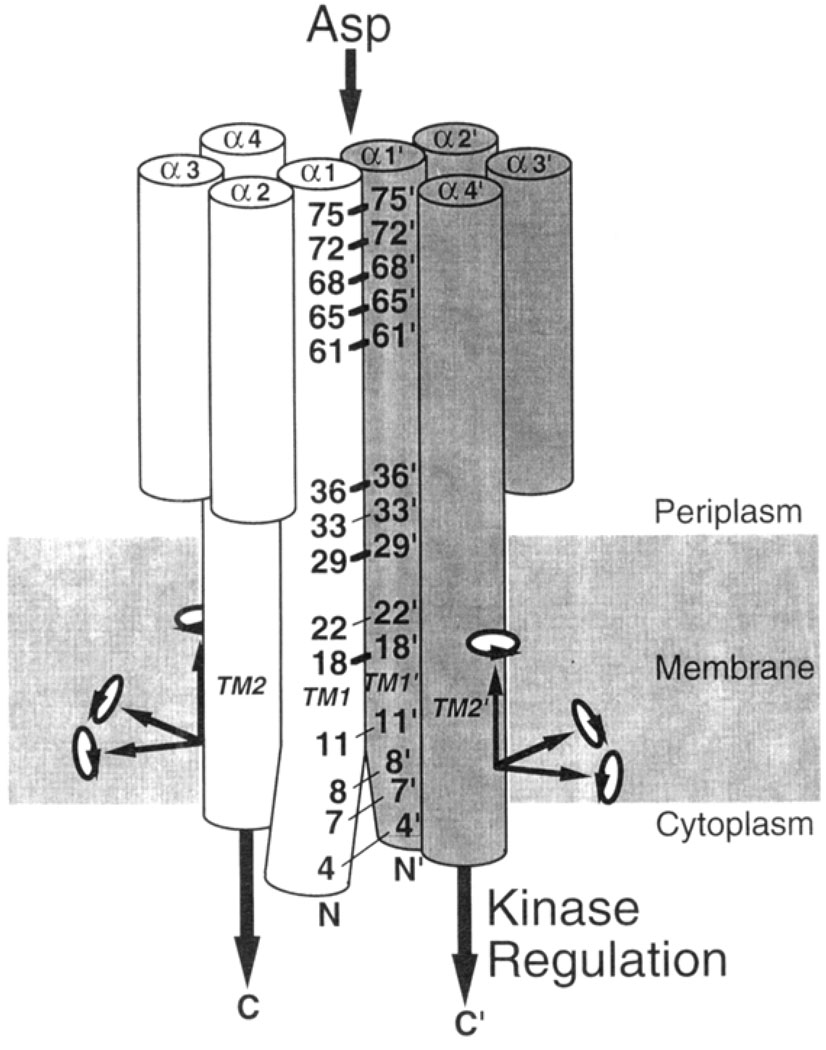
Mechanistic studies of the aspartate receptor are facilitated by the availability of extensive structural information regarding its periplasmic ligand binding and transmembrane domains, which together generate the transmembrane signal. Schematic outlines of both domains are presented in Figure 1.
FIGURE 1.
Schematic structure of the aspartate receptor, including the locations of the engineered cysteines. (A) Schematic representation of the periplasmic helices, looking down their long axes toward the cytoplasm (Milburn et al., 1991). The periplasmic domain consists of two subunits distinguished by shading and primes, each forming a four-helix bundle. The α1 and α1′ helices at the subunit interface form a coiled-coil pair represented by 7-fold helical wheels (Scott et al., 1993), while the remaining standard helices are shown as 18-fold helical wheels. Rotational positions of the engineered cysteines on helix α1 are indicated. (B) Model for the packing of the transmembrane helices (Scott & Stoddard, 1994; Pakula & Simon, 1992: Lynch & Koshland, 1991: Falke et al., 1988), looking toward the cytoplasm. (C) Schematic representation of the periplasmic and transmembrane helices of each subunit, illustrating the positions of the engineered cysteine pairs along the length of helix α1/TM1, No cysteine pairs were placed in the region between positions 36 and 61, where the α1 and α1′ helices separate to create a 6 Å gap in the crystal structure (Milburn et al., 1991). Symbols indicated the degree of motion required for disulfide formation between a given cysteine pair: no motion required (filled square); ≤1.5 Å translation required (open square); undetermined (open circle).
The crystallographic structure of the periplasmic ligand binding domain (Milburn et al., 1991; Yeh et al., 1993) reveals a dimer of two identical subunits, each a four-helix bundle with nearly parallel long axes (Figure 1). Two symmetric aspartate binding sites lie at the interface between the two subunits, near the extreme periplasmic end distal to the membrane. Contacts between the two subunits are primarily between their N-terminal helices α1 and α1′ (the prime distinguishes helices from different subunits), which form a coiled-coil about the C2 rotational symmetry axis at the center of the dimer (Scott et al., 1993).
The transmembrane domain of the receptor consists of four membrane-spanning helices, whose packing has been mapped by engineered disulfide studies (Scott & Stoddard, 1994; Pakula & Simon, 1992b; Lynch & Koshland, 1991; Falke et al., 1988). At the center of the transmembrane domain lie the two N-terminal helices, one from each subunit of the dimer, which begin in the cytoplasm then span the bilayer where they are labeled TM1 and TM1′. These helices emerge in the periplasm where they are believed to be continuous with the α1 and α1′ helices, respectively, lying at the subunit interface in the periplasmic domain. The remaining two transmembrane helices, TM2 and TM2′, are proposed to be extensions of the periplasmic helices α4 and α4′. respectively, and are observed to lie farther from the center of the helix cluster spanning the bilayer (Figure 1).
Two different classes of models have been proposed for the conformational change triggered by aspartate: one class proposes that the two subunits move relative to one another, thereby altering the subunit interface, while the other class suggests that the signal is carried by more localized structural changes within each subunit (Lee et al., 1995; Kim, 1994; Danielson et al., 1994; Scott & Stoddard, 1994; Parkinson, 1993: Pakula & Simon, 1992a; Stoddard et al., 1992; Lynch & Koshland, 1992; Milligan & Koshland, 1991; Mowbray & Koshland, 1987). In order to probe the role of the subunit interface in transmembrane signaling, the present study engineers free cysteines and disulfide bonds into the interface, thereby generating targetted perturbations and covalent links between α1 and α1′ in the perplasmic domain, or between TM1 and TM1′ in the transmembrane domain. These alterations provide a powerful tool with which to resolve different models for the transmembrane signal. For example, models which propose significant structural rearrangements of the subunit interface (a change in the number of subunits, or movement of the subunits relative to one another) predict that the stability conferred to the interface by an engineered disulfide bond will disrupt signal transmission. In contrast, models which propose that the interface is static, such that its structure is unaltered by ligand binding, predict that an intersubunit disulfide may have little or no effect on the transmembrane signal.
An in vitro method is employed to quantitate the effects of engineered cysteines and disulfides on signal transmission across the bilayer. This direct activity assay measures ligand-sensitive, transmembrane regulation of the kinase CheA in the reconstituted receptor–CheW–CheA ternary complex (Borkovich et al., 1989). The results indicate that (i) the periplasmic and transmembrane regions of the subunit interface are effectively static during the transmembrane signal, while (ii) some engineered perturbations of the interface significantly distort the receptor structure. For comparison, the effects of engineered cysteines and disulfides on in vitro methylation of the receptor are also examined. The methylation and phosphorylation results are generally in agreement, although exceptions to this rule indicate that the two assays may monitor different features of receptor structure.
MATERIALS AND METHODS
Materials
The aspartate receptor (Tar), kinase (CheA), coupling protein (CheW), and methyltransferase (CheR) of S. typhimurium, as well as the phosphoacceptor protein (CheY) of E. coli, were all expressed in E. coli. The strain used for expression of plasmids bearing the aspartate receptor gene was E. coli RP3808 [Δ(cheA-cheZ) DE2209 tsr-1 leuB6 his-4 eda-50 rpsL136 [thi-1 Δ (gal-attL) DE99 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx-78] /mks/] while chemotaxis swarm assays were conducted using plasmids expressed in E. coli RP8611 [Δ(tsr)DE7028 A(tar-tap)DE5201 zbd::Tn5 Δ(trg)-DE100 leuB6 his-4 rpsL136 [thi-1 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx-78] /CP362 of G. Hazelbauer via F. Dahlquist, pa/]. Both strains were kindly provided by Sandy Parkinson (University of Utah; Liu & Parkinson, 1989). Strains and plasmids used for expression of CheA (HB101/pM04), CheW (HB101/pME105), and CheR (JM109/pME43) were kindly provided by Jeff Stock (Princeton University; Stock et al., 1987; Simms et al., 1987), while the strain-plasmid combination used for expression of CheY (RBB455/pRBB40) was graciously provided by Bob Bourret (University of North Carolina; Bourret et al., 1993). Radiolabeled chemicals [α-35S]dATP (1000 Ci/mmol) and [3H-methyl]-S-adenosyl-methionine (55–85 mCi/mmol) were purchased from Dupont-NEN. [γ-32P]ATP (6000 Ci/mmol) was purchased from Amersham.
Cloning and Mutagenesis
The gene encoding the wild-type aspartate receptor under the control of its native promoter was subcloned on an EcoRI–HindIII fragment (Falke & Koshland, 1987) into pBluescript KS− (Stratagene, La Jolla, CA) to create pSCF6, the phagemid used for mutagenesis and overexpression of wild-type and engineered receptors. Site-directed mutagenesis was carried out according to Kunkel et al. (1991), with the modifications described by the Muta-Gene phagemid mutagenesis kit (Bio-Rad).
Preparation of Membranes Containing the Aspartate Receptor
Membranes containing wild-type or cysteine-engineered derivatives of the aspartate receptor were prepared according to Foster et al. (1985) with the following modifications, which were designed to minimize oxidation of engineered cysteines during preparation.
E. coli strain RP3808 was transformed with pSCF6. Saturated liquid cultures were grown at 37 °C in Luria Broth (Ausubel et al., 1992) and then diluted 1/250 into minimal growth medium containing Vogel Bonner Citrate components (Vogel & Bonner, 1956) supplemented with 0.75% glycerol (v/v), 40 µg/mL d,l-histidine, 20 µg/mL l-methionine, 20 µg/mL l-leucine, 20 µg/mL l-threonine, 1 µg/mL thiamine, and 100 µg/mL ampicillin, all prepared with deionized water to minimize metal-catalyzed cysteine oxidation in the periplasm or during membrane isolation. Cells were grown for 14–18 h at 30 °C to early stationary phase and then were pelleted by centrifugation (11000g, 8 min) and resuspended in ice-cold low-salt buffer containing 0.1 M NaH2PO4, pH 7.0 with NaOH, 10% (w/v) glycerol, 10 mM EDTA, 1 mM PMSF, 5 mM 1,10-phenanthroline, and 50 mM DTT. Cells were lysed by french press, and unbroken cells were removed by centrifugation at 12000g for 20 min. The membranes in the supernatant were pelleted at 2 °C for 60 min at 186000g in a Ti45 rotor (Beckman), and then the pellet was resuspended in 10 mL of low-salt buffer by extrusion through an 18 guage syringe needle, followed by a 21 guage needle. The membranes were further suspended by sonication, with cooling in an ice–NaCl water bath, for 3 × 20 s intervals using an 1/8 inch microtip set at 50% maximum power (Heat-Systems, Inc.). These membrane pelleting and resuspension steps were repeated using the following buffers: two washes in high-salt buffer containing 20 mM NaH2PO4, pH 7.0 with NaOH, 2 M KCl, 10% (w/v) glycerol, 10 mM EDTA, 5 mM DTT, 1 mM 1,10-phenanthroline, and 0.5 mM PMSF; then one wash in final buffer containing 20 mM NaH2PO4, pH 7.0 with NaOH, 10% (w/v) glycerol, 0.1 mM EDTA, and 1 mM PMSF. Membranes were quick-frozen in liquid N2 and stored at −70 °C for up to 6 months.
The concentration of the aspartate receptor in the resulting membrane preparations was determined as follows. The total membrane protein concentration was measured via the BCA assay (Stoscheck, 1990), in which the color-developing incubation was carried out at 65 °C in the presence of 0.1 % SDS. To ascertain the fraction of this total protein comprised by the receptor, the membrane components were resolved on 7.5% SDS–PAGE gels (Laemmli, 1970), and the intensities of the Coomassie-blue stained protein bands were integrated by laser densitometry (UltroScan XL, LKB).
Preparation of Soluble Chemotaxis Components
CheA, CheW, and CheY were purified to ≥95% homogeneity according to published procedures modified as follows. CheA (Stock et al., 1988) was purified by FPLC on a Q-Sepharose HP 26/10 column (Pharmacia) prior to Affi-Gel blue (Bio-Rad) liquid chromatography and FPLC on a Superdex 200 16/60 column (Pharmacia). CheW (Stock et al., 1987) was isolated by DEAE Sephacel liquid chromatography followed by FPLC on a Superdex 75 16/60 column (Pharmacia). CheY was purified according to the procedure of Bourret et al. (1993). Proteins were quick frozen in liquid N2 and stored at −70 °C in buffer containing 10 mM Tris base, pH 7.5 with HCl, 10% (v/v) glycerol, 0.1 mM EDTA, and 1 mM PMSF. The storage buffer for CheA also contained 25 µM DTT. Total protein concentrations were determined by BCA assays as described above.
Cell lysate containing overexpressed CheR was isolated according to Simms et al. (1987), except that cells were opened by french press in buffer containing 0.1M NaH2PO4, pH 7.0 with NaOH, 10% (w/v) glycerol, 10 mM EDTA, 5 mM DTT, and 1 mM PMSF. The final cell-free supernatant was extensively dialyzed against a buffer containing 50 mM Tris base, pH 7.2 with HCl, 5 mM EDTA, and 10% (w/v) glycerol before storage at −70 °C.
Reduction and Oxidation of Cysteine-Containing Receptors
Formation of disulfide-linked receptors was accomplished using either of two oxidation systems to catalyze disulfide bond formation, while reduction of disulfide-linked receptors was carried out with DTT. Briefly, membrane suspensions containing 10 µM receptor dimer were treated 20 min at 37 °C with (i) 0.2 mM Cu(II)•(1,10-phenanthro-line)3 and ambient O2 [approximately 250 µM; see Careaga and Falke (1992) and Falke and Koshland (1987)], (ii) 1.0 mM I2 (added as a 10 mM stock solution prepared by dissolving iodine crystals in ethanol; Pakula & Simon, 1991), or (iii) 0.1 M DTT. By the end of this incubation the I2 reagent was depleted, while the O2 reaction was inactivated by addition of 0.1 mM sodium persulfate, which depletes the remaining O2. Subsequently the reactions were transferred to ice and used immediately in phosphorylation or methylation assays.
In Vitro Phosphorylation Reactions
The ability of the aspartate receptor to regulate the activity of the CheA kinase was quantitated using a coupled phosphorylation assay, which monitors the production of phospho-CheY by the receptor–CheW–CheA ternary complex (Borkovich & Simon, 1991; Ninfa et al., 1991; Borkovich et al., 1989). Reactions consisted of isolated receptor-containing E. coli membranes (see above) to which purified proteins were added to yield the following monomeric concentrations: ~6 µM receptor (only half of which is likely to be accessible for kinase binding in the sonicated membrane preparation, which has a randomized sidedness), 2 µM CheW, 0.25 µM CheA kinase, and 10 µM. CheY. These concentrations of receptor, CheW, and kinase were well below the KD for ternary complex formation (Gegner et al., 1992). Together these conditions ensured that kinase autophosphorylation in the activated ternary complex varied linearly with both (i) the receptor affinity for CheW and kinase, which controlled the amount of ternary complex formed, and (ii) the Vmax of autophosphorylation in the ternary complex. Moreover, the indicated molar excess of substrate CheY ensured that autophosphorylation of CheA was the rate-limiting step in the reaction. Thus the reconstituted system yielded a phosphorylation rate which was highly sensitive to changes in receptor-mediated kinase regulation.
A relevant feature of the assay was a high molar ratio of receptor/kinase. This was necessary both to stay below the KD for ternary complex formation, as explained above, and to minimize the background signal of the free kinase, which was not regulated by the receptor. Because the kinase molecules coupled to the unliganded receptor have greatly enhanced activity, the phosphorylation rate of the receptor-bound kinase was able to overwhelm that of the free kinase, thereby maximizing sensitivity to receptor regulation.
Due to the high receptor/kinase ratio, however, the assay monitors only a small fraction of the engineered receptor population, which in principle could complicate interpretation of results when the receptor population is heterogeneous. This was not a problem in the studies described herein for the following reasons. The described experiments compared the kinase regulation of two engineered receptor populations: reduced and oxidized. The reduced receptor population was homogeneous, consisting of reduced receptor dimers. The assay directly measured the kinase regulation of this population, enabling straightforward determination of the effect of engineered cysteines on receptor signaling. The oxidized receptor population was heterogeneous because the oxidation reaction only approached completion, yielding both (i) a major population of disulfide-linked oxidized dimers and (ii) a smaller population of uncross-linked dimers containing sulfhydryls in higher oxidation states, or in the reduced state. The behavior of the reduced receptor was independently determined; thus all observed changes in kinase activity must have arisen from receptor disulfide formation or oxidation. Activity changes due to disulfide formation can be positively identified by their reversal when the receptor is reduced. Thus, even though the kinase activity does not probe the entire receptor population, the in vitro assay faithfully reports changes in receptor regulation of the kinase triggered by engineered cysteines or disulfides.
Three control experiments confirmed the usefulness of the in vitro phosphorylation assay in the present application. (i) Addition of 1 mM aspartate to the native receptor yielded a 50-fold reduction in the rate of phospho-CheY formation, providing a large transmembrane signal as previously reported (Borkovich & Simon, 1991; Ninfa et al., 1991; Borkovich et al., 1989). (ii) The rate of phospho-CheY formation varied linearly with the concentration of active receptor, where the receptor was varied either by dilution or by formation of an inhibitory disulfide bond to different degrees of completion (data not shown for the V8C, V8C′ receptor). (iii) Changes in kinase regulation due to disulfide formation in multiple engineered receptors were confirmed by their reversal upon reduction (S. A. Chervitz and J. J. Falke, unpublished results).
Other details of the in vitro phosphorylation reaction were as follows. The reaction buffer contained 50 mM Tris base, pH 7.5 with HCl, 50 mM KCl, and 5 mM MgCl2, with or without 1 mM l-aspartate. Twenty microliter reactions were preincubated at 23 °C for 40–60 min to permit formation of the receptor–kinase complex (Gegner et al., 1992). CheA autophosphorylation, followed by phosphotransfer to CheY, was initiated by the addition of [γ32P]ATP (4000–8000 cpm/pmol) to a final concentration of 0.1 mM. Five microliter aliquots were quenched at 10 and 30 s by mixing with 15 µL of 2 × Laemmli sample buffer containing 25 mM EDTA at room temperature, essentially fixing the level of phospho-CheY. Thirty minutes after initiation, the remaining 10 µL reaction mixture was quenched by mixing with an equal volume of a 2× Laemmli nonreducing sample buffer containing 100 mM EDTA, 2 mM sodium arsenate (to chelate trace DTT), and 200 mM N-ethylmaleimide (added fresh), followed immediately by heating to 95 °C for 1 min. The additional components of the latter quench served to stabilize the oxidation state of the receptor, so that the extent of engineered disulfide formation could be measured directly for each phosphorylation reaction.
Initial phospho-CheY formation rates were analyzed by SDS–PAGE on Laemmli discontinuous gels consisting of (i) a stacking gel of 9.6% (w/v) acrylamide and 0.048% (w/v) bisacrylamide, and (ii) a separatory gel of 16% (w/v) acrylamide and 0.5% bisacrylamide, as well as 22% (w/v) urea and 6.5% (v/v) glycerol. Gels were dried down immediately after electrophoresis and phospho-CheY was quantitated by phosphor-imaging (Molecular Dynamics). The initial phospho-CheY formation rate was determined by averaging the slopes of three pairs of 0 and 10 s time points. The resulting rate was normalized to a fixed receptor concentration (6 µM monomer) as determined by laser densitometric analysis of Coomassie-stained gels. In addition, each rate was multiplied by a correction factor which accounted for the effects of oxidation or reduction treatments on reactions containing the wild-type receptor. The correction factors were as follows: (i) 1.1 for DTT reduction; (ii) 1.2 for O2 oxidation; and (iii) 0.43 for I2 oxidation. The larger correction required for the I2 oxidation stemmed from the superactivation of the ternary complex by the ethanol in which the oxidation agent was dissolved (3% v/v final); this superactivation is not understood but is under study.
The extent of receptor disulfide bond formation during phosphorylation was determined by analysis of the N-ethylmaleimide-quenched reactions on nonreducing Laemmli 7.5% SDS–PAGE gels. The relative receptor monomer and dimer populations were quantitated by a laser densitometric analysis of Coomassie-stained receptor bands.
In Vitro Methylation Reactions
Methyl-esterification of the regulatory glutamate side chains of the aspartate receptor was assayed in reactions containing ~3 µM membrane-bound receptor dimers in 50 mM KH2PO4, pH 7.0 with KOH, 10% (v/v) glycerol, 1 mM EDTA, and 0.5 mM PMSF, with or without 1 mM l-aspartate. Thirty microliter samples were preincubated 23 °C for 20–30 min prior to addition of S-adenosyl-[3H-methyl]-l-methionine (0.6 µM at 60–85 Ci/mmol diluted with 50 µM unlabeled material, final) and cell lysate containing the methyltransferase CheR (0.25 mg/mL total protein, final, of which CheR comprises ~10%). The resulting reactions were incubated at 30 °C. At 15, 30, and 60 s, 9 µL aliquots were quenched by mixing with an equal volume of nonreducing 2× Laemmli sample buffer containing 200 mM N-ethylmaleimide, with heating for 20 s at 95 °C. The oxidation state of the receptor was determined using Laemmli 7.5% SDS–PAGE gels as described for the phophorylation assay. Following that determination, the oxidized and reduced receptor bands were cut out and the extent of receptor methylation was quantitated using the vapor-phase equilibrium method (Chelsky et al., 1984).
Initial methylation rates were calculated by averaging the slopes of three pairs of 0 and 15 s time points and then were normalized to the same receptor concentration and corrected for the effects of reduction or oxidation treatments on the wild type rate. The multiplicative correction factors were as follows: (i) 0.84 for DTT reduction; (ii) 1.1 for O2 oxidation; and (iii) 0.40 for I2 oxidation. As indicated for the phosphorylation reaction, the large increase in methylation activity caused by ethanol in the I2 reaction is not understood but is under study.
Analysis of Engineered Receptors in Vivo
Chemotaxis swarm plate assays (Adler, 1966) were conducted using E. coli strain RP8611 transformed with plasmid pSCF6. Saturated liquid cultures were grown at 37 °C in Luria Broth and spotted onto 0.23% agar minimal plates prepared from deionized water containing Vogel Bonner Citrate medium (Vogel & Bonner, 1956) supplemented with 0.1% (v/v) glycerol, 20 mM lactate, 40 µg/mL d,l-histidine, 20 µg/mL l-leucine, 1 µg/mL thiamine, and 100 µg/mL ampicillin, with or without 0.1 mM l-aspartate (Weiss & Koshland, 1988). Swarm diameters were measured at 3–4 h intervals starting approximately 24 h after spotting onto swarm plates incubated at 30 °C. Swarm rates, which described the increasing diameter of the swarm ring as a function of time, were calculated using the method of linear least-squares.
The fraction of the receptor population containing disulfide bonds in vivo was determined using E. coli strain RP3808 transformed with pSCF6, which together expressed sufficient receptor to assay receptor oxidation state by SDS–PAGE and Coomassie staining. Cells were grown at 30 °C in minimal growth medium as described above for membrane preparation. Cultures were harvested at early stationary phase, pelleted by centrifugation at 7700g for 8 min, and resuspended in ice-cold low-salt buffer containing 0.1 M NaH2PO4, pH 7.0 with NaOH, 10% (w/v) glycerol, and 10 mM EDTA. Immediately prior to lysis by sonication, 1 mM PMSF and 20 mM N-ethylmaleimide were added. The latter reagent served to alkylate the cellular reducing equivalents and free receptor cysteines, thereby preserving the in vivo extent of receptor disulfide formation. Subsequently, the standard membrane preparation was carried out (see above), except that 5 mM N-ethylmaleimide was substituted for reducing agent in all buffers. Membrane proteins were separated and the extent of disulfide formation quantitated on discontinuous 7.5% SDS–PAGE gels as described above.
Protein Graphics
Coordinates of the periplasmic domain containing the C36–C36′ engineered disulfide (Milburn et al., 1991) were displayed on a Silicon Graphics Personal Iris 4D/35 running the Insight II molecular graphics package (BioSym).
Statistics
Error ranges represent the standard deviation or standard error of the mean for n ≥ 3.
RESULTS
Locations of Engineered Cysteine Pairs
In order to introduce local perturbations and covalent linkages into the subunit interface, pairs of engineered cysteines were targetted to the periplasmic and transmembrane regions of the interface. Two considerations were used in choosing positions for substitution: (i) when possible, engineered cysteine pairs were placed in sufficient proximity to enable disulfide formation without large distortions of the protein backbone; and (ii) cysteine pairs were distributed to sample the entire length of the subunit interface from the periplasmic domain through the bilayer. Since the receptor is a homodimer lacking intrinsic cysteines, the incorporation of a single cysteine residue into the linear sequence was sufficient to generate an assembled dimer containing a unique, symmetric cysteine pair. Altogether, 14 engineered receptors were designed, each possessing a single pair of cysteines: R4C,R4C′; V7C,V7C′; V8C,V8C′; L11C,L11C′; F18C,F18C′; Q22C,Q22C′; L29C,L29C′; L33C,L33C′; N36C,N36C′; L61C,L61C′; 165C,I65C′; S68C,S68C′; A72C,A72C′; and M75C,M75C′ (the prime distinguishes substitutions in different subunits). The locations of the cysteine substitutions are schematically summarized in Figure 1A–C.
Seven of the 14 cysteine pairs were targetted to helices α1 and α1′ of the periplasmic domain (Figure 1A,C), making use of the available crystal structures (Milburn et al., 1991) to identify symmetric positions with β-carbon separations satisfying Cβ-Cβ′ ≤ 6.1 Å in both the apo and aspartate-occupied conformations as summarized in Table 1 (L33C,L33C′; N36C,N36C′; L61C,L61C′; 165C,165C′; S68C,S68C′; A72C,A72C′; M75C,M75C′). The angular separation of each pair lies within the range observed for protein disulfides (Careaga & Falke, 1992; Srinivasan et al., 1990; Balaji et al., 1989). indicating that disulfide formation may result from simple translational motions. Moreover, four of the seven resulting cysteine pairs (excepting M75C,-M75C′; I65C, I65C′; and A72C,A72C′) satisfied an even more stringent condition, exhibiting β-carbon distances within the range allowed for protein disulfides, 3.4 Å ≤ Cβ- Cβ′ ≤ 4.6 Å. Thus these four cysteine pairs may form disulfide bonds with little or no perturbation of the backbone structure (Figure 1).
Table 1.
Engineered Cysteine Pairs: Spatial Parameters
| anglesb | ||||||||
|---|---|---|---|---|---|---|---|---|
| engineered cysteine pair |
interatomic distance rβij (Å)a | χβij (deg) | θij (deg) | θij (deg) | ||||
| (−)Asp | (+) Asp | (−) Asp | (+) Asp | (−)Asp | (+) Asp | (−) Asp | (+) Asp | |
| M75C, M75C′ | 5.1 (0.5) | 4.4 (0.0) | 88 (0) | 94 (0) | 142 (0) | 136 (0) | 142 (0) | 148 (0) |
| A72C, A72C′ | 5.9(1.3) | 5.1 (0.5) | −112(0) | −109 (0) | 91(0) | 77 (0) | 91(0) | 78 (0) |
| S68C, S68C′ | 3.6 (0.0) | 3.6 (0.0) | −49 (0) | 86 (0) | 143 (0) | 131 (0) | 143 (0) | 135 (0) |
| I65C, I65C′ | 6.1 (1.5) | 5.7(1.1) | −103 (0) | −102 (0) | 77 (0) | 76 (0) | 77 (0) | 73 (0) |
| L61CL61C′ | 4.3 (0.0) | 4.4 (0.0) | 54 (0) | 80 (0) | 133 (0) | 131 (0) | 133 (0) | 137 (0) |
| N36C,c N36C′ | 3.9 (0.0) | 4.1 (0.0) | 93 (0) | 91 (0) | 139 (0) | 141 (0) | 139 (0) | 135 (0) |
| L33C, L33C′ | 4.5 (0.0) | 4.6 (0.0) | −109(0) | −112(0) | 76 (0) | 76 (0) | 76 (0) | 75 (0) |
Calculated from the crystallographic coordinates of Milburn et al. (1991). Tabulated distances (rβij) are between the β-carbons of residues n and n′ in the two identical subunits of the dimer. Values in parentheses indicate the minimum translation required to bring the engineered cysteines within the range required for disulfide formation (3.4 ≤ rβij ≤ 4.6 Å; Careaga & Falke, 1992; Srinivasan, 1990; Balaji et al., 1989). Parameters for positions in the transmembrane region could not be determined since the transmembrane helices are absent in the crystal structure; thus no parameters are given for the remaining cysteine pairs employed: L29C,L29C′; Q22C,Q22C′; F18C,F18C′; L11C,L11C′; V8C,V8C′; V7C,V7′; R4C,R4C′.
The pseudodihedral angle χβij Was determined using the atomic coordinates for Cαi, Cβi, Cβj, and Cαj. Pseudo-bond angles θij and θij were calculated using the atomic coordinates for Cαi–Cβi-Cβj and Cαj,–Cβj–Cβi, respectively. Values in parentheses indicate the minimum rotation required to bring the engineered cysteines within the range required for disulfide formation (−80° ≥ χβij ≤ −60° and 60° ≥ χβij ≤ 80° are forbidden pseudodihedral angles; 60° ≤ θij, θij, ≤ 180° are allowed pseudobond angles; Careaga & Falke, 1992; Srinivasan, 1990; Balaji et al., 1989).
Residues N36C and N36C′ are linked by a disulfide bond in the crystal structure.
The remaining seven cysteine pairs were engineered into transmembrane helices TM1 and TM1′, specifically targetted to locations along the subunit interface as proposed by the current transmembrane helix packing model (R4C,R4C′; V7C,V7C′; V8C,V8C′; L11C,L11C′; F18C,F18C′; Q22CQ22C′; L29C,L29C′;). Figure 1B,C summarizes the positions of the resulting symmetric cysteine pairs, each of which had previously yielded at least 50% disulfide bond formation in engineered aspartate receptors from S. typhimurium (Lynch & Koshland, 1991) or E. coli (Pakula & Simon, 1992). Due to the lack of high resolution structural information for the transmembrane region, however, the inter-cysteine spatial separations of these pairs are unknown.
Construction, Expression, and in Vivo Activity of Engineered Receptors
Engineered receptors were constructed by oligonucleotide-directed mutagenesis of the receptor gene in the phagemid pSCF6, followed by transformation of the altered phagemid into an E. coli strain lacking the aspartate receptor. Each of the 14 engineered genes was observed to express high levels receptor protein and to correctly incorporate the receptor into the cytoplasmic membrane. In general, despite the oxidizing environment of the periplasm (Zapun et al., 1995), the engineered cysteine pairs were reduced in vivo where no more than 10% of the receptor population was found to be disulfide-linked, as summarized in Table 2. The sole exception was the Cys75,Cys75′ receptor possessing the cysteine pair most exposed to the periplasmic solvent (S. A. Chervitz and J. J. Falke, unpublished results): this receptor yielded 80% disulfide-linked dimer in vivo, which could be fully reduced by DTT prior to in vitro assays.
Table 2.
Effect of Engineered Cysteines on in Vivo Swarming Activity
| receptor | fraction S – S in vivoa | relative swarm rateb |
|---|---|---|
| M75C | 0.8 | 0.9 ± 0.1 |
| A72C | 0.0 | 1.1 ± 0.1 |
| S68C | 0.1 | 1.3 ± 0.1 |
| I65C | 0.0 | 1.0 ± 0.1 |
| L61C | 0.0 | 0.7 ± 0.1 |
| N36C | 0.1 | 1.6 ± 0.1 |
| L33C | 0.02 | 0.5 ± 0.2 |
| L29C | 0.0 | 1.4 ± 0.1 |
| Q22C | 0.0 | 2.4 ± 0.1 |
| F18C | 0.0 | 0.3 ± 0.3 |
| L11C | 0.0 | 1.5 ± 0.1 |
| V8C | 0.0 | 1.6 ± 0.1 |
| V7C | 0.0 | 1.2 ± 0.1 |
| R4C | 0.1 | 0.6 ± 0.1 |
| no Tar | na | 0.1 ± 0.1 |
The ambient level of intersubunit disulfide formation in vivo (see Materials and Methods).
Rate of chemotactic swarming in minimal agar containing aspartate, relative to the swarm rate observed for the wild-type receptor (see Materials and Methods).
Each of the engineered receptors retained significant in vivo signaling activity. Receptors overexpressed by phagemid pSCF6 in E. coli RP8611 restored the ability of this strain to sense aspartate, as measured by the chemotactic swarm-plate assay (Weis & Koshland, 1988; Adler, 1966). The swarm rates observed for engineered receptors ranged from 30% to 240% that of the wild type swarm rate (Table 2). Of the four engineered receptors possessing significantly depressed swarm rates (Cys4,Cys4′; Cys18,Cysl8′; Cys33,-Cys33′; and Cys61,Cys61′), the least active was Cys18,-Cys18′, for which the swarm rate was 30% that of the native receptor. It should be noted that the in vivo swarm experiment is a relatively insensitive assay for receptor defects, since the adaptation branch of the chemosensory pathway is able to compensate for many signaling perturbations.
Isolation of Engineered Receptors for in Vitro Assays
To further examine the effects of engineered cysteines and disulfides on receptor signaling, receptor-containing membranes were isolated for in vitro phosphorylation and methylation assays. Each engineered receptor was overexpressed by phagemid pSCF6 in strain E. coli RP3808, and then bacterial membranes were isolated using standard procedures (Foster et al., 1985). The resulting receptor population comprised 10–15% of the total membrane protein and was homogeneous with respect to the modification state of its regulatory methylation sites, since the expression strain lacked the modification enzymes CheR and CheB.
Effect of Engineered Cysteines on Transmembrane Kinase Regulation in Vitro
Receptor-mediated regulation of the reconstituted phosphorylation pathway provides a sensitive and direct in vitro assay for transmembrane signaling (Ninfa et al., 1991; Borkovich et al., 1989). To reconstitute the pathway, isolated E. coli membranes containing the over-expressed receptor were added to a mixture of purified components including the coupling protein (CheW), kinase (CheA), phosphoacceptor protein (CheY), and ATP (Borkovich & Simon, 1991). The resulting receptor–CheW–CheA ternary complex exhibited a rapid initial rate of phospho-CheY formation which was down-regulated 50-fold by aspartate, thereby providing a direct measure of transmembrane kinase modulation as illustrated in Figure 2. Assay conditions ensured that perturbations of the receptor-kinase interaction would be detected with maximum sensitivity (see Materials and Methods). Such perturbations were detected as either (i) an altered maximum rate of phospho-CheY formation in the absence of aspartate, stemming from a change in the stability, Vmax or KM of the ternary complex, or (ii) modified regulation of the CheY phosphorylation rate in response to asparate, arising from changes in ligand binding or the ensuing transmembrane signal.
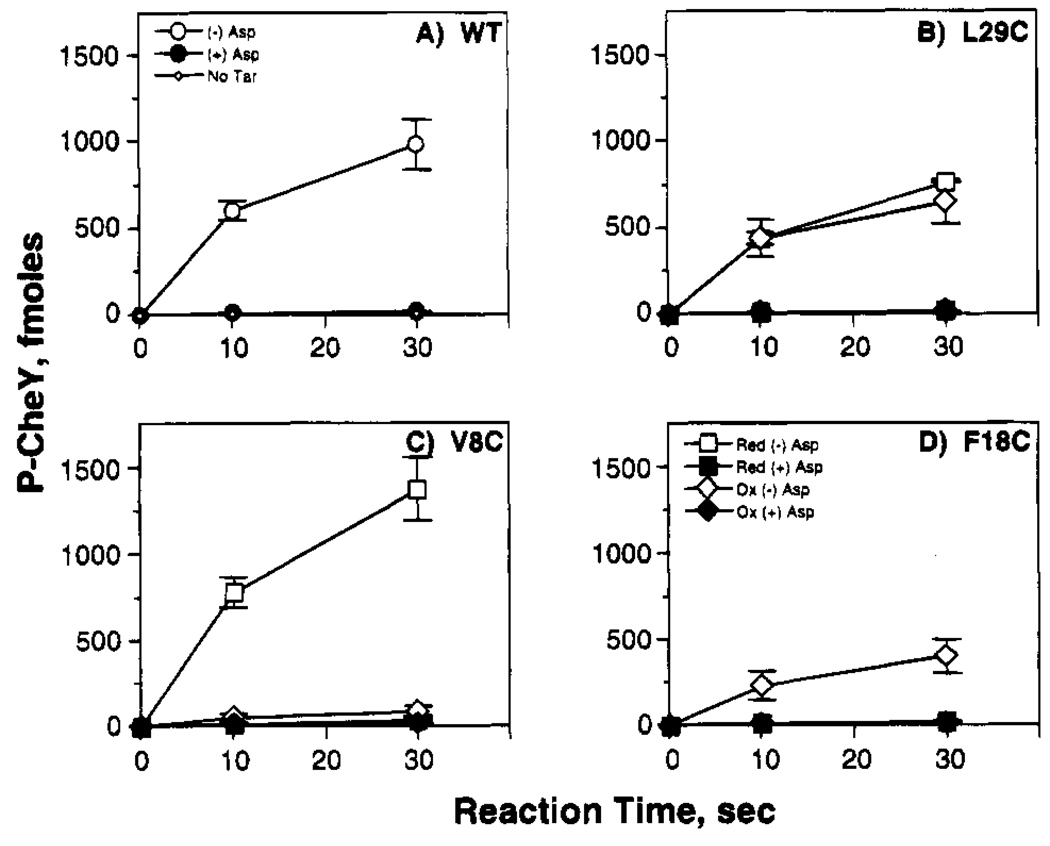
FIGURE 2.
Effect of engineered cysteines and disulfides on the in vitro phosphorylation reaction. Shown is the timecourse of CheY phosphorylation by the reconstituted receptor–kinase complex, illustrating the activities of the wild-type receptor and three classes of engineered receptors. Isolated E. coli membranes containing the wild-type (WT) and indicated engineered cysteine receptors were reduced (red) to eliminate pre-existing disulfide bonds or oxidized (ox) to drive disulfide bond formation linking engineered cysteine pairs. Subsequently, purified CheA, CheW, and CheY were added to reconstitute the receptor–kinase complex, and the effects of 1 mM aspartate (Asp) on CheY phosphorylation were quantitated (see Methods). The illustrated reactions are representative of (A) the wild type receptor or (B–D) receptors in which an engineered disulfide retains, destroys, or restores transmembrane kinase regulation, respectively.
To identify regions of the intersubunit receptor interface which are sensitive to cysteine substitution, the effects of engineered cysteines on the ternary complex were analyzed using receptors in their reduced state as summarized in Figure 2 and Table 3. Eleven of the fourteen cysteine pairs retained ≥20% of the native activation and aspartate regulation of the kinase: these were R4C,R4C′; V7C,V7C′; V8C,V8C′; L11C,L11C′; L29C,L29C′; N36C,N36C′; L61C,L61C′; I65C,I65C′; S68C,S68C′; A72C,A72C′; and M75C,M75C′. Three cysteine pairs, F18C,F18C′, Q22C,Q22C′, and L33C,L33C′, severely damaged or destroyed both kinase activation and aspartate regulation (≤10% that of native). Overall, the transmembrane signal is relatively insensitive to cysteine substitutions in most regions of the interface but is significantly inhibited by substitutions at certain positions.
Table 3.
Effect of Engineered Cysteines and Disulfides on in Vitro Phosphorylation
| relative phosphorylation ratec | ||||||
|---|---|---|---|---|---|---|
| receptor | conditiona | fraction S–Sb | (−) aspartate (/WT (+)Asp) |
(+) aspartate (/WT (+)Asp) |
maximum activityd (/WT (−) Asp) |
aspartate regulatione (/WT) |
| WT | (−) | na | 50 ±5 | 1.0 ±0.2 | 1.0 | 1.0 |
| red | na | 50 ± 5 | 1.0 ±0.2 | 1.0 | 1.0 | |
| ox (O2, I2) | na | 50 ± 5 | 1.0 ±0.2 | 1.0 | 1.0 | |
| Me | na | 50 ± 5 | 1.0 ±0.2 | 1.0 | 1.0 | |
| M75C | red | 38 ±2 | 0.6 ±0.1 | 0.8 | 1.3 | |
| ox (O2) | 0.9 ± 0.1 | 25 ±4 | 0.8 ±0.1 | 0.5 | 0.7 | |
| A72C | red | 61 ± 1 | 1.0 ±0.5 | 1.2 | 1.2 | |
| ox (I2) | 0.8 ± 0.1 | 82 ± 2 | 2.4 ±0.1 | 1.6 | 0.7 | |
| S68C | red | 70 ± 4 | 0.6 ± 0.2 | 1.4 | 2.3 | |
| ox (I2) | 0.9 ± 0.1 | 11 ±2 | 0.5 ±0.1 | 0.2 | 0.4 | |
| I65C | red | 39 ± 5 | 0.8 ±0.5 | 0.8 | 1.0 | |
| ox (O2) | 0.8 ± 0.1 | 12 ± 2 | 1.3 ±0.7 | 0.2 | 0.2 | |
| L61C | red | 34 ± 2 | 0.6 ±0.5 | 0.7 | 1.1 | |
| ox (O2) | 0.6 ± 0.1 | 19 ± 1 | 0.7 ± 0.4 | 0.4 | 0.6 | |
| N36C | red | 76 ± 12 | 1.3 ±0.2 | 1.5 | 1.2 | |
| ox (O2) | 0.9 ± 0.1 | 43 ± 2 | 1.3 ±0.1 | 0.9 | 0.7 | |
| L33C | red | 3 ± 1 | 0.7 ± 0.3 | 0.1 | 0.1 | |
| ox (O2) | 1.0 ± 0.1 | 16 ± 1 | 2.8 ± 0.7 | 0.3 | 0.1 | |
| L29C | red | 37 ± 3 | 0.8 ±0.1 | 0.7 | 0.9 | |
| ox (O2) | 0.9 ± 0.1 | 37 ± 9 | 0.9 ± 0.2 | 0.7 | 0.8 | |
| Q22C | red | 1 ± 0.5 | 0.3 ±0.1 | 0.02 | 0.04 | |
| ox (I2) | 0.9 ± 0.1 | 1 ± 0.2 | 0.2 ±0.1 | 0.02 | 0.04 | |
| red (Me) | 28 ± 3 | 2.8 ±0.3 | 0.6 | 0.2 | ||
| ox (Me,I2) | 1.0 ± 0.1 | 7 ± 1 | 1.0 ±0.1 | 0.1 | 0.1 | |
| F18C | red | 1 ± 0.2 | 0.3 ± 0.2 | 0.02 | 0.04 | |
| ox (I2) | 0.9 ± 0.1 | 19 ± 1 | 0.7 ± 0.3 | 0.4 | 0.5 | |
| L11C | red | 137 ±5 | 1.9 ±0.1 | 2.7 | 1.4 | |
| ox (I2) | 0.7 ± 0.1 | 2 ± 0.4 | 1.0 ±0.3 | 0.04 | 0.03 | |
| V8C | red | 65 ± 7 | 1.1 ±0.2 | 1.3 | 1.2 | |
| ox (O2) | 0.8 ± 0.1 | 5 ± 2 | 0.9 ±0.1 | 0.1 | 0.1 | |
| V7C | red | 8± 2 | 0.3 ±0.1 | 0.2 | 0.5 | |
| ox (O2) | 0.6 ± 0.1 | 1 ± 0.1 | 0.4 ±0.1 | 0.02 | 0.01 | |
| R4C | red | 24 ± 3 | 1.1 ±0.1 | 0.5 | 0.4 | |
| ox (O2) | 0.7 ± 0.1 | 2 ± 0.5 | 1.0 ±0.1 | 0.04 | 0.02 | |
| no Tar | (−) | na | 1.0 ± 0.2 | 1.0 ±0.2 | 0.02 | 0.0 |
Isolated membranes were treated as described in Materials and Methods: no treatment (−); reduction by dithiothreitol (red); oxidation (ox) by molecular iodine (I2) or oxygen (O2); methylation by CheR prior to reduction or oxidation (Me).
Fraction of receptor population linked by disulfide bonds after oxidation.
Rate of CheY phosphorylation by the reconstituted receptor–CheW–CheA ternary kinase complex in the absence or presence of ligand, relative to the rate observed for the aspartate-occupied wild-type complex subjected to the same chemical treatment (1.2 fmol of Pi-CheY s−1). All rates are normalized to the same receptor concentration (3µM dimer). In the presence of ligand, CheY phosphorylation was dominated by the free kinase, yielding the same rate observed in the absence of the receptor (no Tar).
Rate of CheY phosphorylation by the unliganded receptor–CheW–CheA ternary kinase complex, relative to corresponding rate observed for the unliganded wild-type complex subjected to the same chemical treatment (61 fmol of Pj-CheY s−1 for 3 µM receptor dimer).
Effect of aspartate on the rate of CheY phosphorylation by the receptor–CheW–CheA ternary kinase complex, relative to the effect of aspartate on the wild-type complex subjected to the same chemical treatment (50-fold downregulation upon aspartate addition). Calculated as the ratio {([rate (−) Asp]/[rate (+) Asp]) −1}/49, yielding a range between 0 and 1 for no aspartate regulation or wild-type regulation, respectively.
Effect of Engineered Disulfides on Transmembrane Kinase Regulation in Vitro
Each engineered cysteine pair was oxidized to yield an intersubunit disulfide bond, thereby covalently linking the subunit interface. Two oxidation systems were used to drive disulfide formation: (i) oxidation by molecular oxygen (O2) in a reaction catalyzed by the aqueous copper complex Cu(II)•(l,10-phenanthroline)3; or (ii) oxidation by molecular iodine (I2) (Pakula & Simon, 1992a; Careaga & Falke, 1992; Kobashi, 1968). For a given cysteine pair, the oxidation system which yielded the highest final extent of disulfide formation was determined empirically and used in subsequent studies. These disulfide formation reactions proceeded to 60–100% completion, yielding a receptor population consisting predominantly of disulfide-linked dimers as quantitated in Table 3. In general, the O2 system produced higher yields of disulfide-linked product in environments accessible to the aqueous phase, while the I2 system gave higher yields in the membrane phase. In cases where disulfide formation was not complete, further disulfide formation was found to be prevented by a competing reaction which oxidized free cysteine to oxyacid products incapable of disulfide formation (such as R-SO22−and R-SO32−). The chemistry of disulfide formation and the competing oxidation reaction has been discussed elsewhere (Careaga & Falke, 1992; Oae, 1991).
The engineered disulfides were resolved into three classes based on their effect on the transmembrane signal in the reconstituted phosphorylation pathway: those that retained, destroyed, or restored signaling. (i) Seven of the 14 disulfides retained substantial activation and aspartate regulation of the kinase (≥20% that of native): L29C-L29C′, N36C-N36C′, L61C-L61C′, I65C-I65C′, S68C-S68C′, A72C-A72C′, M75C-M75C′. (ii) Four disulfide bonds near the N-terminal end of the interface severely damaged or destroyed transmembrane kinase regulation and activation (≤10% native): R4C-R4C′, V7C-V7C′, V8C-V8C′, L11C-L11C′. (The latter disulfide formation reactions reached only 60–80% completion, but receptor signaling was inhibited over 90%, suggesting that the oxyacid products of the competing oxidation reaction also inactivated the transmembrane signal.) (iii) One disulfide bond in the transmembrane region, F18C-F18C′, restored kinase regulation to this receptor, which exhibited defective kinase activation in the reduced state (<10% native kinase activation when reduced, 40% when oxidized). Thus the cysteine pair F18C,F18C′ perturbed the subunit interface, while formation of the F18C–F18C′ disulfide returned the interface to a more native conformation. It is difficult to classify the effects of the remaining two engineered disulfides, Q22C–Q22C′ and L33C–L33C′, because the corresponding receptors exhibited highly perturbed kinase regulation in both their reduced and oxidized states, although the signaling of the former receptor was partially restored by methylation (Table 3).
Overall, the effects of engineered disulfides on transmembrane signaling indicate that the receptor subunit interface can be perturbed by covalent intersubunit linkages at specific locations, particularly near the N-terminus of the first membrane-spanning helix. However, the majority of the intersubunit disulfides (eight of the total 14) retain or restore transmembrane kinase regulation.
Effect of Engineered Cysteines and Disulfides on Receptor Methylation in Vitro
In order to further analyze the effects of receptor engineering on signaling and structural integrity, the engineered receptors were used as substrates for in vitro methylation by the methyltransferase enzyme, CheR. The methylation reaction modifies four specific glutamate side chains on the cytoplasmic signaling domain of each receptor subunit (Terwilliger et al., 1984). The rate of receptor methylation in vitro is highly sensitive to distortion of receptor structure, and aspartate binding triggers a 2–3-fold increase in the methylation rate (Kleene et al., 1979; Falke & Koshland, 1987). Thus, in principle, measurement of the receptor methylation rate, as well as the sensitivity of this rate to aspartate binding, can help identify engineered receptors which are (i) capable of binding aspartate, or (ii) locked in a native signaling state, or (iii) locked in a nonnative, perturbed conformation.
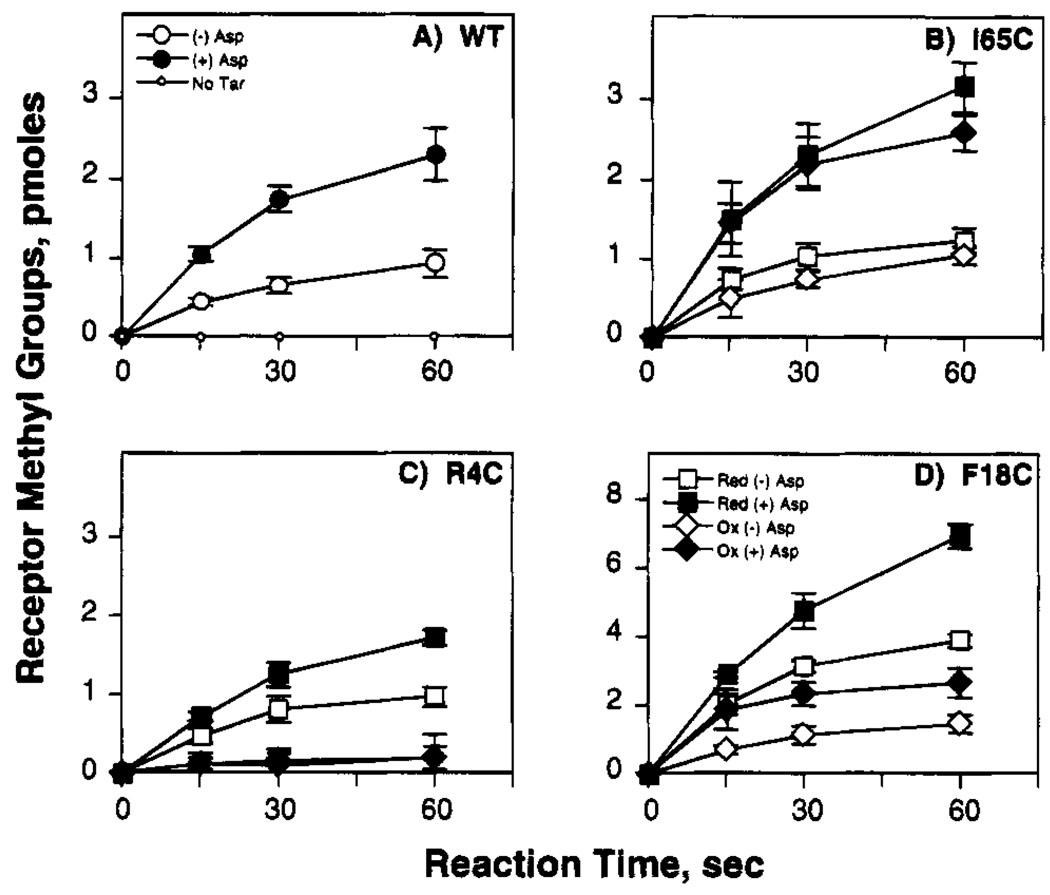
The effect of engineered cysteines and disulfides on the maximum methylation rate achieved in the presence of aspartate illustrates the sensitivity of the in vitro methylation reaction to structural perturbations, as summarized in Figure 3 and Table 4. All 14 of the engineered receptors exhibit altered maximum methylation rates in the reduced state, indicating that cysteine substitutions at the subunit interface generally trigger structural changes which are transmitted to a region of the cytoplasmic domain involved in CheR binding or catalysis. Interestingly, 12 of the 14 reduced receptors possess significantly higher methylation rates than the wild type, suggesting that a simple relationship may exist between intersubunit packing and the in vitro methylation reaction (Table 4). An exception to this simple relationship is the N-terminal end of the interface, where two cysteine substitutions, R4C,R4C′ and V8C,V8C′, yielded decreased methylation rates. Upon formation of the intersubunit disulfide bond, seven of the 14 engineered receptors exhibit methylation rates closer to that of wild type, suggesting that the covalent linkage in these cases restores a more native structure to the interface (Table 4). At the N-terminal region of the interface, however, disulfide formation significantly inhibits the methylation reaction in all four cases examined, R4C-R4C′, V7C-V7C′, V8C-V8C′, and L11C-L11C′, suggesting that intersubunit linkages drastically perturb the structure of this region.
FIGURE 3.
Effect of engineered cysteines and disulfides on the in vitro methylation reaction. Shown is the timecourse of receptor methylation by the methyltransferase CheR, illustrating the activities of the wild-type receptor and three classes of engineered receptors. Isolated E. coli membranes containing the wild type (WT) and indicated engineered cysteine receptors were reduced (red) to eliminate pre-existing disulfide bonds or oxidized (ox) to drive disulfide bond formation linking engineered cysteine pairs. Subsequently, cellular extract containing CheR was added, and the effects of 1 mM aspartate (Asp) on receptor methylation were quantitated (see Materials and Methods). The illustrated reactions are representative of (A) the wild-type receptor or (B–D) receptors in which an engineered disulfide retains, destroys, or restores receptor methylation, respectively (note different vertical scale in panel D).
Table 4.
Effect of Engineered Cysteines and Disulfides on in Vitro Methylation
| relative methylation ratec | ||||||
|---|---|---|---|---|---|---|
| receptor | conditiona | fraction S– Sb | (−) aspartate (/WT (−)Asp) |
(+) aspartate (/WT (−)Asp) |
maximum activityd (/WT (+) Asp) |
aspartate regulatione (/WT) |
| WT | (−) | na | 1.0 ±0.1 | 2.3 ± 0.2 | 1.0 | 1.0 |
| red | na | 1.0 ±0.1 | 2.3 ±0.2 | 1.0 | 1.0 | |
| ox (O2, I2) | na | 1.0 ±0.1 | 2.3 ± 0.2 | 1.0 | 1.0 | |
| M75C | red | 3.2 ± 0.5 | 5.1 ± 1.2 | 2.2 | 0.4 | |
| ox (O2) | 0.9 ±0.1 | 2.4 ± 0.2 | 4.2 ± 0.4 | 1.8 | 0.6 | |
| A72C | red | 2.0 ± 0.4 | 3.4 ± 0.4 | 1.5 | 0.5 | |
| ox (I2) | 0.8 ±0.1 | 1.7 ±0.5 | 3.6 ± 1.1 | 1.6 | 0.9 | |
| S68C | red | 2.2 ± 0.8 | 5.5 ±0.9 | 2.3 | 1.1 | |
| ox (I2) | 0.9 ±0.1 | 2.4 ± 0.8 | 2.8 ±0.5 | 1.2 | 0.1 | |
| I65C | red | 1.6 ±0.3 | 3.4 ± 1.1 | 1.4 | 0.8 | |
| ox (O2) | 0.8 ±0.1 | 1.1 ±0.5 | 3.3 ±0.6 | 1.4 | 1.5 | |
| L61C | red | 2.0 ± 0.6 | 3.5 ± 1.1 | 1.4 | 0.4 | |
| ox (O2) | 0.6 ±0.1 | 1.7 ± 1.0 | 2.6 ±0.6 | 1.1 | 0.4 | |
| N36C | red | 4.1 ± 1.1 | 9.9 ±2.1 | 4.3 | 1.1 | |
| ox (O2) | 0.9 ±0.1 | 5.3 ± 1.5 | 11 ±2.2 | 4.6 | 0.8 | |
| L33C | red | 4.2 ± 0.5 | 7.7 ± 0.6 | 3.3 | 0.6 | |
| ox (O2) | 1.0 ±0.1 | 2.5 ± 0.5 | 3.6 ± 0.8 | 1.6 | 0.3 | |
| L29C | red | 3.7 ± 0.4 | 7.1 ±0.5 | 3.1 | 0.7 | |
| ox (O2) | 0.9 ±0.1 | 3.5 ±0.7 | 7.7 ± 1.1 | 3.3 | 0.9 | |
| Q22C | red | 12 ± 1.8 | 18 ±4.5 | 7.9 | 0.4 | |
| ox (I2) | 0.9 ±0.1 | 13 ± 1.4 | 7.4 ± 1.0 | 3.2 | 0.0 | |
| F18C | red | 4.5 ±0.6 | 6.4 ± 0.2 | 2.8 | 0.3 | |
| ox (I2) | 0.9 ±0.1 | 1.6 ±0.3 | 4.2± 1.3 | 1.8 | 1.3 | |
| L11C | red | 3.7 ± 0.4 | 7.0 ± 0.7 | 3.0 | 0.7 | |
| ox (I2) | 0.7 ±0.1 | 1.0 ±0.1 | 0.6 ± 0.2 | 0.3 | 0.0 | |
| V8C | red | 0.4 ± 0.04 | 0.9 ± 0.2 | 0.4 | 0.9 | |
| ox (O2) | 0.8 ±0.1 | 0.2 ±0.1 | 0.4 ± 0.2 | 0.2 | 0.5 | |
| V7C | red | 2.1 ±0.1 | 3.3 ±0.1 | 1.4 | 0.5 | |
| ox (O2) | 0.6 ±0.1 | 0.1 ±0.1 | 0.2 ±0.1 | 0.1 | 0.9 | |
| R4C | red | 1.1 ±0.2 | 1.6 ±0.1 | 0.7 | 0.4 | |
| ox (O2) | 0.7 ±0.1 | 0.3 ±0.3 | 0.3 ± 0.2 | 0.1 | 0.1 | |
| no Tar | (–) | na | 0 | 0 | 0 | 0 |
Isolated membranes were treated as described in Materials and Methods: no treatment (−); reduction by dithiothreitol (red); oxidation (ox) by molecular iodine (I2) or oxygen (O2).
Fraction of receptor population linked by disulfide bonds after oxidation.
Rate of in vitro receptor methylation by the methyltransferase CheR in the absence or presence of ligand, relative to the rate observed for the unliganded wild-type receptor subjected to the same chemical treatment (1.8 pmol of methyl groups min−1). All rates are normalized to the same receptor concentration (3 µM dimer).
Rate of in vitro receptor methylation by CheR in the presence of aspartate, relative to rate observed for the aspartate-occupied wild-type receptor subjected to the same chemical treatment (4.2 pmol of methyl groups min−1 for 3 µM receptor dimer).
Effect of aspartate on the rate of in vitro receptor methylation by CheR, relative to the effect of aspartate on the wild-type receptor subjected to the same chemical treatment (2.3-fold up-regulation upon aspartate addition). Calculated as the ratio {([rate (+) Asp]/[rate (−) Asp]) −1}/1.3, yielding a range between 0 and 1 for no aspartate regulation or wild-type regulation, respectively.
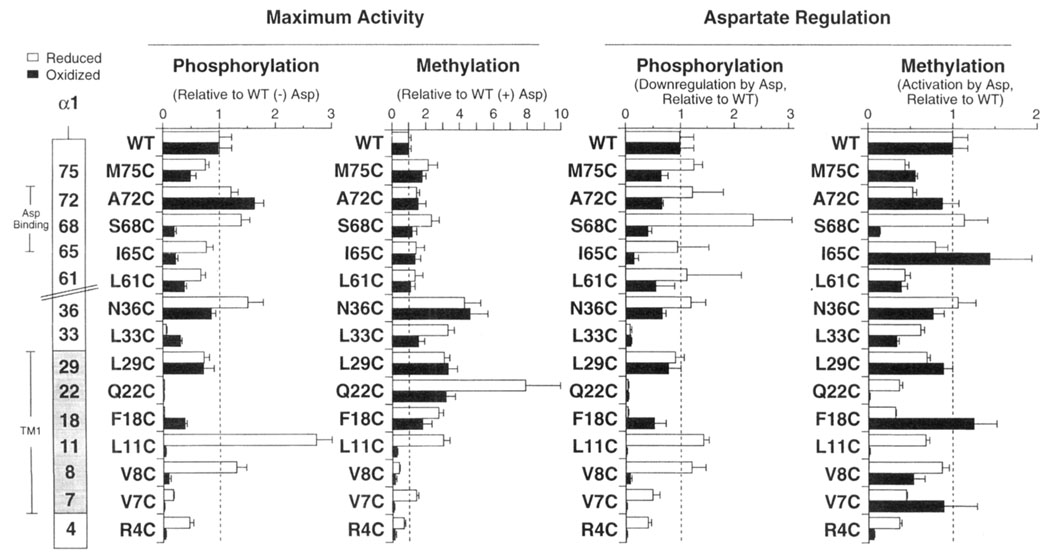
By contrast, the regulation of methylation by aspartate is relatively insensitive to perturbation by cysteine substitution or disulfide formation. Twenty-four of the 28 engineered cysteines and disulfides yield methylation rates retaining 20–130% of the native aspartate-triggered activation. Even four engineered cysteines or disulfides which virtually destroy aspartate regulation of phosphosignaling (≤10% native) are observed to retain aspartate regulation of methylation (30–90% native for oxidized and reduced L33C,L33C′, oxidized V8C,V8C′, and oxidized V7C,V7C′ receptors: compare Table 3 and Table 4). The methylation and phosphorylation rates are directly compared in Figure 4. Overall, the comparison reveals that the in vitro methylation and phosphorylation reactions generally yield a similar picture for the effect of receptor perturbations on the transmembrane signal. Some exceptional perturbations, however, are detected differently by the two assays, indicating that in vitro methylation and phosphorylation may monitor different conformational features.
FIGURE 4.
Effects of engineered cysteines and disulfides on the in vitro phosphorylation and methylation reactions: Summary. The left panels plot the maximum phosphorylation and methylation rates produced by each engineered receptor in its reduced and oxidized states, relative to the activity of the wild-type receptor. The maximum rale of CheY phosphorylation or receptor methylation was obtained in the absence or presence of ligand, respectively. The right panels summarize the ability of aspartate (1.0 mM) to down-regulate CheY phosphorylation, or stimulate receptor methylation, where each aspartate effect is relative to reactions utilizing the wild-type receptor (see Materials and Methods and Tables 3 and 4 for further details).
DISCUSSION
The schematic model presented in Figure 5 summarizes the conclusions of the present study, in which engineered cysteines and disulfides placed at the subunit interface of the aspartate receptor (i) confirm that the dimeric form of the receptor is the functional unit in transmembrane signaling, (ii) confirm the position and orientation of the interfacial first membrane-spanning helix in the dimer, and (iii) reveal that the periplasmic and transmembrane regions of this homodimeric interface do not shift significantly during the transmembrane signal. These deduced characteristics may be general features of other receptors exhibiting the same transmembrane topology and dimeric organization as the aspartate receptor, particularly the large class of structurally and functionally analogous receptors found in other prokaryotic and eukaryotic histidine kinase pathways.
FIGURE 5.
Locations of engineered disulfides which retain or restore the transmembrane signal. Shown is a schematic model of the subunit interface in the periplasmic and transmembrane regions of the receptor dimer (note that perspectives have been altered for clarity), illustrating all 14 engineered disulfides examined in this study. The highlighted eight disulfide bonds (bold lines) retain or restore aspartate-triggered transmembrane regulation of kinase activity, yielding ≥20% native kinase activity and aspartate regulation. In contrast, the remaining six disulfides (fine lines) severely damage or destroy transmembrane kinase regulation, yielding ≤10% native kinase activity and aspartate regulation. The model proposes that the interface between the α1/TM1 and α1′/TM1′ helices is structural, and does not shift significantly during the transmembrane signal. This interface does not include the N-terminal ends of the α1/TM1 and α1′/TM1′ helices, which are proposed to diverge. The transmembrane signal is depicted as an undefined movement of the α4/TM2 helix relative to the subunit interface, which triggers a shift of the cytoplasmic domain into a different signaling state, thereby modulating its associated histidine kinase activity.
The location of the subunit interface, which was predicted by previous crystallographic and disulfide mapping studies (Pakula & Simon, 1992b; Milburn et al., 1991; Lynch & Koshland, 1991; Hazelbauer, et al., 1990; Falke et al., 1988), is largely supported by the new results although the interface is less extensive than previously believed. The interface consists primarily of contacts between the N-terminal helices of the two subunits, designated TM1/α1 and TM1′/α1′, respectively. These contacts extend approximately from Phe 18 in the bilayer to Met 75, the penultimate residue of the N-terminal helix. Within this region, eight of the 14 pairs of engineered cysteines lie within sufficient proximity to form a disulfide covalently linking the two subunits while retaining significant levels of receptor signaling, as assayed by transmembrane activation and regulation of CheA kinase activity (Table 3 and Figure 4). Six of these eight functional disulfides lie at the contact region between helices α1 and α1′ in the crystal structure of the periplasmic domain (N36C–N36C′, L61C–L61C′, I65C–I65C′, S68C–S68C′, A72C–A72C′, M75C–M75C′); the other two lie within the bilayer at the TM1–TM1′ helix–helix interface predicted by disulfide mapping (F18C–F18C′, L29C–L29C′; sec-Figure 5).
The new results suggest that the intersubunit contact does not include the N-terminus of the receptor. The region extending from the Met 1 to Phe 18 is thought to be an extension of the first membrane-spanning helix TM1. All four of the cysteine pairs placed at the N-terminal ends of TM1 and TM1′ form disulfides less efficiently than cysteine pairs within the confirmed interfacial region. Furthermore, these four N-terminal disulfides (R4C–R4C′, V7C–V7C′, V8C–V8C′, L11C–L11C′) each significantly reduce kinase activation, indicating that they distort the native receptor structure even though they are thought to bridge the proximal faces of TM1 and TM1′ (Table 3 and Figure 4). Thus, the model presented in Figure 5 proposes that the TM1 and TM1′ helices diverge at their cytoplasmic N-termini. The kinase inhibition generated by disulfides in this region could result from either reduced kinase binding or a decrease in the Vmax or KM of the ternary complex.
The structural interface between the subunits is observed to be coupled to the transmembrane signal, albeit weakly since receptor regulation by the kinase is perturbed by only three of 14 engineered interfacial cysteine pairs (F18C,F18C′; Q22C,Q22C′; L33C,L33C′). These pairs significantly reduce transmembrane kinase activation, suggesting that they distort the structure of the subunit interaction. Interestingly, signaling is restored by disulfide formation between one of these perturbing pairs (F18C–F18C′), indicating that the covalent link returns the altered interface to a more native structure.
Although receptor signaling is sensitive to specific perturbations of the interface, the subunit interaction appears to remain essentially static during the transmembrane signal. Each of the eight intersubunit disulfide bonds which permits transmembrane regulation of the kinase strongly constrains the conformational freedom of the interface (Figure 5). The magnitude of such a constraint can be estimated from the allowed distance between β-carbons in the disulfide linkage, which ranges 3.4 Å ≤ Cβ-Cβ′ ≤ 4.6 Å in protein crystal structures (Srinivasan et al., 1990). Thus a given intersubunit disulfide allows at most ~1.2 Å relative motion of the two subunits. Models which invoke large relative motions of the two subunits during the transmembrane signal are strongly disfavored by these results; instead, the model presented in Figure 5 proposes that the first membrane-spanning helix (α1/TM1) lying at the subunit interface plays a static structural role in stabilizing the native dimer.
Since the first transmembrane helix appears to be a static structural element, it follows that the transmembrane signal could be carried by a movement of the second membrane-spanning helix (α4/TM2) relative to the subunit interface. Such a hypothesis is supported by existing evidence from X-ray crystallographic studies (Milburn et al., 1991). When the crystal structures of the apo- and aspartate-occupied ligand binding domain are compared, apartate binding is observed to trigger significant changes in the distance between helices α1 and α4′, and between helices α4 and α4′. In contrast, the separation of helices α1 and α1′ is relatively insensitive to aspartate binding (with the exception of the region surrounding the aspartate binding site, residues 67–76, where significant aspartate-induced rearrangements are observed). Similarly, 19F NMR studies of the isolated periplasmic domain in solution (Danielson et al., 1994) have revealed aspartate-triggered structural changes in the vicinity of a fluorine probe near the α4/TM2 junction (Phe 180), while ligand binding yields no detectable perturbations of probes near the α1/TM1 junction (Phe 30, Phe 40; Figure 5). Thus, at least in the isolated periplasmic domain, the aspartate-generated signal appears to bypass α1 while triggering a movement of α4. In the intact receptor such a movement would be transmitted through α4/TM2 to the cytoplasmic domain of the receptor where it would generate a local structural change or regulate subunit interactions, thereby altering the signaling state of the cytoplasmic domain and its associated histidine kinase.
Perturbations of the receptor generated by engineered cysteines and disulfides generally yield similar effects on the in vitro methylation and phosphorylation assays, but exceptions are observed (Table 3 and Table 4; Figure 4). In particular, certain interfacial cysteines perturb the basal methylation rate more than the basal phosphorylation rate, while some interfacial disulfides perturb aspartate regulation of phosphosignaling more than aspartate control of methylation (compare Table 3 and Table 4). It follows that the in vitro methyltransferase and kinase reactions may recognize different conformational features of the receptor. Alternatively, the native transmembrane signal may require the fully assembled receptor–CheW–CheA ternary complex, which is lacking in the in vitro methylation assay. The reconstituted phosphorylation system provides an accurate and sensitive in vitro method for quantitating the transmembrane signal: this system directly monitors CheA autophosphorylation activity, which ultimately controls both the signaling and adaptation branches of the chemotaxis pathway.
The mechanism of transmembrane signaling utilized by the aspartate receptor is likely to be a general feature of structurally and functionally similar receptors which regulate histidine kinase activity. Evidence for such generality is provided by the construction of an active chimeric receptor linking the aspartate receptor’s ligand binding and transmembrane domains to the cytoplasmic histidine kinase domain of the EnvZ receptor, a related bacterial protein responsible for osmosensing (Utsumi et al., 1989). More recently, in vivo disulfide engineering studies of Trg, a bacterial ribose and galactose receptor exhibiting high homology to the aspartate receptor, have yielded results consistent with the signaling role of the second membrane-spanning helix proposed in Figure 5 (Lee et al., 1995, 1994). These mechanistic similarities make it plausible to suspect that the receptors controlling eukaryotic histidine kinase pathways, which possess transmembrane structures similar to those of their prokaryotic counterparts, may well use the same mode of transmembrane signaling.
ACKNOWLEDGMENT
We gratefully acknowledge Dr. Jeffry Stock for E. coli strains and plasmids, Dr. John Parkinson for E. coli strains, Drs. Sung-Hou Kim and Daniel Koshland for crystallographic coordinates, James Cate for expert technical assistance, and the NIH for funding (GM40731 to J.J.F.).
Footnotes
Support provided by NIH Grant GM40731.
REFERENCES
- Adler J. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- Alex LA, Simon MI. Trends Genet. 1994;10:133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Vol. 1. New York: Greene Publishing Association; 1992. p. 1.1.2. [Google Scholar]
- Balaji VN, Mobasser A, Rao SN. Biochem. Biophys. Res. Commun. 1989;160:109–114. doi: 10.1016/0006-291x(89)91627-6. [DOI] [PubMed] [Google Scholar]
- Bogonez E, Koshland DE. Proc. Natl. Acad. Sci. U.S.A. 1985;82:4891–4895. doi: 10.1073/pnas.82.15.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Simon MI. Methods Enzymol. 1991;200:205–214. doi: 10.1016/0076-6879(91)00140-r. [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Kaplan N, Hess JF, Simon MI. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB, Borkovich KA, Simon MI. Annu. Rev. Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Bourret RB, Drake SK, Chervitz SA, Simon MI, Falke JJ. J. Biol. Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- Careaga CL, Falke JJ. J. Mol. Biol. 1992;226:1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleeker AB, Meyerowitz EM. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chelsky D, Gutterson NI, Koshland DE. Anal. Biochem. 1984;141:143–148. doi: 10.1016/0003-2697(84)90437-8. [DOI] [PubMed] [Google Scholar]
- Danielson MA, Biemann HP, Koshland DE, Falke JJ. Biochemistry. 1994;33:6100–6109. doi: 10.1021/bi00186a009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Koshland DE. Science. 1987;237:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Dernburg AF, Sternberg DA, Zalkin N, Milligan DL, Koshland DE. J. Biol. Chem. 1988;263:14850–14858. [PubMed] [Google Scholar]
- Foster DL, Mowbray SL, Jap BK, Koshland DE. J. Biol. Chem. 1985;260:11706–11710. [PubMed] [Google Scholar]
- Gegner JA, Graham DR, Roth AF, Dahlquist FW. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- Hazelbauer GL. Curr. Opin. Struct. Biol. 1992;2:505–510. [Google Scholar]
- Hazelbauer GL, Yaghami R, Burrows GG, Baumgartner JW, Dutton DP, Morgan DG. In: Biology of the Chemotactic Response. Armitage JB, Lackie JM, editors. New York: Cambridge University Press; 1990. pp. 107–134. [Google Scholar]
- Kim SH. Protein Sci. 1994;3:159–165. doi: 10.1002/pro.5560030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene SJ, Hobson AC, Adler J. Proc. Natl. Acad. Sci. U.S.A. 1979;76:6309–6313. doi: 10.1073/pnas.76.12.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashi K. Biochim. Biophys. Acta. 1968;158:239–245. doi: 10.1016/0304-4165(68)90136-0. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K, McClary J. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee GF, Burrows GG, Lebert MR, Dutton DP, Hazelbauer GL. J. Biol. Chem. 1994;269:29920–29927. [PubMed] [Google Scholar]
- Lee GF, Lebert ME, Lilly AA, Hazelbauer GL. Proc. Natl. Acad. Sci. U.S.A. 1995 doi: 10.1073/pnas.92.8.3391. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Parkinson JS. Proc. Natl. Acad. Sci. U.S.A. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch BA, Koshland DE. Proc. Natl. Acad. Sci.U.S.A. 1991;88:10402–10406. doi: 10.1073/pnas.88.23.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch BA, Koshland DE. FEBS Lett. 1992;307:3–9. doi: 10.1016/0014-5793(92)80891-j. [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Kim SH. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Milligan DL, Koshland DE. Science. 1991;254:1651–1654. doi: 10.1126/science.1661030. [DOI] [PubMed] [Google Scholar]
- Mowbray SL, Koshland DE. Cell. 1987;50:171–180. doi: 10.1016/0092-8674(87)90213-3. [DOI] [PubMed] [Google Scholar]
- Ninfa EG, Stock A, Mowbray S, Stock J. J. Biol. Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- Oae S. In: Organic Sulfur Chemistry: Structure and Mechanism. Doi JT, editor. Boca Raton, FL: CRC Press; 1991. pp. 203–281. [Google Scholar]
- Ota IM, Varshavsky A. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- Pakula AA, Simon MI. Methods: A Companion to Methods in Enzymology. 1991;3:175–181. [Google Scholar]
- Pakula A, Simon M. Nature. 1992a;355:496–497. doi: 10.1038/355496a0. [DOI] [PubMed] [Google Scholar]
- Pakula AA, Simon MI. Proc. Natl. Acad. Sci. U.S.A. 1992b;89:4144–4148. doi: 10.1073/pnas.89.9.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Schuster SC, Swanson RV, Alex LA, Bourret RB, Simon MI. Nature. 1993;365:343–347. doi: 10.1038/365343a0. [DOI] [PubMed] [Google Scholar]
- Scott WG, Stoddard BL. Structure. 1994;2:877–887. doi: 10.1016/s0969-2126(94)00088-3. [DOI] [PubMed] [Google Scholar]
- Scott WG, Milligan DL, Milburn MV, Prive GG, Yeh J, Koshland DE, Kim SH. J. Mol. Biol. 1993;232:555–573. doi: 10.1006/jmbi.1993.1411. [DOI] [PubMed] [Google Scholar]
- Simms SA, Stock AM, Stock JB. J. Biol. Chem. 1987;262:8537–4853. [PubMed] [Google Scholar]
- Srinivasan N, Sowdhamini R, Ramakrishnan C, Balaram P. Int. J. Pept. Protein Res. 1990;36:147–155. doi: 10.1111/j.1399-3011.1990.tb00958.x. [DOI] [PubMed] [Google Scholar]
- Stock JB, Surette MG. Escherichia coli and Salmonella typhimurium. 2nd ed. 1995. in press. [Google Scholar]
- Stock A, Mottonen J, Chen T, Stock J. J. Biol. Chem. 1987;262:535–537. [PubMed] [Google Scholar]
- Stock A, Chen T, Welsh D, Stock J. Proc. Natl. Acad. Sci. U.S.A. 1988;85:1403–1407. doi: 10.1073/pnas.85.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JB, Lukat GS, Stock AM. Annu. Rev. Biophys. Biophys. Chem. 1991;20:109–136. doi: 10.1146/annurev.bb.20.060191.000545. [DOI] [PubMed] [Google Scholar]
- Stoddard BL, Bui JD, Koshland DE. Biochemistry. 1992;31:11978–11983. doi: 10.1021/bi00163a004. [DOI] [PubMed] [Google Scholar]
- Stoscheck CM. Methods Enzymol. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Koshland DE. J. Biol. Chem. 1984;259:7719–7725. [PubMed] [Google Scholar]
- Terwilliger TC, Bogonez E, Wang EA, Koshland DE. J. Biol. Chem. 1983;258:9608–9611. [PubMed] [Google Scholar]
- Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- Vogel HJ, Bonner DM. J. Biol. Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- Weis RM, Koshland DE. Proc. Natl. Acad. Sci. U.S.A. 1988;85:83–87. doi: 10.1073/pnas.85.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JI, Biemann HP, Pandit J, Koshland DE, Kim SH. J. Biol. Chem. 1993;268:9787–9792. [PubMed] [Google Scholar]
- Zapun A, Missiakas D, Raina S, Creighton TE. Biochemistry. 1995;34:5075–5089. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]