Abstract
Evolutionary selective pressures have tuned the efficiency of the protein-folding reaction in the crowded complex environment in the cell. Nevertheless, the fidelity of folding is imperfect, leading to off-pathway intermolecular interactions that compete with proper folding and to consequent formation of thermodynamically stable aggregates. Such aggregates constitute the histopathological hallmarks of many neurodegenerative pathologies. Yet, most of the approaches to characterize protein folding and/or misfolding are limited to in vitro conditions. Here, we describe a strategy to directly monitor the behavior of a protein in prokaryotic and eukaryotic cells. The method is based on incorporation of structurally non-perturbing, specific binding motifs for a bis-arsenical fluoroscein dye, FlAsH, in sites that result in distinct dye fluorescence signals for the folded and unfolded states of the protein under study. Our approach has been developed using as a case study the predominantly β-sheet intracellular lipid-binding protein, cellular retinoic acid-binding protein, alone or as a chimera fused to the exon 1-encoded fragment of huntingtin, which harbors a polyglutamine repeat tract. We have designed protocols to label this protein in vivo and to monitor the resulting fluorescence signal, which reports on any misfolding transition and formation of aggregates, yielding quantitatively interpretable data.
I. Introduction
Protein-folding pathways are remarkably complex, and the fidelity of the folding process has been optimized in the highly heterogeneous and crowded cellular environment. Intramolecular factors (e.g., mutations) and/or environmental stresses present additional challenges to the folding landscape and can drastically alter the fidelity of folding. As a result, alternative nonnative, partially folded states can be populated, leading to off-pathway intermolecular interactions. Despite the evolutionarily optimized mechanisms to avoid enrichment of misfolded species, some fraction of misfolded proteins can escape the cellular quality control mechanisms and form thermodynamically stable aggregates. Notably, failure in protein folding and the consequent deposition of metabolically stable aggregates constitute histopathological hallmarks of several neurodegenerative pathologies, including Alzheimer’s, Huntington’s, and Parkinson’s diseases. Cellular protein misfolding also has implications in the research laboratory and biotechnology industry: Off-pathway aggregation limits the yield of recombinant proteins.
Technical challenges make the goal of monitoring protein folding and misfolding in cells extremely difficult; the protein of interest has to be specifically highlighted to allow observation at physiological concentrations in the background of all other cellular constituents. Ideally the approach should provide information on structure formation, and the signal should report on conformational states visited by the target protein. Few groups have interrogated the conformational states a protein can sample in vivo. Thomas and coworkers (Wigley et al., 2001) devised a structural complementation assay with a direct read-out to monitor the formation of the soluble native structure. Similar approaches using structural complementation of a reporter protein provide an easily detectable phenotypic selection for mutants with improved folding efficiency in vivo (Maxwell et al., 1999; Philipps et al., 2003). However, these approaches are restricted to monitoring the end stage of the folding process and cannot readily be used to track various conformational explorations of a protein in vivo.
II. Rationale
Here we describe a fluorescence-based approach that we have recently developed to determine protein stability in vivo (in Escherichia coli cells) and in vitro (Ignatova and Gierasch, 2004) and that may be used to monitor in vivo folding propensities and aggregation kinetics in real time and in a noninvasive manner. The protein of interest is visualized in the context of all cellular macromolecules using the membrane-permeable bis-arsenical fluorescein-based dye “FlAsH” (named originally as a “fluorescein arsenical helix” binder), which specifically ligates to a genetically engineered, highly uncommon tetracysteine motif (Cys-Cys-Xxx-Yyy-Cys-Cys) (Griffin et al., 2000). In addition to its utility in measuring the bulk fluorescence of a population of cells for overall folding data and real-time aggregation assays, the FlAsH-labeling strategy has been very useful for fluorescence microscopy imaging of the state of the expressed protein and in-cell aggregates. We have combined this approach with isolation and electron microscopy of aggregates to garner information about the nature of the aggregates (Ignatova and Gierasch, 2005) and the mechanism of aggregation (Ignatova et al., 2007b). We have also extended our published work using E. coli cells and have developed methods to observe protein aggregation in eukaryotic cells. We describe these protocols here as well.
III. Methods
A. Design of the Tetra-Cys Protein Fluorescence Reporter System
We integrated the tetracysteine sequence (here Cys-Cys-Gly-Pro-Cys-Cys, incorporating the native Gly-Pro present in this loop) into an internal Ω-loop of the 136-amino acid cellular retinoic acid-binding protein I (referred to here as CRABP) (Ignatova and Gierasch, 2004). The Ω-loop shows the lowest sequence conservation among the intracellular lipid-binding protein family (Gunasekaran et al., 2004) and tolerates sequence expansions and deletions (Ignatova and Gierasch, 2004). By this intramolecular insertion of the FlAsH-binding motif, we found that the FlAsH-fluorescence emission signal reports on the conformational state of the protein, with misfolded, denatured, and aggregated states hyperfluorescent compared to the native state (Ignatova and Gierasch, 2004). This system allowed us to measure in vivo stability using a urea titration (Ignatova and Gierasch, 2004; Ignatova et al., 2007a). Mutation of the helix-terminating residue Pro39 to Ala is known to slow down the folding and unfolding of CRABP (Eyles and Gierasch, 2000), and P39A tetra-Cys CRABP shows a high tendency to form inclusion bodies in vivo (Ignatova and Gierasch, 2004) and aggregates in vitro (Ignatova and Gierasch, 2005). The structural integrity and alterations in the native structure of the host protein after the insertion of the tetra-Cys motif must be verified by different methods, for example, circular dichroism spectroscopy (CD), fluorescence, or activity. In their native states, both tetra-Cys CRABP and the aggregation-prone P39A tetra-Cys CRABP variants are indistinguishable in structure and function from their native counterparts, whether FlAsH-labeled or unlabeled, based on far-UV CD or on ligand-binding (here, retinoic acid-binding) activity (Ignatova and Gierasch, 2004).
The FlAsH-labeled tetra-Cys CRABP protein was also useful as a reporter in chimeras in order to assess the impact of other domains, for example the exon 1-encoded fragment of huntingtin (Htt) (Ignatova and Gierasch, 2006), the protein that aggregates in Huntington’s disease (Ross and Poirier, 2004). This chimera yielded insight into the aggregation mechanism of Htt domains and their flanking regions (Ignatova et al., 2007b).
1. FlAsH Labeling in E. coli Cells
Step 1
The FlAsH-binding motif is engineered in the soluble pseudo-wild-type CRABP [carrying an N-terminal His-tag and harboring a stabilizing Arg→Gln mutation at position 131 (Zhang et al., 1992)] and in the slow-folding and aggregation-prone P39A CRABP mutant in three successive mutational steps using the oligonucleotide-directed QuikChange mutagenesis protocol (Stratagene) (Ignatova and Gierasch, 2004). cDNAs encoding tetra-Cys CRABP and P39A tetra-Cys CRABP are cloned in a multi-copy pET16b (AmpR) plasmid; transcription is under the control of the T7 promoter. E. coli BL21(DE3) cells, carrying the DE3 lysogen for high level expression of T7 polymerase, were used as a host. The Htt constructs (tetra-Cys CRABP Htt20, tetra-Cys CRABP Htt40, and tetra-Cys Htt53) are constructed by C-terminal in-frame fusion of the full-length exon 1 of Huntingtin, encoding polyQ tracts of 20, 40, or 53 residues, to the tetra-Cys CRABP. The genetic manipulations led to insertion of two new amino acids (Leu-Glu) at the junction between the C-terminus of tetra-Cys CRABP and the beginning of the exon 1-encoded fragment of Htt (Ignatova and Gierasch, 2006).
Step 2
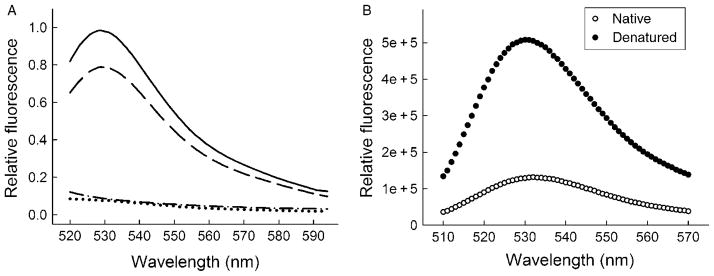
A single colony from freshly transformed cells is inoculated in LB medium containing 100 μg/ml ampicillin and grown overnight at 30 °C by constant shaking at 200 rpm. The cultured cells are sterile-harvested (2060 × g, 15 min, 4 °C) and brought up to 25× the original volume in fresh LB medium containing 100 μg/ml ampicillin. The cultures are grown at 37 °C until the OD600 = 0.5 and then treated with lysozyme (50 ng/ml) for 10 min on ice. This gentle lysozyme pretreatment enhances the permeability of the outer membrane of the bacterial host cell for FlAsH (Fig. 1A) and enables preloading with FlAsH at a rate and concentration sufficient to saturate newly expressed tetra-Cys protein (Ignatova and Gierasch, 2004).
Fig. 1.
FlAsH labeling in E. coli cells and in vitro. (A) FlAsH labeling of E. coli cells expressing tetra-Cys CRABP (dashed line) and P39A tetra-Cys CRABP (solid line). Note that cells expressing wild-type CRABP without the tetra-Cys motif do not fluoresce (dotted line). For comparison, the intrinsic fluorescence of non-labeled cells expressing tetra-Cys CRABP is shown (dash–dot lines). (B) Fluorescence of FlAsH-labeled purified tetra-Cys CRABP in its native and denatured states. Reproduced from Ignatova and Gierasch (2004).
Step 3
The lysozyme-containing medium is removed by centrifugation (2060 × g, 15 min, 4 °C), and the cell pellet is resuspended in the same amount of fresh sterile LB medium containing 100 μg/ml ampicillin. Aliquots of 1 ml are labeled with 0.2 μM FlAsH–EDT2 (marketed by Invitrogen under the name Lumio Green™) and 1 μM ethanedithiol (EDT). FlAsH and EDT remain in the medium during the entire time of culture growth. An excess of EDT suppresses the labeling of endogenous cysteines on other proteins (Griffin et al., 2000).
Usually the dye is preloaded one generation before the induction of protein synthesis with IPTG to circumvent limitations from slow dye entry into the cells and to ensure that ample dye is available upon induction. In addition, excess dye is also present in the nutrient medium throughout the induction period and continues to enter the cells. Dye preloading is a prerequisite to application of labeling to kinetic experiments or to monitor folding and misfolding/aggregation reactions. For some studies of equilibrium processes in the cell (e.g., localization), lysozyme pretreatment and FlAsH preloading before the induction of biosynthesis are not necessary. FlAsH enters E. coli cells, albeit more slowly without the pretreatment, and the dye accumulated in the cells throughout the expression cycle will be sufficient to quantitatively label the entire amount of the expressed protein.
Step 4
Cells are further cultured in the dark by constant shaking at 200 rpm. After one generation at 37 °C, at OD600 = 1.0, protein synthesis is induced by adding 0.4 mM IPTG.
2. Monitoring Protein Aggregation: Bulk Cell Fluorescence
Over the time course of aggregation, 150-μl aliquots are withdrawn from the growing culture and subjected to fluorescence measurements in bulk at 530 nm (excitation 500 nm; excitation bandwidth 2 nm, emission bandwidth 3 nm). Fluorescence of cells transformed with a plasmid bearing the wild-type CRABP without the tetra-Cys motif and labeled with FlAsH is used as a blank, and the value is subtracted from each point. The temperature of the cuvette holder is maintained at 37 °C with a water bath.
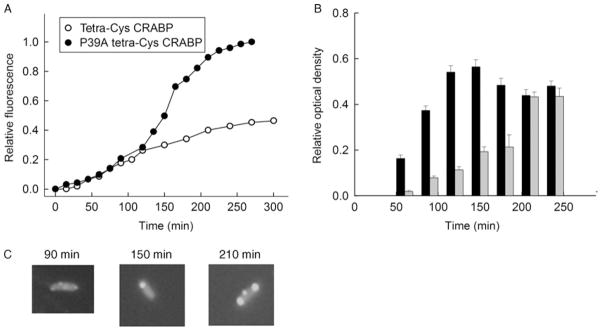
The intensity of fluorescence from the FlAsH-labeled P39A tetra-Cys CRABP cells is higher than that of tetra-Cys CRABP-expressing cells (Fig. 1A), and this higher quantum yield is due to the partial deposition of P39A tetra-Cys CRABP into hyperfluorescent inclusion bodies (Ignatova and Gierasch, 2004, 2005). The quantum yield and spectral properties of the aggregates are reminiscent of the hyperfluorescent denatured state of purified FlAsH-labeled tetra-Cys CRABP protein (Fig. 1B). The time course of bulk fluorescence of FlAsH-labeled P39A tetra-Cys CRABP in cells differs from that of cells expressing soluble tetra-Cys CRABP (Fig. 2A). The steady-state increase in the FlAsH fluorescence of tetra-Cys CRABP reports the synthesis of this protein and plateaus when de novo synthesis ceases and the cells enter stationary phase. The fluorescence curve of the aggregation-prone P39A tetra-Cys CRABP is a cumulative signal of both synthesis and aggregate formation, and the sharp rise in the fluorescent signal between 120 and 150 min reports the enrichment of misfolded and aggregated species.
Fig. 2.
Soluble and aggregation-prone variants of CRABP show different patterns of bulk FlAsH fluorescence. (A) Time evolution of the expression and aggregation of P39A tetra-Cys CRABP (filled symbols) and the expression of tetra-Cys CRABP (open symbols), monitored by fluorescence at 530 nm (excitation 500 nm). (B) Partitioning of P39A tetra-Cys CRABP protein between soluble (black bars) and insoluble (light gray bars) cytoplasmic fractions. (C) Examples of fluorescence microscopic images of the P39A tetra-Cys CRABP-expressing cells, taken at the indicated times after induction of protein synthesis with IPTG. Parts of the figure are reproduced from Ignatova and Gierasch (2004).
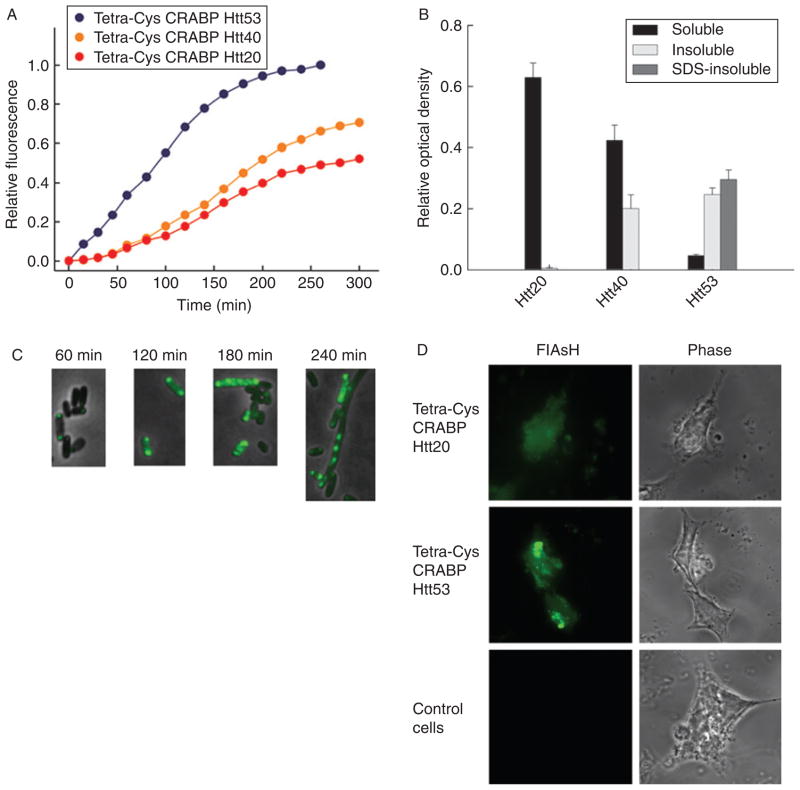
The use of tetra-Cys CRABP as a reporter in chimeras to assess their aggregation propensity is illustrated by fusions of tetra-Cys CRABP with Htt exon 1-encoded domains containing polyQ repeats of varying length (Ignatova and Gierasch, 2006; Ignatova et al., 2007b). Those with shorter polyQ repeats (Q = 20) (i.e., in the non-pathological range) show a fluorescence pattern (Fig. 3A) superimposable on that of tetra-Cys CRABP (Fig. 2A). Increasing the polyQ length beyond the threshold length of 35 consecutive glutamines characteristic of polyQ diseases (Ross and Poirier, 2004; Zoghbi and Orr, 2000) causes an increase in the bulk-fluorescence signal, and reduces the time for onset of the rise in fluorescence. Furthermore, the plateau fluorescence increases in proportion to length of the polyQ tract (Fig. 3A). The sharp increase in the FlAsH fluorescence of the bulk cell suspension expressing the tetra-Cys CRABP Htt53 chimera suggests that the aggregation is favored as soon as expression of protein commences.
Fig. 3.
Chimeras of tetra-Cys CRABP and the exon 1-encoded portion of Htt show polyQ-length-dependent aggregation. (A) Time course of bulk fluorescence (at 530 nm; excitation 500 nm) of FlAsH-labeled E. coli cells expressing tetra-Cys CRABP Htt20 (red), Htt40 (orange), and Htt53 (blue). (B) Partitioning of the chimeras between the soluble (black bars), detergent-labile insoluble (light gray bars), and SDS-resistant insoluble (dark gray bars) cytoplasmic fractions at the conclusion of the experiment shown in Fig. 2A (260 min after induction of the protein synthesis). (C) Tetra-Cys CRABP Htt53 chimera forms hyperfluorescent aggregates in the E. coli host, and the appearance of the SDS-resistant species coincides with the appearance of the filamentous phenotype of the E. coli cells. (D) Fluorescence microscopic images of FlAsH-labeled eukaryotic HEK293T cells transiently transfected with tetra-Cys CRABP Htt chimeras. Tetra-Cys CRABP Htt20 is soluble and yields uniformly distributed FlAsH fluorescence, whereas tetra-Cys CRABP Htt53 generates hyperfluorescent loci. Cells transfected with empty pcDNA3.1 vector serve as control. The localization of the tetra-Cys CRABP Htt chimeras is visualized as indicated (by FlAsH fluorescence or phase contrast). Parts of this figure are reproduced from Ignatova and Gierasch (2006).
3. Monitoring Protein Aggregation: Fractionation of E. coli Cells
Step 1
E. coli BL21(DE3) cells are transformed with either tetra-Cys CRABP or P39A tetra-Cys CRABP, and their expression is induced by adding 0.4 mM IPTG. Ten-milliliter aliquots are withdrawn at different time points after induction, and bacteria are harvested by centrifugation at 2060 × g for 15 min at 4 °C.
Step 2
The cell pellet is resuspended in 1.5 ml of 50 mM phosphate buffer (pH 8) containing 300 mM NaCl. Cells are treated with lysozyme (at a final concentration of 500 μg/ml for 30 min) and DNAse (at a final concentration of 50 μg/ml for 15 min) followed by sonication with 20-s bursts for 3 min (30% duty cycle), all steps on ice. Cell lysates are fractionated into soluble and insoluble fractions by centrifugation at 27000 × g at 5 °C in a tabletop centrifuge for 30 min.
Step 3
The insoluble pellet fraction is resuspended in 1.5 ml of 10 mM Tris–HCl buffer (pH 8) containing 8 M urea. In the case of tetra-Cys Htt chimeras, the insoluble fraction is resuspended in 1.5 ml of 50 mM phosphate buffer (pH 8) containing 8 M urea and 2% SDS and then centrifuged again at 15294 × g at 5 °C in a tabletop centrifuge for 30 min. The detergent-insoluble fraction is solubilized in 100% formic acid for 3 h at 37 °C (Hazeki et al., 2000). Twenty-microliter aliquots of each fraction, mixed with 10 μl 3× SDS-loading buffer and preheated for 3 min at 95 °C, are loaded onto an SDS-PAGE gel (12%), and the tetra-Cys CRABP content in each fraction is quantified by optical densitometry of the Coomassie-stained SDS gel, on which known amounts of purified tetra-Cys CRABP are electrophoresed for comparison.
Fractionation of the cell lysates of P39A tetra-Cys CRABP-expressing cells into soluble and insoluble components confirms that the increase in bulk fluorescence reports on the enrichment of insoluble protein in the cells (Fig. 2B). At t 120 min, both tetra-Cys CRABP and P39A tetra-Cys CRABP are expressed as native proteins in comparable quantity. Subsequently, the slow-folding P39A tetra-Cys CRABP shows a rapidly increasing fluorescence intensity, which corresponds to a growing quantity of partially folded states. When the population of the partially folded intermediates reaches a critical threshold, formation of inclusion bodies ensues (Fig. 2C).
In the set of the polyQ chimeras, the FlAsH-fluorescence intensity correlates with the partitioning of the chimeras between the soluble and insoluble fractions (Fig. 3A and B). The Q40 fusion protein is found equally distributed between the soluble and insoluble fractions, whereas the Q53 protein is almost entirely present in the insoluble fraction with near equal partitioning between the detergent-labile and SDS-resistant fractions. The Htt20 chimera remains soluble over the entire expression period (Fig. 3B).
4. Monitoring Protein Aggregation: Fluorescence Microscopy
During the time course of growth of the labeled cells expressing different proteins, 20-μl aliquots of cell suspension are withdrawn at intervals and concentrated twice by centrifugation [2655 × g for 3 min, resuspension in 10 μl of 50 mM Hepes buffer (pH 7.5)]. Two microliters of this concentrated suspension is immobilized in 1% agarose in LB, and the cells are imaged with a fluorescent microscope (Nikon Eclipse E600) with excitation at 485 nm and a 510-nm emission cutoff filter. The images are processed with the Openlabs software (Improvision, Lexington, MA).
The fluorescence microscopic images of individual E. coli cells (Figs. 2C and 3C) recapitulate the observations made by the FlAsH-fluorescence measurements in bulk and by the centrifugal fractionation studies. In the case of cells containing FlAsH-labeled P39A tetra-Cys CRABP, the fluorescence is initially diffuse throughout the cell, indicative of a soluble protein at early times post-induction. Subsequently, hyperfluorescent aggregates form at the poles of the cell (Fig. 2C), and their appearance coincides with the sharp increase in FlAsH fluorescence (Fig. 2A). The extremely high aggregation propensity of the tetra-Cys CRABP Htt53 chimera is confirmed by fluorescence microscopy; hyperfluorescent polarly localized loci appear at the earliest time points (Fig. 3C). At later time points (>180 min after induction) a fraction of the cells change their morphology into a filamentous phenotype, and this transition parallels the appearance of the detergent-resistant aggregates in the cytoplasm (Fig. 3C).
B. Aggregation Propensity in Eukaryotic Cells
1. Expression and FlAsH Labeling in Eukaryotic Cells
Step 1
For expression in mammalian cells, tetra-Cys CRABP Htt20 and tetra-Cys Htt53 fusions were sub-cloned into the pcDNA3.1 vector (Invitrogen). For transient transfections, 70% confluent human embryonic kidney (HEK) 293T cells are transfected with 20-μg plasmid DNA by electroporation. Transiently transfected cells are grown in 35-mm, 6-well plates for 24 h in 1-ml Dulbecco’s modified Eagle’s medium (Gibco/Invitrogen) supplemented with 10% fetal bovine serum and 100 U/ml penicillin at 37 °C in an atmosphere of 5% CO2. Each well is supplied with a sterile polylysine precoated cover slip.
Step 2
The FlAsH labeling is performed as described (Gaietta et al., 2002). Confluent cells, 24 h after transfection, are washed twice with phenol red-free and bovine-serum-free Dulbecco’s modified Eagle’s medium (D-MEM/F-12, Gibco/Invitrogen) and then incubated in D-MEM/F-12 supplemented with 1 μM FlAsH–EDT2 and 10 μM EDT for 1 h at 37°C.
Step 3
Cells are fixed on cover slips with 4% formaldehyde (from paraformaldehyde) in phosphate-buffered saline for 30 min at room temperature. The cover-slips are washed once for 5 min with relatively high concentrations of EDT (250 μM in PBS pH 7.2, Gibco/Invitrogen) to remove the nonspecifically bound FlAsH dye and twice with phosphate-buffered saline. Fixed samples are mounted in anti-fading solution (Invitrogen) and examined using a Leica inverted microscope equipped with a 63× oil objective. HEK 293T cells transfected with empty pcDNA3.1 vector, otherwise labeled and treated identically, serve as a negative control.
2. Monitoring Protein Aggregation in Eukaryotic Cells
The tetra-Cys CRABP Htt chimeras transiently expressed in HEK 293T cells (Fig. 3D) show the same phenotypes as observed in the E. coli expression system. The tetra-Cys CRABP Htt20 is diffusely distributed in the cytosol, indicative of soluble expression, and tetra-Cys CRABP Htt53 is localized in small hyperfluorescent aggregates that progressively fuse into one large inclusion body.
IV. Summary
We have exploited the specific labeling strategy introduced by Adams and Tsien and coworkers based on a bis-arsenical fluoroscein derivative “FlAsH” (Adams et al., 2002; Gaietta et al., 2002; Griffin et al., 2000) to observe the folding and aggregation of a protein of interest in cells. By suitable incorporation of the FlAsH-binding tetra-Cys motif into an internal loop of a β-barrel intracellular lipid-binding protein, CRABP, we have been able to determine equilibrium stability in cells and to follow the time-course of aggregate formation by a slow-folding mutant and by a chimera containing a portion of Htt, including the polyglutamine-rich tract. The sensitivity of FlAsH fluorescence to conformational changes in the host CRABP protein converts it into an attractive reporter system to monitor misfolding and aggregation. The FlAsH-fluorescence time course measured in a bulk cell suspension upon induction of protein synthesis differs for soluble and aggregation-prone protein, and FlAsH intensity can be used as a read-out to monitor protein aggregation in vivo and in vitro. This general approach is amenable, as reported earlier, to quantitative determination of the size of the nucleus of an aggregating system in vitro (Ignatova and Gierasch, 2005) or to direct in-cell exploration of the impact of highly aggregation-prone sequences on the behavior of an otherwise stably folded protein when chimeras of the two are created (Ignatova and Gierasch, 2006; Ignatova et al., 2007b). This approach has equipped us with a tool to follow fundamental events of major biomedical importance for protein misfolding diseases directly in cells.
Our method may be applied more generally if appropriate care is taken in the design of the FlAsH-labeling site. The sensitivity of the FlAsH quantum yield to the conformational state of the protein of interest is the requisite precondition for the application of this approach to measure in vivo protein folding, stability, and aggregation propensity. We have carried out extensive studies of the relationship of the geometry of the tetra-Cys-binding sites to the FlAsH fluorescence (B. Krishnan and L. M. Gierasch, submitted). To apply this approach to other proteins, a careful structure-based design should be performed with consideration of all sequence-specific and structural constraints so that the incorporated tetra-Cys sequence would be tolerated within the protein of interest and the FlAsH quantum yield would be sensitive to the conformational states of the protein host.
Acknowledgments
The authors acknowledge support from the National Institutes of Health (grants GM027616 and a 2006 NIH Director’s Pioneer Award to LMG), and DFG-project IG73/4-1 and the Heisenberg award IG73 1-1 (to ZI).
References
- Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: Synthesis and biological applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- Eyles SJ, Gierasch LM. Multiple roles of prolyl residues in structure and folding. J Mol Biol. 2000;301:737–747. doi: 10.1006/jmbi.2000.4002. [DOI] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Griffin BA, Adams SR, Jones J, Tsien RY. Fluorescent labeling of recombinant proteins in living cells with FlAsH. Methods Enzymol. 2000;327:565–578. doi: 10.1016/s0076-6879(00)27302-3. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K, Hagler AT, Gierasch LM. Sequence and structural analysis of cellular retinoic acid-binding proteins reveals a network of conserved hydrophobic interactions. Proteins. 2004;54:179–194. doi: 10.1002/prot.10520. [DOI] [PubMed] [Google Scholar]
- Hazeki N, Tukamoto T, Goto J, Kanazawa I. Formic acid dissolves aggregates of an N-terminal huntingtin fragment containing an expanded polyglutamine tract: Applying to quantification of protein components of the aggregates. Biochem Biophys Res Commun. 2000;277:386–393. doi: 10.1006/bbrc.2000.3682. [DOI] [PubMed] [Google Scholar]
- Ignatova Z, Gierasch LM. Monitoring protein stability and aggregation in vivo by real-time fluorescent labeling. Proc Natl Acad Sci USA. 2004;101:523–528. doi: 10.1073/pnas.0304533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatova Z, Gierasch LM. Aggregation of a slow-folding mutant of a β-clam protein proceeds through a monomeric nucleus. Biochemistry. 2005;44:7266–7274. doi: 10.1021/bi047404e. [DOI] [PubMed] [Google Scholar]
- Ignatova Z, Gierasch LM. Extended polyglutamine tracts cause aggregation and structural perturbation of an adjacent beta barrel protein. J Biol Chem. 2006;281:12959–12967. doi: 10.1074/jbc.M511523200. [DOI] [PubMed] [Google Scholar]
- Ignatova Z, Krishnan B, Bombardier JP, Marcelino AM, Hong J, Gierasch LM. From the test tube to the cell: Exploring the folding and aggregation of a β-clam protein. Biopolymers. 2007a;88:157–163. doi: 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatova Z, Thakur AK, Wetzel R, Gierasch LM. In-cell aggregation of a polyglutamine-containing chimera is a multi-step process initiated by the flanking sequence. J Biol Chem. 2007b;282:36736–36743. doi: 10.1074/jbc.M703682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KL, Mittermaier AK, Forman-Kay JD, Davidson AR. A simple in vivo assay for increased protein solubility. Protein Sci. 1999;8:1908–1911. doi: 10.1110/ps.8.9.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps B, Hennecke J, Glockshuber R. FRET-based in vivo screening for protein folding and increased protein stability. J Mol Biol. 2003;327:239–249. doi: 10.1016/s0022-2836(03)00077-9. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10 (Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Wigley WC, Stidham RD, Smith NM, Hunt JF, Thomas PJ. Protein solubility and folding monitored in vivo by structural complementation of a genetic marker protein. Nat Biotechnol. 2001;19:131–136. doi: 10.1038/84389. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu ZP, Jones TA, Gierasch LM, Sambrook JF. Mutating the charged residues in the binding pocket of cellular retinoic acid-binding protein simultaneously reduces its binding affinity to retinoic acid and increases its thermostability. Proteins. 1992;13:87–99. doi: 10.1002/prot.340130202. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]